Abstract
The gastrointestinal (GI) tract harbors a diverse and host-specific gut microbial community. Whereas host-microbe interactions are based on homeostasis and mutualism, the microbiome also contributes to disease development. In this review, we summarize recent findings connecting the GI microbiome with GI disease. Starting with a description of biochemical factors shaping microbial compositions in each gut segment along the longitudinal axis, improved histological techniques enabling high resolution visualization of the spatial microbiome structure are highlighted. Subsequently, inflammatory and neoplastic diseases of the esophagus, stomach, and small and large intestines are discussed and the respective changes in microbiome compositions summarized. Finally, approaches aiming to restore disturbed microbiome compositions thereby promoting health are discussed.
Keywords: Gut microbiome, Spatial microbiome organization, Dysbiosis, Inflammation, Carcinogenesis, Fecal microbiome transplantation
Introduction
All body surfaces being in contact with the environment, like the skin and the gastrointestinal (GI), urogenital, and respiratory tracts are colonized by microorganisms. These microbial consortia, collectively termed “microbiome” or “microbiota,” are now viewed as integral part of our body, being essential for proper organ function. Thus humans are considered holobionts composed of not only “own” (eukaryotic) cells but also microbial cells and understanding the mechanisms underlying health and disease development needs to encounter the microbial part of our body too [1, 2]. Different types of microorganisms are part of the human microbiome, including bacteria (prokaryotes), archea, fungi, protists, and virus. Dependent on the habitat, the composition of the microbiome differs significantly [3]. For instance, the gut microbiota is mainly populated by bacteria (> 99% [4]), whereas the skin harbors also significant amounts of fungi (≈ 10% [5]). The vast majority of commensal microorganisms reside in the colon, with estimates of up to 1013 bacteria followed by the skin, which harbors about 1012 bacteria. Thus, the collective microbiome colonizing our body (approx. 1013 microbial cells) outnumbers our own nucleated body cells (approx. 1012) by the factor of 10, which gives already an estimate of the biological potential of our “second genome” [4, 6].
The biological functions conferred by the microbiome are manifold. The gut microbiome is a major factor involved in metabolism and energy regulation [7]. Up to 10% of our daily consumed calories are provided by the gut microbiota partly via degradation of complex (plant-derived) polysaccharides into short-chain fatty acids (SCFAs; e.g., butyrate), a process called fermentation. Thus, the gut microbiome is a major factor contributing to obesity and its sequels like type II diabetes mellitus [8–10]. Another prominent feature of the microbiome is the education of the immune system. The mucosal immune system needs to tolerate the resident microbiome, whereas it needs to react against pathogens. This homeostasis is achieved by an intricate interplay of the microbiome and the host [11]. Especially the induction of tolerance via induction of anti-inflammatory cells and cytokines (e.g., regulatory T cells, IL-10, TGFβ) is an important trait of the microbiota, conferred by special microbial products directly interacting with the host’s immune system [12–15]. In addition, it has been recognized that CD4+ T cell responses are directed and modulated by specific commensals towards a T helper (Th) cell 1 or Th17 immune reaction, which has major implications not only in mucosal defense but also in autoimmune and autoinflammatory processes beyond the GI tract [16].
Moreover, physiologically colonized body surfaces are intrinsically protected from colonization with pathogens, a highly effective defense mechanism called pathogen exclusion or colonization resistance [17]. Various mechanisms have been described in this context. So it has been shown that a low-fiber diet quickly shapes the structural composition of the microbiome promoting the expansion of a mucus-degrading microbiota. This renders mice more susceptible to colitis elicited by certain intestinal pathogens [18]. On the other hand, a fiber-rich diet reduces the numbers of mucus-degrading commensals and promotes the bloom of fiber-degrading SCFA-producing bacteria. SCFA then support mucosal barrier functions through distinct mechanisms impacting on oxygen consumption by intestinal epithelial cells [19], modulating the threshold of intracellular danger receptors such as inflammasomes [20], or shifting naïve T helper cells towards regulatory T cells [13]. Other mechanisms include less-well investigated mechanisms of microbe-microbe interactions. So it has been shown that by producing iron-binding siderophores certain pathobionts and pathogens acquire a growth advantage during colitis when iron is scarce [21]. Certain protective commensals harness this circumstance by coupling these siderophores with antimicrobial microcins, which then enter and target pathobionts as “Trojan horses” through siderophore-receptor based uptake [22].
Factors shaping the spatial organization of the human gut microbiome
Babies are born sterile. During birth, the body becomes immediately colonized by microbes from the surroundings, which is the main determinant shaping microbiome composition early in life [23]. Consequently, babies born naturally acquire different microbes like Lactobacillus and Prevotella resembling the mother’s vaginal microbiota, whereas babies born via Cesarian section are dominated by “skin”-type bacteria like Staphylococcus, Corynebacterium, and Propionibacterium [24]. Interestingly, differences in early colonization are supposed to contribute also to different susceptibilities to immune-mediated diseases, like asthma and allergies, later in life [25]. The first year of life is signified by an increased variability of the microbiome, which “stabilizes” when adult diet is introduced after weaning [26]. At this time-point (about 1 year of age), the infant microbiome resembles largely an adult microbiome.
The structure of the human microbiome is mainly determined by environmental factors like diet. Consequently, relatives or individuals living in the same household and having the same living habits share more microbes than unrelated individuals [27]. Overall, the gut microbiome appears to be quite stable over time for years, possibly life-long [26]. In addition, there seems to be also a (small) heritable component determining GI microbiome structure [28–30]. For instance, several genotyping studies correlating host genotype with gut microbiome composition have revealed a genetic association of the human lactase gene locus with Bifidobacterium abundance providing evidence of a gene-diet-gut microbiome interaction and giving new clues about pathogenesis of lactose intolerance [31, 32].
The human GI tract could be simply seen as a tube with an input and output. A constant flux of microbes originating from the environment, diet, and the oral cavity exists which potentially facilitates entry of foreign and potentially harmful microbes. Nevertheless, a quite specific and stable microbiota is maintained in each gut segment under healthy conditions. Compositions and densities of the gut microbiome along the GI tract are mainly governed by biochemical factors like pH, oxygen, antimicrobial peptide (AMP) gradients, and presence of bile acids, as well as the speed of transit [33]. As indicated in Fig. 1, pH is lowest in the stomach, gradually increases towards the terminal ileum, decreases in the cecum, and again gradually increases towards the distal colon. An oxygen gradient exists along the length of the gut, with levels highest in the upper GI tract which decrease to anaerobic conditions in the distal colon. However, there is also a radial oxygen gradient in the colon, with anoxic conditions in the lumen and a slight increase in oxygen tension towards the mucosa, which can be consumed by microorganisms living in proximity to the mucosa, also from enteropathogens [35]. Each gut segment harbors a unique repertoire of AMPs, specifically suppressing certain groups of bacteria. Saliva contains high amounts of lysozyme efficiently degrading the murein wall of microbes and also the stomach epithelium is able to produce AMPs [36]. In the small intestine, Paneth cells secrete AMPs like α-defensins, C-type lectins, lysozyme, and phospholipase A2. In the large intestine, enterocytes secrete AMPs like β-defensins, C-type lectins, cathelicidins, galectins, and lipocalin [37]. Interestingly, the microbiome of the large intestine encodes genes providing resistance to specific AMPs conferring resilience to the microbiome during inflammation, when AMP levels are high, allowing faster recovery of a “healthy” microbiome after infection [38].
Fig. 1.
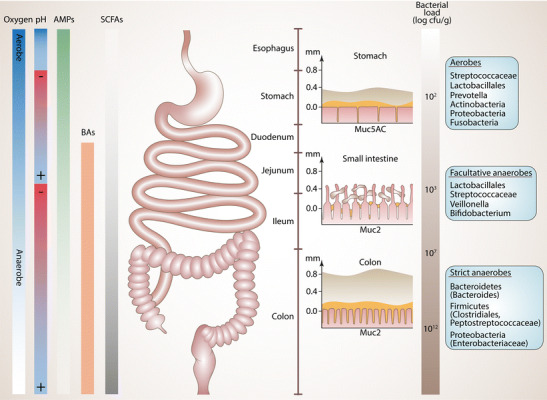
Biogeography and factors shaping the spatial organization of the gut microbiome. Left: factors determining gut segment specific microbiome composition like oxygen, pH, bile acids (BA), antimicrobial peptides (AMPs), and concentration of short-chain fatty acids (SCFAs). Middle: schematic representation of the GI tract and of the segment specific mucus layer architecture (adapted from [33, 34]). The inner solid (amber) and the outer loose mucus layer (gray) are shown. Note that in the stomach and colon the mucus layer is continuous, whereas in the small intestine the layer is discontinuous. Muc5AC and Muc2 denote the dominant mucins produced in the respective gut segment. Right: bacterial load and typical taxonomic compositions of different gut segments
The microbiota is spatially organized along the transverse axis of the GI tract, from the lumen to the mucosa. A major factor driving this transverse organization is the mucus layer covering the GI tract. Mucins are gel-forming glycoproteins, polymerizing into a mesh like structure. There are two discriminable mucus layers in the stomach and colon, an outer “loose” layer which is densely populated by bacteria and an inner “solid” layer, which is enriched in innate and adaptive immune effectors providing a biochemical barrier that segregates the microbiota from direct contact with the epithelium [34, 39]. This inner layer is nearly free of microbes. However, specific microbes like the mucin utilizer Akkermansia muciniphila are able to degrade the layer thus reaching inner areas [40, 41], in addition to various pathogens, like Helicobacter pylori in the stomach or enteropathogenic Escherichia coli, Salmonella, Yersinia, and Campylobacter in the colon. In the small intestine, the mucus layer is discontinuous and less defined; the tips of the villi often not covered by mucin (Fig. 1). Importantly, routine histological preparations do not preserve mucus layer architecture. The mucus layer is heavily hydrated and dehydration, which happens during conventional fixation with formaldehyde, shrinks the mucus layer leading to a very thin film lining the epithelium. Techniques for improved mucus preservation employ fixation of tissue with chloroform, dry methanol, and glacial acetic acid (i.e., Carnoy’s fixative), and processing in water-free solutions before embedding in paraffin, which preserves mucus layer architecture [34, 39, 42]. Moreover, this fixation method allows also for simultaneous staining of bacterial RNA/DNA by fluorescence in situ hybridization (FISH). Application of this technique allows for a high-resolution analysis of gut-microbiome interactions as exemplified in Fig. 2, wherein a mouse colon fixed in Carnoy’s solution was used to track a specific gut pathobiont (Alistipes finegoldii) in the GI tract in situ.
