Abstract
Extended-spectrum beta-lactamase-producing Escherichia coli (ESBL-EC) strains are emerging around the world as a source of resistance to β-lactam antibiotics such as ampicillin, cefotaxime, and ceftazidime. mcr-1 is a novel plasmid-mediated gene conferring resistance to colistin. The aim of this study was to investigate the prevalence of ESBL-EC mcr-1 of chicken origin in the different provinces of China during 2008–2014. Overall, 341 of 821 isolates were determined to be ESBL-EC strains, and the proportion of ESBL-positive strains almost doubled from 2008 to 2014. The findings of our study revealed regional differences, with significantly more ESBL-EC isolates from stockbreeding in concentrated poultry industry areas in Shandong than from the other four provinces. The ESBL type analysis showed that blaCTX-M was the most prevalent ESBL-encoding gene (92.7%). In total, twelve subtypes of CTX-M genes were detected, among which, blaCTX-M-55 (34.3%) and blaCTX-M-65 (17.9%) were the major identified genotypes. In addition, blaTEM and pAmpC genes were carried by 86.0% and 8.5% of isolates, respectively. In this study, we also observed 44 E. coli isolates with multiple ST types (ST46, ST1286, ST10, ST29, ST101, and ST354) carrying mcr-1, and the majority of mcr-1–carrying plasmids were IncI2. The whole-genome sequencing analysis indicated the co-existence of blaCTX-M and mcr-1 in ESBL-EC of both animal and human origin, and phylogenetic analysis further revealed their close relationship, especially several isolates sharing a small number of SNPs, which suggested the increasing trend of co-existence and transmission of ESBL and mcr-1 in both clinical medicine and veterinary medicine.
Introduction
Antibiotic resistance in bacteria has become one of the major threats to public health because of the extensive use of antibiotics in human medicine and animal farming. Global antimicrobial consumption in food animal production may increase by 67% by 2030, driven primarily by BRICS (Brazil, Russia, India, China, and South Africa) countries, as large-scale and intensive farming operations are heavily in demand with the rise in income and meat consumption1. One study predicted that antimicrobial consumption in chickens and pigs in Asia would grow by 129% and 124%, respectively, and the remarkable growth in consumption of chickens is primarily a result of the rapid expansion of the breeding industry in India2. Over 50% of total antibiotic production is used routinely in subtherapeutic doses for growth promotion, and this corresponds to a consumption of antimicrobials per kilogram of animal produced of ~148 mg kg−1 in chicken production3. The rapid increase in the multi-drug resistance of E. coli has been observed not only in clinical medicine4, but also widely in food animal production5, particularly with an increasing prevalence of ESBL and AmpC β-lactamase producing strains, which greatly compromises the therapeutic efficacy and increases morbidity and mortality.
ESBLs are enzymes that confer resistance to most β-lactam antibiotics, especially to third-generation cephalosporins (e.g., ceftazidime, cefotaxime, and ceftriaxone) and aztreonam but not to cephamycins (cefoxitin and cefotetan) and carbapenems6,7. The majority of ESBLs are blaTEM, blaSHV and blaCTX-M types and CTX-M-producing E. coli isolates, which are recognized as a major cause of hospital and community-onset infections8–10. ESBL-EC is usually considered an indicator bacterium to trace the spread of antibiotic resistance genes, as resistance genes transfer between the same species and different species through genetic elements, especially plasmids11. Plasmid-encoded AmpC (pAmpC) β-lactamases or ESBL conferring resistance to penicillins; first, second, and third-generation cephalosporins; and monobactams have been reported both in human and food animal isolates worldwide. CMY-2 is the most common pAmpC in E. coli in several geographical areas, including Asia, North America, and Europe12–15, and has been reported in food animals on all continents except Australia16,17. ESBL and pAmpC genes have been widely detected in food-producing animals, especially in poultry and retail meat18–20.
Moreover, mcr-1, a novel colistin-resistance mechanism, was discovered in 2015 and caught world-wide attention21. Multiple studies have confirmed its wide spread in both clinical settings22 and animal husbandry23. After the discovery of mcr-1, the Chinese government implemented a fast track and banned the use of colistin as a growth promoter, effective since April 1st, 2017, and the European Medicines Agency immediately initiated the re-assessment of colistin use in food animal production and advised member countries to reduce the usage of colistin in these animals24. However, the emergence and prevalence of mcr-1 are still not well understood. Although one specific study revealed that the emerging and elevated spreading of mcr-1 in food animals could be traced back to 2009, details are still lacking. The elevated prevalence of ESBL has been observed since 2009, as revealed in previous studies, and the coexistence of ESBL and mcr-1 in an E. coli strain was first reported in China in 201625. A recent study suggested that cephalosporin resistance genes are mainly disseminated in animals and humans via distinct plasmids26. It is highly likely that food animals have become one of the most important sources for the spread of these resistance-gene-carrying bacteria to humans through horizontal gene transfer. To evaluate the co-existence and prevalence of ESBL and mcr-1 in E. coli of chicken origin, we investigated the trends of ESBL-EC prevalence in chickens in the different provinces of China from 2008 to 2014 and further elucidated the predominant genotype of ESBL and the phylogenetic relationship among ESBL-EC carrying mcr-1.
Results
Prevalence of ESBL-EC
We obtained 341 ESBL-EC strains from 821 isolates of chicken origin (Table 1), and the ESBL-EC increased from 23.8% in 2008 to 57.0% in 2014 (Fig. 1). Although there were slight differences in the resistance rates among the five sampling regions, they all showed elevated resistance from 2008 to 2014 (Table 1 and Fig. 1). Shandong Province had the highest rate, while the lowest rate was in Shanxi Province (Fig. 2). The ESBL-EC also showed considerable multiple drug resistance to various non-β-lactamase antibiotics, including doxycycline, ciprofloxacin, gentamicin, nalidixic acid, sulfamethoxazole, and florfenicol (Table 2). The resistances to tetracycline, nalidixic acid, and sulfamethoxazole were 100% in ESBL-EC isolates. Multiple drug resistance of ESBL-positive strains was significantly higher than that of ESBL-negative isolates for 6 major antibiotic drug classes, except nalidixic acid, as the resistance of both ESBL-EC positive and negative strains peaked (Table 2 and Fig. 3).
