Abstract
Plk2 is a target of p53. Our previous studies demonstrated that with wild-type p53, Plk2 impacts mTOR signaling in the same manner as TSC1, and Plk2-deficient tumors grew larger than control. Other investigators have demonstrated that Plk2 phosphorylates mutant p53 in a positive feedback loop. We investigated Plk2’s tumor suppressor functions in relationship to mTOR signaling. Archival specimens from 12 colorectal adenocarcinomas were stained for markers including Plk2, phosphorylated mTOR (serine 2448) and ribosomal S6 (Serine 235/236). We show that Plk2 is expressed in normal colon, with a punctate staining pattern in supranuclear cytoplasm. In colorectal adenocarcinoma, Plk2 demonstrates complete or partial loss of expression. Strong expression of phosphorylated mTOR is observed in the invasive front. Phosphorylated S6 expression partially correlates with phosphorylated mTOR expression but appears more diffuse in some cases. p53 and Ki67 expression is diffuse, in the subset of cases examined. In order to determine whether Plk2 is lost prior to the development of invasive cancer, 8 colon polyps from 6 patients were evaluated for Plk2 expression. All polyps are positive for Plk2. A Cancer Genome Atlas search identified Plk2 mutations to be infrequent in colorectal adenocarcinomas. Neither Plk2 methylation (in the gene body) nor copy number variations correlated with changes in mRNA expression levels. Loss of Plk2 expression along with accentuated expression of phosphorylated mTOR and phosphorylated S6 at the invasive front in some colorectal carcinomas is consistent with previous findings that an interaction between Plk2 and TSC1 / mTOR signaling molecules plays a role in tumor suppression. Plk2 protein expression is lost at the same stage in colorectal carcinogenesis as p53. The p53 dependence of Plk2 loss and tumor suppressor function in relationship to mTOR signaling may have therapeutic implications.
Abbreviations: Plk2, Polo-like kinase 2; TSC, tuberous sclerosis complex; mTOR, mammalian target of rapamycin; p70S6K/S6K, S6: Ribosomal protein S6 kinase; AMPK, AMP-responsive protein kinase; REDD 1, regulated in DNA damage and development 1; 4EBP1, eukaryotic translation initiation factor 4E binding protein 1; PTEN, phosphatase and tensin homolog
Introduction
Dysregulated cell growth and proliferation can facilitate tumorigenesis. Often activated in neoplasms, the mammalian target of rapamycin (mTOR), is a central promotor of cell growth. Mammalian target of rapamycin complex 1 (mTORC1) and mTORC2 are two multiprotein complexes formed by mTOR [1]. Ribosomal protein S6 kinase (p70S6K/S6K) and eukaryotic translation initiation factor 4E binding protein 1 (4E-BP1), are downstream targets of mTOR that regulate mRNA translation. A heterodimer formed from tuberous sclerosis complex 1 (TSC1) and TSC2 negatively regulates mTOR, preventing the phosphorylation of S6K. Phosphorylation of ribosomal S6 protein, which is a S6K/p70S6K substrate is routinely utilized in both research laboratories and clinics as a biomarker of mTOR activity. While upstream regulators of TSC2 are well-characterized and less is known about the function and regulation of TSC1, mutations in either TSC1 or TSC2 cause the same disease (tuberous sclerosis complex) [1], [2]. TSC1 has been demonstrated to interact with both Polo-like kinase 1 (Plk1) in cell cycle regulated, phosphorylation-dependent manner [3] and Plk2 [4]. Polo-like Kinase 2 (Plk2/Snk) was identified in our laboratory as a direct target for transcriptional regulation by the tumor suppressor p53 protein [5]. Evidence for multiple links between the p53 and mTOR pathways continue to emerge. The p21 accumulation that follows genotoxic stress appears to be mTORC1-dependent [6]. Genotoxic stress may inhibit mTOR activity through up-regulation of known negative regulators PTEN (phosphatase and tensin homolog), TSC2 and AMPKβ1 (AMP-responsive protein kinase) β1 and AMPKα in a p53-dependent manner [7], [8]. Activation of AMPK, which phosphorylates TSC2 and stimulates its GAP activity and subsequent mTOR inhibition [8] is mediated through the products of two p53 target genes, Sestrin 1 and 2.
The polo-like kinases (Plks) comprise a family of serine/threonine kinases that regulate cell cycle progression, mitosis, centrosome duplication, cytokinesis, and the cellular response to DNA damage [9], [10]. The polo-like kinases are characterized by an N-terminal serine/threonine kinase domain and a C-terminal polo-box domain, which regulates Plk localization, activity and function [9]. The five Plks that have been identified in vertebrate cells are Plk1, Plk2/Snk, Plk3/Fnk/Prk, Plk4/Sak3 and Plk5. The best characterized of the polo like kinases, Plk1, plays a role in multiple cell cycle processes including mitosis [10]. Plk2, one of the least well-characterized of the polo like kinases, has been shown to play a role in centrosome duplication [11], mitotic checkpoint [5], S-phase checkpoint [12] and is involved in the modulation of synapses during normal neuronal activity [9]. Both the Plk2 and Plk3 genes were identified as serum-inducible early growth response genes [13], [14]. The primary sequence of Plk4 is divergent while Plk5 has an inactive pseudo-kinase domain [9]. Plk1 and Plk3 genes are over-expressed in Burkitt’s lymphoma cell lines lacking Plk2, consistent with possible functional degeneracy among the polo kinases [15]. The observation that Plk2-null mice survive with phenotypic abnormalities such as delayed S-phase entry may be attributed to functional degeneracy among the polo like kinases [16].
Mouse brain, heart and lung, but not thymus, spleen, liver or kidney are the organ sites where Plk2 was initially detected [17], [18]. Plk2 mRNA levels were found to be high in the testis, mammary gland, and spleen while intermediate in the brain, heart, uterus and trachea in a more comprehensive study [18], [19]. In response to serum stimulation, various fibroblasts demonstrate transient increases in Plk2 mRNA levels while NIH 3T3 cells demonstrated transient increases in Plk2 protein. Although expressed and catalytically active in the G1 phase of the cell cycle, with a short half-life of approximately 15 minutes [18], [20] little is known about the regulation of Plk2 protein stability. Ubiquitylation and degradation of Plk2 by hVPS18, a RING-H2 type ubiquitin ligase has been reported [21].
