Abstract
Grapevine is among the fruit crops with high economic value, and because of the economic losses caused by abiotic stresses, the stress resistance of Vitis vinifera has become an increasingly important research area. Among the mechanisms responding to environmental stresses, the role of miRNA has received much attention recently. qRT-PCR is a powerful method for miRNA quantitation, but the accuracy of the method strongly depends on the appropriate reference genes. To determine the most suitable reference genes for grapevine miRNA qRT-PCR, 15 genes were chosen as candidate reference genes. After eliminating 6 candidate reference genes with unsatisfactory amplification efficiency, the expression stability of the remaining candidate reference genes under salinity, cold and drought was analysed using four algorithms, geNorm, NormFinder, deltaCt and Bestkeeper. The results indicated that U6 snRNA was the most suitable reference gene under salinity and cold stresses; whereas miR168 was the best for drought stress. The best reference gene sets for salinity, cold and drought stresses were miR160e + miR164a, miR160e + miR168 and ACT + UBQ + GAPDH, respectively. The selected reference genes or gene sets were verified using miR319 or miR408 as the target gene.
Introduction
Grapevine is a fruit crop with high economic value that is widely cultivated worldwide1. Nevertheless, grapevine is sensitive to climate change, which often causes a great loss in grape or wine production when unfavourable climate occurs in both vegetative and reproductive growth periods2,3. Salinity, cold and drought are major abiotic stresses in grapevine and in other plants4,5. With the global climate change that has occurred in recent years, the adverse effects caused by these abiotic stresses may be aggravated in the foreseeable future6. Environmental stressors can damage cellular structure and may lead to the physiological function disorders7, which are manifested in impaired membranes, dysfunctional proteins and increased reactive oxygen species (ROS)8,9. As a result, salinity, cold and drought stress can strongly influence the yield and quality of grapevine and sometimes, can even be lethal for grapevine8. Therefore, salinity, cold and drought resistance has always received focused attention of researchers. Epigenetic regulation, such as methylated modification and post-transcriptional regulation, is the stress response mechanism of plants, which can affect the resistance to stresses10,11. Among the post-transcriptional regulators, microRNAs (miRNAs) are likely of great importance12. The miRNAs are a class of small, non-coding RNAs with a length of 20–24 nucleotides13. In plants, miRNAs are produced from pri-miRNAs with hairpin structure, which are processed by a protein complex containing DICER-LIKE1 (DCL1), SERRATE (SE) and HYL114,15. miRNAs regulate the expression of the target gene by cleavage or translational repression14. In the complex gene regulatory network of plants, miRNAs are key regulatory factors that normally influence the expression of downstream genes by regulating their transcription factors16. In this way, miRNAs participate in many physiological activities, such as development, flowering and stress resistance17. Further understanding of the stress-related molecular mechanisms of miRNAs will provide new prospects in grapevine resistance breeding through molecular and genetic engineering methods18.
Currently, various methods of miRNA identification and quantification have been developed, such as northern blot19, microarray20 and high-throughput sequencing21. As a widely used method for gene expression analysis, the qRT-PCR (quantitative reverse transcriptase-polymerase chain reaction) is applied to identify protein-coding gene abundance in many plants. However, using qRT-PCR to detect miRNA abundance due to its small size was a great challenge until stem-loop22 and poly (A)-tailed23 qRT-PCR approaches were developed to characterize mature miRNA abundance. Because of the sensitivity, accuracy and large dynamic range, qRT-PCR soon became the most commonly used technique for miRNA quantification24,25. Although qRT-PCR is a powerful tool for gene expression analysis, normalization is required in the analysis system, because the quantification results can be easily influenced by a few factors, such as the RNA quality, design of reverse transcription primer and the efficiency of reverse transcription enzyme26. Reference genes are widely used as an internal control in qRT-PCR because the expression of the reference gene can be regarded stable under different experimental conditions and in different tissues. To guarantee the reliability of normalization, the use of multiple reference genes is suggested by Vandesompele et al.27 Notable, the reference gene suitable for all experimental conditions does not exist; thus, the selection of appropriate reference genes for qRT-PCR in different plant species or different stress conditions is necessary. In previous studies, various reference genes have been used in plant species. Among these genes, 5.8S ribosomal RNA (5.8S rRNA) and U6 small nuclear RNA (U6 snRNA) are the most commonly used28,29. However, the potential of miRNAs as reference genes has received close attention recently. As reported by Feng et al.30, the expression of miRNAs is more stable than that of protein-coding genes under biotic and abiotic stresses. Research related to the selection of reference genes for miRNA qRT-PCR has been performed in wheat30, soybean31, lettuce32, peach33 and sugarcane28. Nevertheless, until now, no report on miRNA reference gene evaluation in grapevine has been available. Therefore, suitable reference genes for grapevine miRNA study are urgently required, particularly for use in stress resistance research.
In this study, the experiment was designed to select the most suitable reference genes for grapevine miRNA qRT-PCR assays under abiotic stresses (salinity, cold, and drought stress). Fifteen candidate reference genes, including 4 traditional housekeeping genes, Actin (ACT, GenBank Accession: EC969944), Ubiquitin (UBQ, GenBank Accession: EC929411), Glyceraldehyde 3-phosphate dehydrogenase (GAPDH, GenBank Accession: AT1G13440.1), and Elongation factor1-alpha (EF1, GenBank Accession: AT5G60390.1), 9 miRNAs (miR156a, 159a, 160e, 162, 164a, 167a, 168, 169a, and 396a), and 2 non-coding reference genes, 5.8S rRNA (GenBank Accession: KT344661.1) and U6 snRNA (Location: chr6_15577690_15577792_+), were chosen for expression stability evaluation by the stem-loop qRT-PCR method. The amplification characteristics of these candidate reference genes were assessed first, and then, the Cq values of candidates with acceptable amplification efficiency and specificity were used to evaluate the expression stability using the algorithms of geNorm27, NormFinder34, Bestkeeper35 and deltaCt36. Based on the results obtained, the suitable reference genes in salinity, cold, drought stresses were provided, which promoted a more accurate miRNA qRT-PCR assay.
Results
Amplification characteristic and Cq range of candidate reference genes
The candidate reference genes consisted of 2 non-coding RNAs, 4 housekeeping genes and 9 miRNAs. The sequences and stem-loop primers of these selected miRNAs are shown in Table 1. To confirm the efficient amplification of pair-primers, the standard curves were obtained using a set of diluted cDNA templates. Based on the criteria of standard curve27, the regression coefficient (R2) should be more than 0.98 with the amplification efficiency (E) tending to 100%. According to the selecting criteria, miR156a, miR159a, miR162, miR167a, miR169a and miR396a were excluded from the candidates because of an unsatisfactory E value or R2 (Table 2). Among the remaining candidate reference genes, the E values ranged from 1.97 to 2.17, and R2 varied between 0.982 and 0.999. Moreover, the single melting peak during real-time PCR verified the amplification specificity of the final 9 candidate reference genes (Fig. S1).
