Abstract
IDH mutational status is required for proper diagnosis according to the WHO criteria revised in 2016. The single nucleotide polymorphism (SNP) rs11554137 (IDH1105GGT) at codon 105 of IDH1 has been reported in patients with several tumor types, including those with glioma. The aim of this study is to investigate the prevalence of IDH1105GGT in a cohort of brain tumors, and its association with clinicopathologic features and IDH1 and IDH2 missense mutations. Exon 4 of IDH1 and IDH2 was analyzed in a series of brain tumors classified according to current WHO criteria. DNA from control individuals was analyzed to infer the prevalence of IDH1105GGT in the reference population. Analysis was performed using next generation sequencing. IDH1105GGT was three times more frequent in patients with tumors (44/293 cases, 15.0%) vs. population controls (6/109, 5.5%) (p = 0.0102). IDH1105GGT was more frequent in grade III tumors (26.1%) compared to grade II (10.9%, p = 0.038) and grade IV tumors (13.7%, p = 0.041). IDH1105GGT was more frequent in grade II and III tumors without an IDH tumor missense mutation (43.8%) than in those with (11.5%, p = 0.005). The IDH1105GGT SNP likely represents an important genetic marker, worthy of additional investigation to better understand the clinical and biological features of IDH-WT infiltrating gliomas.
Introduction
The isocitrate dehydrogenase (IDH) family includes three isozymes (IDH1, IDH2, IDH3) involved in the Krebs cycle as active participants in NADPH production. These proteins also play an important role in the cellular control of oxidative damage1,2. The IDH1 protein is localized to the cytoplasm and peroxisome, while IDH2 and IDH3 are located in mitochondria3. IDH1 mutations were first implicated in carcinogenesis by a high-throughput study of the mutational landscape of breast and colorectal cancers4. Since then, mutations in IDH1 or IDH2 genes have been detected in many different tumors, primarily gliomas (>80% of grade II and grade III gliomas)5, acute myeloid leukemia (AML, ~15% of cases)6,7 and chondrosarcomas (~50% of cases)8. IDH mutations have been reported, albeit with a lower prevalence, in thyroid carcinoma (5–15% of cases)9,10, cholangiocarcinoma (15–20% of cases)11, and other solid neoplasms12–15. Among brain tumors IDH mutations are identified in over 80% of grade II and grade III gliomas (astrocytomas, oligodendrogliomas)16,17 and in about 5% of glioblastomas (GBM)16. According to the 2016 World Health Organization (WHO) classification of Central Nervous System tumors, establishing whether a brain tumor is IDH mutated or wild-type (WT) is a crucial requisite for the classification of gliomas18.
The large majority of IDH1 cancer-associated mutations affect codon 132, resulting in the amino acidic arginine(R)-to-histidine(H) substitution (p.R132H, c.395 G > A). Mutations other than p.R132H are found with a lower frequency, such as p.R132C (c.394 C > T), p.R132S (c.394 C > A), p.R132G (c.394 C > G) or p.R132L (c.395 G > T)5,17,19–21. However, other mutations not involving codon 132 have also been detected5. IDH-R132 mutations, as well as other IDH1 and IDH2 mutations (such as IDH1-G97D, IDH1-Y139D, IDH2-R172, IDH2-R140) have been shown to produce the 2-hydroxyglutarate (2HG) oncometabolite, while other rare mutations (e.g. IDH1-H133Q, IDH1-I130M, IDH1-G123R, IDH1-I99M, IDH1-V178I, IDH1-V71I) result in decreased IDH activity without a concomitant increase in 2HG production22.