Cancer prognosis has been linked to alterations in the expression of polo-like kinase family members, Plk1 and Plk3 in human tumors [22]. Plk1 has been demonstrated to be elevated in prostate and ovarian cancer, and clearly linked to patient prognosis in prostate [23], ovarian [22], breast [24], lung, head-neck squamous cell [25], esophageal and gastric carcinoma [26]. Plk2 expression is lost in B-cell malignancies [15]. While Plk1 expression decreases following DNA damage, Plk2, Plk3 and Plk5 are activated after DNA damage [18], [27], [28]. Plk2 loss as a result of methylation-dependent silencing of the Plk2 gene has been discovered in patients with B-cell malignancies [15]. Methylation-dependent gene silencing has been reported in a number of cancers [15]. Differences in methylation patterns have been associated with patient outcomes and used to identify novel genes critical to the development of cancer [29], [30]. Loss of Plk2 protein in tumors may result from p53 mutations or alterations in Plk2 stability. The majority of p53 mutations are missense mutations leading to the production of full-length p53. Possible outcomes of p53 mutations include loss of tumor suppressor activities, suppression of the functions of wild-type p53 by either a dominant-negative mechanism or by gain of oncogenic function independent of wild-type p53. An inactive mutant p53 with an intact tetramerization domain may have a dominant-negative effect yielding the loss of the transactivation activity of wild-type p53 [31].
The Plk family members signal with p53. Polo-like Kinase 2 gene (Plk2/Snk) is a direct target for transcriptional regulation by p53 [5]. We subsequently identified a novel connection between the p53 tumor suppressor pathway and the oncogenic mTOR pathway through Plk2 [4]. These recent studies support a role for p53-regulated Plk2 as a candidate tumor suppressor gene whose encoded protein physically associates with TSC proteins to regulate checkpoint signaling and tumor growth [4]. Our overall goal is to further evaluate the function of Plk2 as it relates to mTOR signaling. Our specific objective is to evaluate alterations in Plk2 protein expression and determine at what stage in colon cancer carcinogenesis expression is altered in relationship to mTOR signaling. Archived pathology specimens from colorectal adenocarcinomas and colon polyps were utilized in an IHC analysis to address these questions. In order to guide future studies in the p53-dependence and mechanisms of Plk2 loss in colorectal adenocarcinomas, a TCGA search was conducted to evaluate Plk2 mutations and Plk2 gene methylation in colorectal adenocarcinomas.
Materials and Methods
Immunohistochemistry
Paraffin-embedded colon cancer samples sectioned with adjacent normal tissue were processed using standard protocols including antigen unmasking with citric acid. Antibody was detected using a peroxidase kit with diaminobenzidine (DAB) as the chromogen and counterstained with hematoxylin. The primary antibodies are as follows: Rabbit anti-Plk2 antibody (Abcam; 1:150; ab7133; manual), rabbit antibody anti-mTOR phosphorylated at Serine 2448 (P~mTOR Ser 2448; Cell Signaling #2976; 1:100; manual) and rabbit anti-ribosomal protein S6 phosphorylated at Serine 235/236 (P~S6 Ser 235/236; Cell Signaling #4858; 1:100; manual), Ki67 (Ventana, Clone 30-9; 1:20), and p53 (Cell Marque, Clone D07: 1:200) were utilized for IHC. p53 and Ki67 staining was performed with a Ventana automated stainer. A cut-off of 5% was used for positive versus negative staining of Plk2 and a cut-off of 50% was used for partial versus complete / diffuse Plk2 loss. When less than 5% of the cells were positive for Plk2 expression, the sample was scored as negative or loss of expression. When more than 5% of the cells but less than 50% of the cells were positive, the sample was scored as having partial loss of expression. One representative tumor section (with abundant tumor cells) was examined for each case.
Cancer Genome Atlas (TCGA) Data Analysis
Plk2 mutations were identified amongst 494 colorectal cancer cases obtained from Broad GDAC Firehose site (https://gdacbroadinstitute.org) and TCGA data repository (https://gdc.cancer.gov). Similarly in the case of methylation profiles, 312 colorectal cancer cases were identified of which 305 also had mRNA expression data. Of the 309 cases with Plk2 copy number variation (CNV) (GISTIC 2 values [32] data and 305 cases with Plk2 mRNA data (RSEM values; RSEM=RNA seq by expectation-maximization), 302 cases over-lapped, containing both. For methylation data, beta values for probes mapping to CpG islands located in transcription start sites (TSS), 5′UTR through to first exon and gene body including 3′UTR were compared between tumor and their matching normal colon samples. These comparisons are represented as box plots. All computations were done using wateRmelon package [33] implemented in R Bioconductor environment. These samples were analysed in cbioportal (http://www.cbioportal.org) [34].