Fig. 2.
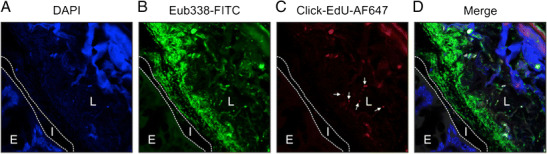
In vivo tracking and improved spatial resolution of gut-microbiome interactions. A mouse colon was fixed in Carnoy’s solution to preserve mucus layer architecture. a The section was counterstained with 4′,6-diamidino-2-phenylindole (DAPI) indicating the colonic epithelium (E) in the left lower corner, the interlaced (I) mucus layer (indicated by the two dotted lines), which is devoid of bacteria, and the colonic lumen (L) on the right side. The structure in the upper right depicts a plant component. b Bacteria were stained by FISH using a fluorescein isothiocyanate (FITC) pan-specific EUB338 probe (green) which covers approximately 90% of the domain bacteria. c In this experiment, mice were gavaged with the gut bacterium Alistipes finegoldii (phylum Bacteroidetes), which were cultured in the presence of the thymidine analog 5-ethynyl-2′-deoxyuridine (EdU). Metabolically active Alistipes was tracked utilizing click chemistry that is based on a copper-catalyzed covalent reaction between an alkyne (within the EdU) and an AlexaFluor® 647-containing azide. Note that the bacterium colonizes the luminal site of the colon, not the mucus layer (arrows). d The right picture shows the merged panels. Cells were imaged on a Zeiss Axioobserver Z1 microscope equipped with a LSM700 confocal unit. Original magnification 400×
The total number of microbes increases from the esophagus to the distal colon, where the microbial load is estimated to be 1012 microbes per gram of feces. The acidic pH and oxygenated environment of the upper GI tract limit microbial colonization to acid- and oxygen-tolerant bacteria (e.g., Lactobacillus, Streptococcus, Veillonella), whereas in the large intestine the flow is slower, and metabolism favors fermentation of complex plant-derived polysaccharides (e.g., fiber) or from host mucus. This results in greater species richness (i.e., number of prevalent taxa), higher microbial densities and dominance of the saccharolytic Bacteroidales and Clostridiales in the large intestine [39].
Dysbiosis—chicken and egg
It is important to note that many studies investigating the microbiome in the disease context describe associations and the reported shifts in microbiome composition (termed “dysbiosis”) are often not proven causal for the respective disease and could just represent epiphenomena, wherein changed habitat factors (e.g., by change of substances produced by the host cells during disease) lead to altered microbial community compositions [43]. However, changed habitat factors and subsequent dysbiosis could contribute to disease as well, especially if these alterations are persistent. Paradigmatic in this context are epithelial tumors of microbially colonized organs like colorectal cancer (CRC) and its precursors (adenomas). Recently, it has been shown that the neoplastic colon epithelium overexpresses a polysaccharide, D-galactose-b(1-3)-N-acetyl-D-galactosamine (Gal-GalNAc), which is selectively bound by a Fusobacterium nucleatum lectin (Fap2) enriching this protumorigenic microbe in CRC tissue [44]. Another impressive example for a direct contribution of the microbiome to disease pathogenesis is exemplified by approaches aiming in restoration of dysbiosis, like bacteriotherapy or fecal microbiome transplantation (FMT). This approach is already regularly used to treat recurrent Clostridium difficile (pseudomembranous) colitis a disease caused by antibiotic-induced dysbiosis and subsequent pathogen overgrowth [45, 46]. Importantly, the restoration of dysbiosis by FMT shows also efficacy in chronic inflammatory GI diseases like ulcerative colitis and even in metabolic diseases like in individuals with metabolic syndrome [47, 48].
Microbiome and inflammatory diseases of the upper GI tract
Esophagus and gastro-esophageal junction
The composition of the esophageal microbiome is heavily influenced by microbes originating from the oral cavity, dominated by taxa like Streptococcus followed by Prevotella, Veillonella, and Fusobacterium, which represent the healthy esophageal core microbiome [49–51]. Chronic exposure of the distal esophagus to gastric acid and duodenal bile salts is thought to be a major factor underlying the pathogenesis of gastro-esophageal reflux disease (GERD), Barrett’s esophagus (BE), and subsequently adenocarcinoma of the gastro-esophageal (GE) junction. As discussed above, changing habitat factors lead also to altered microbiome compositions, which could in turn fuel inflammation and tumorigenesis. Certain studies noted significant taxonomic changes in the esophageal microbiome in GERD, BE, and adenocarcinoma of the GE junction. Dominant taxa like the Gram-positive Streptococcus are depleted, whereas Gram-negative taxa like Veillonella, Prevotella, Campylobacter, Fusobacterium, Haemophilus, and Neisseria are enriched in the diseased states [52, 53]. Notably, the oral taxons Campylobacter concisus and C. rectus have been found enriched in the mucosa of GERD and BE but were depleted in adenocarcinoma of the GE junction [54, 55]. Interestingly, C. concisus seems to be adapted to the harsh (acidic) environment of the upper GI tract. Significantly increased RNA transcripts of C. concisus compared to other stomach microbes were detected in the human gastric juice [56]. These taxa are also increased in individuals with IBD, especially children with Crohn’s disease [57]. In addition, also Fusobacterium nucleatum, a Gram-negative filiform bacterium normally inhabiting the human oral cavity (dental plaque), is overabundant in Crohn’s disease, in addition to its association with adenomas and CRC [58]. Importantly, several molecular studies have shown a proinflammatory and protumorigenic behavior of F. nucleatum, wherein the bacterium was shown to specifically activate epithelial cell proliferation, to induce a protumorigenic immune-microenvironment and inhibits immunological tumor surveillance [59–61].
Another prominent disease of the esophagus is represented by eosinophilic esophagitis (EoE). Although the primary cause of EoE is thought to be a non-IgE-mediated food hypersensitivity [62], also increased levels of Gram-negative bacteria (Neisseria, Corynebacterium, Haemophilus) were reported [63, 64]. If these microbiome changes also contribute to disease pathogenesis needs to be clarified. Recent reviews have summarized the majority of existing studies investigating microbiome compositions in diseases of the upper GI tract [49–51].
Stomach
Helicobacter pylori is a bacterium purely adapted to the human host. This restricted host spectrum has led to a coevolution of the bacterium with humans [65] and has shaped the molecular determinants of host-pathogen interaction manifold [66]. Noteworthy, the intimate relationship between the bacterium and humans has led also to beneficial effects of H. pylori infection aside of the clear pathogenic effects leading to chronic gastritis, ulcers, and subsequently gastric adenocarcinoma and MALT lymphoma [67, 68]. Noteworthy, it has for instance been shown that early life-time infection with H. pylori lowers significantly the risk of developing asthma and celiac disease later in life [69, 70]. Many of this beneficial traits are induced by immune system modulation of H. pylori, due to the induction of a tolerogenic immune-state (e.g., induction of regulatory T cells), which helps the bacterium to persist in the human host. Therefore, H. pylori represents a paradigm “pathobiont,” a term which specifies bacteria with a commensal and pathogenic lifestyle that is determined not just exclusively by bacterial traits but also by host (e.g., age, concomitant microbiome) or environmental factors [71]. The view of a commensal lifestyle of H. pylori is also supported by reports showing low level colonization of asymptomatic individuals [72–76]. Under disease conditions, H. pylori is the dominant stomach bacterium outcompeting the normal resident microbiome and its preferred niche is the gastric mucosal surface (Fig. 3). Interestingly, H. pylori infection also significantly impacts lower gut microbiome composition [77, 78]. That pathogenicity of H. pylori is also determined by non-H. pylori factors is supported by the finding that the concomitant gastric microbiota is also important to drive gastric tumorigenesis. Interestingly, distinct sequential changes in microbiome compositions occur along the gastric metaplasia-dysplasia development [79–81]. In analogy to adenocarcinoma of the GE junction also bacteria originating from the oral cavity seem to be involved in tumorigenesis of the H. pylori-infected stomach [82].