Usually, synonymous single nucleotide polymorphisms (SNPs) do not change protein function, insofar as the amino acid sequence of the protein is not affected by the nucleotide change. Some silent SNPs, however, may lead to a protein defect, for example when they are localized in a splicing site23,24. In the case of the IDH1 gene, Wagner et al. (2010) found a silent SNP in a cohort of cytogenetically normal AML samples, that changes codon 105 of exon 4 from “GGC” (Gly) to “GGT” (Gly)25. The SNP (p.G105G, rs11554137:C > T -IDH1105GGT, minor allele frequency 0.0569) has since been frequently reported in AML and is linked to an adverse prognosis26–28. It has also been reported in brain tumors in a study of patients with gliomas (grade II to IV) from France and Germany29 and in a series of Bulgarian GBM patients30, as well as in thyroid tumors (both carcinomas and adenomas)9,10.
The role and biologic significance of the IDH1105GGT SNP in tumorigenesis is poorly understood, but it appears to be associated with increased IDH1 mRNA levels leading to altered NADPH production25,29.
The aim of the present study was to assess the prevalence of the IDH1105GGT SNP in a cohort of Italian patients with brain tumors classified according to 2016 WHO criteria, and investigate its association with clinicopathologic features and IDH tumor missense mutations.
Results
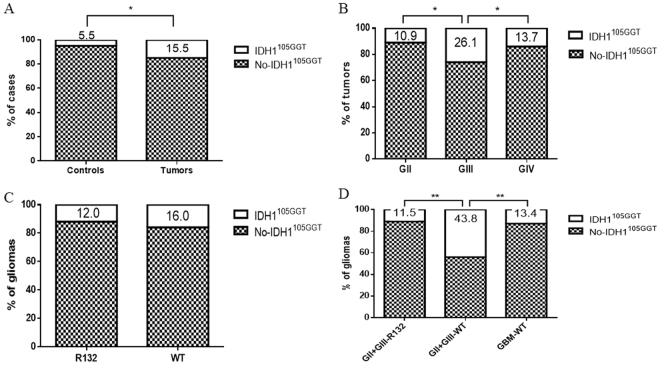
NGS primers allowed a reliable analysis of the nucleotide sequence of codon 105 in all samples. Overall, IDH1105GGT was found in 44 of 293 (15.0%) enrolled tumors (Table 1) and in 6 of 109 (5.5%) control individuals (p = 0.0102) (Fig. 1A).
Table 1.
Histological classification of the tumor samples analyzed and distribution ofIDH1105GGT. All the oligodendrogliomas harboured a mutation in IDH1 or IDH2 genes and showed co-deletion of chromosome arms 1p/19q. WT: Wild Type; °IDH1105GGTwas in the homozygous state.
| Diagnosis | N° of cases | IDH1105GGT (%) |
|---|---|---|
| Grade II tumors | 64 | 7 (10.9) |
| Astrocytomas | 34 | 4 (11.8) |
| Diffuse astrocytoma, IDH-WT | 6 | 2 (33.3) |
| Diffuse astrocytoma, IDH-mutant | 25 | 1 (4) |
| Gemistocytic Astrocytoma, IDH-mutant | 1 | 1 (100) |
| Pleomorphicxanthoastrocytoma, IDH-WT | 2 | 0 (−) |
| Oligodendrogliomas | 25 | 2 (8) |
| Oligodendroglioma, IDH-mutant and 1p/19q codeleted | 25 | 2° (8) |
| Other Grade II brain tumors | 5 | 1 (20) |
| Ependymoma | 4 | 0 (−) |
| Neurocytoma | 1 | 1 (100) |
| Grade III tumors | 46 | 12 (26.1) |
| Anaplastic Astrocytomas | 29 | 9 (31.0) |
| Anaplastic astrocytoma, IDH-WT | 7 | 4 (57.1) |
| Anaplasticastrocytoma, IDH-mutant | 21 | 4 (19) |
| Anaplastic Pleomorphic xanthoastrocytoma, IDH1-WT | 1 | 1 (100) |
| AnaplasticOligodendrogliomas | 15 | 2 (13.3) |
| Anaplastic oligodendroglioma, IDH-mutant and 1p/19q codeleted | 15 | 2 (13.3) |
| Other Grade III brain tumors | 3 | 1 (33.3) |
| Anaplastic ependimoma | 2 | 1 (50.0) |
| Grade IV | 183 | 25 (13.7) |
| Glioblastoma, IDH-WT | 179 | 24° (13.4) |
| Glioblastoma, IDH-mutant | 4 | 1 (25.0) |
Figure 1.