Table 1.
The sequences and stem-loop primers of candidate reference miRNAs.
| Gene | Sequence | Stem-loop primer (5′-3′) |
|---|---|---|
| miR156a | TGACAGAAGAGAGGGAGCAC | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGTGCTCCC |
| miR159a | CTTGGAGTGAAGGGAGCTCTC | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGAGAGCTC |
| miR160e | TGCCTGGCTCCCTGTATGCCA | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTGGCATAC |
| miR162 | TCGATAAACCTCTGCATCCAG | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTGGATGC |
| miR164a | TGGAGAAGCAGGGCACGTGCA | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTGCACGTG |
| miR167a | TGAAGCTGCCAGCATGATCTG | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCAGATCAT |
| miR168 | TCGCTTGGTGCAGGTCGGGAA | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTTCCCGAC |
| miR169a | CAGCCAAGGATGACTTGCCGG | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCGGCAG |
| miR396a | TTCCACAGCTTTCTTGAACTA | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTAGTTCAA |
Table 2.
The primers and amplification characteristics of candidate reference genes.
| Gene | Gene code | Forward primer (5′-3′) | Reverse primer (5′-3′) | Efficiency | R2 |
|---|---|---|---|---|---|
| 5.8S rRNA | KT344661.1 | GCTCTCGCATCGATGAAGAAC | AGACTCGATGATTCACGGGATT | 2.03 | 0.997 |
| U6 snRNA | u6wg_chr6_15577690_15577792_+ | CCGATAAAATTGGAACGATACAGAG | TCGATTTGTGCGTGTCATCCT | 2.13 | 0.995 |
| ACT | EC969944 | GACAATGGATGGACCAGATTCA | CTTGCATCCCTCAGCACCTT | 1.99 | 0.997 |
| UBQ | EC929411 | GTGGGTGCAGACGTGCATAA | CCTTGATGCAATTGGCTAGGA | 2.14 | 0.991 |
| GAPDH | CB973647.1 | AGCCCTCAACGAGAAGTTCTTG | TCGATCACACGGGAGCTGTA | 1.97 | 0.995 |
| EF1 | EC959059 | AACCGAAAGCACCTCGATCA | TGGTTGAGTCCTTTGCTTTTCC | 1.98 | 0.999 |
| miR160e | MIMAT0005656 | GCGGCGGTGCCTGGCTCC | GTGCAGGGTCCGAGGT | 2.10 | 0.982 |
| miR164a | MIMAT0005658 | GCGGCGGTGGAGAAGCAG | GTGCAGGGTCCGAGGT | 2.05 | 0.997 |
| miR168 | MIMAT0005675 | GCGGCGGTCGCTTGGTGC | GTGCAGGGTCCGAGGT | 2.17 | 0.996 |
| miR156a | MIMAT0005640 | GCGGCGGTGACAGAAGAG | GTGCAGGGTCCGAGGT | 2.09 | 0.977 |
| miR159a | MIMAT0005648 | GCGGCGGCTTGGAGTGAA | GTGCAGGGTCCGAGGT | 2.37 | 0.976 |
| miR162 | MIMAT0005657 | GCGGCGGTCGATAAACCT | GTGCAGGGTCCGAGGT | 2.57 | 0.989 |
| miR167a | MIMAT0005670 | GCGGCGGTGAAGCTGCCA | GTGCAGGGTCCGAGGT | 3.31 | 0.911 |
| miR169a | MIMAT0005676 | GCGGCGGCAGCCAAGGAT | GTGCAGGGTCCGAGGT | 2.34 | 0.987 |
| miR396a | MIMAT0005724 | GCGGCGGTTCCACAGCTT | GTGCAGGGTCCGAGGT | 2.46 | 0.958 |
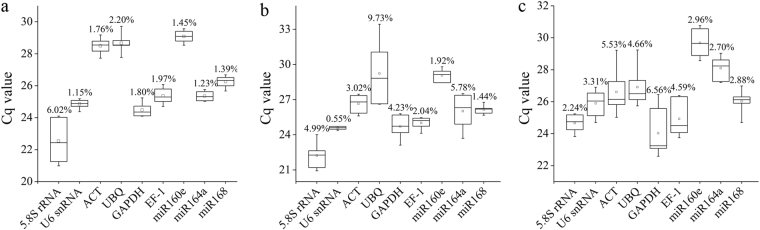
The mean Cq values of candidate reference genes in all 18 samples (six samples under each stress condition) ranged from 23.14 (5.8S rRNA) to 29.28 (miR160e) (Fig. 1). 5.8S rRNA was the most highly expressed gene under salinity and cold stresses, and GADPH showed the highest level of expression under drought stress, whereas miR160e expressed with the lowest level under all three stress conditions. For CV values, the highest and lowest values under the three different treatments were 6.02% (5.8S rRNA) and 1.15% (U6 snRNA) in salinity stress; 9.73% (UBQ) and 0.55% (U6 snRNA) in cold stress; and 6.56% (GAPDH) and 2.24% (5.8S rRNA) in drought stress, respectively (Fig. 1). The rank of mean CV values of candidate reference genes was U6 snRNA < miR168 < miR160e < EF1 < miR164a < ACT < GAPDH < 5.8S rRNA < UBQ, which represented the fluctuation difference of Cq values.
Figure 1.
The expression levels of candidate reference genes. The Cq values generated by qRT-PCR of six time points under different abiotic stresses are shown as box plots. Whisker caps, boxes, lines and square boxes represent maximum/ minimum, 25/75 percentiles, median and mean, respectively. Long boxes and whiskers indicate greater variation. (a) salinity stress, (b) cold stress, (c) drought stress.
Analysis of gene expression stability
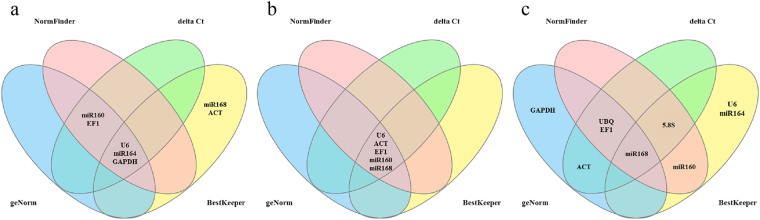
Four algorithms including geNorm, NormFinder, deltaCt and Bestkeeper were used to evaluate the expression stability of the candidate reference genes. geNorm is an algorithm of which the core concept is to compare the average pairwise expression ratio between candidate reference genes. The least stable gene can be excluded by a stepwise approach, and then the M value is provided to evaluate a candidate gene. As the algorithm suggests, a gene with an M value over 1.5 is not suitable to be a reference gene. The NormFinder algorithm uses ANOVA-based mathematical analysis to generate the inter- and intra-group variations of candidate genes. For the Bestkeeper algorithm, the lowest coefficient of variance (CV) and standard deviation (SD) are the evaluation criteria for stably expressed reference genes. The principle of the deltaCt algorithm is similar to that of the algorithm of geNorm; the deltaCt method is used to compare the expression stability of pairs of genes. The expression stability values generated from these algorithms were unified as the stability value (SV), and the SVs of candidate reference genes under salinity, cold and drought stress conditions are shown in Table 3–5, respectively. Additionally, the top 5 most stable reference genes generated by each algorithm are presented as the Venn plot in Fig. 2.