Fig. 3.
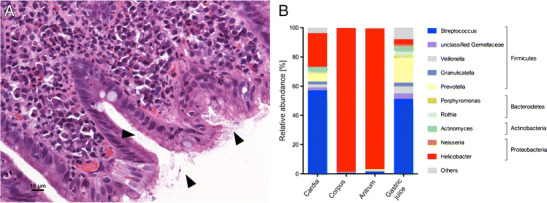
Stomach microbiome in chronic H. pylori gastritis. a Gastric corpus biopsy signifying the preferred mucosal niche of H. pylori (arrow heads). b Microbiome analysis (based on the 16S rRNA gene) indicates the dominance of H. pylori at the mucosal sites, whereas in gastric juice only few H. pylori are prevalent
Another form of chronic gastritis is lymphocytic gastritis (LyG), characterized by increased CD8+ intraepithelial lymphocytes (IELs; ≥ 25 per 100 epithelial cells). In addition to the association with celiac diseases (CeD), a great proportion of LyG has unclarified causes. H. pylori is normally absent in LyG; however, eradication therapy seems to be an effective treatment of LyG, even in the absence of identifiable H. pylori, suggesting an alternative bacterial cause for the disease. We recently identified Propionibacterium acnes as a possible LyG disease trigger inducing the natural killer group 2 member D (NKG2D) system and the proinflammatory cytokine interleukin (IL)-15 in the gastric mucosa [76]. Natural killer (NK) cells, CD8+ T cells, and certain other T cells express the NKG2D receptor. The NKG2D receptor ligands (NKG2DLs) are expressed mainly on epithelial cells at low levels under physiological conditions, but their expression is induced under conditions of cell stress, such as infection, neoplastic transformation, or challenge with specific metabolites (e.g., short-chain fatty acids, SCFAs) or gliadin in the case of CeD. Upon ligand-receptor interaction, NKG2D triggers a cytotoxic response in the receptor-bearing lymphocyte, eliminating the stressed cell that is overexpressing the ligand. This reaction is enhanced by the presence of IL-15 [83–86]. Of note, the NKG2D-NKG2DL system and IL-15 are important for immunological tumor surveillance, which is necessary for the elimination of neoplastic cells. The system has therefore been investigated as a potent target for cancer immunotherapy [87, 88].
Small intestine
In CeD, the NKG2D system is critical for recruitment of CD8+ IELs and subsequent villus atrophy in the duodenum [83, 84]. In addition to the genetic causes (i.e., HLA-DQ), also environmental factors play a role in the development of CeD, including the microbiome [89]. Phenomena like the so-called Swedish CeD epidemic, wherein the incidence of CeD fourfold increased in children within a short period followed by a rapid drop, resembles an infectious disease (i.e., “out-break pattern”) [90]. Recently, it was shown that gut bacteria are able to differentially degrade gluten giving a possible explanation for these phenomena. Specifically, overgrowth of the opportunistic pathogen Pseudomonas aeruginosa was reported in CeD, which is able to produce a specific elastase (lasB). This enzyme degrades gluten in a specific manner enabling the released peptides to better translocate the intestinal barrier, which subsequently leads to the activation of gluten-specific T cells driving the disease [91].
Another inflammatory disease often prevalent in the small intestine, wherein the microbiome plays a role, is graft versus host disease (GvHD). GvHD is caused by the alloactivation of T cells, which recognize host antigens as foreign, causing autoimmune attack to organs such as the GI tract, lungs, liver, and skin [92]. A microbial factor contributing to disease development was already suspected in the 1970s when it was demonstrated that mice kept under germ-free conditions developed less GI GvHD [93, 94]. Although the underlying molecular mechanisms of the contribution of the microbiome to GvHD development are largely unknown, depletion of the resident microbiome, due to the intensive antibiotic and chemotherapeutic treatment regimens, seems to play a pivotal role. Notably, the magnitude of intestinal diversity loss is a risk factor for treatment-related mortality including death from GvHD [95]. The intestinal diversity loss (e.g., depletion of specific Clostridia) leads to impaired microbial fermentation and a lack of SCFAs (e.g., butyrate), the main energy source of gut epithelia. SCFA deprivation has been shown to induce apoptosis in intestinal epithelial cells, the hallmark histological change observed in GvHD [96]. Moreover, a common dysbiotic fecal microbiome signature in GvHD was reported recently, specified by an (over-)abundance of Enterococcus species (E. faecium, E. faecalis). This microbiome type is significantly associated with the risk to develop gut GvHD after hematopoietic stem cell transplantation [97]. Interestingly, treatment strategies which restore a physiological gut microbiome (e.g., FMT) appear to be beneficial in patients with chronic active GvHD [92, 98].
Microbiome and inflammatory diseases of the lower GI tract
Antibiotic-associated colitis
Colitis is a frequent side effect of antibiotic therapy [99]. Besides direct drug-induced toxicity of antibiotics, depletion of the gut microbiome and subsequent pathogen overgrowth are major disease causes, like in Clostridium difficile colitis (CDC [45]). In CDC, the bile acid 7α-dehydroxylating intestinal bacterium Clostridium scidens seems to be depleted which leads to a lack of suppression of C. difficile [100]. Noteworthy, FMT is already an established highly effective treatment for recurrent CDC, indicating the potential of therapies aiming to restore an altered microbiome [46]. Of note, antibiotic-associated colitis could be caused also by other pathogens, like Klebsiella oxytoca, the causative agent of antibiotic-associated hemorrhagic colitis (AAHC). AAHC is usually observed after therapy with penicillins and represents as segmental, often patchy hemorrhagic colitis, typically dominant in the right colon [101, 102]. In AAHC, overgrowing K. oxytoca intrinsically resistant to beta-lactams and producing the enterotoxin tilivalline lead to intestinal epithelial apoptosis and colitis (Fig. 4). In extreme forms of antibiotic-associated colitis microbiome depletion can lead to disease courses resembling severe acute GvHD. We described recently a series of severe apoptotic enterocolitis cases emerging after therapy with antibiotics and steroids, wherein severe microbiome depletion seemed to trigger the disease. Notably, FMT performed in one case restored a normal gut microbiome and was highly effective to dampen epithelial cell death and enterocolitis [103].
Fig. 4.
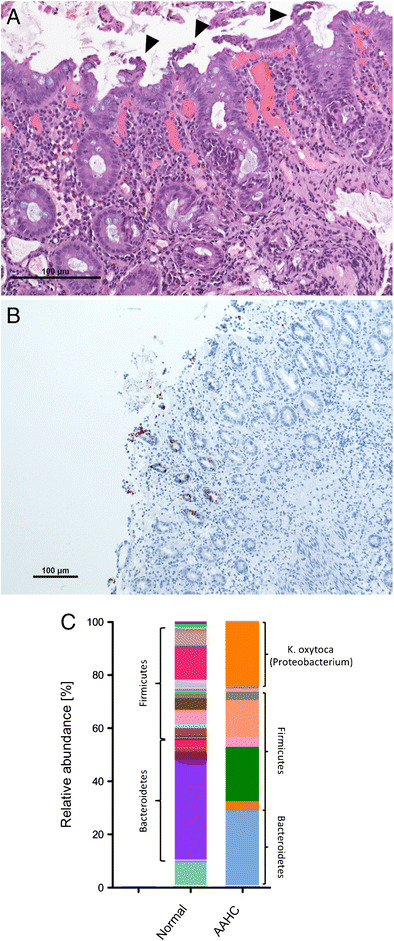
Histology and microbiome representation of antibiotic-associated hemorrhagic colitis (AAHC). a Colon histology with micropapillary epithelial protrusions (arrow heads) indicating the cytotoxic effect of the enterotoxin tilivalline produced by K. oxytoca. b Activated caspase-3 immunohistochemistry signifying epithelial cell apoptosis. c Fecal microbiome composition in AAHC (based on the 16S rRNA gene analysis). A highly reduced overall diversity is evident with the overgrowth of the proteobacterium K. oxytoca. A diverse healthy fecal microbiome is shown on the left
Inflammatory bowel diseases
Crohn’s disease and ulcerative colitis represent the two major clinically defined forms of inflammatory bowel diseases (IBD). Our current mechanistic understanding puts the intestinal epithelial cell as the central orchestrator of the innate immune system into the limelight, integrating genetically based interactions between the intestinal microbiome, the mucosal immune system, and environmental factors [104]. A genetic component in IBD has been postulated early in the process of identifying and understanding disease etiology with evidence from early epidemiological [105, 106] and twin studies [107]. Recent genome-wide association studies (GWAS) identified more than 200 IBD susceptibility loci, with most of them conferring modest disease risk in terms of odds-ratios [108]. Nevertheless GWAS resulted in new insights in IBD biology revealing a substantial overlap between the genetic risks of CD and UC and between other autoimmune and autoinflammatory diseases [109, 110]. Furthermore, these techniques pointed out previously unappreciated pathways in IBD, ahead of autophagy [111].