Prevalence of IDH105GGT. (A) Comparison of IDH105GGT between controls and patients with brain tumors. (B) IDH105GGT in grade II, III and IV brain tumors. (C) IDH105GGTin gliomas with and without IDH tumor missense mutation. (D) IDH105GGT in grade II and III gliomas with and without IDH tumor missense mutation and in grade IV without IDH tumor missense mutation. IDH105GGT: cases with IDH105GGT; No-IDH105GGT: cases without the IDH105GGT; GII: grade II brain tumors; GIII: grade III brain tumors; GIV: grade IV brain tumors; GBM: glioblastoma; R132: cases with IDH-missense mutation; WT: wild type.
In all but two patients harboring IDH1105GGT, the SNP was detected in about 50% of alleles analyzed (range: 45–53%), a frequency that is fully compatible with a heterozygous germline event. In the other two patients, IDH1105GGT was detected in 100% of the alleles analyzed, compatible with a homozygous germline condition.
IDH1105GGTand histological grade
Seven of 64 (10.9%) grade II cases (including 59 gliomas, 4 ependymomas and 1 neurocytoma) harbored IDH1105GGT (Fig. 1B), all but one in the heterozygous state. In one case (oligodendroglioma, IDH1-mutated and 1p/19q co-deleted), the SNP was detected in 100% of the alleles analyzed, compatible with a homozygous condition (Table 1). Among grade II gliomas, 6 of 59 (10.1%) harbored IDH1105GGT.
Twelve of 46 (26.1%) grade III tumors (including 43 gliomas and 3 anaplastic ependymomas) harbored IDH1105GGT, all in the heterozygous condition (Fig. 1B). Among grade III gliomas, 11 of 43 (26.6%) harbored IDH1105GGT (Table 1).
As regards grade IV tumors, 25 of 183 (13.7%) GBM were positive for IDH1105GGT (Fig. 1B), all but one (a GBM-IDH WT) in the heterozygous state.
We found a statistically significant difference in the prevalence of IDH1105GGT between tumor grades, with the highest frequency among tumors belonging to grade III, compared to grades II and IV (p = 0.038 and p = 0.041 respectively, Chi-squared test, Fig. 1B). Also among gliomas, IDH1105GGT is more frequent in grade III than in grade II or IV cases (p = 0.039 and p = 0.046, respectively). No statistically significant difference in prevalence was observed between grades II and IV, for both tumors and gliomas (p = 0.5765 and p = 0.6546, respectively, Chi-squared test).
IDH1105GGT and other IDH mutations
We observed an IDH1 or IDH2 mutation in 51 of 64 (79.7%) grade II tumors (all 51 cases were gliomas and 37 of these harbored the common p.R132H IDH1 mutation), in 36 of 46 (78.3%) grade III tumors (all 36 cases were gliomas and 31 harbored p.R132H), and in 4 of 183 (2.2%) grade IV tumors (all p.R132H).
Among 91 IDH-mutated gliomas, 11 (12.1%) also carried IDH1105GGT. In 31 gliomas, IDH1105GGT was detected in the absence of any IDH missense mutation. In accordance with data previously reported29, we found no correlation between the presence of IDH missense mutations in the tumor and the presence of IDH1105GGT (p = 0.4749, Fisher’s exact test) (Fig. 1C). However, IDH1105GGT was more frequent in grade II and III gliomas without than with IDH missense tumor mutations (43.8% vs 11.5% respectively – p = 0.005, Fisher’s exact test) (Fig. 1D). The SNP was also more frequent in grade II and III gliomas lacking IDH missense mutations than in GBM lacking IDH missense mutations (43.8% vs. 13.4%, p = 0.005, Fisher’s exact test) (Fig. 1D).