Table 1.
No. of strains, ESBL-EC strains and ESBL-EC positive rates across different provinces from 2008 to 2014
| Year | Province | Total | ||||
|---|---|---|---|---|---|---|
| Guangdong | Shanghai | Shanxi | Sichuan | Shandong | ||
| 2008 | 22a(4b,18.2c) | 12(2,16.7) | – | 58(15, 25.9) | 13(4, 30.8) | 105d(25e, 23.8f) |
| 2009 | 23(5, 21.7) | 45(11, 24.4) | 25(6, 24.0) | 27(7, 25.9) | 20(7,35) | 140(36, 25.7) |
| 2010 | 23(8, 34.8) | 20(8, 40) | 12(4, 33.3) | 34(15, 44.1) | 13(7, 53.8) | 102(42, 41.2) |
| 2011 | 22(9, 40.9) | 18(8, 44.4) | 39(15, 38.5) | 28(14, 50.0) | 20(11, 55.0) | 127(57, 44.9) |
| 2012 | 25(11,44.0) | 20(9, 45.0) | 25(11, 44.0) | 23(12, 54.5) | 22(14, 56.5) | 115(57, 49.6) |
| 2013 | 25(12, 48.0) | 20(9, 45.0) | 25(9, 36.0) | 28(16, 57.1) | 20(13, 65.0) | 118(59, 50.0) |
| 2014 | 36(19, 52.8) | 30(16, 53.3) | – | 28(16, 57.1) | 20(14, 70.0) | 114(65, 57.0) |
| Total | 176g | 165 | 126 | 226 | 128 | 821(341) |
a Number of strains
b Number of positive strains
c ESBL-EC positive rate
d, g Total number of strains
e Total number of positive strains
f Total positive rate of ESBL-EC
Fig. 1.

Positive rate of ESBL and polymyxin E resistance from 2008 to 2014
Fig. 2.

Positive rate of ESBL-EC in five broiler production regions of China from 2008 to 2014
Table 2.
Elevated multiple drug resistance rate in the E. coli of chicken origin from 2008 to 2014
| Antibiotics | Species | Drug resistance rate by year (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| 2008 | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | ||
| GEN | ESBL+ | 52.9 | 54.8 | 63.0 | 65.7 | 72.2 | 76.04 | 77.8 |
| ESBL− | 41.9 | 42.3 | 48.7 | 50.3 | 52.9 | 56.7 | 56.9 | |
| DOX | ESBL+ | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| ESBL− | 67.6 | 71.4 | 74.1 | 78.9 | 83.3 | 85.3 | 88.9 | |
| FFC | ESBL+ | 67.6 | 71.4 | 74.1 | 78.9 | 83.3 | 85.3 | 88.9 |
| ESBL− | 50.3 | 51.4 | 53.3 | 53.4 | 54.9 | 56.1 | 58.4 | |
| NAL | ESBL+ | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| ESBL− | 93.9 | 98.6 | 98.7 | 100 | 100 | 100 | 100 | |
| CIP | ESBL+ | 52.9 | 59.2 | 75.9 | 80.3 | 81.9 | 84.0 | 86.4 |
| ESBL− | 43.6 | 45.0 | 45.3 | 47.1 | 46.4 | 48.4 | 52.6 | |
| P E | ESBL+ | 0.0 | 2.0 | 5.6 | 5.6 | 6.9 | 8.0 | 8.6 |
| ESBL− | 0.0 | 0.0 | 1.3 | 1.9 | 2.1 | 2.6 | 2.9 | |
| SXT | ESBL+ | 88.2 | 96.0 | 98.1 | 100 | 100 | 100 | 100 |
| ESBL− | 57.0 | 60.5 | 68.0 | 68.3 | 71.2 | 73.2 | 75.9 | |
GEN gentamicin, DOX doxycycline, FFC florfenicol, NAL nalidixic acid, CIP ciprofloxacin, PE polymyxin E, SXT compound sulfamethoxazole
Fig. 3.
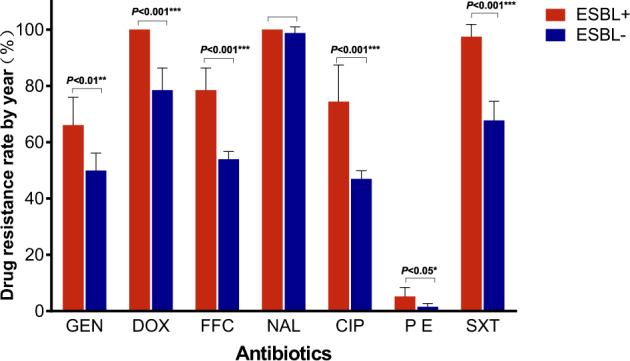
Multiple drug resistance rate between ESBL-EC and non-ESBL-EC
Characterization of the ESBLs genes in the E. coli isolates
In this study, 92.7% (316/341) of ESBL-EC isolates contained blaCTX-M genes, among which, blaCTX-M-55 (34.3%), blaCTX-M-65 (17.9%), and blaCTX-M-101 (7.0%) were the most dominant blaCTX-M types (Fig. 4a)27–32. To the best of our knowledge, for the first time, we identified blaCTX-M-98 in E. coli of food animals in China, which has been detected in humans previously33,34. We also analyzed over 6000 whole genome sequences of E. coli isolates submitted to the NCBI database. The distribution of all CTX-M-producing E. coli isolates (n = 660) from the NCBI database were also analyzed and presented (Fig. 4b). As over 80% of them were from human isolates, blaCTX-M-15 (41.1%) was the dominant type in the human isolates.
Fig. 4. The distribution of CTX-M types of ESBL-EC.
