Summary
Amyotrophic Lateral Sclerosis (ALS) is a muscle-bone degenerative disease, which lacks a specific index for diagnosis. In our previous studies, we found that exosomes mediated the interaction mechanism between muscle and bone at the cellular level, and myoblast exosomes can transfer miR-27a-3p to promote osteoblast mineralization. Therefore, we suppose that the expression of miR-27a-3p in the serum exosomes of ALS patients also changes. In this study, we used healthy human serum as a sample to find out the conditions and methods for extraction and detection. Then through comparison of the expression of miR-27a-3p in the serum exosomes of 10 ALS patients and healthy subjects, we found that in the ALS patients miR-27a-3p was down-regulated, and may be involved in the development of ALS, and therefore has potential as a reference for the diagnosis of ALS in the clinic.
Keywords: Amyotrophic Lateral Sclerosis (ALS), exosome, miR-27a-3p, serum, stem loop
1. Introduction
Amyotrophic Lateral Sclerosis (ALS) is a progressive and fatal motor neuron degenerative disease, in which the main manifestation is progressive muscle weakness and atrophy, and gradually develops into systemic muscle atrophy and paralysis, with death finally due to respiratory failure. The cause is not yet clear, the early symptoms are not obvious, and lack of specific biological indicators makes clinical diagnosis difficult with a high misdiagnosis rate (1). Once diagnosed, the average survival time of patients are three to five years. The disease develops rapidly, and there is a significant difference between patients, with no definite diagnosis and cure method.
Exosomes are double lipid vesicles with a diameter about 40–100 nm, which are secreted by a variety of cells. Exosomes are widespread in the peripheral body fluid, can carry and enrich various biologically active molecules, such as proteins, nucleic acids and so on, and have high stability, agility and specificity (2). Exosomes can reflect the physiological and pathological changes of the source cells, including proteins and nucleic acids, which have a potential biomarker function, and make exosomes have potential to be a new biomarker for disease diagnosis (3). MicroRNA (miRNA) is a class of endogenous non-encoding small RNA, with a length of 18-22 nt. MiRNAs are enriched in exosomes, and the exosome membrane structure can protect miRNA from degradation by RNA enzymes, this has made exosome miRNA become a hotspot in recent years (4,5).
In this experiment, we used the serum of healthy subjects, and compared and analyzed different extraction methods for serum exosomes and exosome miRNAs. Detection of miRNA expression was by polyadenylate poly A tail and stem loop methods, in order to find the most suitable method for extraction and detection of serum exosomes and exosome miRNAs. Then we compared the expression level of miR-27a-3p in serum exosomes of ALS patients and healthy subjects, which laid a foundation for further research on the role of exosomes in the early diagnosis of ALS (Figure 1).
Figure 1.
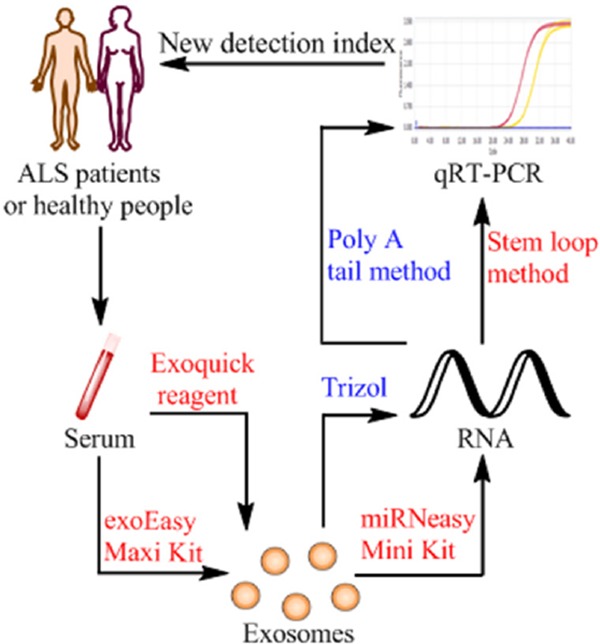
Extraction and identification methods of exosome miRNA in the study of ALS patients. A diagram showing the conditions and methods for extraction and detection and their applications. Exosome were isolated from ALS patients and healthy subject's serum by Exoquick reagent or exoEasy Maxi Kit, then using Qiagen miRNeasy Mini Kit to isolate RNA from serum exosomes, finally using stem loop method to detect the expression of miR-27a-3p in two groups of RNA samples. The results of the different expressions can be used for the clinical diagnosis of ALS patients.