Results
Plk2 Protein Expression is Lost or Partially Lost in Colorectal Tumors
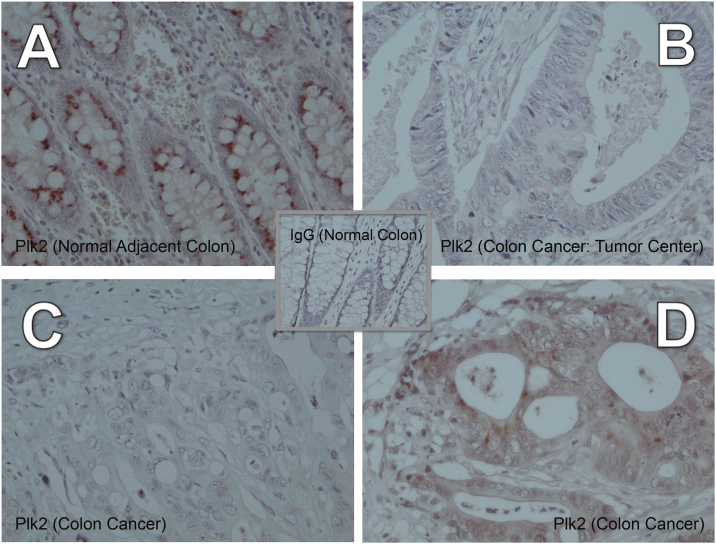
Our previous studies demonstrated that in the wild-type p53 context, human tumor cells deficient in Plk2 grew significantly larger than control tumor cells. If p53 target Plk2 has a tumor suppressor role, Plk2 protein expression may be lost during colon cancer progression. After a recent screening of various commercially available Plk2 antibodies in cancer cells and human tissue samples, optimization of the Abcam Plk2 antibody has yielded promising results in IHC analysis of colorectal cancer specimens (Figure 1). Archived pathology specimens from 12 colorectal adenocarcinomas (5 early stage and 7 advanced stage) were retrieved, and IHC staining for Plk2 was conducted. Plk2 is expressed in adjacent normal colon epithelium, with a punctate staining pattern in the supranuclear region (Figure 1A & Figure S1). In colorectal adenocarcinoma, Plk2 demonstrates partial loss of expression or complete loss of expression (Figure 1, B–D; Table 1). Disrupted Plk2 expression manifested as irregular or abnormal localization was also observed. Loss of Plk2 expression is more pronounced in the invasive front in one case (Table 1; patient 12). In a stage IV colorectal cancer patient’s tumor, significant loss of Plk2 protein was observed (Figure 1B compared to Figure 1A; Table 1, patient 1). A stage IIIB colorectal cancer patient’s tumor demonstrating partial loss of Plk2 is shown in Figure 1, C–D & Table 1 (patient 11). Except for one patient with a mucinous carcinoma, all patients had either loss (5 patients) or partial loss (6 patients) of Plk2 protein expression.
Figure 1.
Plk2 loss in colorectal adenocarcinomas. Plk2 expression is punctuated in distinct areas / intracellular structures adjacent to the nucleus (arrows). A) Normal adjacent colon (patient 1), 400×), B) Colorectal adenocarcinoma: center of the tumor, the area with the greatest Plk2 loss (stage IV, patient 1), 400×. Plk2 loss was more prominent in C) some areas of the tumor than D) other areas (stage IIIB, patient 11). Panels A-B are from one patient with stage IV colorectal adenocarcinoma shown in Figure 2, A–D. Proteins detected by antibody: brown color; Hematoxylin nuclear counterstain: blue.
Table 1.
Plk2 expression is frequently lost and mTOR activated in colorectal adenocarcinomas
| Pt | Stage | K-Ras | Plk2 | P~mTOR | P~S6 | p53 | Ki67 |
|---|---|---|---|---|---|---|---|
| 1 | IV | NT | Loss | IF | IF | Diffuse, S | Diffuse |
| 2 | IIIB | NT | Partial Loss | IF | IF,L | Diffuse, S | Diffuse |
| 3 | I | NT | Partial Loss | IF | IF | Diffuse, S | Diffuse, S |
| 4 | I | NT | Pos (Muc. carc) | IF | Diffuse | W | Diffuse, S |
| 5 | IV | NT | Partial Loss | IF | IF | Diffuse, S | Diffuse |
| 6 | I | NT | Loss | Neg | Neg | Diffuse, W | Diffuse, S |
| 7 | IIIB | G12D | Loss | IF, L | IF, L | NE | NE |
| 8 | IIIB | WT | Loss | IF,L | Neg/L | NE | NE |
| 9 | IIB | G12D | Loss | F/Neg | Neg | NE | NE |
| 10 | IIIB | WT | Partial Loss | IF | Diffuse | NE | NE |
| 11 | IIIB | WT | Partial LossΨ | IF | L | NE | NE |
| 12 | I | NT | Partial Loss-IF | IF | Diffuse | NE | NE |
| IF, Invasive front | L, Luminal | C, Center | NT Not tested for K-ras mutation | NE not evaluated |
| S, Strong/high levels | Patients with high levels (strong) of p53ere predicted to have mutant p53 | |||
| W, Weak/low levels | Patients with low levels (weak) of p53 were predicted to likely have wi-typep53 | |||
| F, Focal | G12D Mutation in Codon 12 of K-ras | |||
| Neg Most or all pf the tumor is negative for expression of this protein | ||||
Plk2 expression: adjacent normal tissue/ staining did not allow for internal comparison with tumor.
The mTOR Pathway Activated and Plk2 Lost in Colorectal Adenocarcinomas
Our previous studies demonstrated that in the wild-type p53 context, Plk2 restrains mTOR signaling in the same manner as TSC1. In order to identify a possible inverse relationship between Plk2 protein expression and mTOR signaling, an immunohistochemical (IHC) analysis of colorectal adenocarcinomas was conducted. Ribosomal protein S6 is phosphorylated by p70S6 Kinase during activation of the mTOR pathway. In the same 12 archived pathology specimens of colorectal adenocarcinomas IHC staining for phosphorylated mTOR (serine 2448) [35] and phosphorylated ribosomal protein S6 (Serine 235/236) was performed. Given tumor heterogeneity, attention was paid to invasive front expression of these proteins as compared to expression closer to the surface.
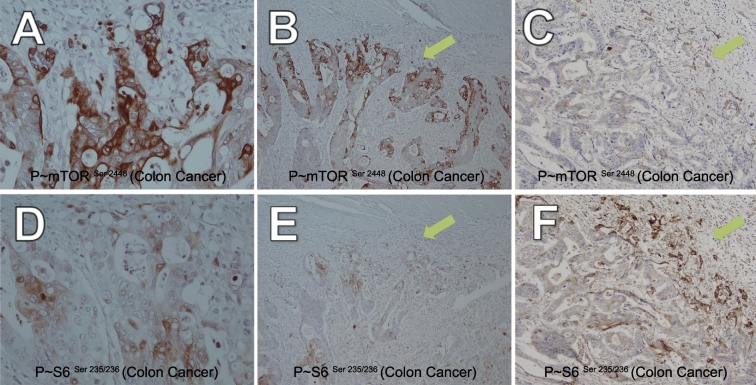
We observed that in the majority of tumors, mTOR signaling is high in the colorectal invasive front, consistent with the literature [36]. This is exemplified by two representative cases colorectal adenocarcinoma cases demonstrating elevation of phospho-mTOR Ser2448 in the tumor invasive front (Figure 2, A and B, stage IIIB; Figure 2C, stage IV) and elevation of phospho-S6 Ser234/235 in the invasive front of the same stage IIIB patient (Figure 2, D and E) and stage IV (Figure 2F) patients. This stage IV patient was described above as having loss of Plk2 (Figure 1B; Table 1, patient 1) and this stage IIIB patient described as having partial loss of Plk2 expression (Figure 1C and D; Table 1, patient 11). In 10 of the 12 cases examined, invasive front phosphorylation (ser 2448) of mTOR was observed (Table 1). Phosphorylated S6 expression partially correlates with upstream phosphorylated mTOR expression [35] in some cases but appears more diffuse in other cases (Table 1). The specificity of these high quality antibodies is further demonstrated by the high expression of these phospho-proteins in the luminal cells as compared to the basal cells of normal breast ducts for phospho-mTOR (Figure S2A) and in the breast ducts as compared to the surrounding tissue for phospho-S6 (Figure S2B).