Nevertheless, the incidence of both Crohn’s disease and ulcerative colitis is increasing dramatically worldwide, which is of course hardly explained by changes in the genetic landscape [112]. IBD was among the first diseases in which the microbiome has been studied intensively. These studies identified not only significant compositional alterations of the microbiome with reduction of potentially-protective commensals such as Faecalibacterium prausnitzii [113] but also a less stable microbiome, an increased adherence of microbes to the epithelial surface, however, no clear signature with specific pathobionts. Of note, many of the early studies included patients under medication which potentially influenced some of the results. More recently, Gevers and colleagues studied the treatment-naïve microbiome including 447 children and adolescents with newly diagnosed Crohn’s disease and 221 controls. They identified a strong correlation of disease with increased abundance of Enterobacteriaceae, Pasteurellacaea, Veillonellaceae, and Fusobacteriaceae and a decreased abundance of Erysipelotrichales, Bacteroidales, and Clostridiales [58]. This study included several other interesting aspects showing that composition of the gut microbiome may be predictive for an individual disease course and that in early-stage and lower grade inflammation the mucosa-associated microbiome, e.g., from rectal biopsies, may be superior to fecal microbiome analysis. Noteworthy, more recent studies implied relevant alterations in the structural composition of the gut “virome” in IBD [114], and two recent clinical trials showed that blocking IL-17A with secukinumab or IL-17RA with brodalumab worsened Crohn’s disease, with some patients developing mucocutaneous candidiasis ventilating a role for the gut mycobiome in IBD, which has been implicated in intestinal inflammation as well [115, 116].
Are aberrant immune responses to the intestinal microbiome indeed causally related to IBD? Evidence from clinical studies showed that mucosal inflammation recurs quickly after reinfusion of ileal content from a protective proximal loop ileostomy, which certainly proved that the “enemy” lies within the “fecal stream” [117]. Various experimental models implicated that an altered dysbiotic microbiota causes transmissible disease, both in the ileum and the colon. Schaubeck and coworkers demonstrated that only a certain proportion of TNFΔARE mice, which overproduce TNFα and develop spontaneous ileitis, will develop high-grade histological inflammation, while others show no inflammation at all, despite identical genetic background and a comparable environment. Using a reductionist approach, they transplanted the stool from inflamed and non-inflamed TNFΔARE mice into germ-free animals and demonstrated that the microbiome was indeed the driving force for inflammation in this model [118]. Our own studies indicated a transplantable dysbiotic microbiome in mice double-deficient of IL-10 and lipocalin 2 (Lcn2) driving colonic inflammation. In cross-foster experiments, IL-10 pups raised by double-deficient nursing mothers developed the same phenotype irrespectively of their own genotype [119].
However, there are additional relevant environmental modulators of the intestinal microbiome. “We are, what we eat” and the axis food-microbiome represents another important player particularly in IBD [120]. From clinical trials, we have learned that an elemental or polymeric diet is highly effective in the treatment of Crohn’s disease in children [121]; however, the underlying mechanisms remain elusive as recent studies suggest that an exclusive enteral nutrition further aggravates dysbiosis [122]. Moreover, a western diet, enriched in animal protein and fat, reduced in dietary fiber, and the intake of processed foods has been shown to promote intestinal inflammation through different mechanisms [15, 123, 124].
Colorectal cancer
Colorectal cancer (CRC) represents the third most commonly diagnosed malignancy and the fourth leading cause of cancer-related deaths worldwide. CRC burden is expected to increase by 60% until 2030. CRC is one of the clearest markers of cancer transition and incidence is growing fastest in low- and middle-income countries and is associated with adoption of a western lifestyle [125]. Mechanistically, malignant transformation of intestinal epithelial cells and the development of CRC includes at least three relevant steps namely (i) the induction of oncogenic mutations within the Lgr5+ intestinal stem cells (SC), (ii) an altered beta-catenin/Wnt signaling, and (iii) proinflammatory cascades such as TNFα-NFκB and IL6-STAT3 that catalyze CRC development [126].
CRC has been associated with specific changes in gut microbiome composition [127–129]. Recently, we studied microbial alterations along the adenoma-carcinoma sequence, collecting stools from healthy controls, patients with advanced adenomas and patients with CRC [127]. A metagenome-wide association study (MGWAS) was performed and found, that certain Bacteroides spp. (e.g., B. dorei, B. vulgatus, B. massilensis) and E. coli were associated with systemic inflammation and tumor stage. In line with others, Parvimonas, Bilophila wadsworthia, Fusobacterium nucleatum, and Alistipes spp. were also overrepresented in CRC patients. Importantly, the presence of SCFA-producing and bile acid-metabolizing bacteria was clearly positively related to consumption of meat and negatively related to the intake of fruits and vegetables again indicating the important role of a western lifestyle in development of CRC [127].
There is emerging evidence regarding a causal role of certain bacteria in CRC evolution such as F. nucleatum, colibactin-producing E. coli and toxigenic Bacteroidis fragilis. Interestingly, little is known about bacteria and mechanisms that protect from CRC development. F. nucleatum was one of first bacteria associated with human CRC, shown to be enriched in tumors [59, 130]. Furthermore, F. nucleatum has been strongly associated with certain tumor types, such as microsatellite instable CRC and cancers with BRAF mutations [131, 132]. Mechanistically, it has been shown that the F. nucleatum FadA antigen is a ligand for E-cadherin on intestinal epithelial cells that activates the β-catenin signaling pathway, thereby promoting uncontrolled cell growth, acquisition of a stem cell-like phenotype of epithelia and loss of cell polarity [61]. Also, mucosa-associated E. coli are overrepresented in CRC, which correlates with tumor stage and prognosis [133]. Moreover, some E. coli strains harbor a genomic polyketide synthase (pks) island that encodes for the genotoxin colibactin capable of inducing DNA damage and mutations in epithelial cells [134].
Finally, it is increasingly recognized that the microbiome contributes to the efficacy of cancer therapies. Several recent papers demonstrated convincingly that bacterial nucleotide metabolism genes affect efficacy of 5-fluoruracil and camptothecin antineoplastic therapy [135]. Again, F. nucleatum promoted colorectal cancer resistance to chemotherapy by a complex network of mechanisms including toll-like receptor signaling, microRNAs and induction of autophagy [136]. The intestinal microbiome and associated intestinal immune mechanisms seem particularly relevant for response to checkpoint inhibitors such as anti-PD1 and anti-PDL1 therapies. This was first recognized in experimental models, wherein response to anti-PD-L1 antibodies was associated with the presence of Bifidobacterium and oral administration of Bifidobacterium boosted the efficacy of such therapies [137]. Gopalakrishnan and coworkers recently demonstrated that the efficacy of anti-PD1 therapy was strongly affected by concomitant antibiotic therapy, as well. Mechanistically, the response was strongly dependent on the presence of the mucin degrader A. muciniphila. Response to anti-PD1 therapy in mice that received the stool from non-responders could be restored by administration of A. muciniphila and was dependent on Akkermansia-induced, IL-12-dependent Th1 responses [138].
Conclusion
It is now evident that the human gut microbiome significantly contributes not only to the maintenance of GI health but also to disease development. Recent scientific findings support the view that the gut microbiome might serve as a future diagnostic and therapeutic target for inflammatory and also neoplastic GI diseases.
Acknowledgements
Open access funding provided by Medical University of Graz.
Funding
The work in the laboratory of GG is supported by the Austrian Science Foundation (FWF W1241-B18) and BioTechMed Graz. The work in the laboratory of AM is supported by the Christian Doppler research foundation. The financial support by the Austrian Federal Ministry of Science, Research and Economy and the National Foundation for Research, Technology and Development is gratefully acknowledged.
Compliance with ethical standards
Does not apply.
Conflict of interest
The authors declare that they have no conflicts of interest.
Contributor Information
Gregor Gorkiewicz, Email: gregor.gorkiewicz@medunigraz.at.
Alexander Moschen, Email: alexander.moschen@i-med.ac.at.