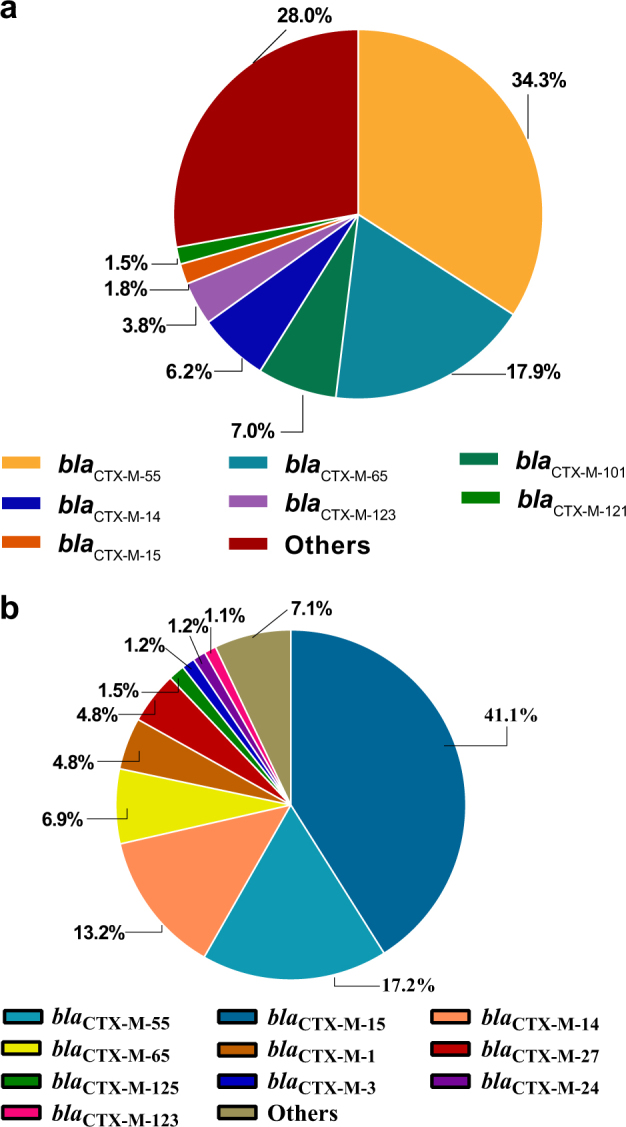
a The distribution of CTX-M types of ESBL-EC of chicken origin in China. b The distribution of CTX-M types of ESBL-EC from the NCBI whole-genome sequencing database
Of the isolates from this study, 86.0% were detected as harboring blaTEM genes at a proportion >46.5%; blaTEM-1 accounted for 80.0% of all blaTEM genes, a remarkable prevalence in food animals, and the remaining were blaTEM-52 and blaTEM-116 genes. pAmpC encoding genes, which were found in 37 tested strains, included CMY2 (19) and OXA10 (18) but not SHV, as previously reported11,16,34. ESBL or AmpC-producing E. coli were further investigated for genetic relationship using PFGE (Supplementary Figure S1). The results illustrated that most of the strains isolated in this study were clonally unrelated, similar to previous reports35.
Co-existence of ESBL and mcr-1 genes and whole-genome sequencing analysis
From this study, the emergence of resistance to polymyxins has also been observed since 2009, and it has been increasing in the last few years (Table 1 and Fig. 1). To further understand the co-existence of mcr-1 and ESBL, all colistin-resistant isolates were selected for the screening of mcr-1 and other mcr genes (mcr-2, mcr-3, mcr-4, and mcr-5) using a PCR method. Only mcr-1 was identified; none of other mcr genes was identified. All 44 mcr-1–carrying isolates were analyzed using whole genome sequencing to retrieve the complete antibiotic resistance gene profiles. Through the whole genome sequencing, multiple MLST types have been identified, such as ST46 (9), ST1286 (4), ST10 (3), ST29 (3), ST101 (3), and ST354 (3) (Table 3). Furthermore, information on plasmids carrying mcr-1 was also revealed (Table 3). The Inc type of the majority of these mcr-1 carrying plasmids was IncI2, and the plasmid size ranged from 58 to 62 kb. Several mcr-1–carrying contigs were not sufficiently long to identify the plasmid size and Inc type. The insert sequence ISApl1 surrounding mcr-1 was identified in the most IncI2 plasmids. Two isolates were identified as carrying Tn6330 (R26 and R46). The whole-genome sequence data revealed that 77.3% of mcr-1 positive isolates (34/44) carried at least one ESBL gene, including blaCTX-M (n = 25), blaOXA (n = 11), and blaTEM (n = 25). blaTEM-198 (7/25) is the dominant type in the blaTEM-1, and blaCTX-M-55 (6/44), blaCTX-M-14 (6/44), blaCTX-M-65 (4/44) are the three major blaCTX-M types. Eleven isolates carry both blaCTX-M and blaTEM genes; one isolate carries blaCTX-M-15, blaCTX-M-65, blaOXA-1, blaOXA-10, and blaTEM-141; and six isolates contain multiple ESBL genes (blaTEM-blaCMY-blaOXA, blaTEM- blaCTX-M-blaOXA, blaTEM-blaCTX-M-blaCMY).
Table 3.