IDH1105GGT and histological subtypes
In grade II and III tumors, IDH1105GGT was more frequent in astrocytomas (13 of 63 cases −20.6%) than in oligodendrogliomas (4 of 40 cases −10%), but the difference did not reach statistical significance (p = 0.1837, Fisher’s exact test). Even after the inclusion of GBMs in the astrocytoma group (38 of 246–15.4%), the prevalence of IDH1105GGT was not statistically different between the oligodendroglial and astrocytic lineages (p = 0.4743, Fisher’s exact test).
IDH1105GGT and age
The age of patients harboring IDH1105GGT ranged from 26 to 74 years (mean 51.7ys). These patients were slightly younger than those without the SNP (mean age 53.2ys; age range: 17–84ys), but the difference was not statistically significant (p = 0.4476, Mann Whitney test). Figure 2 summarizes the statistical relationship between age, IDH1105GGT and IDH missense tumor mutations. There are significant differences among patient age and the distribution of IDH1105GGT and that of IDH missense tumor mutations. In particular, patients bearing only the SNP (mean age 55.7 years) were older that patients bearing both the SNP and IDH missense tumor mutations (mean 45.6ys, p < 0.05, Tukey’s multiple comparisons test), or those bearing only missense mutations (mean 42.3ys, p < 0.001, Tukey’s multiple comparisons test).
Figure 2.
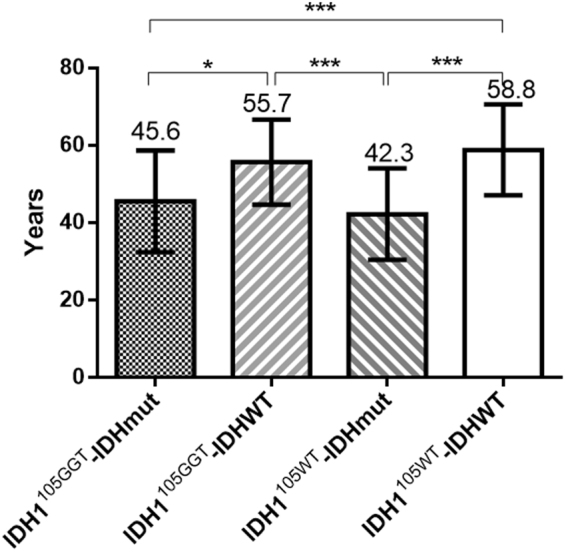
Age distribution of patients according to the presence of IDH1105GGT and IDH missense tumor mutations. IDH1105GGT: patients with tumor with IDH1105GGT; IDH1105WT: patients with tumor without IDH1105GGT; IDHmut: patients with tumor with IDH missense mutation; IDHWT: patients with tumor without IDH missense mutation. *p < 0.05 (Tukey’s multiple comparisons test); ***p < 0.001 (Tukey’s multiple comparisons test). Bars represent standard deviation (SD).
Discussion
In our Italian cohort, the prevalence of the IDH1105GGT SNP was considerably higher in patients with brain tumors compared to the control population (15.5% vs 5.5%, respectively).
Few studies have analyzed IDH1105GGT in brain tumors, likely because routine molecular pathology methods do not always allow its identification. IDH1105GGT status cannot be inferred by immunohistochemical methods, or by the mutation-specific PCR assays commonly used to diagnose p.R132H. Furthermore, sequencing requires the design of specific primers to include codon 105. Our NGS primers allowed us to reliably diagnose the SNP genotype in all samples.
Wang et al. in a cohort of French and German patients with gliomas, did not find a statistical correlation between IDH1105GGT and tumor histological grade29. In our cohort of Italian patients, we found a statistical association of IDH1105GGT with grade III gliomas, in particular with grade III astrocytomas. Importantly, among grade II and III gliomas, IDH1105GGT was more frequent in those cases without IDH missense tumor mutations (Fig. 1D). Previous studies reported this polymorphism as an adverse prognostic factor in patients with acute myeloid leukemia25; findings in the series of Wang et al. suggested a strong association with adverse outcome in patients with malignant glioma29. No association of the IDH1105GGT SNP with survival was found in the GBM series of Stancheva et al.30.