Figure 2.
The mTOR pathway is activated in colorectal adenocarcinomas. A) P~mTOR Ser 2448 in the colorectal adenocarcinoma invasive front (stage IIIB, patient 11), 400×. B) Lower power version of A, 100×. C) P~mTOR Ser 2448 in the colorectal adenocarcinoma invasive front (stage IV, patient 1), 200×. D) P~S6 Ser 235/236 in the colorectal adenocarcinoma invasive front (stage IIIB, patient 11), 400×. E) Lower power version of D, 100×. F) P~S6 Ser 235/236 in the colorectal adenocarcinoma invasive front (stage IV, patient 1), 200×. Phospho-protein detected by antibody: reddish-brown color; Hematoxylin counterstain of nuclei: blue. Arrows point to invasive front.
Colorectal Adenocarcinomas Demonstrate Diffuse Expression of p53 and Ki67
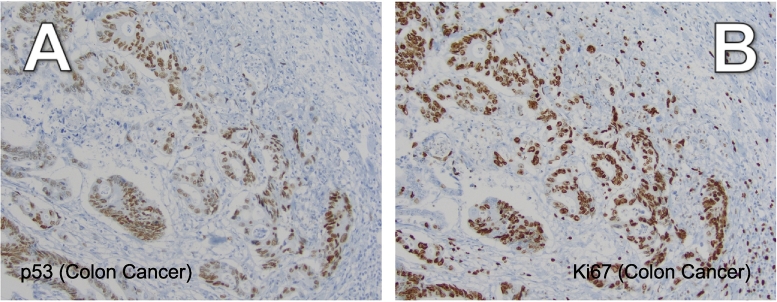
In the same archived pathology specimens from 12 colorectal adenocarcinomas, IHC staining for p53 and Ki67 was performed. p53 and Ki67 expression is diffuse, rather than specific to the tumor invasive front in the subset of cases examined (Figure 3; Table 1; Figures S3 & S4). The Ki67 proliferation index was 85% (Figure 3B), 90%, 90%, 85% and 90% respectively (Figure S3, A–D). It should be noted that in wild-type p53 cells, p53 levels are expected to be low at baseline. As contrasted to the patient tumors shown in Figure S4, A and B with low p53 protein expression, the tumors in the lower panels (Figure S4, C and D) exhibiting robust p53 protein expression are likely comprised of cells with mutant p53 (Table 1). Colorectal adenocarcinomas with the no p53 protein expression are expected to be p53 mutant, but were not encountered in this study.
Figure 3.
Ki67 and p53 expression are diffuse in colorectal adenocarcinomas. A) p53 protein expression, 200×, B) Ki67 protein expression (Ki67 proliferation index= 85%), 200×. Tissue sections are from the same representative stage IV colorectal adenocarcinoma tumor (patient 1).
Pre-Malignant Colorectal Lesions and Adjacent Normal Tissue Demonstrate Robust Plk2 Expression
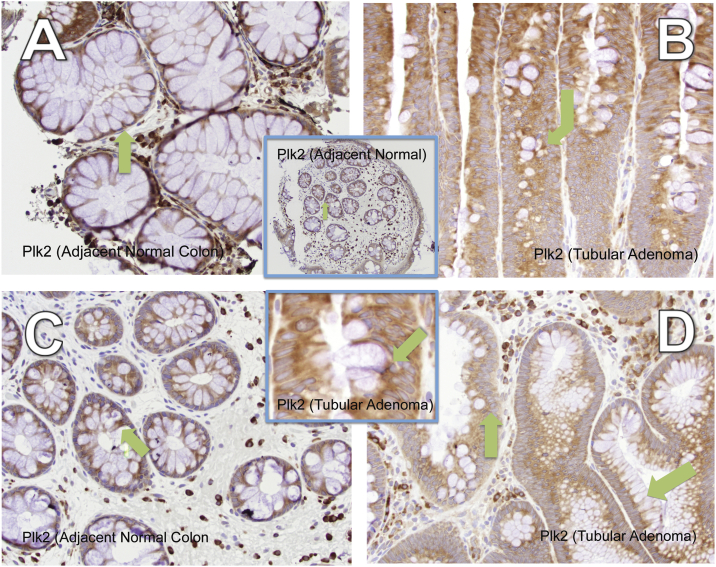
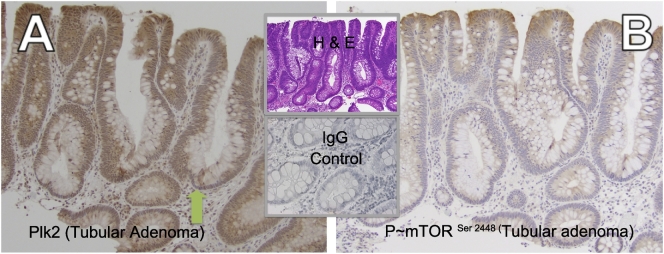
In the eight different pre-malignant lesions (4 tubular adenomas, 1 tubulovillous adenoma, 2 sessile serrated polyp and 1 hyperplastic polyp) from six patients, Plk2 was present (Figure 4, Table 2 and Figure S5). Robust cytoplasmic Plk2 expression was observed in the enterocytes and goblet cells of the tubular adenomas and the adjacent normal tissue. Plk2 was observed in the basal portion of all goblet cells.
Figure 4.