The information regarding mcr-1 positive E. coli and mcr-1–carrying plasmids
| E. coli Isolate ID | BioSample ID | Year | Area | MLST Type | Plasmid Inc type | mcr-1 contig size (bp) | No. of contigs |
|---|---|---|---|---|---|---|---|
| R02 | SAMN07983251 | 2011 | Chongqing | 10 | − | 2943 | 74 |
| R24 | SAMN07983271 | 2011 | Sichuan | 10 | IncI2 | 60,207 | 28 |
| R46 | SAMN07983287 | 2010 | Shanghai | 10 | IncI2 | 61,831 | 21 |
| R26 | SAMN07983273 | 2013 | Shanghai | 29 | IncI2 | 60,873 | 31 |
| R37 | SAMN07983280 | 2010 | Guangzhou | 29 | IncI2 | 60,955 | 32 |
| R95 | SAMN07983324 | 2013 | Shanghai | 29 | IncI2 | 60,178 | 34 |
| R10 | SAMN07983257 | 2013 | Shanghai | 46 | IncI2 | 59,879 | 27 |
| R12 | SAMN07983261 | 2012 | Shanghai | 46 | IncI2 | 61,943 | 25 |
| R13 | SAMN07983262 | 2013 | Shanghai | 46 | IncI2 | 62,380 | 25 |
| R19 | SAMN07983267 | 2013 | Shanghai | 46 | IncI2 | 61,022 | 27 |
| R35 | SAMN07983279 | 2012 | Shanghai | 46 | – | 11,027 | 84 |
| R40 | SAMN07983283 | 2012 | Shanghai | 46 | IncI2 | 58,887 | 29 |
| R53 | SAMN07983292 | 2013 | Shanghai | 46 | IncI2 | 63,724 | 26 |
| R56 | SAMN07983295 | 2012 | Shanghai | 46 | IncI2 | 60,445 | 28 |
| R64 | SAMN07983303 | 2010 | Shanghai | 46 | IncI2 | 61,022 | 27 |
| R34 | SAMN07983278 | 2011 | Chongqing | 48 | IncI2 | 60,203 | 32 |
| R59 | SAMN07983298 | 2012 | Guangzhou | 93 | – | 2863 | 164 |
| R61 | SAMN07983300 | 2013 | Shandong | 93 | IncI2 | 60,872 | 27 |
| R14 | SAMN07983263 | 2014 | Sichuan | 101 | IncI2 | 60,872 | 35 |
| R28 | SAMN07983275 | 2011 | Guangzhou | 101 | IncI2 | 60,872 | 34 |
| R41 | SAMN07983284 | 2012 | Sichuan | 101 | IncHI2A + IncHI2 + IncI2 | 257,133 | 6 |
| R44 | SAMN07983286 | 2014 | Shandong | 156 | IncI2 | 58,932 | 28 |
| R47 | SAMN07983288 | 2014 | Sichuan | 165 | IncI2 | 62,807 | 26 |
| R07 | SAMN07983254 | 2011 | Shanghai | 354 | IncI2 | 59,541 | 28 |
| R48 | SAMN07983289 | 2011 | Shanghai | 354 | – | 9910 | 65 |
| R51 | SAMN07983291 | 2011 | Shanghai | 354 | – | 20,901 | 46 |
| R75 | SAMN07983308 | 2011 | Shanghai | 354 | IncI2 | 59,294 | 29 |
| R09 | SAMN07983256 | 2009 | Guangzhou | 533 | − | 11,573 | 43 |
| R57 | SAMN07983296 | 2009 | Guangzhou | 533 | − | 11,525 | 45 |
| R74 | SAMN07983307 | 2009 | Guangzhou | 533 | − | 11,022 | 44 |
| R25 | SAMN07983272 | 2013 | Shanghai | 542 | − | 2943 | 94 |
| R39 | SAMN07983282 | 2013 | Shanghai | 542 | − | 2943 | 90 |
| R38 | SAMN07983281 | 2014 | Shandong | 617 | IncI2 | 61,088 | 25 |
| R27 | SAMN07983274 | 2014 | Shandong | 648 | IncI2 | 60,752 | 24 |
| R01 | SAMN07983250 | 2012 | Unknown | 1286 | IncI2 | 61,068 | 27 |
| R134 | SAMN08180604 | 2012 | Shanghai | 1286 | – | 11,029 | 49 |
| R62 | SAMN07983301 | 2014 | Sichuan | 1286 | IncI2 | 64,482 | 29 |
| R78 | SAMN07983311 | 2012 | Shanghai | 1286 | – | 12,112 | 45 |
| R86 | SAMN07983317 | 2012 | Shanghai | 1286 | IncI2 | 61,931 | 27 |
| R96 | SAMN07983325 | 2012 | Shanghai | 1286 | – | 10,915 | 48 |
| R50 | SAMN07983290 | 2010 | Shanghai | 1564 | – | 2946 | 82 |
| R58 | SAMN07983297 | 2012 | Shanghai | 1589 | IncI2 | 62,044 | 29 |
| R31 | SAMN07983276 | 2012 | Sichuan | 3014 | IncI2 | 60,874 | 27 |
| R32 | SAMN08238410 | 2014 | Shandong | 5229 | – | 4631 | 67 |
In total 39 blaCTX-M-55–carrying isolates of different origin from four countries (China, Germany, United States, Vietnam) from the NCBI database were identified, combined with 6 ESBL-EC isolates carrying both blaCTX-M-55 and mcr-1 from this study and subjected to a core genome phylogenetic analysis (Fig. 5). In total 3 of 6 isolates from this study had blaCTX-M-55 surrounded by ISEcp1. A total of 122,307 SNPs were used to calculate the phylogenetic relationship among these isolates; the number of SNPs among closely related isolates ranged from 0 to 105 SNPs. Interestingly, 34 of 45 isolates were identified as carrying both blaCTX-M-55 and mcr-1, and ESBL-EC of animal origin had a higher positive rate (83.3%, 25 of 30) compared with human origin (60%, 9 of 15). Several isolates carrying both blaCTX-M-55 and mcr-1 from animal and human origin were clustered very closely and belonged to geographically distinct regions, e.g., China and Vietnam.
Fig. 5.
The phylogenetic tree of ESBL-EC carrying both blaCTX-M-55 and mcr-1. * Red, ESBL-EC from China; Green, ESBL-EC from Vietnam; Blue, ESBL-EC from Germany
Discussion
China has the second largest broiler production and the largest consumption of antibiotics in the world36, and the overall antibiotic consumption has continued rising, which could potentially facilitate the spread of antibiotic resistance. The prevalence of ESBL-EC of chicken origin in China has been increasing since 2006, with slight differences among geographical locations and animal species5. Recently, mcr-1, mcr-2, and mcr-3, a group of genes mediating colistin resistance through plasmids, have been widely discovered around the world21,37,38. The mechanism of colistin resistance related to mcr-1 and mcr-2 has also been revealed39,40. However, there are few reports studying the prevalence of the co-existence of mcr-1 and ESBL in the broiler production. Herein, we screened over 800 E. coli isolates from several major broiler production areas of China during 2008-2014 and found that the fast rise of ESBL and emergence of mcr-1–positive EC (MCRPEC) in the chicken were consistent with previous reports5,41. This investigation is the first to reveal the co-rising of ESBL and mcr-1 in E. coli isolates in a longitudinal study.