Although the functional effects of this polymorphism are still unclear, prediction analysis has shown that nucleotide 315 of the IDH1 gene may be within a putative Exonic Splicing Silencer (ESS) motif (ESRsearch Tool, http://esrsearch.tau.ac.il/)31. A nucleotide substitution in this region could lead to a protein defect due to incorrect regulation of constitutive or alternative splicing. Moreover, IDH1105GGT may be in linkage disequilibrium with other “tumor predisposing” variants.
Current opinion favors the existence of two major glioma groups: IDH-mutant gliomas, that are typically grade II and III tumors with a relatively favorable prognosis and IDH-WT tumors with a worse prognosis. As the large majority of IDH-WT tumors are grade IV, some authors have suggested that IDH-WT astrocytomas are in fact under-sampled IDH-WT GBMs and that they should be treated accordingly32. However, some subsets of IDH-WT low-grade gliomas do not have the molecular characteristics of GBM. These tumors likely represent other entities on a biological level. Some IDH-WT astrocytomas correspond to so called “pediatric type” tumors, sharing genetic and epigenetic features with pilocytic astrocytomas33.
In this context, the IDH1105GGT SNP may represent an important marker to further dissect and understand the clinical and biological features of IDH-WT infiltrating gliomas. Additional studies are warranted to clearly define the genetic profile and clinical outcome of patients with the IDH1105GGT SNP.
Methods
Case selection
A total of 293 consecutive cases of primitive brain tumors (64 grade II tumors, 46 grade III tumors, 183 grade IV tumors) were retrieved from the archives of Anatomic Pathology of Bellaria Hospital (Bologna, Italy). Samples were diagnosed and reclassified according to 2016 WHO criteria18. Patients were 181 males (61.8%) and 112 females (38.2%), aged from 17 to 84 years (mean age 52.9ys). Control DNA samples were analyzed from the peripheral blood of 109 individuals who underwent blood testing at the same institution to infer the prevalence of IDH1105GGT in the reference population. None of the controls was affected by brain tumor or other neoplastic diseases. The study was approved by Ethic Committee of Azienda Sanitaria Locale di Bologna (protocol number CE09113 of 29th September 2013, Bologna, Italy). All information regarding the human material was managed using anonymous numerical codes and all samples were handled in compliance with the Helsinki Declaration (https://www.wma.net/policies-post/wma-declaration-of-helsinki-ethical-principles-for-medical-research-involving-human-subjects/).
IDH1 and IDH2 analysis
All analyses were performed on DNA from formalin fixed and paraffin embedded (FFPE) specimens, extracted with the QuickExtract FFPE DNA Extraction Kit (Epicentre, Madison, WI, U.S.A.). Control DNA from blood specimens were extracted using the MasterPure DNA Purification Kit (Epincentre, Madison WI, USA). IDH1 (exon 4, codons 96–138) and IDH2 (exon 4, codons 151–178) amplicons were generated using the following primers: IDH1 Fw 5′-GAAACAAATGTGGAAATCACCA-3′, IDH1 Rv 5′-TCACATTATTGCCAACATGACT-3′; IDH2 Fw 5′-AGCCCATCATCTGCAAAAA-3′, IDH2 Rv 5′-TGTGGCCTTGTACTGCAGA-3′. The IDH1105GGT SNP (rs11554137) is 27 codons (81 nucleotides) upstream of the IDH1 hot spot codon (p.R132), well within the DNA region amplified by our set of primers.
Sequencing was performed using the 454 GS-Junior next generation sequencer (NGS) (Roche Diagnostic, Mannheim, Germany) according to established protocols (http://www.454.com/)34.
Categorical variables were compared using the Chi-square test or Fisher’s exact test. Continuous variables were compared using the Mann-Whitney test. Statistical comparison among IDH1 alterations and age was determined by the one-way analysis of variance (ANOVA) with Tukey’s multiple comparison test. A p-value < 0.05 was considered as statistically significant. Statistical analyses were performed using GraphPad Prism 6.01 (GraphPad Software).