Plk2 is present in tubular adenomas. Plk2 expression in A) Adjacent normal and B) Tubular adenoma (patient E), 400×, C) Adjacent normal and D) Tubular Adenoma of a different patient than panels A & B (patient F), 400×. Center top panel: Adjacent normal (patient F), 200×; Center bottom panel: zoom section of Panel B.
Table 2.
Plk2 expression is preserved and mTOR inactive: polyps and tubular adenomas
| Pt | Type | Plk2 | P~mTOR |
|---|---|---|---|
| A-1 | TA / SSP | Pos | Neg |
| A-2 | SSP | Pos | Neg |
| B | HP | Pos | Neg |
| C | TA | Pos | Neg |
| D | TVA | Pos | Neg |
| E | TA | Pos | NE |
| F | TA | Pos | Neg |
Pos, Most or all of the section is positive for this protein; Neg, Most or all of the section is negative for this protein; TA, Tubular adenoma; TVA, Tubulovillous adenoma; SSP, Sessile serrated polyp; HP, Hyperplastic polyp; NE, Not evaluated.
Pre-Malignant Colon Lesions Express Plk2 but mTOR Remains Inactivated
A possible inverse relationship between Plk2 and mTOR signaling during progression to colon carcinoma was evaluated in the pre-malignant lesions described above. It was determined that the Plk2 expression coincided with absence of mTOR activation as demonstrated by the absent or very low phosphorylation of mTOR at serine 2448 in the evaluated pre-malignant lesions (Table 2). This is exemplified by a representative tubular adenoma with Plk2 expression (Figure 5A), but no P~mTOR Ser2448 in the same tumor area (Figure 5B). The increased stain at the luminal edge of the section is most likely due to edge effect and thus considered non-specific.
Figure 5.
Plk2 presence in tubular adenomas coincides with the absence of activated mTOR. A) Plk2 expression in tubular adenoma (patient C), 200×, B) Phosphorylated mTOR ser2448 is negative in the same tumor area as panel A, 200×.
Plk2 Gene Body Methylation is Present in Both Colon Cancer Tumors and Normal Colon
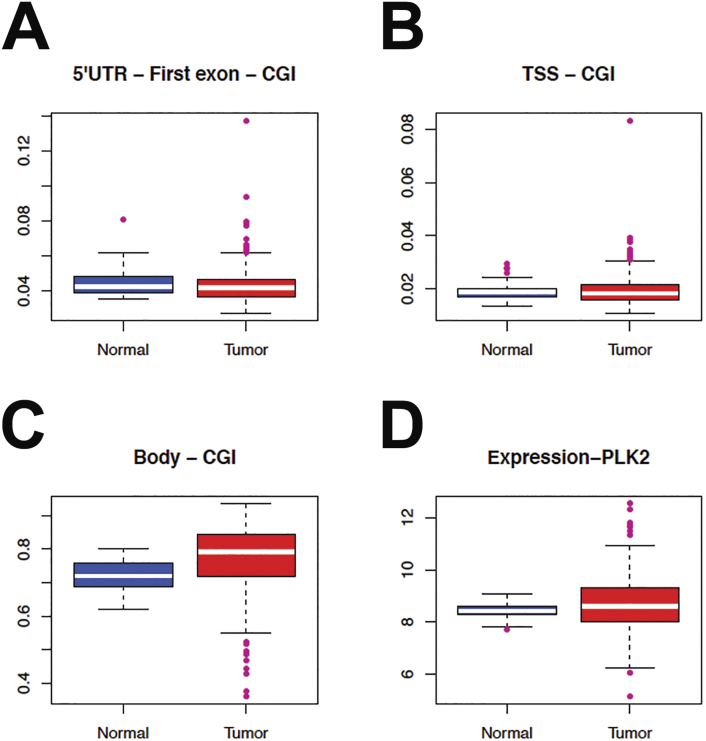
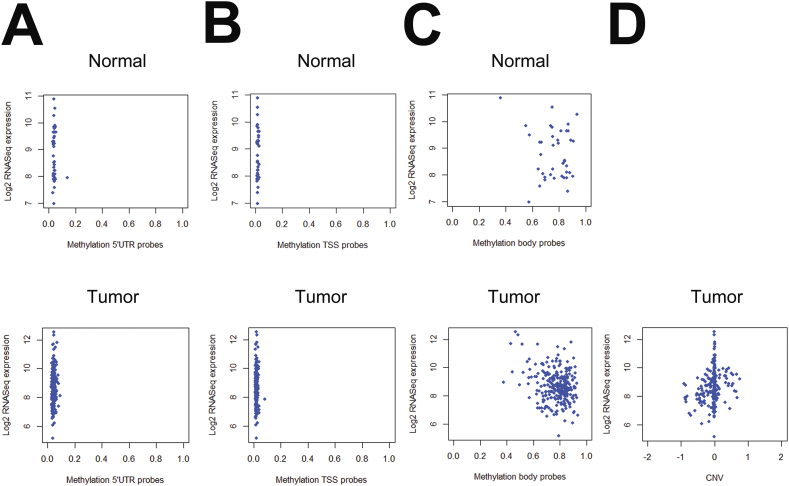
Many important tumor suppressors are suppressed by methylation of promotor CpG islands [30]. Plk2 has been demonstrated to be methylation silenced in Burkitt’s lymphoma and other B-cell and hematological malignancies [37] as well as ovarian cancer [15]. We conducted a Cancer Genome Atlas (TCGA) search of Plk2 methylation in colorectal adenocarcinomas (312 cases with methylation data) as compared to normal. The Plk2 gene body is methylated in both colon cancer and normal (Figure 6C; beta value above 0.4) with median beta values above 0.7. No methylation was reported in the regulatory region CpG islands (CGIs) and shores of the Plk2 gene as shown in the 5′UTR to first exon CGI and the transcription start site (TSS) CGI (Figure 6, A and B). CpG islands promotor methylation is associated with gene repression whereas exonic and genic methylation is associated with active gene expression [38]. Median Plk2 mRNA expression in normal versus colorectal cancer were similar (Figure 6D). Neither Plk2 regulatory site methylation nor copy number variations (CNV) correlated (Spearman rank correlations 0.3) with mRNA expression changes (Figure 7) and were independent of p53 status. Of the 610 TCGA colorectal cancer samples for which stage information is available, there are 109 stage I, 230 stage II, 181 stage III and 90 stage IV cases.