Table 3.
The expression stability of candidate reference genes under salinity stress.
| Rank | geNorm | NormFinder | deltaCt | Bestkeeper | ||||
|---|---|---|---|---|---|---|---|---|
| Gene | SV | Gene | SV | Gene | SV | Gene | SV | |
| 1 | miR160e | 0.216 | U6 snRNA | 0.012 | U6 snRNA | 0.617 | U6 snRNA | 0.208 |
| 2 | miR164a | 0.216 | miR164a | 0.012 | miR164a | 0.633 | miR164a | 0.269 |
| 3 | U6 snRNA | 0.228 | EF1 | 0.014 | miR160e | 0.685 | miR168 | 0.280 |
| 4 | GAPDH | 0.425 | GAPDH | 0.015 | GAPDH | 0.701 | GAPDH | 0.333 |
| 5 | EF1 | 0.475 | miR160e | 0.016 | EF1 | 0.711 | ACT | 0.363 |
| 6 | miR168 | 0.531 | ACT | 0.018 | ACT | 0.732 | miR160e | 0.370 |
| 7 | ACT | 0.578 | miR168 | 0.023 | miR168 | 0.791 | EF1 | 0.379 |
| 8 | UBQ | 0.620 | UBQ | 0.024 | UBQ | 0.842 | UBQ | 0.410 |
| 9 | 5.8S rRNA | 0.788 | 5.8S rRNA | 0.058 | 5.8S rRNA | 1.375 | 5.8S rRNA | 1.142 |
Table 5.
The expression stability of candidate reference genes under drought stress.
| Rank | geNorm | NormFinder | deltaCt | Bestkeeper | ||||
|---|---|---|---|---|---|---|---|---|
| Gene | SV | Gene | SV | Gene | SV | Gene | SV | |
| 1 | ACT | 0.255 | miR168 | 0.024 | EF1 | 1.120 | 5.8S rRNA | 0.427 |
| 2 | UBQ | 0.255 | UBQ | 0.027 | UBQ | 1.129 | miR168 | 0.484 |
| 3 | GAPDH | 0.451 | EF1 | 0.027 | miR168 | 1.140 | miR164a | 0.635 |
| 4 | EF1 | 0.519 | miR160e | 0.030 | ACT | 1.236 | miR160e | 0.690 |
| 5 | miR168 | 0.845 | 5.8S rRNA | 0.035 | 5.8S rRNA | 1.259 | U6 snRNA | 0.703 |
| 6 | miR160e | 0.990 | ACT | 0.036 | miR160e | 1.260 | UBQ | 0.914 |
| 7 | 5.8S rRNA | 1.104 | miR164a | 0.042 | GAPDH | 1.370 | EF1 | 0.952 |
| 8 | miR164a | 1.194 | GAPDH | 0.050 | miR164a | 1.427 | ACT | 1.092 |
| 9 | U6 snRNA | 1.281 | U6 snRNA | 0.053 | U6 snRNA | 1.584 | GAPDH | 1.320 |
Figure 2.
The top 5 most stable reference genes generated by geNorm, NormFinder, deltaCt and Bestkeeper. The blue, pink, green and yellow circles each contain the top 5 most stable reference genes of geNorm, NormFinder, deltaCt and Bestkeeper, respectively. The genes in the overlap area are the ones confirmed as the top 5 most stable reference genes by more than one algorithm. (a) salinity stress, (b) cold stress, (c) drought stress.
Three stress treatments were conducted in this experiment, and H2O2 (Hydrogen Peroxide) content, MDA (Malondialdehyde) content and SOD (Superoxide Dismutase) activity were measured to reflect the stress severity (Fig. S2). In salinity-treated samples, U6 snRNA and miR164a ranked at the top 3 in all algorithms, indicating their stable expression under the condition of salinity stress. However, the least stable of all candidate genes calculated by the four algorithms under salinity stress were 5.8S rRNA and UBQ. For other candidate genes, the rankings were variable among different algorithms. For example, miR168 was the 6th, 7th, 7th, and 3rd most stable gene evaluated by geNorm, NormFinder, deltaCt and Bestkeeper, respectively (Table 3). Under the cold stress condition, the most stable reference genes were similar across the four algorithms (Table 4), and collectively, U6 snRNA and miR168 were the first and second most stable genes according to three algorithms, whereas U6 snRNA and ACT were the top two stable genes evaluated by NormFinder. For the least stable genes, the results given by geNorm, NormFinder, deltaCt and Bestkeeper were consistent, which confirmed that miR164a and UBQ were unstable under cold stress. As noted above, candidate genes with a SV (M) over 1.5 are not suitable to be a reference gene according to the algorithm of geNorm. Among all samples under different treatments, the SVs of 9 candidate genes were all below 1.5, except for UBQ in cold-treated samples. UBQ had a SV of 1.525 under cold stress, which indicated that UBQ was not a suitable reference gene for cold-treated grapevine. For drought stress, the SV rankings of the candidate genes were variable among algorithms (Table 5). However, UBQ and miR168 were indicated as the top 2 most stable genes by at least two algorithms. For the genes with low stability, U6 snRNA was the least stable gene from geNorm, NormFinder and deltaCt, whereas GAPDH was evaluated as the least stable gene by Bestkeeper. Furthermore, miR164a showed an unstable expression trait evaluated by most algorithms under drought stress.
Table 4.
The expression stability of candidate reference genes under cold stress.
| Rank | geNorm | NormFinder | deltaCt | Bestkeeper | ||||
|---|---|---|---|---|---|---|---|---|
| Gene | SV | Gene | SV | Gene | SV | Gene | SV | |
| 1 | miR160e | 0.258 | U6 snRNA | 0.007 | U6 snRNA | 1.071 | U6 snRNA | 0.106 |
| 2 | miR168 | 0.258 | ACT | 0.011 | miR168 | 1.169 | miR168 | 0.259 |
| 3 | U6 snRNA | 0.363 | miR160e | 0.021 | ACT | 1.208 | EF1 | 0.396 |
| 4 | EF1 | 0.593 | miR168 | 0.021 | miR160e | 1.217 | miR160e | 0.458 |
| 5 | ACT | 0.709 | EF1 | 0.022 | EF1 | 1.247 | ACT | 0.692 |
| 6 | GAPDH | 0.824 | GAPDH | 0.032 | GAPDH | 1.373 | 5.8S rRNA | 0.796 |
| 7 | 5.8S rRNA | 0.939 | 5.8S rRNA | 0.047 | 5.8S rRNA | 1.538 | GAPDH | 0.867 |
| 8 | miR164a | 1.164 | miR164a | 0.076 | miR164a | 2.114 | miR164a | 1.205 |
| 9 | UBQ | 1.525 | UBQ | 0.090 | UBQ | 2.789 | UBQ | 2.426 |
Considering the limits and biases of different algorithms, four algorithms were performed to evaluate the expression stability of candidate genes. To reach a consensus on the rankings of optimal reference genes, the results generated by the four algorithms were aggregated by the Cross-Entropy Monte Carlo approach using Rpackage RankAggreg36. Based on the aggregated rankings (Fig. 3), the most and least stable candidate genes under different treatments were almost consistent with the results of ranking orders provided by each algorithm (Tables 3–5). U6 snRNA and miR164a, U6 snRNA and miR160e, and miR168 and UBQ were the candidate genes ranked at the top 2 under salinity, cold and drought stress, respectively, whereas the least stable candidate genes under these stresses were UBQ and 5.8S rRNA, miR164a and UBQ, and miR164a and U6 snRNA, respectively.