In this study, the positive rate of ESBL-EC of chicken origin in China was higher than that in some previous reports28–30,32,42 but lower than that in other reports6,28,43,44 for the same period, in which the sampling area and total number of isolates were relatively limited and did not cover major poultry production areas. This study surveyed three major broiler production areas, namely, Guangdong, Shandong, and Sichuan Provinces, accounting for ~40% of the total broiler production in China45, which should give a more comprehensive estimation of major poultry production areas compared with previous studies. One report from PNAS also confirmed that an antibiotic consumption per 10 km2 in chickens exceeding 250 kg in these areas has the highest density throughout China2. Although the positive rate of ESBL-EC differed among geographical locations, the resistance rates all doubled in a short period of time (Table 2 and Fig. 2) and have continued rising steadily. Shandong Province ranks first in poultry production in China; the province’s annual poultry production, including chickens and ducks, has reached 1.87 billion. The highly concentrated poultry industry in Shandong Province may also facilitate the expansion of ESBLs, causing this province to have the highest ESBL rate. These results were similar to those obtained in previous investigations9,12,28,46. A very recent study reported that the overall prevalence of ESBLs from India in the broiler chicken was 87.0%, which was much higher than that in China (57.0%, 2014)1. In this study, not only has the resistance rate of ESBL-EC strains been increasing at an alarming rate but also ESBL-EC has shown significant resistance to other major antibiotics, such as doxycycline, nalidixic acid, and sulfamethoxazole, with the resistance even reaching 100% in several classes of antibiotics. This was also observed in an ESBL study of India chicken production with overwhelming multiple drug resistance to nalidixic acid, ciprofloxacin, and chloramphenicol1. Such phenomena may be attributed to the co-selection of antibiotic resistance caused by improper combination usage of multiple classes of broad-spectrum antibiotics. The presence of these multiple drug resistance phenotypes further indicated that the co-existence of ESBLs with other resistance genes in E. coli strains results in an expansion of their resistance spectrum to β-lactam antibiotics or to other antibiotics, which poses a serious challenge to the application of antibiotics in the poultry industry. Therefore, ESBL-EC may be used as an indicator for the surveillance of multiple-drug-resistant E. coli in food animal production.
blaCTX-M type was the dominant ESBL-encoding gene in the ESBL-EC6,11,28,30,32. Of the 341 ESBL-EC isolates in this study, 92.7% had blaCTX-M genes, slightly higher than the previously reported 86% in chickens in China6,11 but lower than those (96.9%) in human isolates47. The predominant blaCTX-M genotype, blaCTX-M-55, was consistent with the previous report5,48–51. Furthermore, the prevalence of blaCTX-M-55 was even higher than those of blaCTX-M-14 and blaCTX-M-65 combined, and the second-most dominant blaCTX-M was blaCTX-M-65, which has surpassed blaCTX-M-14, compared with previous reports5. Overall, the blaCTX-M-1 group (47%) has replaced the blaCTX-M-9 (26%) group as the dominant blaCTX-M genotype. blaCTX-M-55 was not reported in human clinical bacteria in China before 201033, but several studies have recently reported the emergence of blaCTX-M-55 in human isolates, which has become the second-most common blaCTX-M enzyme in China, surpassing blaCTX-M-5528.
Interestingly, the prevalence of mcr-1 was higher in the ESBL-EC than in the non-ESBL-EC (p < 0.001, 77.3% vs 22.7%), and the fast rise of ESBL apparently also increased the selective pressure of colistin resistance. Within the mcr-1 positive E. coli, more than 75% of the isolates had at least one ESBL gene, and several isolates carried three or more ESBL genes, suggesting that the ESBL-EC are more likely to recruit the mcr-1 gene than are non-ESBL-EC. Both β-lactams and colistin could damage bacteria cell walls by inhibiting the synthesis of peptidoglycan and disrupting the outer membrane, respectively. Maintaining the cell wall integrity has become the top priority for bacteria to survive in the battle against antibiotics, leading to the high prevalence of mcr-1 in ESBL-EC isolates. Recently, from the whole-genome sequence data of ESBL-EC submitted to the NCBI database, over 75% of isolates carried mcr-1 within the CTX-M-55–positive ESBL-EC, which was much higher than other CTX-M type isolates. According to the whole-genome sequence data of ESBL-EC submitted to the NCBI database, a similar pattern was also observed. Considering that blaCTX-M-55 has become the dominant blaCTX-M type in the ESBL-EC of animal origin in the last decade but was still very rare in the ESBL-EC of human origin; this finding might suggest that blaCTX-M-55 and mcr-1 emerged and rose under the heavy selective pressure of antibiotic usage in the animal husbandry in the last decade. Interestingly, one strain isolated from Vietnam in 2012 was clustered together with three other strains isolated in the Shanghai area, all carrying both blaCTX-M-55 and mcr-1 and sharing 1440 SNPs (1.2%) of 122,307 total SNPs (Fig. 5). In addition, several ESBL and mcr-1-carrying isolates from Vietnam were closely related to the ESBL-positive and mcr-1–negative isolates of human origin from Zhejiang Province and shared 566 SNPs. The ST types also confirmed the closely phylogenetic relationship among these isolates (Fig. 5), while a recent study revealed the co-existence of blaCTX-M-15 and mcr-1 in the ESBL-EC of human origin25. However, these mcr-1–negative isolates have the potential capability to eventually acquire the mcr-1 gene because of its high resemblance of genetic context to the ESBL-EC of animal origin23,41,52,53. The whole-genome sequencing also revealed the genetic context of plasmids carrying mcr-1 in the most mcr-1–positive ESBL-EC isolates, which are highly similar to the plasmid pHNSHP45 identified in the previous study21. The Inc type of the majority of the mcr-1–carrying plasmids was IncI2, which was similar to that of the E. coli isolates of pig origin from a recent study54. The multiple ST types suggested that the IncI2 plasmid could be adapted to various E. coli hosts. Several ST types were the predominant ST types within the prevalence regions, such as ST46, ST354, and ST1286 in Shanghai; however, the same ST type could also be found from different regions, such as ST10. ST354 with an IncI2 plasmid has been reported in the previous study55,56.