2. Materials and Methods
2.1. Materials
Serum from 20 healthy persons and 10 ALS patients were provided by Qilu Hospital and Ji'nan infectious diseases hospital, and stored at -80oC. Exoquick reagent and Anti CD63 were purchased from System Biosciences (Mountain View, CA, USA). ExoEasy Maxi Kit and miRNeasy Mini Kit were purchased from Qiagen (Valencia, CA, USA). TRIzol reagent was purchased from Invitrogen (Carlsbad, CA, USA). MiDETECT A Track™ miRNA qRT-PCR Starter Kit was purchased from RiboBio (Guangzhou, China). Reverse transcriptase kit was from Toyobo (Osaka, Japan).
2.2. Extraction and identification of serum exosome
The serum exosome was extracted by Exoquick reagent (System Biosciences, Mountain View, CA, USA) according to the manufacter's protocol with minor modifications. The specific steps were as follows: the serum was centrifuged at 3,000 × g for 30 min (4°C), to obtain the supernatant. We used a 1 mL syringe and repeatedly sucked 5 times. Exoquick reagent was added in a proportion of 4:1, and mix thoroughly by pipetting. It was stored at 4°C for 30 min, centrifuged at 13,000 × g for 2 min, discarded the supernatant; centrifuged again and discarded the supernatant, then 100 µL phosphate-buffered saline (PBS) was added for resuspension, the exosome resuspension named exo-A was obtained.
The serum exosome was extracted by exoEasy Maxi Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's protocol with minor modifications. Briefly, serum filtration was used to remove diameters greater than 0.8 µm particles, an equal volume of buffer XBP was added mixed well immediately, and transfered into the exoEasy spin column and centrifuged 1 min at 500 × g, the flow-through was discarded; 10 ml Buffer XWP was added and centrifuged 5 min at 5,000 × g, the flow-through was discarded; the spin column was transfered to a new collection tube, 100 uL Buffer XE was added and centrifuged 5 min at 5,000 × g, the flow-through was collected as exosome resuspension B, named exo-B.
Western-blot for detection of exosome specific marker proteins: Exosome protein content was measured using the BCA protein assay kit (Beyotime Institute of Biotechnology, Shanghai, China). Equal amounts (20 µg) of exosome resuspension were taken respectively, subjected to sodium dodecyl sulphate polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride (PVDF) membrane. After blocking, membrane was washed with TBST and then incubated overnight using antibodies against CD63 (1:1,000, System Biosciences, Mountain View, CA, USA). The blots signals were developed using ECL Plus (Millipore, Billerica, MA, USA) and visualized with Fusion SOLO S (Vilber, Collégien, France).
2.3. Isolation of total RNA from serum exosomes
Combined with the extraction of serum exosomes, we used two different methods to isolate total RNA from serum exosomes.
Using the traditional TRIzol reagent method to isolate total RNA from serum exosomes: The extracted exosome was mixed with a proper volume of TRIzol reagent (Invitrogen, Carlsbad, CA, USA), stored at room temperature for 5 min; chloroform was added according to a proportion of 1:5 in order to separated the lysate, transfer the upper aqueous phase to a new collection tube, and add isopropyl alcohol to precipitate the RNA, finally the RNA precipitation was cleaned using 75% ethanol, and then RNA could be precipitated after centrifugation.
Using Qiagen miRNeasy Mini Kit (Qiagen, Valencia, CA, USA) to isolate total RNA from serum exosomes: the extracted exosomes were washed and centrifuged using Buffer XBP and XWP, and mixed with a proper volume of QIAzol lysate, stored at room temperature for 5 min; and chloroform was added according to the proper proportion to separate the lysate, the upper aqueous phase was transferred to a new collection tube, after addition of 2 volumes of 100% ethanol, the mixture was transferred into an RNeasy MinElute spin column in a 2 ml collection tube, after centrifugation, RNA was combined on the column membrane, and then washed with Buffer RWT and RPE, DNase/RNase-Free water was added and centrifuged 1 min at 12,000 × g, the flow-through collected was the exosome total RNA.