Figure 3.
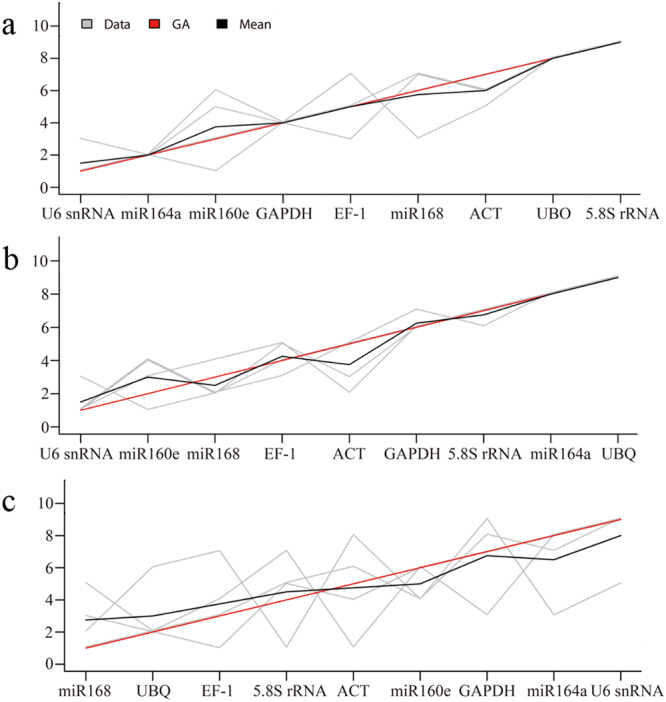
Consensus stability ranking generated by a Monte Carlo algorithm. The R package RankAggreg (Version 0.5) was used to integrate ranking lists of four stability algorithms (geNorm, NormFinder, deltaCt and Bestkeeper) by a Monte Carlo model. Each grey line represents the ranking of an individual algorithm; the black and red lines represent the mean ranking and consensus ranking generated by the Monte Carlo algorithm, respectively. (a) salinity stress, (b) cold stress, (c) drought stress.
Optimal combinations of reference genes under salinity, cold and drought stresses
As noted above, one reference gene may result in bias during qRT-PCR. Using a reference gene combination can obtain more accurate and reliable normalization of gene expression data compared with that of a single reference gene for qRT-PCR26. The optimal number of multiple reference genes was determined by geNorm through calculating the V value (pairwise variation) ratio (V = Vn/Vn + 1) between normalization factors (NF). The value of 0.15 was set as the threshold of the V value to determine whether an additional reference gene was required in the reference gene set. When Vn/Vn + 1 ≤ 0.15, the n most stable genes were sufficient to create a stable reference gene combination. As shown in Fig. 4, the V2/3 of candidate genes under salinity and cold stress was 0.069 and 0.133, respectively, which were below 0.15. Therefore, the optimal reference gene sets under salinity and cold stress were miR160e + miR164a and miR160e + miR168, respectively. For drought stress, because V2/3 = 0.179 was more than 0.15, whereas V3/4 = 0.130 was less than 0.15, the top three candidate genes ACT, UBQ and GAPDH were the best genes to normalize the qRT-PCR results.
Figure 4.
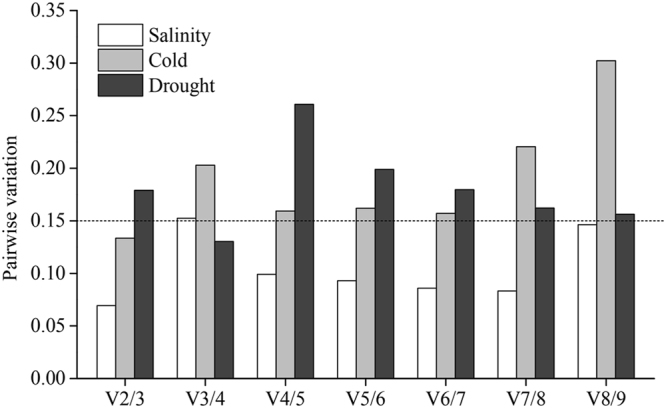
Determination of the optimal number of candidate reference genes by geNorm analysis. Pairwise variation (Vn/n + 1) between the normalization factors NFn and NFn+1 was used to determine the optimal number of multiple reference genes. When the value of Vn/n + 1 is ≤0.15, the addition of one or more reference genes into the reference gene combination would not be necessary.
Verification of reference genes
For each of the three stress conditions, the 3 most stable single reference genes and a gene set were selected to validate the effectiveness of reference genes. Based on the rankings and V values, which were generated by RankAggreg and geNorm, respectively, the reference genes and gene set were selected as follow: U6 snRNA, miR164a, miR160e, and miR160e + miR164a for salinity stress; U6 snRNA, miR160e, miR168, and miR160e + miR168 for cold stress; miR168, UBQ, EF1, and ACT + UBQ + GAPDH for drought stress. According to previous studies, miR319 responds to salinity and cold stresses, whereas miR408 is associated with drought stress in different plant species37. Thus, miR319 and miR408 were chosen as target genes to perform qRT-PCR to validate the normalization efficiency of selected reference genes. When normalized by U6 snRNA, miR164a, miR160e and miR160e + miR164a, the expression of miR319 under salinity stress increased through time until the peak was reached at 24 h, followed by a slight decrease at 48 h (Fig. 5a). The expression patterns normalized by different reference genes or gene set showed consistency. However, at the time points of 12 and 24 h, a significant difference was observed between U6 snRNA and the other normalization factors. Under cold stress, miR319 expression decreased at time points of 8, 12 and 24 h. The expression levels of miR319 normalized by different reference genes were not exactly the same (Fig. 5b); although the normalization results of U6 snRNA and miR168 were similar, the expression normalized by miR160e and miR160e + miR168 showed no significant differences at most time points. For drought stress, the expression patterns of miR408 with different normalization factors were not as consistent as those under the salinity and cold stresses (Fig. 5c). The expression of miR408 increased at the time points of 3 and 9 d, followed by a gradual decrease. Significant differences occurred at all time points except for d 0 and were primarily between miR168 and the other reference genes.
