Abstract
Genome editing using CRISPR/Cas9 is considered the best instrument for genome engineering in plants. This methodology is based on the nuclease activity of Cas9 that is guided to specific genome sequences by single guide RNAs (sgRNAs) thus enabling researchers to engineer simple mutations or large chromosomal deletions. Current methodologies for targeted genome editing in plants using CRISPR/Cas9 are however largely inefficient, mostly due to low Cas9 activity, variable sgRNA efficiency and low heritability of genetic lesions. Here, we describe a newly developed strategy to enhance CRISPR/Cas9 efficiency in Arabidopsis thaliana focusing on the design of novel binary vectors (pUbiCAS9-Red and pEciCAS9-Red), the selection of highly efficient sgRNAs, and the use of direct plant regeneration from induced cell cultures. Our work demonstrates that by combining these three independent developments, heritable targeted chromosomal deletions of large gene clusters and intergenic regulatory sequences can be engineered at a high efficiency. Our results demonstrate that this improved CRISPR/Cas9 methodology can provide a fast, efficient and cost-effective tool to engineer targeted heritable chromosomal deletions, which will be instrumental for future high-throughput functional genomics studies in plants.
Introduction
Phenotypic alterations caused by genetic lesions are the most widely used approach to study gene function in eukaryotes. Over the last century, forward mutagenesis screens have been conducted in several plant species taking advantage of the mutagenic activity of chemicals, ionizing radiation and the mobility of transposable elements1–3. However, over the last three decades significant advances in plant transformation technologies have led to the majority of mutagenesis studies exploiting the random integration of foreign transfer DNA (T-DNA)4,5. Using the latter approach, extensive mutant libraries have been generated in different plant species6–8. However, the major limitations of T-DNA and other traditional mutagenesis strategies are that they are time-consuming to confirm the insertion, collections are largely incomplete due to not all genes in the genome being disrupted, and combining multiple mutations is difficult for those genes that are genetically linked. To overcome some of these limitations, plant researchers have employed reverse genetic screens using transgenes that induce gene silencing by RNA interference (RNAi) or viruses causing viral-induced gene silencing (VIGS)9. These methodologies are rapid and efficient in down-regulating gene expression irrespective of gene copy number or chromosomal position, however they normally cause a wide range of hypomorphic phenotypes that can complicate functional studies10.
The recent development of targeted gene editing technologies offers significant advantages over the aforementioned methods. Targeted mutations in plants have been generated using zinc-finger-nucleases (ZFN)11 and transcription activator-like effector nucleases (TALEN)12. Both TALENs and ZFNs encode in their amino acid sequence the specific DNA binding motifs, thus their sequence recognition domains must be designed to target each specific sequence13,14. In the last decade, the discovery of a microbial adapted immune system based on the clustered regularly interspaced short palindromic repeats-Cas (CRISPR/Cas9)15–19 has revolutionised genome editing technology by offering a user-friendly tool for use in microbial, plant and animal species20–22. The CRISPR/Cas9 system functions by recognizing genomic target sites through a single chimeric guide RNA (sgRNA)21. Chimeric sgRNAs commonly used for CRISPR/Cas9 contain a 17- to 20-nt long variable region, the protospacer, which is complementary to the targeted genomic sequence23. The simplicity of designing sgRNAs to recognize different target sequences is one of the biggest advantages of the CRISPR/Cas9 system over previous gene targeting methodologies. The feasibility of CRISPR/Cas9 mediated genome editing has been demonstrated in several plant species, primarily for the generation of targeted loss-of-function mutants through point mutations or small deletions at genes of known function24–33. However, the efficiency and heritability of CRISPR/Cas9 genetic lesions in plants is far from optimal with regards to high throughput functional studies34.
In the present study, we show how current methodologies for CRISPR-mediated genome targeted deletions in Arabidopsis can be inefficient due to variable sgRNA activity and low heritability. We further demonstrate how the standard CRISPR/Cas9 methodology could be improved to overcome these caveats and aid the rapid and cost-effective generation of targeted heritable chromosomal deletions, thus enhancing further the potential scope for functional genomic analyses in plants.
Results
A new methodology for the rapid generation of targeted chromosomal deletions in Arabidopsis
To generate heritable CRISPR/Cas9 chromosomal deletions in plants we created two novel binary vectors named pUbiCAS9-Red and pEciCAS9-Red. For both vectors we used the backbone of pDE-CAS929 (Fig. 1A). Both contain a codon optimized Cas9 driven by the Petroselinum crispum Ubiquitin4–2 promoter (PcUbi4–2) or by the synthetic promoter comprising the EC1.2 enhancer fused to the EC1.1 promoter35. To aid the rapid insertion of multiple synthetic Cas9 sgRNAs we included an attR1-attR2 Gateway cassette and generated an entry vector containing two Arabidopsis U6–26 RNA polymerase III promoters flanked by a Cas9 small guide RNA (sgRNA) scaffold flanked by a type IIS restriction enzyme site. This design allows for a rapid and PCR-free direct cloning of two or more chemically synthesized sgRNAs by Golden-Gate assembly36 (Fig. 1A). To facilitate positive/negative selection of Cas9-containing plants we included, additionally to the existing bialaphos resistance (BAR) gene in the vector, which confers tolerance to glufosinate37, a seed-specific fluorescent reporter38 (Fig. 1A). These reporter allows a fast and non-destructive selection of Cas9-negative plants without the need for molecular genotyping. However, it is possible that some lines may carry partial integrations of the transgene.
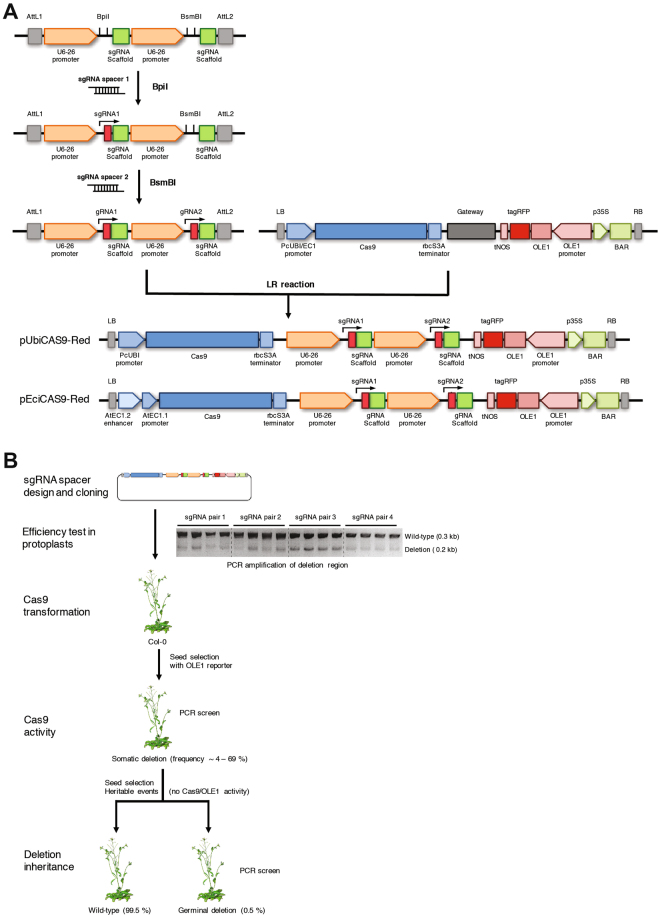
Figure 1.