The concentration and purity of RNA samples were detected by Thermo micro spectrophotometer NanoDrop2000, and values were recorded.
2.4. Quantitative real-time PCR (qRT-PCR)
The expression level of miR-27a-3p in serum exosomes was detected by fluorescence quantitative polyadenylate poly A tail and stem loop methods, with miR-16 as endogenous exosome control. We compared the difference in expression of miR-27a-3p between ALS patients and healthy subjects.
Polyadenylate poly A tail method: mature miRNA sequences were provided (http://www.mirbase.org), the primers were designed by RiboBio company, and included reverse transcription, forward and reverse primers. The qRT-PCR was performed with miDETECT A Track™ miRNA qRT-PCR Starter Kit (RiboBio, Guangzhou, China), specific steps and the reaction system was conducted according to the manufacturer's protocol.
Stem loop method: Based on the miRNA mature sequences and the stem loop frame, we designed specific RT primers with a stem loop structure and forward-reverse primers, the sequence of the primers for miR-27a-3p and miR-16 are listed in the Table 1. RNA samples were premodified at 65°C for 5 min, and a reverse transcriptase kit (Toyobo, Osaka, Japan) was used for the RT reactions. After cDNA and diluted 20 times, the qRT-PCR reaction was performed according to the following reaction system: Realtime PCR Master Mix 5 µL, forward primer (10 µmol/L) 1 µL, reverse primer (10 µmol/L) 1 µL, and 3 µL cDNA dilution solution. The conditions of qRT-PCR reaction were: 95°C 10 min; 95°C 2 s, 60°C 20 s, 70°C 10 s, 45 cycles. After the end of the cycle, the melting curve was analyzed immediately.
Table 1. Sequences of primers used in this study.
| Gene | Primers | |
|---|---|---|
| miR-27a-3p | RT: | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCGGAA |
| Forward: | CGGCGGTTTCACAGTGGCTAAG | |
| Reverse: | CCAGTGCAGGGTCCGAGGTAT | |
| miR-16 | RT: | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCGCCAAT |
| Forward: | CGCGCTAGCAGCACGTAAAT | |
| Reverse: | GTGCAGGGTCCGAGGT |
2.5. Statistical analysis
All data were analyzed statistically by SPSS software (version 19.0). The measurement data are expressed as mean ± standard deviation (S.D.), the mean values between groups were compared by one-way analysis of variance (ANOVA), and p < 0.05 was considered statistically significant.
3. Results
3.1. Identification of exosome
Western-blots were used to detect the specific marker proteins (6) of serum exosomes extracted by the two methods, and CD63 is one of the most commonly used for identification of exosome protein molecules. The results showed that CD63 proteins were all expressed in two groups (Figure 2).
Figure 2.
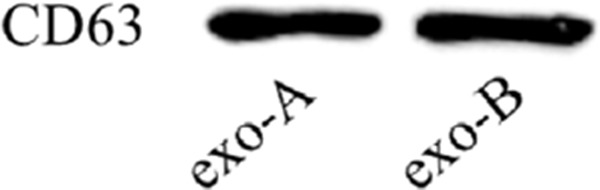
Identification of surface marker proteins of exosomes. Using Exoquick and exoEasy Maxi Kit extract exosomes from serum, respectively. CD63 level was monitored by Western blot.
3.2. Comparison of the concentration and purity of RNA were extracted by 4 different methods
In order to facilitate the description, the RNA samples obtained by Exoquick-Trizol method were named group A, those obtained by Exoquick- miRNeasy Mini Kit were named group B, the exoEasy Maxi Kit – Trizol method used to obtain the RNA samples were named group C, and the exoRNeasy Serum/Plasma Midi Kit method for the RNA samples were named group D.