The co-rising of the ESBL-EC and MECPEC can be traced back to 2008 from this study, which is highly synchronized with the rapid growth of veterinary drug consumption, especially of amoxicillin/clavulanic acid and colistin, in the poultry industry57. By 2014, the use of amoxicillin and colistin reached ~7,000,000 kg and 22,000,000 kg, respectively, in food animal production in China57. According to the United States FDA, the overall consumption of all penicillin class drugs, including amoxicillin, ampicillin, cloxacillin and penicillin, was only 885,975 kg in 201458. However, from the National Antimicrobial Resistance Monitoring System (NARMS) Integrated Report of 2014, the cephalosporin resistance rate in E. coli isolates from retail chicken even declined from a peak of 13% in 2011 to 6.6% in 2014, while the usage of penicillins and cephalosporins increased by 57.0% and 28.0%, respectively59. This may suggest that the proper usage of antibiotics during food animal production is crucial to controlling the spread of antibiotic resistance. In the last decade, amoxicillin has been widely used in drinking water for chicks (<7 days) as a preventive measure, and colistin is used as feed additive for the entire production period of chicken. This usage pattern may contribute a significant portion of selective pressure on the bacteria. Although colistin has been banned as a feed additive since April 1st, 2017, it will take some time to observe the decline in colistin resistance in food animals. Taken together, the wide spread of ESBL-EC and mcr-1 in animal husbandry in China further indicates the extensive use of antibiotics in food animal production, which may facilitate the transmission of antibiotic resistance to humans and the environment and cause a serious threat to health. The phylogenetic analysis also showed the close relationship of ESBL- and mcr-1–carrying E. coli between the animal isolates and clinical isolates, which suggested that we should pay more attention to monitoring the prevalence of ESBL- and mcr-1–carrying E. coli in both clinical medicine and food animal production.
Materials and methods
Bacterial isolation and antimicrobial susceptibility testing
A total of 821 E. coli strains, isolated from cloacal swabbing of chickens, were provided by the China Institute of Veterinary Drug Control. The strains were isolated from different regions between 2008 and 2014 (see Table 1). Screening and confirmation of ESBL producers were conducted according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI, 2013). The minimum inhibitory concentrations (MICs) of cefotaxime or ceftazidime for E. coli isolates were initially performed using the broth microdilution method. For isolates with an MIC of cefotaxime or ceftazidime >1 mg/l, the double disk synergy test was further utilized to confirm the ESBL-EC. Seven other antibiotics, namely, doxycycline, gentamicin, ciprofloxacin, compound sulfamethoxazole, polymyxin E, nalidixic acid, and florfenicol, were also tested for MIC using the broth microdilution method. E. coli ATCC25922 was used as the quality control strain.
Detection of the ESBL and pAmpC, and mcr genes
ESBL-producing strains were screened for β-lactamase genes, ESBL genes (blaCTX-M, blaTEM and blaSHV), pAmpC genes (blaCMY, blaOXA), and mcr 1–5 using PCR testing. All the primers used are listed in Table S1. The positive products were sent for Sanger sequencing. The DNA sequences obtained were further checked for ESBL and pAmpC gene subtypes using BLAST analysis (http://www.ncbi.nlm.nih.gov/) or the β-lactamase classification system (http://www.lahey.org/studies/webt.asp).
Whole genome sequencing analysis and genome typing
All isolates containing mcr-1 were subjected to whole genome sequencing. Total DNA from the E. coli isolates was extracted using a Wizard® Genomic DNA Purification kit (Promega, WI, USA) and then subjected to whole genome sequencing. The library was constructed using a Next® Ultra™ DNA Library Prep kit (New England Biolabs, Ipswich, UK) according to the manufacturer’s protocol, and 250-bp paired-end reads were obtained from an Illumina Hiseq2500 platform (Bionova Biotech Co.). For each E. coli isolate analyzed by whole-genome sequencing, at least 100-fold coverage of raw reads were yielded and collected. The clean reads were searched against the ResFinder and PubMLST databases using the SRST2 program to retrieve all the resistance genes and the MLST types as previous study23. A draft assembly of the sequences was generated using SPAdes as previous study23. A total of 6320 E. coli isolate whole-genome sequences (by July 31st, 2017) were download from the NCBI database for the screening of ESBL genes, and all the ESBL-EC isolates were analyzed to retrieve the ESBL genotypes for further analysis. Draft genome sequences were aligned and then applied for phylogenetic tree construction by Parsnp in the Harvest package, and the phylogenetic tree was visualized with FigTree as previously described. Sixty ESBL-EC strains were randomly selected to conduct the PFGE analysis using the methods as described. All whole genome sequences have been submitted to GenBank under BioProject (PRJNA417344).
Electronic supplementary material
Acknowledgements
This work was supported in part by the National Key Research and Development Program of China (2016YFD0501301, 2016YFD0501304, and 2016YFD0501305), the National Natural Science Foundation of China (31572568, 81661138002).
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Juchun Lin, Email: juchunlin@126.com.
Shaolin Wang, Email: shaolinwang@cau.edu.cn.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41426-018-0033-1).