Figure 5.
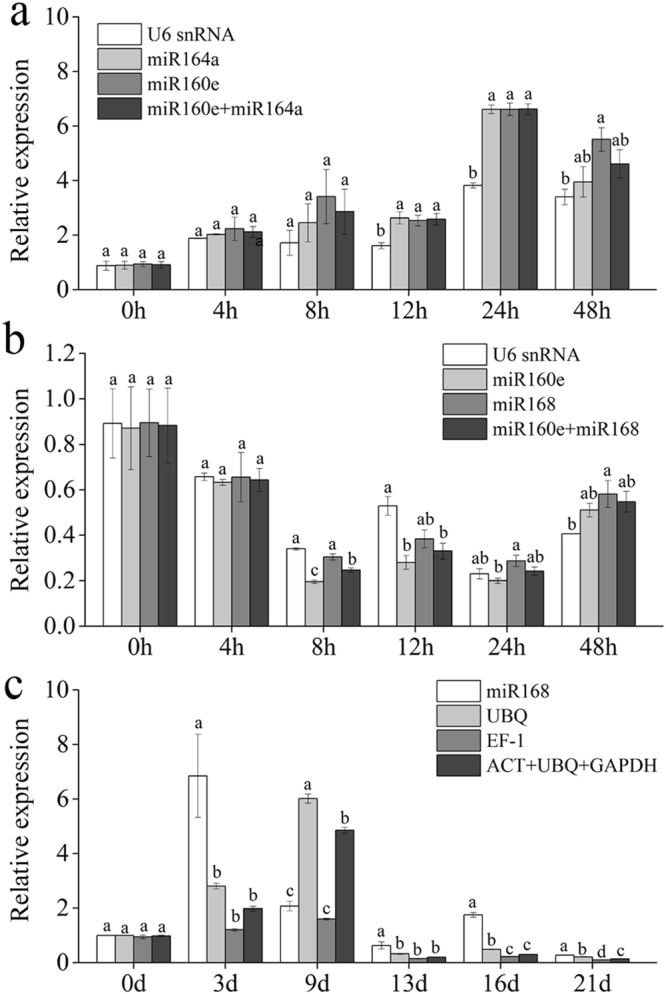
Relative expression of target genes. Different miRNAs were chosen to be target genes for different abiotic stresses. Selected reference genes or gene combination was used to normalize the expression results. (a) salinity stress. The columns represent the relative expression of miR319. (b) cold stress. The columns represent the relative expression of miR319. (c) drought stress. The columns represent the relative expression of miR408.
Discussion
qRT-PCR is a widely used method for gene expression analysis because of the high sensitivity and accuracy21,38. However, the final result of qRT-PCR can be easily influenced by technical error because of the advantage of high sensitivity39, and such bias may be caused by RNA extraction, cDNA reverse transcription, or the qRT-PCR process27. To obtain a more reliable result, the most common approach is to introduce one or more reference genes as the normalization factors25,40. According to previous research, the most suitable reference genes for a given experimental condition, such as a biotic or abiotic stress, may be different from the suggested reference genes for other conditions41,42. The optimal reference genes should be validated for different species, treatments or specific tissues.
Several studies have conducted reference gene validation experiments for grapevine mRNA qRT-PCR but not for grapevine miRNA qRT-PCR43,44. In these studies, protein-coding genes (such as ACT and UBQ) or ribosome RNAs (such as 5.8S rRNA and 18S rRNA) are always considered the most appropriate normalization factors for mRNA quantification. However, whether these selected reference genes are also suitable for grapevine miRNA qRT-PCR remains unclear. Because of the lack of supporting evidence, the widely used reference genes in other species, such as U6 snRNA and 5.8S rRNA, are usually chosen for normalizing grapevine miRNA quantitation data28. In addition to mRNAs, rRNAs and snRNAs, the potential of miRNAs as reference genes has received much attention recently. In some plant species, such as lettuce31, wheat29 and peach32, miRNAs are more stable than the currently used reference genes under specific conditions. Furthermore, compared with other types of RNAs, miRNA is much shorter in length, and therefore, miRNA extraction and reverse transcription are different from the protocols for longer RNAs28. Using miRNAs as reference genes could avoid the bias caused by different RNA extraction and reverse transcription processes between reference genes and target genes45. Therefore, to guarantee a comprehensive comparison, the candidate reference genes of this research included traditional housekeeping genes (ACT, UBQ, GAPDH and EF1), widely used reference genes in miRNA qRT-PCR (5.8S rRNA and U6 snRNA), and conserved miRNAs (miR160e, miR164a and miR168). The genes miR156a, miR159a, miR162, miR167a, miR169a, and miR396a were excluded from the candidate list because of unsatisfactory amplification efficiency or regression coefficient of the standard curve.
Because of no consensus about the most reliable gene expression stability evaluation algorithm, four algorithms (geNorm, NormFinder, deltaCt, Bestkeeper) were used to minimize the bias46. U6 snRNA was the most stable reference gene under salinity and cold stress in this research, which ranked first in NormFinder, deltaCt, Bestkeeper and in the comprehensive ranking. U6 snRNA is one of the most commonly used reference genes in miRNA qRT-PCR27 and is reported as the most stable gene in reference gene selection research of citrus45 and tea47. According to previous research, U6 snRNA is highly conserved among species48 and functions as a primary component of the RNA spliceosome, participating in the process of mRNA precursors49,50. However, U6 snRNA ranked at the bottom under drought stress, as evaluated by geNorm, NormFinder and deltaCt, which could be because drought stress is lethal for plants5. Under drought stress, maintaining normal physiology processes in leaves is difficult, which leads to the decrease of mRNA content and that of U6 snRNA in the late stage of drought stress51. Another traditional miRNA reference gene 5.8S rRNA was less stable than U6 snRNA under salinity and cold stresses but was more stable than U6 snRNA under drought stress. This result indicates that no reference gene is suitable for every experimental condition. The test of expression stability under a particular experimental condition is always required, when not previously confirmed.
miR160e, miR164a and miR168 were ranked at the top one or top two under cold, salinity and drought stresses, indicating these genes were more stable than protein-coding genes and 5.8S rRNA. This result is consistent with previous studies29,31,32, indicating miRNAs are qualified to be miRNA qRT-PCR reference genes for grapevine under abiotic stresses. These 3 miRNAs all belong to conserved miRNA families and undertake important physiological functions. The target genes of miR160e are ARF family members that respond to auxin and associate with the development of multiple organs, such as seeds, leaves, roots and flowers52. For miR168, the function is regulation of the ARGONAUTE1 protein53, which is the primary component of the RNA-induced slicing complex (RISC) and influences plant growth and development54,55. Because of the involvement in fundamental physiological processes, the stable expression of miR160e and miR168 was a reasonable result. miR164a reportedly negatively regulates the expression of NAC, a transcription factor that responds to salinity stress. However, in this research, miR164a showed relatively high stability under salinity stress, which is consistent with a previous report in maize56. In the maize experiment, NAC showed a more dramatic response to salinity stress after treatment, whereas miR164a only changed moderately, decreasing to approximately 90% relative expression on average compared with that of the control.