References
- 1.Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. 2016;375(24):2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 2.Bordenstein SR, Theis KR. Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol. 2015;13(8):e1002226. doi: 10.1371/journal.pbio.1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, MetaHIT Consortium. Bork P, Ehrlich SD, Wang J. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halwachs B, Madhusudhan N, Krause R, Nilsson RH, Moissl-Eichinger C, Högenauer C, Thallinger GG, Gorkiewicz G. Critical issues in mycobiota analysis. Front Microbiol. 2017;8:180. doi: 10.3389/fmicb.2017.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell. 2016;164(3):337–340. doi: 10.1016/j.cell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 7.Nieuwdorp M, Gilijamse PW, Pai N, Kaplan LM. Role of the microbiome in energy regulation and metabolism. Gastroenterology. 2014;146(6):1525–1533. doi: 10.1053/j.gastro.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 8.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 9.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 10.Sonnenburg JL, Bäckhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535(7610):56–64. doi: 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489(7415):231–241. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453(7195):620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 13.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504(7480):446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 14.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, Xavier RJ, Teixeira MM, Mackay CR. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461(7268):1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341(6145):569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL, Alonzo F, Ng C, Chen A, Lin X, Sczesnak A, Liao JJ, Torres VJ, Jenkins MK, Lafaille JJ, Littman DR. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature. 2014;510(7503):152–156. doi: 10.1038/nature13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawley TD, Walker AW. Intestinal colonization resistance. Immunology. 2013;138(1):1–11. doi: 10.1111/j.1365-2567.2012.03616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, Young VB, Henrissat B, Wilmes P, Stappenbeck TS, Núñez G, Martens EC. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell. 2016;167(5):1339–1353.e21. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ, Wilson KE, Glover LE, Kominsky DJ, Magnuson A, Weir TL, Ehrentraut SF, Pickel C, Kuhn KA, Lanis JM, Nguyen V, Taylor CT, Colgan SP. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe. 2015;17(5):662–671. doi: 10.1016/j.chom.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macia L, Tan J, Vieira AT, Leach K, Stanley D, Luong S, Maruya M, Ian McKenzie C, Hijikata A, Wong C, Binge L, Thorburn AN, Chevalier N, Ang C, Marino E, Robert R, Offermanns S, Teixeira MM, Moore RJ, Flavell RA, Fagarasan S, Mackay CR. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat Commun. 2015;6:6734. doi: 10.1038/ncomms7734. [DOI] [PubMed] [Google Scholar]
- 21.Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio SP, Paixao TA, Butler BP, Chu H, Santos RL, Berger T, Mak TW, Tsolis RM, Bevins CL, Solnick JV, Dandekar S, Bäumler AJ. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe. 2009;5(5):476–486. doi: 10.1016/j.chom.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sassone-Corsi M, Nuccio SP, Liu H, Hernandez D, Vu CT, Takahashi AA, Edwards RA, Raffatellu M. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature. 2016;540(7632):280–283. doi: 10.1038/nature20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, D Lieber A, Wu F, Perez-Perez GI, Chen Y, Schweizer W, Zheng X, Contreras M, Dominguez-Bello MG, Blaser MJ. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016;8(343):343ra82. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107(26):11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dominguez-Bello MG, De Jesus-Laboy KM, Shen N, Cox LM, Amir A, Gonzalez A, Bokulich NA, Song SJ, Hoashi M, Rivera-Vinas JI, Mendez K, Knight R, Clemente JC. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat Med. 2016;22(3):250–253. doi: 10.1038/nm.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, Khan MT, Zhang J, Li J, Xiao L, Al-Aama J, Zhang D, Lee YS, Kotowska D, Colding C, Tremaroli V, Yin Y, Bergman S, Xu X, Madsen L, Kristiansen K, Dahlgren J, Wang J. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17(6):852. doi: 10.1016/j.chom.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 27.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, Spector TD, Clark AG, Ley RE. Human genetics shape the gut microbiome. Cell. 2014;159(4):789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turpin W, Espin-Garcia O, Xu W, Silverberg MS, Kevans D, Smith MI, Guttman DS, Griffiths A, Panaccione R, Otley A, Xu L, Shestopaloff K, Moreno-Hagelsieb G, GEM Project Research Consortium. Paterson AD, Croitoru K. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat Genet. 2016;48(11):1413–1417. doi: 10.1038/ng.3693. [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Thingholm LB, Skiecevičienė J, Rausch P, Kummen M, Hov JR, Degenhardt F, Heinsen FA, Rühlemann MC, Szymczak S, Holm K, Esko T, Sun J, Pricop-Jeckstadt M, Al-Dury S, Bohov P, Bethune J, Sommer F, Ellinghaus D, Berge RK, Hübenthal M, Koch M, Schwarz K, Rimbach G, Hübbe P, Pan WH, Sheibani-Tezerji R, Häsler R, Rosenstiel P, D'Amato M, Cloppenborg-Schmidt K, Künzel S, Laudes M, Marschall HU, Lieb W, Nöthlings U, Karlsen TH, Baines JF, Franke A. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat Genet. 2016;48(11):1396–1406. doi: 10.1038/ng.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, Spector TD, Bell JT, Clark AG, Ley RE. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe. 2016;19(5):731–743. doi: 10.1016/j.chom.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonder MJ, Kurilshikov A, Tigchelaar EF, Mujagic Z, Imhann F, Vila AV, Deelen P, Vatanen T, Schirmer M, Smeekens SP, Zhernakova DV, Jankipersadsing SA, Jaeger M, Oosting M, Cenit MC, Masclee AA, Swertz MA, Li Y, Kumar V, Joosten L, Harmsen H, Weersma RK, Franke L, Hofker MH, Xavier RJ, Jonkers D, Netea MG, Wijmenga C, Fu J, Zhernakova A. The effect of host genetics on the gut microbiome. Nat Genet. 2016;48(11):1407–1412. doi: 10.1038/ng.3663. [DOI] [PubMed] [Google Scholar]
- 33.Wlodarska M, Kostic AD, Xavier RJ. An integrative view of microbiome-host interactions in inflammatory bowel diseases. Cell Host Microbe. 2015;17(5):577–591. doi: 10.1016/j.chom.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johansson ME, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Albenberg L, Esipova TV, Judge CP, Bittinger K, Chen J, Laughlin A, Grunberg S, Baldassano RN, Lewis JD, Li H, Thom SR, Bushman FD, Vinogradov SA, Wu GD. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 2014;147(5):1055–63.e8. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jäger S, Stange EF, Wehkamp J. Antimicrobial peptides in gastrointestinal inflammation. Int J Inflam. 2010;2010:910283–910211. doi: 10.4061/2010/910283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol. 2012;12(7):503–516. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cullen TW, Schofield WB, Barry NA, Putnam EE, Rundell EA, Trent MS, Degnan PH, Booth CJ, Yu H, Goodman AL. Gut microbiota. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science. 2015;347(6218):170–175. doi: 10.1126/science.1260580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tropini C, Earle KA, Huang KC, Sonnenburg JL. The gut microbiome: connecting spatial organization to function. Cell Host Microbe. 2017;21(4):433–442. doi: 10.1016/j.chom.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berry D, Stecher B, Schintlmeister A, Reichert J, Brugiroux S, Wild B, Wanek W, Richter A, Rauch I, Decker T, Loy A, Wagner M. Host-compound foraging by intestinal microbiota revealed by single-cell stable isotope probing. Proc Natl Acad Sci U S A. 2013;110(12):4720–4725. doi: 10.1073/pnas.1219247110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pédron T, Mulet C, Dauga C, Frangeul L, Chervaux C, Grompone G, Sansonetti PJ (2012) A crypt-specific core microbiota resides in the mouse colon. MBio 3(3). 10.1128/mBio.00116-12 [DOI] [PMC free article] [PubMed]
- 42.Matsuo K, Ota H, Akamatsu T, Sugiyama A, Katsuyama T. Histochemistry of the surface mucous gel layer of the human colon. Gut. 1997;40(6):782–789. doi: 10.1136/gut.40.6.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gorkiewicz G, Thallinger GG, Trajanoski S, Lackner S, Stocker G, Hinterleitner T, Gülly C, Högenauer C. Alterations in the colonic microbiota in response to osmotic diarrhea. PLoS One. 2013;8(2):e55817. doi: 10.1371/journal.pone.0055817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abed J, Emgård JE, Zamir G, Faroja M, Almogy G, Grenov A, Sol A, Naor R, Pikarsky E, Atlan KA, Mellul A, Chaushu S, Manson AL, Earl AM, Ou N, Brennan CA, Garrett WS, Bachrach G. Fap2 mediates fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed gal-GalNAc. Cell Host Microbe. 2016;20(2):215–225. doi: 10.1016/j.chom.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leffler DA, Lamont JT. Clostridium difficile infection. N Engl J Med. 2015;372(16):1539–1548. doi: 10.1056/NEJMra1403772. [DOI] [PubMed] [Google Scholar]