Methodology for the generation of targeted chromosomal deletions in Arabidopsis. (A) Schematic assembly of the CRISPR/Cas9 vectors. Briefly, 24 nt DNA oligos containing the sequences of the two sgRNAs were used to generate a double-stranded DNA molecule and cloned into the entry vector with the two restriction enzyme sites BpiI and BsmBI, respectively. A LR reaction between the entry vector containing the sgRNAs and the binary vector generated the final Cas9 expression vector. (B) The efficiency of the Cas9 expression vector was determined by transfection of Arabidopsis protoplast. Selected vectors were integrated using floral dipping. Positive transformants were selected using a seed-specific fluorescent reporter. Cas9 nuclease activity in independent transgenic lines was confirmed by PCR using oligos flanking the deletion. Heritable deletion events were identified in plants lacking Cas9 activity after selection for seeds lacking the seed-specific fluorescent reporter. PCR was carried out using primers JD460 and JD461.
To assess the efficiency of CRISPR/Cas9 in generating genomic deletions, Arabidopsis plants were transformed with the pUbiCAS9-Red vector containing sgRNA pairs targeting twelve distinct genomic regions. Transgenic plants (T1) were selected based on both bialaphos resistance and the presence of RFP fluorescence in mature seeds (Fig. 1B). To assess the efficiency of the constructs, we grew ~100 independent T1 transgenic lines for each construct and extracted DNA from flower buds to PCR-amplify the targeted genomic region (Supplementary Table 1). This analysis revealed that the targeting efficiency of our Cas9 system in somatic tissues differed dramatically between constructs (4–69%, n = 1,200) (Supplementary Table 1), suggesting that the design of efficient RNA guides is a critical parameter for obtaining high efficiency CRISPR/Cas9 genomic deletions. Because of this, we introduced an additional step to first assess the efficiency of sgRNAs by transfecting binary constructs carrying Cas9 and different combinations of in silico designed sgRNAs into Arabidopsis mesophyll protoplasts. We monitored RNA guide efficiency by semi-quantitative PCR amplification of targeted genomic regions using flanking oligonucleotides (Fig. 1B) and determined the extent of the lesions by sequencing. Using this strategy we identified highly efficient sgRNAs, to target discrete chromosomal regions containing large gene clusters, or non-coding intergenic genome regions.
A significant issue with genome editing using CRISPR/Cas9 in plants is the limited heritability of genetic lesions. To circumvent this problem, we selected pUbiCAS9-Red transgenic T1 plants that displayed a high frequency of somatic deletions (Fig. 1B). Seed progenies from these plants were screened for lack of RFP signal in dry seeds to identify those lacking Cas9 activity. Around 100–300 plants for each line were grown in soil and genotyped by PCR to confirm the inheritance of deletions (Fig. 1B). We found that the frequency of these heritable deletions oscillated between 0.5–5% and we were able to create heritable lines only for two out of the twelve regions targeted (Supplementary Table 1). Collectively, our data show that although CRISPR/Cas9 is a suitable tool for generating heritable genomic deletions in Arabidopsis, its efficiency is very low. Recent reports suggest that the efficiency of single mutations created by Cas9 in plants can be improved by the use of promoters that are highly active during early stages of sexual reproduction35,39, We therefore modified our vector to drive Cas9 expression using a previously described synthetic EC1 promoter that confers expression in egg cells35 and named this vector pEciCAS9-Red. We found that when using this vector the efficiency of heritable deletions increased dramatically (6–100%) for some of the regions targeted. Intriguinly, in some cases we were able to generate offspring carrying homozygous genome deletions, (Supplementary Table 1). These data demonstrate that the use of pEciCAS9-Red vector enhances the efficiency of CRISPR/Cas9 for targeted genome deletions in Arabidopsis.
Targeted deletions of gene clusters and intergenic regulatory sequences to aid functional analyses in Arabidopsis
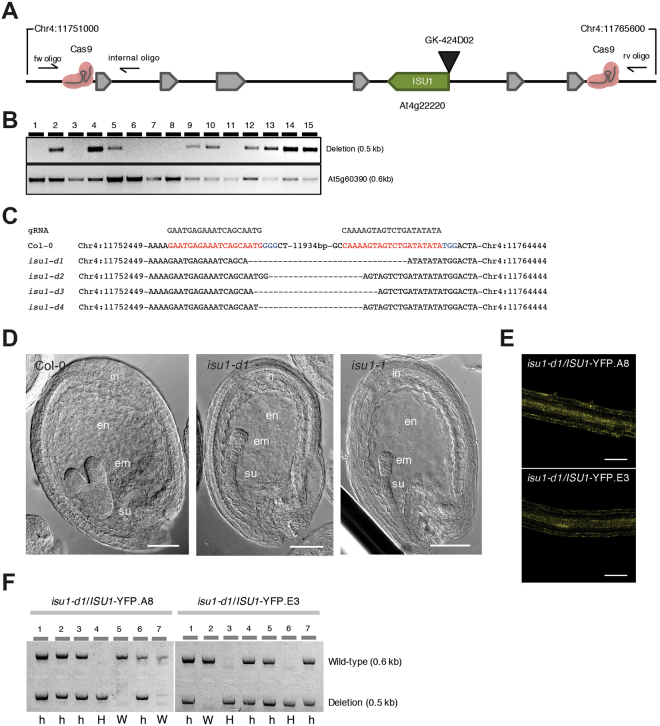
To demonstrate the efficacy of CRISPR-mediated genomic deletions for functional genomic studies, we first selected a genomic region flanking the ISU1 locus, which also contains a large cluster of defensin-like genes (Fig. 2A). ISU1 encodes a scaffold protein targeted to mitochondria that is involved in iron-sulfur cluster biogenesis40,41. It has been previously shown that RNAi downregulation and T-DNA promoter insertions in ISU1 cause a wide range of developmental phenotypes42. We decided to target the full ISU1 locus by removing a ~12 kb segment that includes coding and regulatory sequences (Fig. 2A). We designed different sgRNA pairs that were tested in Arabidopsis protoplast transfections and selected pairs based on their targeting efficiency to generate two ISU1-Cas9 binary constructs using our pUbiCAS9-Red binary vector. We grew 152 independent transgenic lines, genotyped these lines with oligos flanking the expected deletion region and found that 77 plants displayed targeted deletions (51%) (Fig. 2B and Supplementary Table 1), which we further confirmed by sequencing (Fig. 2C). We then selected 396 Cas9-negative seed progenies from nine independent lines and identified two independent heritable events, which we named isu1-d. When comparing T-DNA exonic insertional mutant plants (isu1–1) and Cas9 deletion plants (isu1-d) we found that we could not recover homozygous plants from either population. We then analyzed mature siliques and found that a proportion of the seeds (23.5%, n = 336) displayed a significant reduction in size and in some cases abnormal morphology. To assess the nature of these defects we analyzed developing seeds in heterozygous insertional and targeted deletion mutants, and found that for both mutant lines early globular embryo development was arrested in approximately a quarter of the seeds (24.6%, n = 455) indicating that these mutations are recessive and cause post-zygotic defects during seed development. In particular, globular embryos possessed disorganized apical embryonic cells while extra-embryonic suspensor cells developed normally (Fig. 2D). To confirm that these phenotypes where directly caused by the loss of ISU1 function, we complemented isu1-d plants with a translational ISU1-GFP reporter. We found that out of 22 independent lines tested, six fully restored embryo development and seed viability, which then allowed us to recover homozygous isu1-d/ISU1-GFP plants (Fig. 2E and F).