References
- 1.Brower CH, et al. The prevalence of extended-spectrum beta-lactamase-producing multidrug-resistant Escherichia coli in poultry chickens and variation according to farming practices in Punjab, India. Environ. Health Perspect. 2017;125:077015. doi: 10.1289/EHP292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Boeckel TP, et al. Global trends in antimicrobial use in food animals. Proc. Natl Acad. Sci. USA. 2015;112:5649–5654. doi: 10.1073/pnas.1503141112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Boeckel TP, et al. Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. Lancet Infect. Dis. 2014;14:742–750. doi: 10.1016/S1473-3099(14)70780-7. [DOI] [PubMed] [Google Scholar]
- 4.Chen J, et al. Prevalence and characterization of integrons in multidrug resistant Acinetobacter baumannii in Eastern China: a multiple-hospital study. Int. J. Environ. Res Public Health. 2015;12:10093–10105. doi: 10.3390/ijerph120810093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rao L, et al. Increasing prevalence of extended-spectrum cephalosporin-resistant Escherichia coli in food animals and the diversity of CTX-M genotypes during 2003–2012. Vet. Microbiol. 2014;172:534–541. doi: 10.1016/j.vetmic.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, et al. Prevalence of ESBLs and PMQR genes in fecal Escherichia coli isolated from the non-human primates in six zoos in China. Vet. Microbiol. 2012;159:53–59. doi: 10.1016/j.vetmic.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Canton R, Gonzalez-Alba JM, Galan JC. CTX-M enzymes: origin and diffusion. Front. Microbiol. 2012;3:110. doi: 10.3389/fmicb.2012.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pitout JD, Nordmann P, Laupland KB, Poirel L. Emergence of Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs) in the community. J. Antimicrob. Chemother. 2005;56:52–59. doi: 10.1093/jac/dki166. [DOI] [PubMed] [Google Scholar]
- 9.Li LM, et al. Characterization of integrons among Escherichia coli in a region at high incidence of ESBL-EC. Pak. J. Med. Sci. 2014;30:177–180. doi: 10.12669/pjms.301.4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng VC, Wong SC, Ho PL, Yuen KY. Strategic measures for the control of surging antimicrobial resistance in Hong Kong and mainland of China. Emerg. Microbes Infect. 2015;4:e8. doi: 10.1038/emi.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao, L. et al. Application of swine manure on agricultural fields contributes to extended-spectrum β-lactamase-producing Escherichia coli spread in Tai’an, China. Front. Microbiol.6, 313 (2015). [DOI] [PMC free article] [PubMed]
- 12.Alvarez M, Tran J, Chow N, Jacoby G. Epidemiology of conjugative plasmid-mediated AmpC β-lactamases in the United States. Antimicrob. Agents Chemother. 2004;48:533–537. doi: 10.1128/AAC.48.2.533-537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mulvey MR, et al. Molecular characterization of cefoxitin-resistant Escherichia coli from Canadian hospitals. Antimicrob. Agents Chemother. 2005;49:358–365. doi: 10.1128/AAC.49.1.358-365.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, et al. Prevalence of plasmid-mediated AmpC β-lactamases in a Chinese university hospital from 2003 to 2005: first report of CMY-2-type AmpC β-lactamase resistance in China. J. Clin. Microbiol. 2008;46:1317–1321. doi: 10.1128/JCM.00073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woodford N, et al. Wide geographic spread of diverse acquired AmpC β-lactamases among Escherichia coli and Klebsiella spp. in the UK and Ireland. J. Antimicrob. Chemother. 2007;59:102–105. doi: 10.1093/jac/dkl456. [DOI] [PubMed] [Google Scholar]
- 16.Guo YF, et al. IncA/C plasmid-mediated spread of CMY-2 in multidrug-resistant Escherichia coli from food animals in China. PLoS ONE. 2014;9:e96738. doi: 10.1371/journal.pone.0096738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trott D. β-lactam resistance in gram-negative pathogens isolated from animals. Curr. Pharm. Des. 2013;19:239–249. doi: 10.2174/138161213804070339. [DOI] [PubMed] [Google Scholar]
- 18.Smet A, et al. Diversity of extended-spectrum β-lactamases and class C β-lactamases among cloacal Escherichia coli isolates in Belgian broiler farms. Antimicrob. Agents Chemother. 2008;52:1238–1243. doi: 10.1128/AAC.01285-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Machado E, Coque TM, Cantón R, Sousa JC, Peixe L. Antibiotic resistance integrons and extended-spectrum β-lactamases among Enterobacteriaceae isolates recovered from chickens and swine in Portugal. J. Antimicrob. Chemother. 2008;62:296–302. doi: 10.1093/jac/dkn179. [DOI] [PubMed] [Google Scholar]
- 20.Paterson D, et al. Extended‐spectrum and CMY‐type β‐lactamase‐producing Escherichia coli in clinical samples and retail meat from Pittsburgh, USA and Seville, Spain. Clin. Microbiol Infect. 2010;16:33–38. doi: 10.1111/j.1469-0691.2009.03001.x. [DOI] [PubMed] [Google Scholar]
- 21.Liu YY, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, et al. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect. Dis. 2017;17:390–399. doi: 10.1016/S1473-3099(16)30527-8. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, et al. Comprehensive resistome analysis reveals the prevalence of NDM and mcr -1 in Chinese poultry production. Nat. Microbiol. 2017;2:16260. doi: 10.1038/nmicrobiol.2016.260. [DOI] [PubMed] [Google Scholar]
- 24.EMA updates its advice on the use of colistin in animals. Veterinary Record. 179, 131–132 (2016).
- 25.Zhang H, Seward CH, Wu Z, Ye H, Feng Y. Genomic insights into the ESBL and mcr-1-producing ST648 Escherichiacoli with multi-drug resistance. Sci. Bull. 2016;61:875–878. doi: 10.1007/s11434-016-1086-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Been M, et al. Dissemination of cephalosporin resistance genes between Escherichia coli strains from farm animals and humans by specific plasmid lineages. PLoS Genet. 2014;10:e100477627. doi: 10.1371/journal.pgen.1004776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu YY, et al. Molecular typing of CTX-M-producing Escherichia coli isolates from environmental water, swine feces, specimens from healthy humans, and human patients. Appl. Environ. Microbiol. 2013;79:5988–5996. doi: 10.1128/AEM.01740-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rao L, et al. Increasing prevalence of extended-spectrum cephalosporin-resistant Escherichia coli in food animals and the diversity of CTX-M genotypes during 2003–2012. Vet. Microbiol. 2014;172:534–541. doi: 10.1016/j.vetmic.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 29.Li L, et al. Characterization of antimicrobial resistance and molecular determinants of beta-lactamase in Escherichia coli isolated from chickens in China during 1970–2007. Vet. Microbiol. 2010;144:505–510. doi: 10.1016/j.vetmic.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Liu BT, et al. Dissemination and characterization of plasmids carrying oqxAB-bla CTX-M genes in Escherichia coli isolates from food-producing animals. PLoS ONE. 2013;8:e73947. doi: 10.1371/journal.pone.0073947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Y, et al. High prevalence of blaCTX‐M extended‐spectrum β‐lactamase genes in Escherichia coli isolates from pets and emergence of CTX‐M‐64 in China. Clin. Microbiol. Infect. 2010;16:1475–1481. doi: 10.1111/j.1469-0691.2010.03127.x. [DOI] [PubMed] [Google Scholar]