- 46.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, Speelman P, Dijkgraaf MG, Keller JJ. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368(5):407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 47.Kump P, Wurm P, Gröchenig HP, Wenzl H, Petritsch W, Halwachs B, Wagner M, Stadlbauer V, Eherer A, Hoffmann KM, Deutschmann A, Reicht G, Reiter L, Slawitsch P, Gorkiewicz G, Högenauer C (2017) The taxonomic composition of the donor intestinal microbiota is a major factor influencing the efficacy of faecal microbiota transplantation in therapy refractory ulcerative colitis. Aliment Pharmacol Ther. 10.1111/apt.14387 [DOI] [PMC free article] [PubMed]
- 48.Vrieze A, Van Nood E, Holleman F, Salojärvi J, Kootte RS, Bartelsman JF, Dallinga-Thie GM, Ackermans MT, Serlie MJ, Oozeer R, Derrien M, Druesne A, Van Hylckama Vlieg JE, Bloks VW, Groen AK, Heilig HG, Zoetendal EG, Stroes ES, de Vos WM, Hoekstra JB, Nieuwdorp M. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143(4):913–6.e7. doi: 10.1053/j.gastro.2012.06.031. [DOI] [PubMed] [Google Scholar]
- 49.Nardone G, Compare D, Rocco A. A microbiota-centric view of diseases of the upper gastrointestinal tract. Lancet Gastroenterol Hepatol. 2017;2(4):298–312. doi: 10.1016/S2468-1253(16)30108-X. [DOI] [PubMed] [Google Scholar]
- 50.Hunt RH, Yaghoobi M. The esophageal and gastric microbiome in health and disease. Gastroenterol Clin N Am. 2017;46(1):121–141. doi: 10.1016/j.gtc.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 51.Di Pilato V, Freschi G, Ringressi MN, Pallecchi L, Rossolini GM, Bechi P. The esophageal microbiota in health and disease. Ann N Y Acad Sci. 2016;1381(1):21–33. doi: 10.1111/nyas.13127. [DOI] [PubMed] [Google Scholar]
- 52.Yang L, Lu X, Nossa CW, Francois F, Peek RM, Pei Z. Inflammation and intestinal metaplasia of the distal esophagus are associated with alterations in the microbiome. Gastroenterology. 2009;137(2):588–597. doi: 10.1053/j.gastro.2009.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu N, Ando T, Ishiguro K, Maeda O, Watanabe O, Funasaka K, Nakamura M, Miyahara R, Ohmiya N, Goto H. Characterization of bacterial biota in the distal esophagus of Japanese patients with reflux esophagitis and Barrett’s esophagus. BMC Infect Dis. 2013;13:130. doi: 10.1186/1471-2334-13-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blackett KL, Siddhi SS, Cleary S, Steed H, Miller MH, Macfarlane S, Macfarlane GT, Dillon JF. Oesophageal bacterial biofilm changes in gastro-oesophageal reflux disease, Barrett’s and oesophageal carcinoma: association or causality? Aliment Pharmacol Ther. 2013;37(11):1084–1092. doi: 10.1111/apt.12317. [DOI] [PubMed] [Google Scholar]
- 55.Macfarlane S, Furrie E, Macfarlane GT, Dillon JF. Microbial colonization of the upper gastrointestinal tract in patients with Barrett’s esophagus. Clin Infect Dis. 2007;45(1):29–38. doi: 10.1086/518578. [DOI] [PubMed] [Google Scholar]
- 56.von Rosenvinge EC, Song Y, White JR, Maddox C, Blanchard T, Fricke WF. Immune status, antibiotic medication and pH are associated with changes in the stomach fluid microbiota. ISME J. 2013;7(7):1354–1366. doi: 10.1038/ismej.2013.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Castaño-Rodríguez N, Kaakoush NO, Lee WS, Mitchell HM. Dual role of Helicobacter and Campylobacter species in IBD: a systematic review and meta-analysis. Gut. 2017;66(2):235–249. doi: 10.1136/gutjnl-2015-310545. [DOI] [PubMed] [Google Scholar]
- 58.Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, Morgan XC, Kostic AD, Luo C, González A, McDonald D, Haberman Y, Walters T, Baker S, Rosh J, Stephens M, Heyman M, Markowitz J, Baldassano R, Griffiths A, Sylvester F, Mack D, Kim S, Crandall W, Hyams J, Huttenhower C, Knight R, Xavier RJ. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15(3):382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, El-Omar EM, Brenner D, Fuchs CS, Meyerson M, Garrett WS. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S, Shussman N, Almogy G, Cuapio A, Hofer E, Mevorach D, Tabib A, Ortenberg R, Markel G, Miklić K, Jonjic S, Brennan CA, Garrett WS, Bachrach G, Mandelboim O. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42(2):344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14(2):195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simon D, Cianferoni A, Spergel JM, Aceves S, Holbreich M, Venter C, Rothenberg ME, Terreehorst I, Muraro A, Lucendo AJ, Schoepfer A, Straumann A, Simon HU. Eosinophilic esophagitis is characterized by a non-IgE-mediated food hypersensitivity. Allergy. 2016;71(5):611–620. doi: 10.1111/all.12846. [DOI] [PubMed] [Google Scholar]
- 63.Benitez AJ, Hoffmann C, Muir AB, Dods KK, Spergel JM, Bushman FD, Wang ML. Inflammation-associated microbiota in pediatric eosinophilic esophagitis. Microbiome. 2015;3(1):23. doi: 10.1186/s40168-015-0085-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harris JK, Fang R, Wagner BD, Choe HN, Kelly CJ, Schroeder S, Moore W, Stevens MJ, Yeckes A, Amsden K, Kagalwalla AF, Zalewski A, Hirano I, Gonsalves N, Henry LN, Masterson JC, Robertson CE, Leung DY, Pace NR, Ackerman SJ, Furuta GT, Fillon SA. Esophageal microbiome in eosinophilic esophagitis. PLoS One. 2015;10(5):e0128346. doi: 10.1371/journal.pone.0128346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Falush D, Wirth T, Linz B, Pritchard JK, Stephens M, Kidd M, Blaser MJ, Graham DY, Vacher S, Perez-Perez GI, Yamaoka Y, Mégraud F, Otto K, Reichard U, Katzowitsch E, Wang X, Achtman M, Suerbaum S. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299(5612):1582–1585. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- 66.Atherton JC, Blaser MJ. Coadaptation of Helicobacter pylori and humans: ancient history, modern implications. J Clin Invest. 2009;119(9):2475–2487. doi: 10.1172/JCI38605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Atherton JC. The pathogenesis of Helicobacter pylori-induced gastro-duodenal diseases. Annu Rev Pathol. 2006;1(1):63–96. doi: 10.1146/annurev.pathol.1.110304.100125. [DOI] [PubMed] [Google Scholar]
- 68.Kusters JG, van Vliet AHM, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19(3):449–490. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arnold IC, Dehzad N, Reuter S, Martin H, Becher B, Taube C, Müller A. Helicobacter pylori infection prevents allergic asthma in mouse models through the induction of regulatory T cells. J Clin Invest. 2011;121(8):3088–3093. doi: 10.1172/JCI45041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lebwohl B, Blaser MJ, Ludvigsson JF, Green PH, Rundle A, Sonnenberg A, Genta RM. Decreased risk of celiac disease in patients with Helicobacter pylori colonization. Am J Epidemiol. 2013;178(12):1721–1730. doi: 10.1093/aje/kwt234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chow J, Tang H, Mazmanian SK. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr Opin Immunol. 2011;23(4):473–480. doi: 10.1016/j.coi.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Monstein HJ, Tiveljung A, Kraft CH, Borch K, Jonasson J. Profiling of bacterial flora in gastric biopsies from patients with Helicobacter pylori-associated gastritis and histologically normal control individuals by temperature gradient gel electrophoresis and 16S rDNA sequence analysis. J Med Microbiol. 2000;49(9):817–822. doi: 10.1099/0022-1317-49-9-817. [DOI] [PubMed] [Google Scholar]
- 73.Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Relman DA. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A. 2006;103(3):732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maldonado-Contreras A, Goldfarb KC, Godoy-Vitorino F, Karaoz U, Contreras M, Blaser MJ, Brodie EL, Dominguez-Bello MG. Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME J. 2011;5(4):574–579. doi: 10.1038/ismej.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li XX, Wong GL, To KF. Wong VW, Lai LH, Chow DK, Lau JY, Sung JJ, Ding C. Bacterial microbiota profiling in gastritis without Helicobacter pylori infection or non-steroidal anti-inflammatory drug use. PLoS One. 2009;4(11):e7985. doi: 10.1371/journal.pone.0007985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Montalban-Arques A, Wurm P, Trajanoski S, Schauer S, Kienesberger S, Halwachs B, Högenauer C, Langner C, Gorkiewicz G. Propionibacterium acnes overabundance and natural killer group 2 member D system activation in corpus-dominant lymphocytic gastritis. J Pathol. 2016;240(4):425–436. doi: 10.1002/path.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kienesberger S, Cox LM, Livanos A, Zhang XS, Chung J, Perez-Perez GI, Gorkiewicz G, Zechner EL, Blaser MJ. Gastric Helicobacter pylori infection affects local and distant microbial populations and host responses. Cell Rep. 2016;14(6):1395–1407. doi: 10.1016/j.celrep.2016.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang I, Nell S, Suerbaum S. Survival in hostile territory: the microbiota of the stomach. FEMS Microbiol Rev. 2013;37(5):736–761. doi: 10.1111/1574-6976.12027. [DOI] [PubMed] [Google Scholar]