Figure 2.
CRISPR/Cas9 targeted deletions of the ISU1 locus and genetic complementation with an ISU1-YFP reporter. (A) Schematic representation of the ISU1 locus and the Cas9 binding sites. (B) PCR analysis of the ISU1 locus with oligonucleotides flanking the deletion site. The expected PCR size containing a deletion is 500 bp. Primers JD157 with JD158 and JD125 with JD126 were used for the ISU1 deletion and At5g60390 control, respectively. (C) Sanger sequencing results of four independent deletions (T1) (sgRNA sequence in red and PAM sequence in blue). (D) Defects in early embryo development associated with isu1-d1 and isu1–1 (GK424D02) mutations. Scale bar: 50 μm. in, integuments, en, endosperm, em, embryo, su, suspensor (E) Confocal microscopy images of roots from two independent ISU1-YFP transgenic lines that complement the isu1-d1 mutant. Scale bar: 50 μm. (F) Genotype analysis of plants from two independent isu1-d1/ISU1-YFP complemented lines. Deletions of the ISU1 locus was determined by PCR using oligonucleotides flanking the deletion site. The expected PCR product size for the deletion is 500 bp and the expected PCR size for the wild-type ISU1 allele is 600 bp. Primers JD157 with JD159 and JD157 with JD158 were used for the ISU1 wild-type allele and ISU1 deletion allele, respectively. W, wild-type; H, homozygous; h, heterozygous.
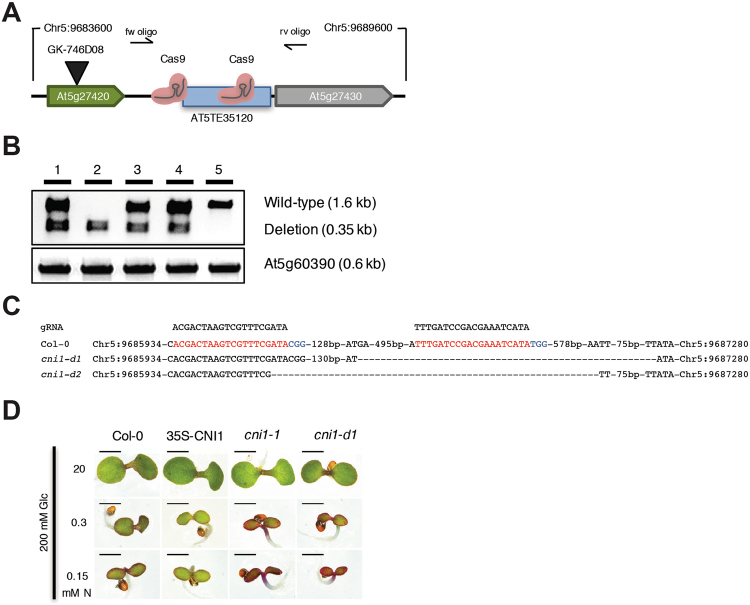
We next tested the application of our CRISPR/Cas9 genome targeting approach to dissect the function of a non-coding regulatory region. For this analysis we selected the intergenic non-coding regulatory region of CARBON/NITROGEN INSENSITIVE 1 (CNI1)- a plant specific ubiquitin ligase critical for carbon and nitrogen homeostasis, defense response and leaf senescence43–45. The region targeted comprises a cis regulatory element located in a transposable-related element, which is sensitive to epigenetic modification in response to stress46. We selected sgRNA pairs by Arabidopsis protoplast transfection and generated two distinct pUbiCAS9-Red binary constructs to target this genomic region (Fig. 3A). We found that both constructs were highly efficient in generating targeted deletions in somatic tissues (Supplementary Table 1) and allow us to identify two independent heritable deletions that we named cni1-d (Fig. 3B). These two deletion lines showed the expected genetic segregation and sequence analysis confirmed that these lines carried independent genomic lesions (Fig. 3C). Interestingly, both heritable lines show different larger additonal deletions adding to a deletion size of ~1.2 kB compared with the expected 0.66 kb. Because CNI1 participates in carbon and nitrogen sensing, we grew homozygous cni1-d1 plants in media containing varying concentrations of nitrogen and glucose. We also included in our analysis a homozygous T-DNA insertion mutant line (cni1–1) and a transgenic line that constitutively express CNI1 (35S-CNI)44. We found that under limiting nitrogen and high glucose growth conditions, both cni1-d1 and cni1–1 plants accumulated strong purple pigmentation and displayed arrested growth phenotypes. On the other hand, wild-type and 35S-CNI1 plants did not show strong accumulation of pigments and/or growth arrest. Collectively, our experiments demonstrate that highly efficient CRISPR/Cas9 targeted genome-editing methodology can enhance significantly functional genomic studies in plants.
Figure 3.
CRISPR/Cas9 targeted deletion of the CNI1 intergenic regulatory region. (A) Schematic representation of the CNI1 locus and sequences targeted by Cas9. (B) PCR analysis of plants segregating for heritable deletion line cni1-d1 (T2) (cni1-d) with oligonucleotides flanking the deletion site. The expected PCR size of the deletion allele is 350 bp, the expected PCR size for the wild-type allele is 1.6 kb. Primers JD271 with JD272 and JD125 with JD126 were used for the CNI1 deletion and At5g60390 control, respectively. (C) Sanger sequencing results of two independent heritable cni1-d lines (T2) (sgRNA sequence in red and PAM sequence in blue). (D) Post-germinative growth phenotype of cni1-d1 compared to 35S-CNI1, cni1–1 mutants and WT after growth (4 days after germination) on medium containing different concentrations of nitrogen (0.15–20 mM) with 200 mM glucose. Scale bar: 1 mm.