Figure 6.
Plk2 gene body methylation and mRNA expression remain unaltered in colorectal cancer as compared to normal: TCGA. Plk2 Gene methylation normal versus colorectal tumors: A) 5′UTR to the First Exon (P=0.2), B) Transcription Start Site (TSS)(P=0.9), C) Gene Body (P<0.001, and D) Plk2 mRNA expression (RSEM values) in normal versus colorectal tumors (P=0.003). Panels A-C: RNA seq by expectation-maximization (RSEM). Beta-values above 0.4 were considered methylated. Cancer Genome Atlas (TCGA) search yielded 312 colorectal cancers with methylation data of which 305 cases also had mRNA data.
Figure 7.
Plk2 mRNA expression does not correlate with Plk2 gene methylation or copy number variation: TCGA. A) Plk2 mRNA expression (RSEM values) versus 5′UTR methylation (Beta values), B) Plk2 mRNA expression (RSEM values) versus transcription start site (TSS) methylation and C) Plk2 mRNA expression (RSEM values) versus gene body methylation. D) Plk2 mRNA expression versus copy number variation (CNV). Top panels: normal; Bottom panels: colorectal cancer. Cancer Genome Atlas (TCGA) search identified 312 colorectal cancers with methylation data of which 305 cases also had mRNA expression data. Of the 309 cases with copy number variation (CNV) data and 305 cases with mRNA data, 302 cases over-lapped. A-C: 309 patients. D: 302 patients. RNA seq by expectation-maximization (RSEM); Gene methylation expressed as Beta-values.
Plk2 Mutations Occur Infrequently in Colorectal Cancer
Based on a TCGA/Broad Firehose analysis Plk2 mutations occur infrequently in colorectal cancer. Amongst the 494 colorectal cancer patients, 13 patients had Plk2 mutations in their tumors of which five also contained p53 mutation(s). One rectal cancer patient had 3 different mutations and one colon cancer patient had two different mutations to equal 16 individual mutations among the 13 patients. These 16 included 9 missense, 3 frame shift deletions, 1 frameshift insertion, 2 nonsense mutations and 1 silent mutation (Table 3). Mutation assessor and polyphen predictions of the impact of these mutations (Table S1) are provided for comparison with our own findings/predictions.
Table 3.
Plk2 gene mutations occur infrequently in colorectal cancer
| Patient | Organ Site | Variation Type | cDNA | Protein | p53-Mutation |
|---|---|---|---|---|---|
| I | Colon | Missense Mutation | c.A1699G | p.S567G | |
| II | Rectal | Nonsense Mutation | c.C2014T | p.R672* | |
| Missense Mutation | c.C1336T | p.R446W | |||
| Missense Mutation | c.C1089A | p.F363L | |||
| III | Colon | Nonsense Mutation | c.1171G>T | p.E391* | |
| IV | Colon | Missense Mutation | c.274G>T | p.G92C | p.D186G;p.R1 75H;p.S95P |
| V | Colon | Missense Mutation | c.346A>C | p.S116R | p.I255T;p.V218A |
| Colon | Missense Mutation | c.274G>T | p.G92C | ||
| VI | Colon | Missense Mutation | c.274G>T | p.G92C | |
| VII | Colon | Silent | c.276C>T | p.G92G | p.R1 75H |
| VIII | Colon | Frame Shift Insertion | c.1004_1005insT | p.L335fs | |
| IX | Colon | Frame Shift Deletion | c.1005delG | p.L335fs | p.P3 01fs |
| X | Colon | Missense Mutation | c.G1856T | p.G619V | p.R273C |
| XI | Rectal | Missense Mutation | c.C1089A | p.F363L | |
| XII | Colon | Frame Shift Deletion | c.1004delT | p.L335fs | |
| XIII | Colon | Frame Shift Deletion | c.1004delT | p.L335fs |
Total Number of Colorectal Cancer Patients = 494 cases The Plk2 gene is on chromosome 5.
Patients I t o III I are common to Broad Firehose an d TCGA.
Patients IX to XIII are not in Broad Firehose but present in TCGA.
Five patients unique to Broad Firehose.
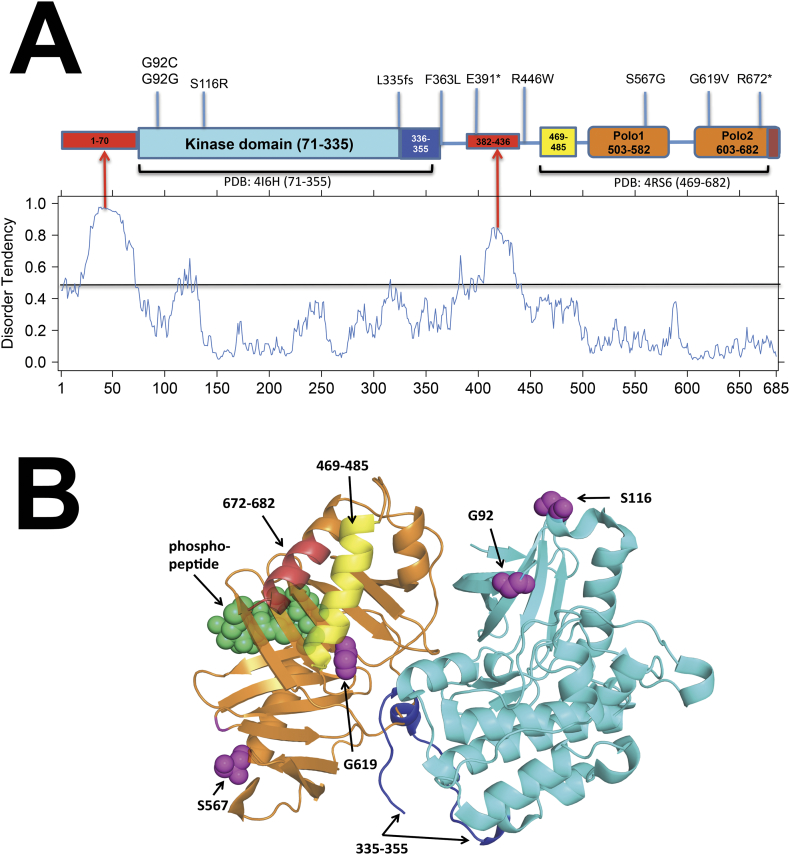
Plk2 Structure and Mutations
We analyzed the sequence and structures of PLK2 domains in order to understand the possible effects of mutations on protein function for variants found in colorectal cancers in TCGA (Table 3). A schematic of the domains and linkers of PLK2 is shown in Figure 8A. The IUPred server [39] predicts two regions of intrinsic disorder in PLK2, residues 1-70 and 382-436 (see red regions, and disorder graph below the protein domain diagram). Experimental X-ray crystallographic structures are available for the kinase domain and the Polo-box domains of PLK2 in the Protein Data Bank (PDB). The structure of the kinase domain (PDB entries: 4I5P, 4I5M, 4I6B, 4I6F, 4I6H) [40] reveal a typical kinase fold in an active DFG-in state comprising residues 71 to 335 (see cyan domain) followed by an ordered C-terminal tail that is folded against the kinase domain (residues 336-355, see dark blue segment). Residues 57 to 70 are present in the crystals but disordered and absent from the coordinates, consistent with the disorder predictions.