The A group was extremely unstable, the concentration range is wide and distributed from 3.6–120.4 ng/µL, the mean value of OD260/280 was 1.42 ± 0.02 and quality was bad. The concentration and purity of group B were improved compared with group A, and the mean value of OD260/280 was 1.54 ± 0.051. The concentration of group C tended to be stable, but the purity of the group changed a little. Group D used a Qiagen kit to extracted exosomes from the serum, we continued to use the kit to isolate RNA, the concentration was about 10 ng/µL and the purity was obviously improved, the mean value of OD260/280 was 1.70 ± 0.05, it basically met the requirements of the experiment, and compared with the other groups, the difference was statistically significant (p < 0.05) (Figure 3).
Figure 3.
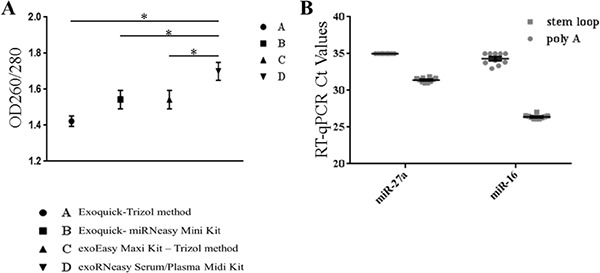
Comparing the different extraction and identification methods. The exosome RNA were extracted by 4 different methods and identificated by poly A tail or stem loop method, respectively. (A) The purity of Exosome RNA were extracted by 4 different methods, and group D was extracted by exoRNeasy Serum/Plasma Midi Kit compared with the other groups, the difference was statistically significant (p < 0.05); (B) The Ct values of miR-27a-3p and miR-16 were detected by poly A tail or stem loop method.
3.3. Comparison of the difference in expression of miRNA by qRT-PCR
The exoRNeasy Serum/Plasma Midi Kit isolated RNA from the serum exosomes, expression level of miR-27a-3p was detected by fluorescence quantitative polyadenylate poly A tail and stem loop methods respectively, the internal control selected miR-16 which is a serum exosome and was relatively stable.
Comparing the specificity of PCR products detected by the two methods, dissolution curves were all a single peak, which proved that the designed primers had effectiveness and specificity.
When the poly A tail method was used to detect miR-27a-3p and miR-16, the Ct values of the partially complex hole is greater than 35, the test results were negative, and the Ct values were unstable, and did not have reproducibility. The qRT-PCR again after changing the concentration of cDNA samples, the relative results are the same as before. The results of the stem loop method showed that the Ct values of miR-27a-3p ranged from 31.31 to 31.71, the Ct values of miR-16 were from 25.5–26.1, which are less than 35 (Figure 3). It was proved that the stem loop method is more sensitive than the polyadenylate poly A tail method, the repeatability of the compound holes is good and the stability is strong.
3.4. Comparing the difference in expression of miR-27a-3p between ALS patients and healthy subjects
We used the exoRNeasy Serum/Plasma Midi Kit to extract RNA from 10 ALS patients and healthy subject's serum exosomes, respectively. The difference in expression of miR-27a-3p in the two groups of RNA samples was detected by the fluorescence quantitative stem loop method. The Ct values of all the samples were less than 35, the dissolution curves were all a single peak, which proved that the amplification of data was effective and the specificity was good. Compared with the control group, the expression of miR-27a-3p in the ALS group was down-regulated, and the difference was statistically significant (p < 0.05) (Figure 4).
Figure 4.
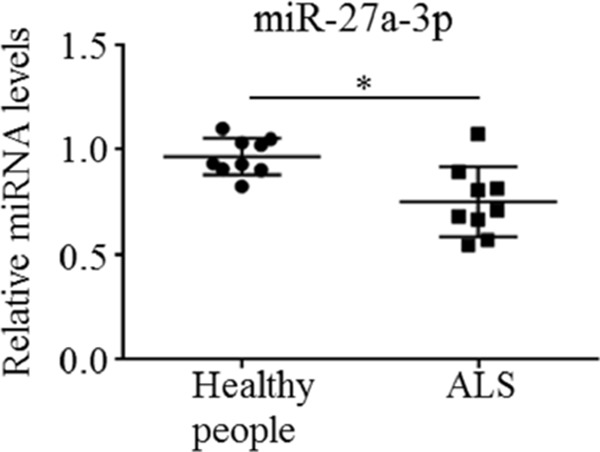
Comparing the difference in expression of miR-27a-3p between ALS patients and healthy subjects. Using stem loop method to detected the miR-27a-3p in 10 ALS patients and healthy subject's serum exosomes, respectively. Compared with the control group, the expression of miR-27a-3p in the ALS group was down-regulated, and the difference was statistically significant (p < 0.05).