A new strategy to increase the heritability rates of targeted CRISPR/Cas9 genomic deletions in Arabidopsis
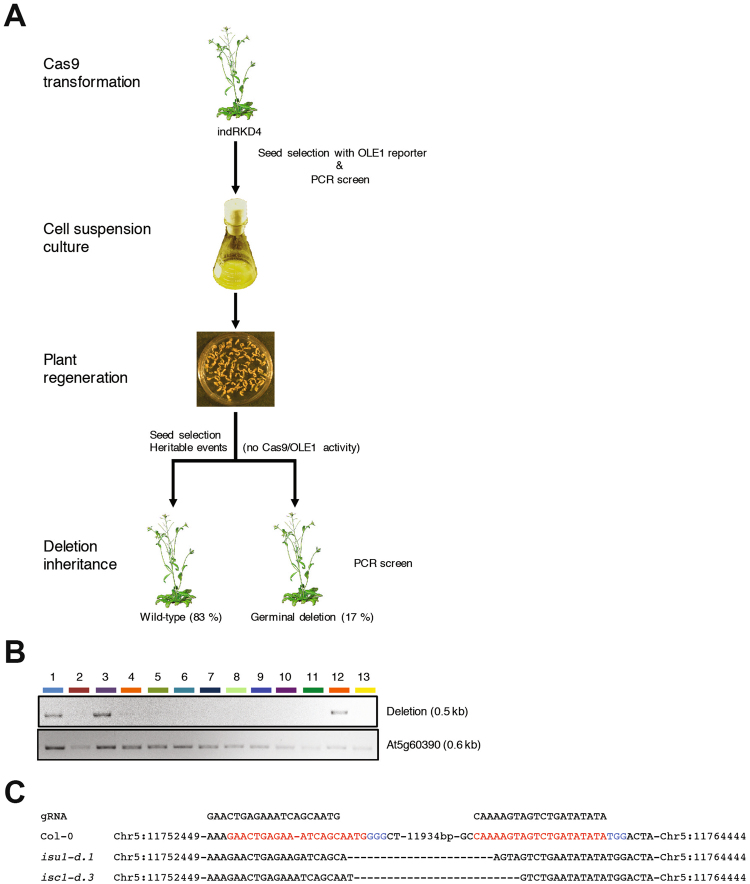
Our work has revealed that it is possible to generate heritable targeted genomic deletions in Arabidopsis using CRISPR/Cas9, although its efficiency is limited by the efficacy of the selected sgRNA, optimal Cas9 activity, and multigenerational analysis of modified plants (Fig. 1). Another major limiting factor of this approach is the reduced transmission of genetic lesions to the offspring. Because chromosomal deletions require the dual targeting of Cas9 to two distant sequences on the same chromatin, this could be particularly problematic in meristematic cells, which are known to have active DNA repair mechanisms that could affect targeting efficiency47. To test this hypothesis, we generated CRISPR/Cas9 lines to induce ISU1 genomic deletions in somatic cells and then recovered these lesions by induced plant regeneration. To facilitate the rapid and cost-effective regeneration of plants from somatic cells we generated a transgenic line expressing the zygotic transcription factor RKD448,49 under the regulation of a chemically inducible system, which we named indRKD4. We transformed indRKD4 plants with a pUbiCas9-Red construct designed to target the ISU1 locus with high efficiency in somatic cells (Fig. 4A). We identified forty primary transformants and selected five for analysis based on their ability to generate ISU1 somatic deletions. From each of the lines we selected 100 Cas9 positive seeds that were placed in liquid media (containing dexamethasone as a chemical inducer) to generate somatic cell suspension cultures. Two weeks after culture, cell suspensions where transferred to solid medium (devoid of dexamethasone) to induce plant regeneration (Fig. 4A). We selected five independent regenerants per line from where we collected ten Cas9-negative dry seeds (10/line). We grew plants from these seeds to identify those carrying heritable ISU1 deletions (Fig. 4B). Almost one fifth of these plants (17%, n = 250) carried independent ISU1 chromosomal deletions, which we confirmed by sequencing (Fig. 4C). Thus through this approach of combining our pUbiCAS9-Red vector with direct plant regeneration from indRKD4 somatic cell cultures we could improve the efficiency of targeted genomic deletions and generate a large number of independent heritable genetic lesions.
Figure 4.
A novel strategy for highly efficient CRISPR/Cas9 targeted genomic deletions in Arabidopsis. (A) The Cas9 expression vector was integrated into indRKD4 plants using floral dipping. Positive transformants were selected using the fluorescent seed-specific reporter. Positive seeds were grown in liquid medium containing dexamethasome. After two weeks cell suspensions were transferred to plates containing standard medium. Regenerated plants were tested for Cas9 nuclease activity by PCR and selection of heritable deletion was determined in plants lacking Cas9 activity. Primers JD157 with JD158 and JD125 with JD126 were used for the ISU1 deletion and At5g60390 control, respectively. (B) PCR analysis of the ISU1 locus to identify targeted deletions. (C) Sanger sequencing results of plants containing independent ISU1 heritable deletions.
Discussion
A highly efficient method for targeted chromosome deletions using CRISPR/Cas9 in plants
Newly developed targeted genomics tools, such as ZFN, TALEN and Cas9, have opened the possibility of rapidly generating precise, customized genetic lesions in plants. These methodologies can be applied to genes of unknown function but also to genes for which the function is not fully understood because mutants generated using classic mutagenesis may contain unwanted genetic lesions or chromosomal rearrangements that could interfere with their functional analyses50,51. For instance, CRISPR-generated loss-of-function alleles of the AUXIN BINDING PROTEIN 1 (ABP1) in Arabidopsis have revealed that phenotypes initially described in T-DNA alleles were inaccurate52. The CRISPR/Cas9 system has also a great potential for promoting functional studies in plants, as it can be easily adapted to generate true loss-of-function mutations for single genes, large gene clusters and also non-coding regulatory sequences. In mammals, single Cas9–sgRNA tiling approaches have been developed to dissect the activity of regulatory elements demonstrating the function of enhancer sequences and their role in gene expression regulation53–55. However, the use of tiling Cas9-sgRNA in plant functional analysis is not straightforward. We instead opted for an alternative approach based on the capacity to create stable targeted deletions of regulatory elements using CRISPR/Cas9. Our work demonstrates for the first time that this approach is suitable for the functional characterization of distantly acting regulatory sequences in plants. This methodology could be further adapted to engineer targeted epigenetic alterations, such as histone modifications or DNA methylation, using catalytically inactive Cas9 (dCas9)56–60.