Figure 8.
Structural analysis of human Polo-like kinase 2. A). Domain schematic of PLK2. Experimental structures are denoted covering the kinase domain (residues 71-335) and its ordered C-terminal tail (dark blue, residues 336-355) and the Polo-box domains and adjacent ordered N-terminal tail and linkers (residues 469-682). The graph shows the prediction of disordered regions with the program IUPred. Values over 0.5 are predicted to be disordered and are marked in red in the domain schematic. Mutations in TCGA colorectal tumors are marked. B). Modeled structure of full length PLK2 based on structural superposition of the kinase domain (PDB: 4I6H) and the Polo-box domains (PDB: 4R6S) onto the experimental structure of zebrafish PLK1 (PDB: 4J7B). The zebrafish structure provides the relative orientation of the kinase and Polo-box domains of Polo-like kinases. In addition, the structure of the phosphopeptide bound Polo-box domains of human PLK1 (PDB: 1Q4K) was superimposed in order to locate the likely position of phosphopeptides bound to the Polo-box domains of human PLK2. The phosphopeptide is shown in green spheres. The N and C-terminal helices of the Polo-box region are indicated in yellow and dark-red respectively, adjacent to the second Polo-box domain.
The crystal structures of the Polo-box domains of PLK2 contain coordinates for residues 469-682 [PDB: 4RS6 [41], 4XB0 [42]. In these structures, an N-terminal helix (residues 469-485, see yellow segment) folds against the second Polo-box domain followed by a linker (residues 486-502) and the first Polo-box domain (residues 503-582, see orange domains), a linker between the two Polo-box domains (residues 583-602), and the second Polo-box domain (residues 603-682). To determine if any structure was available for Polo-box domains in complex with a kinase domain, we utilized the Protein Common Interface Database (ProtCID, http://dunbrack2.fccc.edu/protcid) [43] which helped us to identify a structure of zebrafish PLK1 containing a complex of the kinase domain and the Polo-box domains expressed as two separate proteins (PDB: 4J7B (44)). This structure is in an autoinhibited state, which can be relieved by phosphorylation within the kinase domain or by binding of phosphopeptides to the Polo-box domains [44]. We superimposed the experimental structures of the kinase domain and the Polo-box domains of PLK2 as well as a phosphopeptide-bound structure of human PLK1 onto the zebrafish PLK1 complex structure in order to produce a model of full-length human PLK2 (excluding the disordered regions) bound to a phosphopeptide. This complex is shown in Figure 8B.
The TCGA mutations can be interpreted in terms of their placement on the modeled structure of full-length PLK2. The L335fs frameshift mutation truncates the protein at the end of the kinase domain, prior to a C-terminal tail that is folded against the kinase domain. It is possible that such a construct is thermodynamically unstable and leads to degradation of the resulting protein. Alternatively, such a kinase domain might be constitutively active, since it lacks the C-terminal autoinhibitory Polo-box domains. Similarly, the E391* truncation mutation produces a full-length kinase domain which may be more stable since it includes the C-terminal tail (residues 336-355) plus a portion of the linker between the kinase and Polo-box domains. Sample TCGA-AG-A002 contains a frameshift at residue 672, which truncates the protein by removing the last 14 residues of the full length protein (residues 672-685). Residues 672-682 form the C-terminal helix (see orange-red helix in Figure 8B) of the second Polo-box domain and their removal may result in an unstable protein subject to degradation. The same sample contains two missense mutations, F363L and R446W, in the linker between the kinase domain and the Polo-box domains. While these are in a flexible region of the protein where it is difficult to predict their effect, it is an interesting possibility that inactivation of both alleles may play a role in progression.
Several of the remaining samples contain missense mutations that can be mapped to the modeled three-dimensional structure of PLK2. The G92C mutation is in the Gly-rich loop of the N-terminal domain of the kinase, which is critical to the activity of protein kinases. The mutation is likely to change the conformation of the backbone of this loop, resulting in an inactive kinase. S116R is in the B-helix, a short helix that precedes the C-helix of the N-terminal domain of the kinase. The functional effects of such a mutation are difficult to predict, although the mutation is very near the activation loop and may adversely affect kinase activity. S567G and G619V are in the Polo-box domains. The first mutation, S567G, is on the surface of the first Polo-box domain and is not in contact with either the kinase domain or phosphopeptides bound to the Polo-box domains. This mutation may therefore be functionally neutral. The G619V mutation is in a β turn in the β sheet of the second Polo-box domain and sits adjacent to the kinase domain. The mutation may be disruptive to the fold of the second Polo-box domain since it is likely to disturb the structure of the β turn.
Discussion
Plk2 is a target of p53. Our previous studies demonstrated that in the context of wild-type p53, Plk2 restrains mTOR signaling in the same manner as TSC1, and human tumor cells deficient in Plk2 grew significantly larger than control tumor cells. Other investigators have demonstrated that Plk2 phosphorylates mutant p53 in a positive feedback loop [45]; Plk2 expression may therefore be transcriptionally regulated by either wild-type [5] or mutant p53 [45]. During this study, it was determined that Plk2 protein expression is completely or partially lost in all colorectal tumors examined with the exception of a mucinous carcinoma. Intriguingly, Plk2 loss was found to occur at the same stage in carcinogenesis as p53 mutational loss, between adenoma and carcinoma. Specifically, all colon polyps and adjacent normal colon evaluated demonstrated robust Plk2 expression.