Data availability
The datasets analysed during the current study are available from the corresponding author on reasonable request.
Acknowledgements
This work was supported in part by an Italian Government-Ministero Della Salute Grant No. RF-2011-02350857.
Author Contributions
Conception or design of the work: G.A., M.V., D.d.B., A.A.B., G.T., Data collection: G.A., M.V., G.M., E.F., A.T., Data analysis and interpretation: G.A., M.V., D.d.B., G.M., E.F., A.T., A.A.B., K.J.R., A.P., G.T., Drafting the article: G.A., M.V., D.d.B., G.M., K.J.R., G.T., Critical revision of the article: G.A., M.V., D.d.B., G.M., E.F., A.T., A.A.B., K.J.R., A.P., G.T., Final approval of the version to be published: G.A., M.V., D.d.B., G.M., E.F., A.T., A.A.B., K.J.R., A.P., G.T.
Competing Interests
The authors declare no competing interests.
Footnotes
Giorgia Acquaviva and Michela Visani contributed equally to this work.
Annalisa Pession and Giovanni Tallini jointly supervised this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Thompson CB. Metabolic enzymes as oncogenes or tumor suppressors. The New England journal of medicine. 2009;360:813–815. doi: 10.1056/NEJMe0810213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clark O, Yen K, Mellinghoff IK. Molecular Pathways: Isocitrate Dehydrogenase Mutations in Cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2016;22:1837–1842. doi: 10.1158/1078-0432.CCR-13-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yen KE, Bittinger MA, Su SM, Fantin VR. Cancer-associated IDH mutations: biomarker and therapeutic opportunities. Oncogene. 2010;29:6409–6417. doi: 10.1038/onc.2010.444. [DOI] [PubMed] [Google Scholar]
- 4.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 5.Forbes SA, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic acids research. 2015;43:D805–811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paschka P, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28:3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 7.Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. The New England journal of medicine. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amary MF, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. The Journal of pathology. 2011;224:334–343. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- 9.Hemerly JP, Bastos AU, Cerutti JM. Identification of several novel non-p.R132 IDH1 variants in thyroid carcinomas. European journal of endocrinology. 2010;163:747–755. doi: 10.1530/EJE-10-0473. [DOI] [PubMed] [Google Scholar]
- 10.Murugan AK, Bojdani E, Xing M. Identification and functional characterization of isocitrate dehydrogenase 1 (IDH1) mutations in thyroid cancer. Biochemical and biophysical research communications. 2010;393:555–559. doi: 10.1016/j.bbrc.2010.02.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borger DR, et al. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. The oncologist. 2012;17:72–79. doi: 10.1634/theoncologist.2011-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fathi AT, et al. Isocitrate dehydrogenase 1 (IDH1) mutation in breast adenocarcinoma is associated with elevated levels of serum and urine 2-hydroxyglutarate. The oncologist. 2014;19:602–607. doi: 10.1634/theoncologist.2013-0417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato Kaneko M, et al. Isocitrate dehydrogenase mutation is frequently observed in giant cell tumor of bone. Cancer science. 2014;105:744–748. doi: 10.1111/cas.12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mauzo SH, et al. Immunohistochemical demonstration of isocitrate dehydrogenase 1 (IDH1) mutation in a small subset of prostatic carcinomas. Applied immunohistochemistry & molecular morphology: AIMM. 2014;22:284–287. doi: 10.1097/PAI.0b013e3182649d1c. [DOI] [PubMed] [Google Scholar]
- 15.Ang D, et al. Biphasic papillary and lobular breast carcinoma with PIK3CA and IDH1 mutations. Diagnostic molecular pathology: the American journal of surgical pathology, part B. 2012;21:221–224. doi: 10.1097/PDM.0b013e31826ddbd1. [DOI] [PubMed] [Google Scholar]