A major limitation for the use of CRISPR/Cas9 in functional genomic studies in plants is the difficulty to generate meiotically stable genetic lesions with high efficiency. Our work has revealed that the generation of chromosomal deletions using current methodologies is highly inefficient. The low efficiency of this technique in Arabidopsis could be attributed to several factors; an important factor being the unpredictable efficiency of the in silico designed sgRNAs. Although there are several computational resources that aid the design of highly specific sgRNAs to minimize off-target effects in plants61–63 their efficiency in vivo cannot be predicted. Recent studies in animals have shown that the efficiency of the sgRNA depends on the nucleotide composition of the protospacer sequence64–67. In contrast, a recent study in rice was unable to detect nucleotide preferences for a collection of sgRNA designed for Cas9 gene targeting68. Although protospacer sequences with high GC-content have been associated with higher Cas9 editing efficiencies69, no direct link to nucleotide preference could be drawn in plants. By first testing the efficiency of sRNAs in vivo using protoplast transfection, our strategy offers a fast, simple and high-throughput method for CRISPR/Cas9 functional genomic analysis in plants. A second parameter that influences the efficiency of heritable genomic deletions in Arabidopsis is the promoter used to drive Cas9 expression. We found that a promoter that drives high expression in somatic cells is highly efficient in generating genomic deletions, however these lesions often exhibit low heritability. Our data show that the presence of Cas9-derived genomic deletions in somatic tissues is not a good proxy for strong germline transmission. This may be attributable to differences in DNA repair for somatic and meristematic cells, because heritable Cas9 genome deletions require simultaneous editing activity of the two sgRNAs. These results may be linked to the reported mechanisms that plants have developed to safeguard the genetic integrity of meristematic cells47. By generating Arabidopsis lines expressing the zygotic factor RKD4, we were able to combine Cas9-metiated genome editing in somatic cells and direct plant regeneration, and found that the efficiency of Cas9-mediated genome deletions increased. We foresee that this methodology could make possible CRISPR/Cas9 high-throughput functional studies in crop plants where plant regeneration from cell cultures has been already demonstrated70–74. Our methodology could also be potentially enhanced using transfection or viral infection as a transgene-free approach for the delivery of sgRNA/Cas9 in plants75–77. Several studies have shown that CRISPR/Cas9 editing efficiency can be improved when using promoters active during different stages of plant sexual reproduction35,78–80. We therefore adapted our system to drive Cas9 expression using a synthetic promoter that is preferentially expressed, although not exclusively, in Arabidopsis egg cells. We found that combining the use of a synthetic egg cell promoter with the in vivo sgRNAs screening enhances the efficiency of Cas9 targeted genome deletions. Moreover, we found that some plants produced progenies that were homozygous for the targeted deletion, which indicates that the promoter is active throughout embryo development thus minimizing the formation of chimeric mutations. Since both of the promoters tested - the U6–26 and the synthetic EC1 promoter - are active in Arabidopsis and other dicotyledonous plants27,81–83, we anticipate that our genome editing strategy can be directly applied to other plant species.
One widely reported disadvantage of CRISPR/Cas9 genome editing for functional genomic studies is the occurrence of off-target mutations22,84,85, with the frequency of off-targets in mammals being higher than plants86–88. Various effort has been made to overcome such limitations, ranging from the identification of new CRISPR/Cas systems89,90 to the development of newly engineered Cas9s91,92 Although we have not determined the frequency of off-targets in our CRISPR/Cas9 genome deletions, the generation of independent genetic lesions using different sgRNA pairs coupled to multigenerational propagation by selfing or backcrossing should minimize the impact of off-targets.
In summary, we have established an efficient and cost-effective platform for the generation of heritable targeted chromosomal deletions in Arabidopsis using CRISPR/Cas9. We demonstrate that the high efficiency of our system is based on (i) an improved methodology for initially selecting highly efficient sgRNAs, (ii) expression of Cas9 during early stages of sexual reproduction and (iii) propagating deletions occurring in somatic cells by direct plant regeneration. The optimization of these methodologies will be fundamental to facilitate high-throughput functional studies in crops and plants with complex genomes.
Material and Methods
Plant lines and growth conditions
All plant material used in this study was derived from the wild-type Columbia (Col-0) accession. The isu1–1 mutant (GK_424D02) was obtained from GABI-Kat collection93. The cni1–1 mutant and CNI1 overexpression lines have been reported previously46. Mutant and wild type (Col-0) seeds were sown on soil (John Innes and Perlite mix), stratified for 2 days at 4 °C in the dark, and grown at 20 °C under long photoperiod conditions (150 µmol/m2/s and 16 h/8 h light/dark cycles) to induce flowering. Seeds were surface sterilized for 3 min in 70% ethanol, followed by a 2 min treatment in 1% NaOCl. Seeds were then washed six times in sterile H2O, dispersed in sterile 0.1% agarose and placed on half-strength MS medium (Murashige & Skoog, Duchefa), solidified with 0.8% Phytagar.
Plant transformation and protoplast
Arabidopsis plants were transformed via Agrobacterium-mediated transformation as described previously5. Arabidopsis mesophyll protoplasts for transient expression of the CRISPR/Cas9 construct were prepared as described previously94 with minor modifications. Aproximately 80,000 protoplasts were transformed with 16 μg of plasmid (pUbiCAS9-Red) and incubated for 48 h at 22 °C under long photoperiod conditions (150 µmol/m2/s and 16 h/8 h light/dark cycles). DNA was isolated and concentrations adjusted before performing a semi-quantitative PCR using oligonucleotides flanking the region targeted for deletion (Fig. 1B and Supplementary Table 1).
T-DNA constructs
The binary vectors used in this study were derived from pDE-CAS929,35. The OLE1-RFP reporter was amplified by PCR using the pFAST-R03 vector38 as a template and oligos JD08 and JD09 (Supplementary Table 2). The OLE1-RFP reporter cassette was then cloned in pDE-CAS9 using SpeI and PacI restriction sites, to generate the vector pUbiCAS9-Red. We replaced the UbiCAS9 cassette with the EciCAS9 fragment from pHEE401E (Wang et al.35) to generate the vector pEciCAS9-Red. We generated a customizable entry vector by modifying the pEN-Chimera vector29. The first guide RNA was amplified by PCR using JD01 and JD02 and cloned with SacII and PstI restriction enzymes. A second guide RNA was synthesized by Integrated DNA Technologies, amplified with JD03 and JD04 (Supplementary Table 2). PstI and SacI were used to create the pEN-2xChimera vector. The two protospacer sequences that act as guides to specific target sequence were designed using CRISPR-PLANT62, chemically synthetized and cloned into pEN-2xChimera using BpiI and BsmBI restriction enzymes, respectively. The customized guide RNAs were then transferred into pUbiCAS9-Red or pEciCAS9-Red by Gateway® single-site LR recombination-mediated cloning.
The ISU1 gene including the promoter region was amplified from A. thaliana genomic DNA using the oligos JD335 and JD336. The PCR fragment was cloned into pDONR207 using Gateway BP clonase (Thermo Fisher Scientific). The insert of the entry clone was them integrated into pGWB44095 by LR clonase (Thermo Fisher Scientific).