- 32.Zheng H, et al. Prevalence and characterisation of CTX-M β-lactamases amongst Escherichia coli isolates from healthy food animals in China. Int J. Antimicrob. Agents. 2012;39:305–310. doi: 10.1016/j.ijantimicag.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 33.Liu W, et al. Novel CTX-M β-lactamase genotype distribution and spread into multiple species of Enterobacteriaceae in Changsha, Southern China. J. Antimicrob. Chemother. 2009;63:895–900. doi: 10.1093/jac/dkp068. [DOI] [PubMed] [Google Scholar]
- 34.Liu BT, et al. Dissemination and characterization of plasmids carrying oqxAB-bla CTX-M genes in Escherichia coli isolates from food-producing animals. PLoS ONE. 2013;8:e73947. doi: 10.1371/journal.pone.0073947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Denisuik AJ, et al. Molecular epidemiology of extended-spectrum β-lactamase-, AmpC β-lactamase-and carbapenemase-producing Escherichia coli and Klebsiella pneumoniae isolated from Canadian hospitals over a 5 year period: CANWARD 2007–11. J. Antimicrob. Chemother. 2013;68(Suppl 1):i57–i65. doi: 10.1093/jac/dkt027. [DOI] [PubMed] [Google Scholar]
- 36.Collignon P, Voss A. China, what antibiotics and what volumes are used in food production animals? Antimicrob. Resist Infect. Control. 2015;4:16. doi: 10.1186/s13756-015-0056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xavier, B. B. et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill21, 161 (2016). [DOI] [PubMed]
- 38.Yin W, et al. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. MBio. 2017;8:e00543–17. doi: 10.1128/mBio.00543-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun J, et al. Deciphering MCR-2 colistin resistance. MBio. 2017;8:e00625–17. doi: 10.1128/mBio.00625-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao R, et al. Dissemination and mechanism for the MCR-1 colistin resistance. PLoS Pathog. 2016;12:e1005957. doi: 10.1371/journal.ppat.1005957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen Z, Wang Y, Shen Y, Shen J, Wu C. Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect. Dis. 2016;16:293. doi: 10.1016/S1473-3099(16)00061-X. [DOI] [PubMed] [Google Scholar]
- 42.Gao L, et al. Application of swine manure on agricultural fields contributes to extended-spectrum beta-lactamase-producing Escherichia coli spread in Tai’an, China. Front. Microbiol. 2015;6:313. doi: 10.3389/fmicb.2015.00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lo WU, et al. Highly conjugative IncX4 plasmids carrying blaCTX-M in Escherichia coli from humans and food animals. J. Med. Microbiol. 2014;63:835–840. doi: 10.1099/jmm.0.074021-0. [DOI] [PubMed] [Google Scholar]
- 44.Ma J, et al. Characterization of extended-spectrum β-lactamase genes found among Escherichia coli isolates from duck and environmental samples obtained on a duck farm. Appl. Environ. Microbiol. 2012;78:3668–3673. doi: 10.1128/AEM.07507-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chinese Livestock Industry Almanac for the year 2016; 2016.
- 46.Liebana E, et al. Public health risks of enterobacterial isolates producing extended-spectrum beta-lactamases or AmpC beta-lactamases in food and food-producing animals: an EU perspective of epidemiology, analytical methods, risk factors, and control options. Clin. Infect. Dis. 2013;56:1030–1037. doi: 10.1093/cid/cis1043. [DOI] [PubMed] [Google Scholar]
- 47.Shi H, et al. Epidemiology of CTX-M-type extended-spectrum beta-lactamase (ESBL)-producing nosocomial-Escherichia coli infection in China. Ann. Clin. Microbiol. Antimicrob. 2015;14:1. doi: 10.1186/s12941-015-0063-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu YY, et al. Molecular typing of CTX-M-producing Escherichia coli isolates from environmental water, swine feces, specimens from healthy humans, and human patients. Appl. Environ. Microbiol. 2013;79:5988–5996. doi: 10.1128/AEM.01740-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng H, et al. Prevalence and characterisation of CTX-M beta-lactamases amongst Escherichia coli isolates from healthy food animals in China. Int J. Antimicrob. Agents. 2012;39:305–310. doi: 10.1016/j.ijantimicag.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 50.Sun Y, et al. High prevalence of bla(CTX-M) extended-spectrum beta-lactamase genes in Escherichia coli isolates from pets and emergence of CTX-M-64 in China. Clin. Microbiol. Infect. 2010;16:1475–1481. doi: 10.1111/j.1469-0691.2010.03127.x. [DOI] [PubMed] [Google Scholar]
- 51.Li J, et al. Dissemination of cefotaxime-M-producing Escherichia coli isolates in poultry farms, but not swine farms, in China. Foodborne Pathog. Dis. 2010;7:1387–1392. doi: 10.1089/fpd.2010.0581. [DOI] [PubMed] [Google Scholar]
- 52.Wang Q, et al. Expanding landscapes of the diversified mcr-1-bearing plasmid reservoirs. Microbiome. 2017;5:70. doi: 10.1186/s40168-017-0288-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ye H, et al. Diversified mcr-1-harbouring plasmid reservoirs confer resistance to colistin in human gut microbiota. MBio. 2016;7:e00177. doi: 10.1128/mBio.00177-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li R, et al. Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J. Antimicrob. Chemother. 2017;72:393–401. doi: 10.1093/jac/dkw411. [DOI] [PubMed] [Google Scholar]
- 55.Zhang X, et al. Possible transmission of mcr-1-Harboring Escherichia coli between companion animals and human. Emerg. Infect. Dis. 2016;22:1679–1681. doi: 10.3201/eid2209.160464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mohsin M, Raza S, Roschanski N, Schaufler K, Guenther S. First description of plasmid-mediated colistin-resistant extended-spectrum β-lactamase-producing Escherichia coli in a wild migratory bird from Asia. Int J. Antimicrob. Agents. 2016;48:463–464. doi: 10.1016/j.ijantimicag.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 57.Association CVD. Annual Report on Development of Veterinary Medicine Industry in China (Beijing, 2014).
- 58.Summary Report on Antimicrobials Sold or Distributed for Use in Food-Producing Animals; 2015.
- 59.NARMS Integrated Report. https://www.fda.gov/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/ucm059103.htm (2014).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