- 79.Lertpiriyapong K, Whary MT, Muthupalani S, Lofgren JL, Gamazon ER, Feng Y, Ge Z, Wang TC, Fox JG. Gastric colonisation with a restricted commensal microbiota replicates the promotion of neoplastic lesions by diverse intestinal microbiota in the Helicobacter pylori INS-GAS mouse model of gastric carcinogenesis. Gut. 2014;63(1):54–63. doi: 10.1136/gutjnl-2013-305178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lofgren JL, Whary MT, Ge Z, Muthupalani S, Taylor NS, Mobley M, Potter A, Varro A, Eibach D, Suerbaum S, Wang TC, Fox JG. Lack of commensal flora in Helicobacter pylori-infected INS-GAS mice reduces gastritis and delays intraepithelial neoplasia. Gastroenterology. 2011;140(1):210–220. doi: 10.1053/j.gastro.2010.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Noto JM, Peek RM., Jr The gastric microbiome, its interaction with Helicobacter pylori, and its potential role in the progression to stomach cancer. PLoS Pathog. 2017;13(10):e1006573. doi: 10.1371/journal.ppat.1006573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Coker OO, Dai Z, Nie Y, Zhao G, Cao L, Nakatsu G, Wu WK, Wong SH, Chen Z, Sung JJY, Yu J (2017) Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut. 10.1136/gutjnl-2017-314281 [DOI] [PMC free article] [PubMed]
- 83.Meresse B, Chen Z, Ciszewski C, Tretiakova M, Bhagat G, Krausz TN, Raulet DH, Lanier LL, Groh V, Spies T, Ebert EC, Green PH, Jabri B. Coordinated induction by IL15 of a TCR-independent NKG2D signaling pathway converts CTL into lymphokine-activated killer cells in celiac disease. Immunity. 2004;21(3):357–366. doi: 10.1016/j.immuni.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 84.Hüe S, Mention JJ, Monteiro RC, Zhang S, Cellier C, Schmitz J, Verkarre V, Fodil N, Bahram S, Cerf-Bensussan N, Caillat-Zucman S. A direct role for NKG2D/MICA interaction in villous atrophy during celiac disease. Immunity. 2004;21(3):367–377. doi: 10.1016/j.immuni.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 85.Raulet DH, Gasser S, Gowen BG, Deng W, Jung H. Regulation of ligands for the NKG2D activating receptor. Annu Rev Immunol. 2013;31(1):413–441. doi: 10.1146/annurev-immunol-032712-095951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guerra N, Pestal K, Juarez T, Beck J, Tkach K, Wang L, Raulet DH. A selective role of NKG2D in inflammatory and autoimmune diseases. Clin Immunol. 2013;149(3):432–439. doi: 10.1016/j.clim.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.López-Soto A, Huergo-Zapico L, Acebes-Huerta A, Villa-Alvarez M, Gonzalez S. NKG2D signaling in cancer immunosurveillance. Int J Cancer. 2015;136(8):1741–1750. doi: 10.1002/ijc.28775. [DOI] [PubMed] [Google Scholar]
- 88.El-Gazzar A, Groh V, Spies T. Immunobiology and con icting roles of the human NKG2D lymphocyte receptor and its ligands in cancer. J Immunol. 2013;191(4):1509–1515. doi: 10.4049/jimmunol.1301071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sjöberg V, Sandström O, Hedberg M, Hammarström S, Hernell O, Hammarström ML. Intestinal T-cell responses in celiac disease—impact of celiac disease associated bacteria. PLoS One. 2013;8(1):e53414. doi: 10.1371/journal.pone.0053414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ivarsson A, Persson LA, Nyström L, Ascher H, Cavell B, Danielsson L, Dannaeus A, Lindberg T, Lindquist B, Stenhammar L, Hernell O. Epidemic of coeliac disease in Swedish children. Acta Paediatr. 2000;89(2):165–171. doi: 10.1080/080352500750028771. [DOI] [PubMed] [Google Scholar]
- 91.Caminero A, Galipeau HJ, McCarville JL, Johnston CW, Bernier SP, Russell AK, Jury J, Herran AR, Casqueiro J, Tye-Din JA, Surette MG, Magarvey NA, Schuppan D, Verdu EF. Duodenal bacteria from patients with celiac disease and healthy subjects distinctly affect gluten breakdown and immunogenicity. Gastroenterology. 2016;151(4):670–683. doi: 10.1053/j.gastro.2016.06.041. [DOI] [PubMed] [Google Scholar]
- 92.Staffas A, Burgos da Silva M, van den Brink MR. The intestinal microbiota in allogeneic hematopoietic cell transplant and graft-versus-host disease. Blood. 2017;129(8):927–933. doi: 10.1182/blood-2016-09-691394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.van Bekkum DW, Roodenburg J, Heidt PJ, van der Waaij D. Mitigation of secondary disease of allogeneic mouse radiation chimeras by modification of the intestinal microflora. J Natl Cancer Inst. 1974;52(2):401–404. doi: 10.1093/jnci/52.2.401. [DOI] [PubMed] [Google Scholar]
- 94.Jones JM, Wilson R, Bealmear PM. Mortality and gross pathology of secondary disease in germfree mouse radiation chimeras. Radiat Res. 1971;45(3):577–588. [PubMed] [Google Scholar]
- 95.Taur Y, Jenq RR, Perales MA, Littmann ER, Morjaria S, Ling L, No D, Gobourne A, Viale A, Dahi PB, Ponce DM, Barker JN, Giralt S, van den Brink M, Pamer EG. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood. 2014;124(7):1174–1182. doi: 10.1182/blood-2014-02-554725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Luciano L, Hass R, Busche R, von Engelhardt W, Reale E. Withdrawal of butyrate from the colonic mucosa triggers “mass apoptosis” primarily in the G0/G1 phase of the cell cycle. Cell Tissue Res. 1996;286(1):81–92. doi: 10.1007/s004410050677. [DOI] [PubMed] [Google Scholar]
- 97.Holler E, Butzhammer P, Schmid K, Hundsrucker C, Koestler J, Peter K, Zhu W, Sporrer D, Hehlgans T, Kreutz M, Holler B, Wolff D, Edinger M, Andreesen R, Levine JE, Ferrara JL, Gessner A, Spang R, Oefner PJ. Metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation: loss of diversity is associated with use of systemic antibiotics and more pronounced in gastrointestinal graft-versus-host disease. Biol Blood Marrow Transplant. 2014;20(5):640–645. doi: 10.1016/j.bbmt.2014.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Spindelboeck W, Schulz E, Uhl B, Kashofer K, Aigelsreiter A, Zinke-Cerwenka W, Mulabecirovic A, Kump PK, Halwachs B, Gorkiewicz G, Sill H, Greinix H, Högenauer C, Neumeister P. Repeated fecal microbiota transplantations attenuate diarrhea and lead to sustained changes in the fecal microbiota in acute, refractory gastrointestinal graft-versus-host-disease. Haematologica. 2017;102(5):e210–e213. doi: 10.3324/haematol.2016.154351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Högenauer C, Hammer HF, Krejs GJ, Reisinger EC. Mechanisms and management of antibiotic-associated diarrhea. Clin Infect Dis. 1998;27(4):702–710. doi: 10.1086/514958. [DOI] [PubMed] [Google Scholar]
- 100.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MR, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517(7533):205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Högenauer C, Langner C, Beubler E, Lippe IT, Schicho R, Gorkiewicz G, Krause R, Gerstgrasser N, Krejs GJ, Hinterleitner TA. Klebsiella oxytoca as a causative organism of antibiotic-associated hemorrhagic colitis. N Engl J Med. 2006;355(23):2418–2426. doi: 10.1056/NEJMoa054765. [DOI] [PubMed] [Google Scholar]
- 102.Schneditz G, Rentner J, Roier S, Pletz J, Herzog KA, Bücker R, Troeger H, Schild S, Weber H, Breinbauer R, Gorkiewicz G, Högenauer C, Zechner EL. Enterotoxicity of a nonribosomal peptide causes antibiotic-associated colitis. Proc Natl Acad Sci U S A. 2014;111(36):13181–13186. doi: 10.1073/pnas.1403274111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wurm P, Spindelboeck W, Krause R, Plank J, Fuchs G, Bashir M, Petritsch W, Halwachs B, Langner C, Högenauer C, Gorkiewicz G. Antibiotic-associated apoptotic enterocolitis in the absence of a defined pathogen: the role of intestinal microbiota depletion. Crit Care Med. 2017;45(6):e600–e606. doi: 10.1097/CCM.0000000000002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Ann Rev Immunol. 2010;28(1):573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Orholm M, Munkholm P, Langholz E, Nielsen OH, Sørensen TI, Binder V. Familial occurrence of inflammatory bowel disease. N Engl J Med. 1991;324(2):84–88. doi: 10.1056/NEJM199101103240203. [DOI] [PubMed] [Google Scholar]
- 106.Meucci G, Vecchi M, Torgano G, Arrigoni M, Prada A, Rocca F, Curzio M, Pera A, de Franchis R. Familial aggregation of inflammatory bowel disease in northern Italy: a multicenter study. The Gruppo di Studio per le Malattie Infiammatorie Intestinali (IBD Study Group) Gastroenterology. 1992;103(2):514–519. doi: 10.1016/0016-5085(92)90841-l. [DOI] [PubMed] [Google Scholar]
- 107.Halme L, Paavola-Sakki P, Turunen U, Lappalainen M, Farkkila M, Kontula K. Family and twin studies in inflammatory bowel disease. World J Gastroenterol. 2006;12(23):3668–3672. doi: 10.3748/wjg.v12.i23.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, Bitton A, Dassopoulos T, Datta LW, Green T, Griffiths AM, Kistner EO, Murtha MT, Regueiro MD, Rotter JI, Schumm LP, Steinhart AH, Targan SR, Xavier RJ, Genetics Consortium NIDDKIBD, Libioulle C, Sandor C, Lathrop M, Belaiche J, Dewit O, Gut I, Heath S, Laukens D, Mni M, Rutgeerts P, Van Gossum A, Zelenika D, Franchimont D, Hugot JP, de Vos M, Vermeire S, Louis E, Belgian-French IBD Consortium. Wellcome Trust Case Control Consortium. Cardon LR, Anderson CA, Drummond H, Nimmo E, Ahmad T, Prescott NJ, Onnie CM, Fisher SA, Marchini J, Ghori J, Bumpstead S, Gwilliam R, Tremelling M, Deloukas P, Mansfield J, Jewell D, Satsangi J, Mathew CG, Parkes M, Georges M, Daly MJ. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40(8):955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lees CW, Barrett JC, Parkes M, Satsangi J. New IBD genetics: common pathways with other diseases. Gut. 2011;60(12):1739–1753. doi: 10.1136/gut.2009.199679. [DOI] [PubMed] [Google Scholar]