Selection of germinal deletions
Primary transformants were selected with the seed specific RFP reporter using a Leica MZ-FL III stereomicroscope (Leica Camera AG). Plants were grown on soil and genomic DNA was extracted form approximately 4 week old plants. For genotyping oligos JD157/JDF158 (deletion) and JD157/JD160 (WT) were used. Progeny were checked for a 3:1 segregation. RFP negative seeds of single locus lines were sown on soil and genotyped for deletion events. Deletions were confirmed using Sanger sequencing.
Generation of RKD4 lines and direct plant regeneration from cell cultures
A DNA fragment spanning the entire protein-coding and intronic seqences of RKD4 (gRKD4) was amplified from the genomic DNA prepared from A. thaliana accession Col-0 plants with the primers Apa-RKD4–5′ (5′-ttggggccccagagactatatATGAGTTCGTC-3′) and Cla-Spe-RKD4-CtR (5′-aaatcgatactagTCAATAATAATCATCACC -3′). This fragment was inserted into the pBI-Bar-UAS-NosT-35S-ADH5′-GVG plasmid that contained DNA fragments of pNos-Bar96 5xUAS-NosT (Waki et al. 48), CaMV35S promoter, tobacco ADH 5′ leader97 and Gal4-VP16-GR (GVG)98 in the pBIN19 backbone99 The resulting plasmid (pBI-Bar-UAS-gRKD4–35S-GVG) was introduced into wild-type plants by the floral dip method, and transgenic plants (named indRKD4) were selected with 8 mg/L of bialaphos. Homozygous indRKD4 plants were transformed by floral dip with pUbiCAS9-Red_ISU1 vector and seeds carrying this transgene were isolated by fluorescent seed selection. Seeds were surface sterilized and germinated in inducing medium containing Gamborg’s B-5 Basal Salt Mixture (Sigma-Aldrich), 2% sucrose, 0.05% MES/KOH (pH 5.7), 10 µg/ml thiamine, 1 µg/ml pyridoxine, 1 µg/ml nicotinic acid and containing 30 µM dexamethasone. After two weeks in culture the cell suspension was spread on solid MS medium (without dexamethasone) to induce the formation of somatic embryos and allowing direct plant regeneration. Regenerated plants were transferred to soil and genotyped by PCR to identify those carrying targeted chromosomal deletions.
Low nitrogen/High sugar growth assay
Plants were grown on medium containing 0.15 or 0.3 mM KNO3, 1.5 mM CaCl2, 0.75 mM MgSO4, 0.625 mM KH2PO4 and 200 mM glucose, supplemented with Murashige and Skoog basal salt micronutrient solution (Sigma-Aldrich). ½ MS with 200 mM glucose was used as mock control. Images were taken 5 days after germination. Images were taken using a Zeiss Discovery.V12 stereomicroscope equipped with a Zeiss AxioCam HRc.
Microscopy analysis of developing seeds
Developing ovules, 3 days after pollination (DAP), were mounted in Hoyer’s solution (3.75 g gun arabic, 25 g chloral hydrate, 1.25 ml glycerol and 15 ml H2O). Differential interference contrast (DIC) images were obtained with a Zeiss Axiovert 200 M inverted microscope equipped with a confocal laser-scanning module (Zeiss LSM 710). Image processing was done using Fiji (fiji.sc).
Data availability
The nucleotide sequences reported in this paper have been submitted to NCBI under GenBank accession numbers KY489664 (pEN-2xChimera), KY489665 (pUbiCas9-Red) and KY489666 (pEciCas9-Red).
Electronic supplementary material
Acknowledgements
We are grateful to Miss Shanon Easterlow for technical assistance and to Prof. Holger Puchta (Karlsruhe Institute of Technology, Germany), Dr. Qi-Jun Chen (China Agricultural University, China), Prof. Junji Yamaguchi (Hokkaido University, Japan), Prof. Sofien Kamoun and Dr. Vladimir Nekrasov (Sainsbury Laboratory, UK), and Dr. Janneke Balk (John Innes Centre, UK) for providing reagents and advice. This work was funded by BBSRC awards BB/L003023/1, BB/N005279/1 and BB/N00194X/1.
Author Contributions
J.G.-M. and J.D. designed experiments. J.D. and R.P. conducted experiments and analyzed data. K.N. contributed with reagents and J.G.-M. wrote the manuscript with input from J.D.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-22667-1.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dooner HK, Belachew A. Transposition Pattern of the Maize Element Ac from the Bz-M2(ac) Allele. Genetics. 1989;122:447–457. doi: 10.1093/genetics/122.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Long D, et al. The maize transposable element system Ac/Ds as a mutagen in Arabidopsis: identification of an albino mutation induced by Ds insertion. Proc Natl Acad Sci USA. 1993;90:10370–10374. doi: 10.1073/pnas.90.21.10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Veleminský, J., Gichner, T. & Čsav., Ú. e. b. Induction of mutations and the mutation process: proceedings. (Pub. House of the Czechoslovak Academy of Sciences, 1965).