4. Discussion
Exosomes are a vesicular structure secreted by a variety of cells, and has become a hotspot issue in the field of biomedical research. It has the characteristics of easy to collect and noninvasive or minimally invasive. Noninvasive detection can be realized through the identification of tumor exosomes, or detection of individual biological characteristics used for individualized diagnosis and treatment (7,8). It has been found that exosomes can reflect the phenotype of the source of tumor cells through their molecular characteristics, and the tumor specific antigen or miRNA carried by them can be used as tumor diagnostic markers. Thompson et al. studies have shown that the exosome source of the central nervous system is related to neurodegenerative diseases, and the exosomes in cerebrospinal fluid and serum is an early diagnostic marker for some neurodegenerative diseases by carrying proteins and nucleic acids (9). According to blood tests, researchers from Irish University found that miRNA imbalance appeared in the exosomes of breast cancer, so breast cancer cells can be monitored through miRNA changes in the exosome, and it has proved that exosome miRNA has potential as a marker for breast cancer diagnosis (10). All these suggested that miRNA has the potential to be used as a tumor marker for early diagnosis, evaluation of curative effect, and metastasis or recurrence of the tumor.
The extraction of exosomes by overspeed centrifugation or sucrose gradient centrifugation are the most common methods (11). However, the operation is time-consuming, has a low recovery rate, has an important influence on exosomes, and it is not suitable for extracting sample volumes from a small initial volume. However, Exoquick reagent and Qiagen exoEasy Maxi Kit have obtained exosomes by compound polymerization precipitation or membrane affinity centrifugation columns. The operation is simple and fast, and it is suitable for extraction of serum exosomes (12,13).
In isolation of serum derived exosome RNA, two different principles including traditional TRIzol and miRNeasy Mini Kits were used in this study. The isolation of RNA by TRIzol is a method of splitting decomposition of phenol lysate, as chloroform makes a two-phase separation with a final alcohol precipitation. MiRNeasy Mini Kit was adopted for the phenol type lysate combined with an affinity filtration column, use of a membrane for particle adsorption, and the purified RNA was obtained by different buffer multiple purification.
The current methods of qRT-PCR mainly include a TaqMan probe method and fluorescent dye method. The TaqMan probe method specificity is strong, but the cost is high. Therefore, in this experiment, the qRT-PCR was carried out by combining the poly A tail method or the stem loop method with the fluorescent dye. The polyA tail method was added to miRNA under the action of poly A polymerase, the template chain was obtained by reverse transcription through specific primers, and the fluorescence quantitative PCR was detected at last. Stem loop method used primers with a folded structure like stem loop, directly complementary miRNA to get reverse transcriptional template chains (14,15). Experiments have shown that the stem loop method has high sensitivity, specificity and less sample demand for miRNA. It can effectively detect the expression of miRNA in the samples, and laid a foundation for the next step to effectively detect the expression of miRNA in ALS patients.
As a fatal neurodegenerative disease, ALS seriously influences and endangers the quality of daily life of middle-aged and elderly people. The specific pathogenesis is complex. Now, more detailed studies are being carried out in order to find out the related mechanisms of skeletal degenerative diseases, so as to develop new preventive strategies and therapies. Our previous study found that myoblast exosomes can stimulate mineralization of osteoblasts through miR-27a-3p regulation of the Wnt signaling pathway, which proves that exosomes can regulate the interaction between myoblasts and osteoblasts through miR-27a-3p, and there may be a positive feedback mechanism.