As reported previously, using a reference gene set instead of a single reference gene can provide more reliable normalized results25,26. According to geNorm, miR160e + miR164a, miR160e + miR168, and ACT + UBQ + GAPDH were selected as the most suitable reference gene sets for salinity, cold and drought stresses, respectively. To validate the selected reference genes or gene sets, the top 3 genes of comprehensive ranking under each experimental treatment with the gene sets mentioned above were used to normalize the expression of target genes. Note that the genes with a low or middle ranking also have the potential to be reference genes as long as their SVs meet the requirements of algorithms in some other stress conditions42. As shown in Fig. 5, under salinity and cold stresses, the quantification results of target gene miR319 exhibited similar patterns when normalized by the selected reference genes or gene sets, demonstrating that the tested reference gene or gene set was mutually validated. For drought stress, when normalized by miR168 and the others, the expression of target gene miR408 showed disagreement. The phenomenon that different reference genes lead to different expression patterns is also observed in other research, further indicating the importance of selecting the proper reference gene31,57.
In conclusion, in grapevine, U6 snRNA, miR164a, miR160e and miR160e + miR164a are all suitable reference genes or gene set under salinity stress, whereas U6 snRNA, miR160e, miR168 and miR160e + miR168 can be used for qRT-PCR normalization under cold stress. Considering the target gene was miRNA, and miR168 was the top 1 reference gene of comprehensive ranking under drought stress, we suggest miR168 as the reference gene for miRNA qRT-PCR, rather than the others.
Methods
Plant materials and treatments
This study was performed on one-year-old Vitis vinifera L. cv. Muscat Hamburg grown in the Fruit Experimental Greenhouse of Shanghai Jiao Tong University. These self-rooted grapevines were potted into a soil mixture (garden soil: organic fertilizer: perlite = 1: 1: 1) in 15 L pots. Fifty-four plants were divided equally into three groups to conduct stress treatments after cultivating under greenhouse conditions for 10 weeks. For salinity stress, plants were treated with 200 mM NaCl water solution for several days (sampling times are listed below); for cold stress, plants were exposed to low temperature in an incubator (4 °C, 16 h light/8 h dark); for drought stress, plants were maintained with the water supply withheld. Leaves were collected at 0, 4, 8, 12, 24, and 48 h after salinity and cold treatments and at 0, 3, 9, 13, 16, and 21 d after drought treatment. Three biological replications were included at each time point. All samples were immediately frozen in liquid nitrogen and stored at −80 °C until used. H2O2 content, MDA content and SOD activity were measured using Hydrogen Peroxide assay kit (Nanjing Jiancheng, Nanjing, Jiangsu, China), Malondialdehyde assay kit (Nanjing Jiancheng, Nanjing, Jiangsu, China) and Total Superoxide Dismutase assay kit (Nanjing Jiancheng, Nanjing, Jiangsu, China), respectively.
RNA extraction and cDNA synthesis
Total RNA was isolated using RNAplant Plus Reagent (TIANGEN, Beijing, China), and genomic DNA was removed using Recombinant DNase I (TAKARA Biotechnology, Dalian, Liaoning, China). RNA integrity was evaluated by 2% agarose gel electrophoresis; whereas a NanoDrop 2000 measured RNA concentration and quality (Thermo Scientific, Waltham, Massachusetts, USA). After analysis, samples with an OD 260/280 ratio from 1.9 to 2.1 and an OD 260/230 ratio from 1.8 to 2.2 were chosen for the next step.
The first-strand cDNA was synthesized from 1 μg of total RNA according to the instructions of a FastKing RT Kit (TIANGEN, Beijing, China). The 20 μL reaction system was performed at 42 °C for 15 min and 95 °C for 3 min. For miRNA amplification, cDNA synthesis was conducted using the same protocol described above, except stem-loop reverse transcription primers were used instead of oligo (dT).
Selection of reference genes and primer design
A total of 15 candidate reference genes were selected. The sequences of 5.8S rRNA and U6 snRNA, the two most commonly used reference genes for miRNA qRT-PCR, were obtained from NCBI (GenBank Accession: KT344661.1) and reported by Kullan et al.58, respectively. The sequences of 4 traditional housekeeping genes, ACT, UBQ, GAPDH and EF1, were reported by Reid et al.59. According to the miRNA high-throughput sequencing results in grapevine28,60 and reference gene selection research for miRNA qRT-PCR in other plant species27,29,32, miR156a, miR159a, miR160e, miR162, miR164a, miR167a, miR168, miR169a, and miR396a were chosen as potential normalization factors for miRNA qRT-PCR in grapevine.
The primers of 5.8S rRNA, ACT, UBQ, GAPDH and EF1 were designed by Primer Express (3.0.1), whereas the primers of U6 snRNA were obtained from Kullan et al.58. For miRNA cDNA synthesis, the stem-loop primers were designed according to Chen21, which consisted of 44 conserved and 8 variable nucleotides that were specific to the 3′ end of the miRNA sequences. For miRNA qRT-PCR, the forward primers were designed using 1–11th nucleotides of miRNA sequences, with 7 nucleotides (GCGGCGG) added to the 5′ end to increase the length of primers; whereas the reverse primers were universal.
qRT-PCR analysis
qRT-PCR was performed on a LightCycler480 System (Roche, New York, USA). The 10 μL reaction system contained 5 μL of SYBR® Premix Ex Taq (TAKARA Biotechnology, Dalian, China), 0.3 μM each primer, 2 μL of diluted cDNA (miRNA cDNA and other cDNA diluted 25-fold and 50-fold, respectively) and 2.4 μL of ddH2O. The procedure was conducted as follows: 95 °C for 10 min, amplification for 40 cycles (95 °C for 20 s, 60 °C for 20 s, 75 °C for 20 s). Negative controls were set to test for possible contamination, and the melting curves were evaluated in each reaction to ensure the specificity of amplified action. Each sample was evaluated in three technical triplicates.
Using a series of 5-fold diluted cDNA as templates, the standard curves were generated for each candidate reference gene. The correlation coefficient (R2) and slope were obtained from the linear regression model created by the LightCycle 480 system, and the PCR efficiency (E) was calculated using the following formula:
Data analyses
The expression stability of candidate reference genes was calculated using geNorm, NormFinder, deltaCt and Bestkeeper. As the algorithms required, Cq values of all samples (six time points under salinity, cold and drought stress) were transformed into Q values using the formula Q = EΔCq. ΔCq indicates the difference between the sample Cq and the lowest Cq among the samples of six time points. Bestkeeper and deltaCt were performed by the online analysis tool RefFinder (http://fulxie.0fees.us/?type=reference). To integrate the ranks of the four algorithms, RankAggreg Rpackage (Version 0.5) was used to generate a consensus ranking of candidate reference genes.