- 4.An G, Watson BD, Chiang CC. Transformation of Tobacco, Tomato, Potato, and Arabidopsis thaliana Using a Binary Ti Vector System. Plant Physiol. 1986;81:301–305. doi: 10.1104/pp.81.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 6.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 7.Sessions A, et al. A high-throughput Arabidopsis reverse genetics system. Plant Cell. 2002;14:2985–2994. doi: 10.1105/tpc.004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Rosso MG, Strizhov N, Viehoever P, Weisshaar B. GABI-Kat SimpleSearch: a flanking sequence tag (FST) database for the identification of T-DNA insertion mutants in Arabidopsis thaliana. Bioinformatics. 2003;19:1441–1442. doi: 10.1093/bioinformatics/btg170. [DOI] [PubMed] [Google Scholar]
- 9.Ruiz MT, Voinnet O, Baulcombe DC. Initiation and maintenance of virus-induced gene silencing. Plant Cell. 1998;10:937–946. doi: 10.1105/tpc.10.6.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ostergaard L, Yanofsky MF. Establishing gene function by mutagenesis in Arabidopsis thaliana. Plant J. 2004;39:682–696. doi: 10.1111/j.1365-313X.2004.02149.x. [DOI] [PubMed] [Google Scholar]
- 11.Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujikawa T, Ishihara H, Leach JE, Tsuyumu S. Suppression of defense response in plants by the avrBs3/pthA gene family of Xanthomonas spp. Mol Plant Microbe Interact. 2006;19:342–349. doi: 10.1094/MPMI-19-0342. [DOI] [PubMed] [Google Scholar]
- 13.Boettcher M, McManus MT. Choosing the Right Tool for the Job: RNAi, TALEN, or CRISPR. Mol Cell. 2015;58:575–585. doi: 10.1016/j.molcel.2015.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osakabe Y, Osakabe K. Genome editing with engineered nucleases in plants. Plant Cell Physiol. 2015;56:389–400. doi: 10.1093/pcp/pcu170. [DOI] [PubMed] [Google Scholar]
- 15.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 16.Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169:5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 18.Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sapranauskas R, et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Upadhyay SK, Kumar J, Alok A, Tuli R. RNA-guided genome editing for target gene mutations in wheat. G3 (Bethesda) 2013;3:2233–2238. doi: 10.1534/g3.113.008847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li JF, et al. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol. 2013;31:688–691. doi: 10.1038/nbt.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang W, et al. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013;41:e188. doi: 10.1093/nar/gkt780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nekrasov V, Staskawicz B, Weigel D, Jones JD, Kamoun S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:691–693. doi: 10.1038/nbt.2655. [DOI] [PubMed] [Google Scholar]
- 28.Liang Z, Zhang K, Chen K, Gao C. Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J Genet Genomics. 2014;41:63–68. doi: 10.1016/j.jgg.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 29.Fauser F, Schiml S, Puchta H. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 2014;79:348–359. doi: 10.1111/tpj.12554. [DOI] [PubMed] [Google Scholar]
- 30.Feng Z, et al. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013;23:1229–1232. doi: 10.1038/cr.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mao Y, et al. Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol Plant. 2013;6:2008–2011. doi: 10.1093/mp/sst121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bortesi L, Fischer R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv. 2015;33:41–52. doi: 10.1016/j.biotechadv.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 33.Sugano SS, et al. CRISPR/Cas9-mediated targeted mutagenesis in the liverwort Marchantia polymorpha L. Plant Cell Physiol. 2014;55:475–481. doi: 10.1093/pcp/pcu014. [DOI] [PubMed] [Google Scholar]
- 34.Belhaj K, Chaparro-Garcia A, Kamoun S, Patron NJ, Nekrasov V. Editing plant genomes with CRISPR/Cas9. Curr Opin Biotechnol. 2015;32:76–84. doi: 10.1016/j.copbio.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 35.Wang ZP, et al. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 2015;16:144. doi: 10.1186/s13059-015-0715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Engler C, Gruetzner R, Kandzia R, Marillonnet S. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One. 2009;4:e5553. doi: 10.1371/journal.pone.0005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson CJ, et al. Characterization of the herbicide-resistance gene bar from Streptomyces hygroscopicus. EMBO J. 1987;6:2519–2523. doi: 10.1002/j.1460-2075.1987.tb02538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimada TL, Shimada T, Hara-Nishimura I. A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J. 2010;61:519–528. doi: 10.1111/j.1365-313X.2009.04060.x. [DOI] [PubMed] [Google Scholar]
- 39.Mao Y, et al. Development of germ-line-specific CRISPR-Cas9 systems to improve the production of heritable gene modifications in Arabidopsis. Plant Biotechnol J. 2016;14:519–532. doi: 10.1111/pbi.12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leon S, Touraine B, Briat JF, Lobreaux S. Mitochondrial localization of Arabidopsis thaliana Isu Fe-S scaffold proteins. FEBS Lett. 2005;579:1930–1934. doi: 10.1016/j.febslet.2005.02.038. [DOI] [PubMed] [Google Scholar]
- 41.Leaden L, Busi MV, Gomez-Casati DF. The mitochondrial proteins AtHscB and AtIsu1 involved in Fe-S cluster assembly interact with the Hsp70-type chaperon AtHscA2 and modulate its catalytic activity. Mitochondrion. 2014;19 Pt B:375–381. doi: 10.1016/j.mito.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 42.Frazzon AP, et al. Functional analysis of Arabidopsis genes involved in mitochondrial iron-sulfur cluster assembly. Plant Mol Biol. 2007;64:225–240. doi: 10.1007/s11103-007-9147-x. [DOI] [PubMed] [Google Scholar]
- 43.Aoyama S, et al. Ubiquitin ligase ATL31 functions in leaf senescence in response to the balance between atmospheric CO2 and nitrogen availability in Arabidopsis. Plant Cell Physiol. 2014;55:293–305. doi: 10.1093/pcp/pcu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sato T, et al. CNI1/ATL31, a RING-type ubiquitin ligase that functions in the carbon/nitrogen response for growth phase transition in Arabidopsis seedlings. Plant J. 2009;60:852–864. doi: 10.1111/j.1365-313X.2009.04006.x. [DOI] [PubMed] [Google Scholar]
- 45.Maekawa S, et al. The Arabidopsis ubiquitin ligases ATL31 and ATL6 control the defense response as well as the carbon/nitrogen response. Plant Mol Biol. 2012;79:217–227. doi: 10.1007/s11103-012-9907-0. [DOI] [PubMed] [Google Scholar]
- 46.Wibowo, A. et al. Hyperosmotic stress memory in Arabidopsis is mediated by distinct epigenetically labile sites in the genome and is restricted in the male germline by DNA glycosylase activity. Elife5, 10.7554/eLife.13546 (2016). [DOI] [PMC free article] [PubMed]
- 47.Fulcher N, Sablowski R. Hypersensitivity to DNA damage in plant stem cell niches. Proc Natl Acad Sci USA. 2009;106:20984–20988. doi: 10.1073/pnas.0909218106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waki T, Hiki T, Watanabe R, Hashimoto T, Nakajima K. The Arabidopsis RWP-RK protein RKD4 triggers gene expression and pattern formation in early embryogenesis. Curr Biol. 2011;21:1277–1281. doi: 10.1016/j.cub.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 49.Jeong S, Palmer TM, Lukowitz W. The RWP-RK factor GROUNDED promotes embryonic polarity by facilitating YODA MAP kinase signaling. Curr Biol. 2011;21:1268–1276. doi: 10.1016/j.cub.2011.06.049. [DOI] [PubMed] [Google Scholar]