So in this experiment, we compared the expression of miR-27a-3p in patients with ALS and healthy human serum exosomes, found compared with the healthy group, miR-27a-3p expression in ALS patients was down-regulated, and suggested that exosome miR-27a-3p may act as a detection index of ALS, but the specific mechanism is unclear and needs further study.
Acknowledgements
This study was supported by National Natural Science Foundation of China (81772300, 81371909) and The Innovation Project of Shandong Academy of Medical Sciences.
References
- 1. Worms PM. The epidemiology of motor neuron diseases: a review of recent studies. J Neurol Sci. 2001; 191:3-9. [DOI] [PubMed] [Google Scholar]
- 2. Tkach M, Théry C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell. 2016; 164:1226-1232. [DOI] [PubMed] [Google Scholar]
- 3. Bang C, Thum T. Exosomes: new players in cell-cell communication. Int J Biochem Cell Biol. 2012; 44:2060-2064. [DOI] [PubMed] [Google Scholar]
- 4. Koga Y, Yasunaga M, Moriya Y, Akasu T, Fujita S, Yamamoto S, Matsumura Y. Exosome can prevent RNase from degrading microRNA in feces. J Gastrointest Oncol. 2011; 2:215-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007; 9:654-659. [DOI] [PubMed] [Google Scholar]
- 6. Khatun Z, Bhat A, Sharma S, Sharma A. Elucidating diversity of exosomes: biophysical and molecular characterization methods. Nanomedicine. 2016; 11:2359- 2377. [DOI] [PubMed] [Google Scholar]
- 7. Tan KH, Tan SS, Sze SK, Lee WK, Ng MJ, Lim SK. Plasma biomarker discovery in preeclampsia using a novel differential isolation technology for circulating extracellular vesicles. Am J Obstet Gynecol. 2014; 211:380.. [DOI] [PubMed] [Google Scholar]
- 8. Liu WH, Ren LN, Wang X, Wang T, Zhang N, Gao Y, Luo H, Navarro-Alvarez N, Tang LJ. Combination of exosomes and circulating microRNAs may serve as a promising tumor marker complementary to alpha-fetoprotein for early-stage hepatocellular carcinoma diagnosis in rats. J Cancer Res Clin Oncol. 2015; 141:1767-1778. [DOI] [PubMed] [Google Scholar]
- 9. Thompson AG, Gray E, Heman-Ackah SM, Mäger I, Talbot K, Andaloussi SE, Wood MJ, Turner MR. Extracellular vesicles in neurodegenerative disease - pathogenesis to biomarkers. Nat Rev Neurol. 2016; 12:346-357. [DOI] [PubMed] [Google Scholar]
- 10. Joyce DP, Kerin MJ, Dwyer RM. Exosome-encapsulated microRNAs as circulating biomarkers for breast cancer. Int J Cancer. 2016; 139:1443-1448. [DOI] [PubMed] [Google Scholar]
- 11. Théry C, Amigorena S, Raposo G, Clayton A. Isolation and Characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006; Unit 3.22. [DOI] [PubMed] [Google Scholar]
- 12. Cui Y, Luan J, Li H, Zhou X, Han J. Exosomes derived from mineralizing osteoblasts promote ST2 cell osteogenic differentiation by alteration of microRNA expression. FEBS Lett. 2016; 590:185-192. [DOI] [PubMed] [Google Scholar]
- 13. Enderle D, Spiel A, Coticchia CM, Berghoff E, Mueller R, Schlumpberger M, Sprenger-Haussels M, Shaffer JM, Lader E, Skog J, Noerholm M. Characterization of RNA from Exosomes and Other Extracellular Vesicles Isolated by a Novel Spin Column-Based Method. PLoS One. 2015; 10:e0136133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu RM, Wood M, Thrush A, Walton EF, Varkonyi-Gasic E. Real-time PCR quantification of plant miRNA using universal probeLibrary technology. Biochemica. 2007; 202:12-15. [Google Scholar]
- 15. Wu Y, Xing X, You T, Liang R, Liu J. RT-qPCR with chimeric dU stem-loop primer is efficient for the detection of bacterial small RNAs. Appl Microbiol Biotechnol. 2017; 101:4561-4568. [DOI] [PubMed] [Google Scholar]