Reference gene validation
As reported by Thiebaut et al.61, Zhou et al.62 and Ma et al.37, miR319 is responsive to salt and cold stress, whereas miR408 is responsive to drought stress. Therefore, miR319 (salinity and cold stress) and miR408 (drought stress) were chosen as the target genes to validate the stability of selected reference genes. The relative expression of the target genes at six time points was calculated using the 2−ΔΔCt method, and the significant difference analysis was performed using the SPSS statistical software package (19.0).
Electronic supplementary material
Acknowledgements
The Shanghai Pujiang Program (No. 16PJ1404900) and the Shanghai Natural Science Foundation (Grant No. 17ZR1448800) sponsored this work.
Author Contributions
C. M. and S. W. designed the research. M. L. performed the research. M. L., Z. G., H. L., Q. L., C. Z., W. X., S. S., C. M. and S. W. analysed the data. M. L. wrote the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-22743-6.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chao Ma, Email: chaoma2015@sjtu.edu.cn.
Shiping Wang, Email: fruit@sjtu.edu.cn.
References
- 1.Rocheta M, et al. Transcriptomic comparison between two Vitis vinifera L. varieties (Trincadeira and Touriga Nacional) in abiotic stress conditions. Bmc Plant Biol. 2016;16:224. doi: 10.1186/s12870-016-0911-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones GV, White MA, Cooper OR, Storchmann K. Climate Change and Global Wine Quality. Climatic Change. 2005;73:319–343. doi: 10.1007/s10584-005-4704-2. [DOI] [Google Scholar]
- 3.Mosedale JR, Abernethy KE, Smart RE, Wilson RJ, Maclean IM. Climate change impacts and adaptive strategies: lessons from the grapevine. Global change biology. 2016;22:3814–3828. doi: 10.1111/gcb.13406. [DOI] [PubMed] [Google Scholar]
- 4.Shinozaki K, Yamaguchi-Shinozaki K. Molecular responses to dehydration and low temperature differences and cross talk between two stress signaling pathways. Current Opinion in Plant Biology. 2000;3:217–223. doi: 10.1016/S1369-5266(00)00067-4. [DOI] [PubMed] [Google Scholar]
- 5.Zhu JK. Abiotic Stress Signaling and Responses in Plants. Cell. 2016;167:313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fedoroff NV, et al. Radically rethinking agriculture for the 21st century. Science. 2010;327:833–834. doi: 10.1126/science.1186834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larcher, W. Physiological Plant Ecology 4th edtion. (Springer, 2003).
- 8.Krasensky J, Jonak C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J Exp Bot. 2012;63:1593–1608. doi: 10.1093/jxb/err460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhargava S, Sawant K, Tuberosa R. Drought stress adaptation: metabolic adjustment and regulation of gene expression. Plant Breeding. 2013;132:21–32. doi: 10.1111/pbr.12004. [DOI] [Google Scholar]
- 10.Chinnusamy V, Zhu JK. Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol. 2009;12:133–139. doi: 10.1016/j.pbi.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang, S. et al. PASmiR:a literature-curated database for miRNA molecular regulation in plant response to abiotic stress. Bmc Plant Biol (2013). [DOI] [PMC free article] [PubMed]
- 12.Hackenberg M, Gustafson P, Langridge P, Shi BJ. Differential expression of microRNAs and other small RNAs in barley between water and drought conditions. Plant Biotechnol J. 2015;13:2–13. doi: 10.1111/pbi.12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.Covarrubias AA, Reyes JL. Post-transcriptional gene regulation of salinity and drought responses by plant microRNAs. Plant Cell Environ. 2010;33:481–489. doi: 10.1111/j.1365-3040.2009.02048.x. [DOI] [PubMed] [Google Scholar]
- 15.Kidner CA, Martienssen RA. The developmental role of microRNA in plants. Curr Opin Plant Biol. 2005;8:38–44. doi: 10.1016/j.pbi.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 16.Jones-Rhoades, M. W., Bartel, D. P. & Bartel, B. MicroRNAs and Their Regulatory Roles in Plants. Annual Review of Plant Biology57, 19-53, 10.1146/ (2006). [DOI] [PubMed]
- 17.Sreenivasulu N, Sopory SK, Kavi Kishor PB. Deciphering the regulatory mechanisms of abiotic stress tolerance in plants by genomic approaches. Gene. 2007;388:1–13. doi: 10.1016/j.gene.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 18.Schwarzkopf M, Pierce NA. Multiplexed miRNA northern blots via hybridization chain reaction. Nucleic Acids Res. 2016;44:e129. doi: 10.1093/nar/gkw503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu D, et al. The use of miRNA microarrays for the analysis of cancer samples with global miRNA decrease. Rna. 2013;19:876–888. doi: 10.1261/rna.035055.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han X, et al. Integration of small RNAs, degradome and transcriptome sequencing in hyperaccumulator Sedum alfredii uncovers a complex regulatory network and provides insights into cadmium phytoremediation. Plant Biotechnol J. 2016;14:1470–1483. doi: 10.1111/pbi.12512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen C, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi R, Chiang V. Facile means for quantifying microRNA expression by real-time PCR. BioTechniques. 2005;39:519–525. doi: 10.2144/000112010. [DOI] [PubMed] [Google Scholar]
- 23.Omidvar V, Mohorianu I, Dalmay T, Fellner M. MicroRNA Regulation of Abiotic Stress Response in Male-Sterile Tomato Mutant. The Plant Genome. 2015;8:0. doi: 10.3835/plantgenome2015.02.0008. [DOI] [PubMed] [Google Scholar]
- 24.Akdogan G, Tufekci ED, Uranbey S, Unver T. miRNA-based drought regulation in wheat. Funct Integr Genomics. 2016;16:221–233. doi: 10.1007/s10142-015-0452-1. [DOI] [PubMed] [Google Scholar]
- 25.Bustin SA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical chemistry. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 26.Vandesompele, J., De Preter, K. & Pattyn, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. (2002). [DOI] [PMC free article] [PubMed]
- 27.Yang Y, et al. Selection of Reference Genes for Normalization of MicroRNA Expression by RT-qPCR in Sugarcane Buds under Cold Stress. Front Plant Sci. 2016;7:86. doi: 10.3389/fpls.2016.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pantaleo V, et al. Novel functional microRNAs from virus-free and infected Vitis vinifera plants under water stress. Scientific reports. 2016;6:20167. doi: 10.1038/srep20167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng H, et al. Selection of suitable inner reference genes for relative quantification expression of microRNA in wheat. Plant physiology and biochemistry: PPB / Societe francaise de physiologie vegetale. 2012;51:116–122. doi: 10.1016/j.plaphy.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 30.Kulcheski FR, Marcelino-Guimaraes FC, Nepomuceno AL, Abdelnoor RV, Margis R. The use of microRNAs as reference genes for quantitative polymerase chain reaction in soybean. Analytical biochemistry. 2010;406:185–192. doi: 10.1016/j.ab.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 31.Borowski JM, et al. Selection of candidate reference genes for real-time PCR studies in lettuce under abiotic stresses. Planta. 2014;239:1187–1200. doi: 10.1007/s00425-014-2041-2. [DOI] [PubMed] [Google Scholar]