- 50.Veluthambi K, Ream W, Gelvin SB. Virulence genes, borders, and overdrive generate single-stranded T-DNA molecules from the A6 Ti plasmid of Agrobacterium tumefaciens. J Bacteriol. 1988;170:1523–1532. doi: 10.1128/jb.170.4.1523-1532.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tax FE, Vernon DM. T-DNA-associated duplication/translocations in Arabidopsis. Implications for mutant analysis and functional genomics. Plant Physiol. 2001;126:1527–1538. doi: 10.1104/pp.126.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao Y, et al. Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proc Natl Acad Sci USA. 2015;112:2275–2280. doi: 10.1073/pnas.1500365112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lopes R, Korkmaz G, Agami R. Applying CRISPR-Cas9 tools to identify and characterize transcriptional enhancers. Nat Rev Mol Cell Biol. 2016;17:597–604. doi: 10.1038/nrm.2016.79. [DOI] [PubMed] [Google Scholar]
- 54.Canver MC, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Korkmaz G, et al. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat Biotechnol. 2016;34:192–198. doi: 10.1038/nbt.3450. [DOI] [PubMed] [Google Scholar]
- 56.Hilton IB, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kearns NA, et al. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods. 2015;12:401–403. doi: 10.1038/nmeth.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu X, et al. A CRISPR-based approach for targeted DNA demethylation. Cell Discov. 2016;2:16009. doi: 10.1038/celldisc.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McDonald JI, et al. Reprogrammable CRISPR/Cas9-based system for inducing site-specific DNA methylation. Biol Open. 2016;5:866–874. doi: 10.1242/bio.019067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vojta A, et al. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 2016;44:5615–5628. doi: 10.1093/nar/gkw159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lei Y, et al. CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant. 2014;7:1494–1496. doi: 10.1093/mp/ssu044. [DOI] [PubMed] [Google Scholar]
- 62.Xie K, Zhang J, Yang Y. Genome-wide prediction of highly specific guide RNA spacers for CRISPR-Cas9-mediated genome editing in model plants and major crops. Mol Plant. 2014;7:923–926. doi: 10.1093/mp/ssu009. [DOI] [PubMed] [Google Scholar]
- 63.Oliveros JC, et al. Breaking-Cas-interactive design of guide RNAs for CRISPR-Cas experiments for ENSEMBL genomes. Nucleic Acids Res. 2016;44:W267–271. doi: 10.1093/nar/gkw407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Doench JG, et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu H, et al. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 2015;25:1147–1157. doi: 10.1101/gr.191452.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gagnon JA, et al. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One. 2014;9:e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liang G, Zhang H, Lou D, Yu D. Selection of highly efficient sgRNAs for CRISPR/Cas9-based plant genome editing. Sci Rep. 2016;6:21451. doi: 10.1038/srep21451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ma X, et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol Plant. 2015;8:1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 70.Zhou H, Liu B, Weeks DP, Spalding MH, Yang B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014;42:10903–10914. doi: 10.1093/nar/gku806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Y, et al. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol. 2014;32:947–951. doi: 10.1038/nbt.2969. [DOI] [PubMed] [Google Scholar]
- 72.Xing HL, et al. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014;14:327. doi: 10.1186/s12870-014-0327-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu J, et al. Efficiency and Inheritance of Targeted Mutagenesis in Maize Using CRISPR-Cas9. J Genet Genomics. 2016;43:25–36. doi: 10.1016/j.jgg.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 74.Lawrenson T, et al. Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 2015;16:258. doi: 10.1186/s13059-015-0826-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ali Z, et al. Efficient Virus-Mediated Genome Editing in Plants Using the CRISPR/Cas9 System. Mol Plant. 2015;8:1288–1291. doi: 10.1016/j.molp.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 76.Ali Z, Abul-Faraj A, Piatek M, Mahfouz MM. Activity and specificity of TRV-mediated gene editing in plants. Plant Signal Behav. 2015;10:e1044191. doi: 10.1080/15592324.2015.1044191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Woo JW, et al. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol. 2015;33:1162–1164. doi: 10.1038/nbt.3389. [DOI] [PubMed] [Google Scholar]
- 78.Eid A, Ali Z, Mahfouz MM. High efficiency of targeted mutagenesis in arabidopsis via meiotic promoter-driven expression of Cas9 endonuclease. Plant Cell Rep. 2016 doi: 10.1007/s00299-016-2000-4. [DOI] [PubMed] [Google Scholar]
- 79.Yan L, et al. High-Efficiency Genome Editing in Arabidopsis Using YAO Promoter-Driven CRISPR/Cas9 System. Mol Plant. 2015;8:1820–1823. doi: 10.1016/j.molp.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 80.Tsutsui, H. & Higashiyama, T. pKAMA-ITACHI vectors for highly efficient CRISPR/Cas9-mediated gene knockout in Arabidopsis thaliana. Plant Cell Physiol, 10.1093/pcp/pcw191 (2016). [DOI] [PMC free article] [PubMed]
- 81.Xu R, et al. Gene targeting using the Agrobacterium tumefaciens-mediated CRISPR-Cas system in rice. Rice (N Y) 2014;7:5. doi: 10.1186/s12284-014-0005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gao J, et al. CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol Biol. 2015;87:99–110. doi: 10.1007/s11103-014-0263-0. [DOI] [PubMed] [Google Scholar]
- 83.Ron M, et al. Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model. Plant Physiol. 2014;166:455–469. doi: 10.1104/pp.114.239392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Peterson BA, et al. Genome-Wide Assessment of Efficiency and Specificity in CRISPR/Cas9 Mediated Multiple Site Targeting in Arabidopsis. PLoS One. 2016;11:e0162169. doi: 10.1371/journal.pone.0162169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang H, et al. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J. 2014;12:797–807. doi: 10.1111/pbi.12200. [DOI] [PubMed] [Google Scholar]
- 88.Endo M, Mikami M, Toki S. Multigene knockout utilizing off-target mutations of the CRISPR/Cas9 system in rice. Plant Cell Physiol. 2015;56:41–47. doi: 10.1093/pcp/pcu154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Esvelt KM, et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hou Z, et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci USA. 2013;110:15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kleinstiver BP, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Slaymaker IM, et al. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kleinboelting, N., Huep, G. & Weisshaar, B. Enhancing the GABI-Kat Arabidopsis thaliana T-DNA Insertion Mutant Database by Incorporating Araport11 Annotation. Plant Cell Physiol, 10.1093/pcp/pcw205 (2016). [DOI] [PMC free article] [PubMed]
- 94.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 95.Nakagawa T, et al. Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem. 2007;71:2095–2100. doi: 10.1271/bbb.70216. [DOI] [PubMed] [Google Scholar]
- 96.Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol. 2000;42:819–832. doi: 10.1023/A:1006496308160. [DOI] [PubMed] [Google Scholar]
- 97.Satoh J, Kato K, Shinmyo A. The 5′-untranslated region of the tobacco alcohol dehydrogenase gene functions as an effective translational enhancer in plant. J Biosci Bioeng. 2004;98:1–8. doi: 10.1016/S1389-1723(04)70234-0. [DOI] [PubMed] [Google Scholar]
- 98.Miyashima S, Koi S, Hashimoto T, Nakajima K. Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development. 2011;138:2303–2313. doi: 10.1242/dev.060491. [DOI] [PubMed] [Google Scholar]
- 99.Frisch DA, et al. Complete sequence of the binary vector Bin 19. Plant Mol Biol. 1995;27:405–409. doi: 10.1007/BF00020193. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The nucleotide sequences reported in this paper have been submitted to NCBI under GenBank accession numbers KY489664 (pEN-2xChimera), KY489665 (pUbiCas9-Red) and KY489666 (pEciCas9-Red).