- 110.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar JP, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A, Boucher G, Brand S, Büning C, Cohain A, Cichon S, D'Amato M, De Jong D, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Georges M, Gieger C, Glas J, Haritunians T, Hart A, Hawkey C, Hedl M, Hu X, Karlsen TH, Kupcinskas L, Kugathasan S, Latiano A, Laukens D, Lawrance IC, Lees CW, Louis E, Mahy G, Mansfield J, Morgan AR, Mowat C, Newman W, Palmieri O, Ponsioen CY, Potocnik U, Prescott NJ, Regueiro M, Rotter JI, Russell RK, Sanderson JD, Sans M, Satsangi J, Schreiber S, Simms LA, Sventoraityte J, Targan SR, Taylor KD, Tremelling M, Verspaget HW, De Vos M, Wijmenga C, Wilson DC, Winkelmann J, Xavier RJ, Zeissig S, Zhang B, Zhang CK, Zhao H, International IBD Genetics Consortium (IIBDGC) Silverberg MS, Annese V, Hakonarson H, Brant SR, Radford-Smith G, Mathew CG, Rioux JD, Schadt EE, Daly MJ, Franke A, Parkes M, Vermeire S, Barrett JC, Cho JH. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361(21):2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, Benchimol EI, Panaccione R, Ghosh S, Barkema HW, Kaplan GG. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142(1):46–54.e42. doi: 10.1053/j.gastro.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 113.Becker C, Neurath MF, Wirtz S. The intestinal microbiota in inflammatory bowel disease. ILAR J. 2015;56(2):192–204. doi: 10.1093/ilar/ilv030. [DOI] [PubMed] [Google Scholar]
- 114.Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC, Kambal A, Monaco CL, Zhao G, Fleshner P, Stappenbeck TS, McGovern DP, Keshavarzian A, Mutlu EA, Sauk J, Gevers D, Xavier RJ, Wang D, Parkes M, Virgin HW. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160(3):447–460. doi: 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hueber W, Sands BE, Lewitzky S, Vandemeulebroecke M, Reinisch W, Higgins PD, Wehkamp J, Feagan BG, Yao MD, Karczewski M, Karczewski J, Pezous N, Bek S, Bruin G, Mellgard B, Berger C, Londei M, Bertolino AP, Tougas G, Travis SP, Secukinumab in Crohn’s Disease Study Group Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn’s disease: unexpected results of a randomised, double-blind placebo-controlled trial. Gut. 2012;61(12):1693–1700. doi: 10.1136/gutjnl-2011-301668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Targan SR, Feagan B, Vermeire S, Panaccione R, Melmed GY, Landers C, Li D, Russell C, Newmark R, Zhang N, Chon Y, Hsu YH, Lin SL, Klekotka P. A randomized, double-blind, placebo-controlled phase 2 study of brodalumab in patients with moderate-to-severe Crohn’s disease. Am J Gastroenterol. 2016;111(11):1599–1607. doi: 10.1038/ajg.2016.298. [DOI] [PubMed] [Google Scholar]
- 117.D'Haens GR, Geboes K, Peeters M, Baert F, Penninckx F, Rutgeerts P. Early lesions of recurrent Crohn’s disease caused by infusion of intestinal contents in excluded ileum. Gastroenterology. 1998;114(2):262–267. doi: 10.1016/s0016-5085(98)70476-7. [DOI] [PubMed] [Google Scholar]
- 118.Schaubeck M, Clavel T, Calasan J, Lagkouvardos I, Haange SB, Jehmlich N, Basic M, Dupont A, Hornef M, von Bergen M, Bleich A, Haller D. Dysbiotic gut microbiota causes transmissible Crohn’s disease-like ileitis independent of failure in antimicrobial defence. Gut. 2016;65(2):225–237. doi: 10.1136/gutjnl-2015-309333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Moschen AR, Gerner RR, Wang J, Klepsch V, Adolph TE, Reider SJ, Hackl H, Pfister A, Schilling J, Moser PL, Kempster SL, Swidsinski A, Orth Höller D, Weiss G, Baines JF, Kaser A, Tilg H. Lipocalin 2 protects from inflammation and tumorigenesis associated with gut microbiota alterations. Cell Host Microbe. 2016;19(4):455–469. doi: 10.1016/j.chom.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 120.Tilg H, Moschen AR. Food, immunity, and the microbiome. Gastroenterology. 2015;148(6):1107–1119. doi: 10.1053/j.gastro.2014.12.036. [DOI] [PubMed] [Google Scholar]
- 121.Borrelli O, Cordischi L, Cirulli M, Paganelli M, Labalestra V, Uccini S, Russo PM, Cucchiara S. Polymeric diet alone versus corticosteroids in the treatment of active pediatric Crohn’s disease: a randomized controlled open-label trial. Clin Gastroenterol Hepatol. 2006;4(6):744–753. doi: 10.1016/j.cgh.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 122.Quince C, Ijaz UZ, Loman N, Eren AM, Saulnier D, Russell J, Haig SJ, Calus ST, Quick J, Barclay A, Bertz M, Blaut M, Hansen R, McGrogan P, Russell RK, Edwards CA, Gerasimidis K. Extensive modulation of the fecal metagenome in children with Crohn’s disease during exclusive enteral nutrition. Am J Gastroenterol. 2015;110(12):1718–1729. doi: 10.1038/ajg.2015.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature. 2012;487(7405):104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chassaing B, Koren O, Goodrich JK, Poole AC, Srinivasan S, Ley RE, Gewirtz AT. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature. 2015;519(7541):92–96. doi: 10.1038/nature14232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66(4):683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 126.Garrett WS. Cancer and the microbiota. Science. 2015;348(6230):80–86. doi: 10.1126/science.aaa4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, Zhang D, Xia H, Xu X, Jie Z, Su L, Li X, Li X, Li J, Xiao L, Huber-Schönauer U, Niederseer D, Xu X, Al-Aama JY, Yang H, Wang J, Kristiansen K, Arumugam M, Tilg H, Datz C, Wang J. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun. 2015;6:6528. doi: 10.1038/ncomms7528. [DOI] [PubMed] [Google Scholar]
- 128.Nakatsu G, Li X, Zhou H, Sheng J, Wong SH, Wu WK, Ng SC, Tsoi H, Dong Y, Zhang N, He Y, Kang Q, Cao L, Wang K, Zhang J, Liang Q, Yu J, Sung JJ. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun. 2015;6:8727. doi: 10.1038/ncomms9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liang Q, Chiu J, Chen Y, Huang Y, Higashimori A, Fang J, Brim H, Ashktorab H, Ng SC, Ng SSM, Zheng S, Chan FKL, Sung JJY, Yu J. Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clin Cancer Res. 2017;23(8):2061–2070. doi: 10.1158/1078-0432.CCR-16-1599. [DOI] [PubMed] [Google Scholar]
- 130.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song M, Kostic AD, Giannakis M, Bullman S, Milner DA, Baba H, Giovannucci EL, Garraway LA, Freeman GJ, Dranoff G, Garrett WS, Huttenhower C, Meyerson M, Meyerhardt JA, Chan AT, Fuchs CS, Ogino S. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65(12):1973–1980. doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Park HE, Kim JH, Cho NY, Lee HS, Kang GH. Intratumoral Fusobacterium nucleatum abundance correlates with macrophage infiltration and CDKN2A methylation in microsatellite-unstable colorectal carcinoma. Virchows Arch. 2017;471(3):329–336. doi: 10.1007/s00428-017-2171-6. [DOI] [PubMed] [Google Scholar]
- 133.Bonnet M, Buc E, Sauvanet P, Darcha C, Dubois D, Pereira B, Déchelotte P, Bonnet R, Pezet D, Darfeuille-Michaud A. Colonization of the human gut by E. coli and colorectal cancer risk. Clin Cancer Res. 2014;20(4):859–867. doi: 10.1158/1078-0432.CCR-13-1343. [DOI] [PubMed] [Google Scholar]
- 134.Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, Rhodes JM, Stintzi A, Simpson KW, Hansen JJ, Keku TO, Fodor AA, Jobin C. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338(6103):120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.García-González AP, Ritter AD, Shrestha S, Andersen EC, Yilmaz LS, Walhout AJM. Bacterial metabolism affects the C. elegans response to cancer chemotherapeutics. Cell. 2017;169(3):431–441.e8. doi: 10.1016/j.cell.2017.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, Chen Y, Chen H, Hong J, Zou W, Fang JY. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170(3):548–563.e16. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre ML, Chang EB, Gajewski TF. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350(6264):1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, Cogdill AP, Zhao L, Hudgens CW, Hutchinson DS, Manzo T, Petaccia de Macedo M, Cotechini T, Kumar T, Chen WS, Reddy SM, Sloane RS, Galloway-Pena J, Jiang H, Chen PL, Shpall EJ, Rezvani K, Alousi AM, Chemaly RF, Shelburne S, Vence LM, Okhuysen PC, Jensen VB, Swennes AG, McAllister F, Sanchez EMR, Zhang Y, Le Chatelier E, Zitvogel L, Pons N, Austin-Breneman JL, Haydu LE, Burton EM, Gardner JM, Sirmans E, Hu J, Lazar AJ, Tsujikawa T, Diab A, Tawbi H, Glitza IC, Hwu WJ, Patel SP, Woodman SE, Amaria RN, Davies MA, Gershenwald JE, Hwu P, Lee JE, Zhang J, Coussens LM, Cooper ZA, Futreal PA, Daniel CR, Ajami NJ, Petrosino JF, Tetzlaff MT, Sharma P, Allison JP, Jenq RR, Wargo JA (2017) Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 10.1126/science.aan4236