- 32.Luo X, et al. Selection of suitable inner reference genes for normalisation of microRNA expression response to abiotic stresses by RT-qPCR in leaves, flowers and young stems of peach. Sci Hortic-Amsterdam. 2014;165:281–287. doi: 10.1016/j.scienta.2013.10.030. [DOI] [Google Scholar]
- 33.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 34.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 35.Silver N, Best S, Jiang J, Thein SL. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC molecular biology. 2006;7:33. doi: 10.1186/1471-2199-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pihur V, Datta S, Datta S. RankAggreg, an R package for weighted rank aggregation. BMC bioinformatics. 2009;10:62. doi: 10.1186/1471-2105-10-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma C, Burd S, Lers A. miR408 is involved in abiotic stress responses in Arabidopsis. Plant J. 2015;84:169–187. doi: 10.1111/tpj.12999. [DOI] [PubMed] [Google Scholar]
- 38.Ginzinger DG. Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Experimental Hematology. 2002;30:503–512. doi: 10.1016/S0301-472X(02)00806-8. [DOI] [PubMed] [Google Scholar]
- 39.Radonić A, et al. Guideline to reference gene selection for quantitative real-time PCR. Biochem Bioph Res Co. 2004;313:856–862. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- 40.Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes and immunity. 2005;6:279–284. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- 41.Guenin S, et al. Normalization of qRT-PCR data: the necessity of adopting a systematic, experimental conditions-specific, validation of references. J Exp Bot. 2009;60:487–493. doi: 10.1093/jxb/ern305. [DOI] [PubMed] [Google Scholar]
- 42.Niu, X. et al. Reference Gene Selection for qRT-PCR Normalization Analysis in kenaf (Hibiscus cannabinus L.) under Abiotic Stress and Hormonal Stimuli. Front Plant Sci8, 10.3389/fpls.2017.00771 (2017). [DOI] [PMC free article] [PubMed]
- 43.Borges AF, Fonseca C, Ferreira RB, Lourenco AM, Monteiro S. Reference gene validation for quantitative RT-PCR during biotic and abiotic stresses in Vitis vinifera. Plos One. 2014;9:e111399. doi: 10.1371/journal.pone.0111399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coito JL, Rocheta M, Carvalho L, Amancio S. Microarray-based uncovering reference genes for quantitative real time PCR in grapevine under abiotic stress. BMC research notes. 2012;5:220. doi: 10.1186/1756-0500-5-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kou SJ, et al. Selection and validation of suitable reference genes for miRNA expression normalization by quantitative RT-PCR in citrus somatic embryogenic and adult tissues. Plant Cell Rep. 2012;31:2151–2163. doi: 10.1007/s00299-012-1325-x. [DOI] [PubMed] [Google Scholar]
- 46.Guo J, Ling H, Wu Q, Xu L, Que Y. The choice of reference genes for assessing gene expression in sugarcane under salinity and drought stresses. Scientific reports. 2014;4:7042. doi: 10.1038/srep07042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song, H. et al. Selection and Verification of Candidate Reference Genes for Mature MicroRNA Expression by Quantitative RT-PCR in the Tea Plant (Camellia sinensis). Genes7, 10.3390/genes7060025 (2016). [DOI] [PMC free article] [PubMed]
- 48.Brow DA, Guthrie C. Splicesomal RNA U6 is remarkably conserved from yeast to mammals. Nature. 1988;334:213–218. doi: 10.1038/334213a0. [DOI] [PubMed] [Google Scholar]
- 49.Bessonov S, Anokhina M, Will CL, Urlaub H, Luhrmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846–850. doi: 10.1038/nature06842. [DOI] [PubMed] [Google Scholar]
- 50.Mroczek S, Dziembowski A. U6 RNA biogenesis and disease association. Wiley interdisciplinary reviews. RNA. 2013;4:581–592. doi: 10.1002/wrna.1181. [DOI] [PubMed] [Google Scholar]
- 51.Lenka SK, Katiyar A, Chinnusamy V, Bansal KC. Comparative analysis of drought-responsive transcriptome in Indica rice genotypes with contrasting drought tolerance. Plant Biotechnol J. 2011;9:315–327. doi: 10.1111/j.1467-7652.2010.00560.x. [DOI] [PubMed] [Google Scholar]
- 52.Lin Y, et al. Endogenous target mimics down-regulate miR160 mediation of ARF10, -16, and -17 cleavage during somatic embryogenesis in Dimocarpus longan Lour. Front Plant Sci. 2015;6:956. doi: 10.3389/fpls.2015.00956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li W, et al. Transcriptional regulation of Arabidopsis MIR168a and argonaute1 homeostasis in abscisic acid and abiotic stress responses. Plant Physiol. 2012;158:1279–1292. doi: 10.1104/pp.111.188789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang L, et al. Characterizations of a hypomorphic argonaute1 mutant reveal novel AGO1 functions in Arabidopsis lateral organ development. Plant Mol Biol. 2006;61:63–78. doi: 10.1007/s11103-005-5992-7. [DOI] [PubMed] [Google Scholar]
- 55.Xian Z, et al. miR168 influences phase transition, leaf epinasty, and fruit development via SlAGO1s in tomato. J Exp Bot. 2014;65:6655–6666. doi: 10.1093/jxb/eru387. [DOI] [PubMed] [Google Scholar]
- 56.Kong Y, Elling AA, Chen B, Deng X. Differential Expression of microRNAs in Maize Inbred and Hybrid Lines during Salt and DroughtStress. American Journal of Plant Sciences. 2010;01:69–76. doi: 10.4236/ajps.2010.12009. [DOI] [Google Scholar]
- 57.Wu ZJ, Tian C, Jiang Q, Li XH, Zhuang J. Selection of suitable reference genes for qRT-PCR normalization during leaf development and hormonal stimuli in tea plant (Camellia sinensis) Scientific reports. 2016;6:19748. doi: 10.1038/srep19748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kullan JB, et al. miRVine: a microRNA expression atlas of grapevine based on small RNA sequencing. Bmc Genomics. 2015;16:393. doi: 10.1186/s12864-015-1610-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reid KE, Olsson N, Schlosser J, Peng F, Lund ST. An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. Bmc Plant Biol. 2006;6:27. doi: 10.1186/1471-2229-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pantaleo, V. et al. Identification of grapevine microRNAs and their targets using high-throughput sequencing and degradome analysis. The Plant Journal, 960–976, 10.1111/j.1365-313X.2010.04208.x (2010). [DOI] [PubMed]
- 61.Thiebaut F, et al. Regulation of miR319 during cold stress in sugarcane. Plant Cell Environ. 2012;35:502–512. doi: 10.1111/j.1365-3040.2011.02430.x. [DOI] [PubMed] [Google Scholar]
- 62.Zhou M, et al. Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol. 2013;161:1375–1391. doi: 10.1104/pp.112.208702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.