- 16.Ohgaki H, Kleihues P. The definition of primary and secondary glioblastoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19:764–772. doi: 10.1158/1078-0432.CCR-12-3002. [DOI] [PubMed] [Google Scholar]
- 17.Yan H, et al. IDH1 and IDH2 mutations in gliomas. The New England journal of medicine. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Louis DN, et al. The2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta neuropathologica. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 19.Hartmann C, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta neuropathologica. 2009;118:469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 20.Yang B, Zhong C, Peng Y, Lai Z, Ding J. Molecular mechanisms of “off-on switch” of activities of human IDH1 by tumor-associated mutation R132H. Cell research. 2010;20:1188–1200. doi: 10.1038/cr.2010.145. [DOI] [PubMed] [Google Scholar]
- 21.von Deimling A, Korshunov A, Hartmann C. The next generation of glioma biomarkers: MGMT methylation, BRAF fusions and IDH1 mutations. Brain Pathol. 2011;21:74–87. doi: 10.1111/j.1750-3639.2010.00454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ward PS, et al. Identification of additional IDH mutations associated with oncometabolite R(-)−2-hydroxyglutarate production. Oncogene. 2012;31:2491–2498. doi: 10.1038/onc.2011.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mooney SD, Krishnan VG, Evani US. Bioinformatic tools for identifying disease gene and SNP candidates. Methods Mol Biol. 2010;628:307–319. doi: 10.1007/978-1-60327-367-1_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Vito L, et al. Brachial amyotrophic diplegia associated with the a140a superoxide dismutase 1 mutation. Neurogenetics. 2013;14:255–256. doi: 10.1007/s10048-013-0369-6. [DOI] [PubMed] [Google Scholar]
- 25.Wagner K, et al. Impact of IDH1 R132 mutations and an IDH1 single nucleotide polymorphism in cytogenetically normal acute myeloid leukemia: SNP rs11554137 is an adverse prognostic factor. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28:2356–2364. doi: 10.1200/JCO.2009.27.6899. [DOI] [PubMed] [Google Scholar]
- 26.Damm F, et al. Prevalence and prognostic value of IDH1 and IDH2 mutations in childhood AML: a study of the AML-BFM and DCOG study groups. Leukemia. 2011;25:1704–1710. doi: 10.1038/leu.2011.142. [DOI] [PubMed] [Google Scholar]
- 27.Ho PA, et al. Prognostic implications of the IDH1 synonymous SNP rs11554137 in pediatric and adult AML: a report from the Children’s Oncology Group and SWOG. Blood. 2011;118:4561–4566. doi: 10.1182/blood-2011-04-348888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Willander K, et al. Mutations in the isocitrate dehydrogenase 2 gene and IDH1 SNP 105C > T have a prognostic value in acute myeloid leukemia. Biomarker research. 2014;2:18. doi: 10.1186/2050-7771-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang XW, et al. Prognostic impact of the isocitrate dehydrogenase 1 single-nucleotide polymorphism rs11554137 in malignant gliomas. Cancer. 2013;119:806–813. doi: 10.1002/cncr.27798. [DOI] [PubMed] [Google Scholar]
- 30.Stancheva G, et al. IDH1/IDH2 but not TP53 mutations predict prognosis in Bulgarian glioblastoma patients. BioMed research international. 2014;2014:654727. doi: 10.1155/2014/654727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 32.van den Bent, M. J. et al. A clinical perspective on the 2016 WHO brain tumor classification and routine molecular diagnostics. Neuro-oncology, 10.1093/neuonc/now277 (2017). [DOI] [PMC free article] [PubMed]
- 33.Ceccarelli M, et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell. 2016;164:550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Biase D, et al. Next generation sequencing improves the accuracy of KRAS mutation analysis in endoscopic ultrasound fine needle aspiration pancreatic lesions. PloS one. 2014;9:e87651. doi: 10.1371/journal.pone.0087651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analysed during the current study are available from the corresponding author on reasonable request.