Activation of the mTOR pathway leads to increased protein translation, cell proliferation and growth. Components of the mTOR pathway including phospho-mTOR Ser 2448 and phospho-p70S6KThr389 have been demonstrated to be highly over-expressed in colorectal cancer [46]. In this study, activation of the mTOR pathway as demonstrated by phosphorylation of both mTOR (serine 2448) [47] and down-stream ribosomal S6 (serines 234/235) was observed in the colorectal carcinomas with various types / degrees of Plk2 loss. The tumor invasive front of carcinomas is an important area for tumor prognosis, which is characterized by increased proliferation, increased gain and loss of adhesion molecules, secretion of proteolytic enzymes, and initiation of angiogenesis [48]. Phosphorylation of mTOR at the invasive front was observed in a large percentage (10 out of 12; Table 1) of the colon carcinomas. In some tumors, Plk2 loss was significant in the invasive front. Through IHC of the same tumor area from individual patients, we determined that as with the loss of Plk2, phosphorylation of mTOR (ser2448) increased between adenoma and carcinoma. These findings raise the possibility that the corresponding loss of putative tumor suppressor Plk2 and activation of oncogenic mTOR play a role in progression to invasive colorectal carcinoma.
A role for mTOR in colon carcinogenesis has been previously suggested [36], [49]. The appearance of mTOR ser 2448 phosphoryation in colorectal adenomas with high compared to low intra-epithelial neoplasias (HIN vs LIN) has been reported. These investigators found mTOR signaling in CRC to be stronger than in noncancerous mucosa and LIN [50]. Other studies have reported increased mTOR signaling in colorectal carcinomas compared to matched normal [51]. mTORC1 phosphorylates and activates p70S6K at T389 to activate the ribosomal protein S6 via phosphorylation at S235/236 and S240/244. An analysis of S6 phosphorylation demonstrates p70S6K and mTORC1 kinase activity whereas phospho-AKT S475 indicates mTORC2 kinase activity. The mTOR serine 2448 site is a target of phosphorylation by both AKT and after mitogen and nutrient-derived stimuli by p70 S6 kinase (S6K) [35]. As seen in renal clear cell carcinoma, we expected increased phospho-mTOR Ser 2448 at the invasive front to correlate with phospho-S6 at the invasive front. In some of the advanced colon cancer specimens, the increased phospho-mTOR was not accompanied by increased phospho-S6 indicating that a factor other than p70S6K may be regulating S6 phosphorylation in colon cancer. All the colorectal adenocarcinomas demonstrated high proliferation index based on extensive expression of Ki67 throughout the tumors. There have been reports of regional differences in Ki67 expression in different tumor types [52], [53], [54], [55].
Epigenetic alterations are one of the early events in colon tumorigenesis [30], [56], [57]. In normal cells, promotor CpG islands commonly remain unmethylated and during differentiation are associated with active gene expression. However, methylated CpG islands are associated with gene repression and many important tumor suppressors are suppressed by methylation of promotor CpG islands [30]. Exonic or genic CpG methylation is typically associated with active gene expression [30]. Although Plk2 is methylation silenced in B-cell malignancies and ovarian cancer, our TCGA analysis determined that this is not the case in colon cancer and would therefore not explain the Plk2 protein loss we observed in the colorectal cancer cases evaluated. TCGA analysis further identified Plk2 mutations to be infrequent in colorectal adenocarcinomas. Some of these Plk2 mutations may lead to the Plk2 protein loss in some cases through decreased Plk2 protein stability. Nevertheless, the mechanism behind the vast majority of cases of Plk2 loss in colorectal cancer we observed remains to be elucidated.
Conclusions
Inhibitors of mTOR are currently in use (renal and other cancer types) with new clinical trials being conducted. Plk1 inhibitors are now in the clinic [58]. Promising Plk1 inhibitors such as Volasertib are currently in clinical trials. A Plk4 inhibitor is in early phase trials for advanced cancers. Understanding Plk2 function in the mTOR pathway is important for the design of treatments targeting mTOR and other Plk family members. As a result of the similarity in catalytic domain between Plk1-3, Plk2 and Plk3 are in the range of inhibition by Plk1 inhibitors [58]. Inhibition of tumor suppressors Plk2 and Plk3 decreases the effectiveness of Plk1 inhibitors and spotlights the importance of a better understanding these less well-characterized family members. Plk1 inhibitors that instead target the polo-box domain and thereby inhibit Plk1 protein interactions are being explored as an alternative approach. This study demonstrates Plk2 loss in colorectal carcinomas but not adenomas. Moreover, a reciprocal relationship between Plk2 expression and mTOR signaling was observed during progression from adenomas to carcinomas. An evaluation of data from a large set of colorectal carcinomas demonstrates that the Plk2 gene is not methylated in regions associated with gene repression and Plk2 mutations are infrequent indicating a different mechanism of Plk2 loss in the vast majority of colorectal carcinomas. In the future we will investigate the mechanism of Plk2 loss in the context of p53 status, including changes in mRNA and protein stability and correlate these changes with changes in mTOR signaling. Plk2 loss is a potential marker of progression to invasive colorectal cancer.
Conflict of Interests
The authors have no conflicts of interest.
Funding
This research was funded by the American Cancer Society.
Contributor Information
Elizabeth M. Matthew, Email: mattheeh@yahoo.com.
Zhaohai Yang, Email: zyang2@pennstatehealth.psu.edu.
Suraj Peri, Email: suraj.peri@fccc.edu.
Mark Andrake, Email: mandrake@fccc.edu.
Roland Dunbrack, Email: roland.dunbrack@fccc.edu.
Eric Ross, Email: eric.ross@fccc.edu.
Wafik S. El-Deiry, Email: wafik.eldeiry@fccc.edu.
References
- 1.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12(1):9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 2.Crino PB, Nathanson KL, Henske EP. The tuberous sclerosis complex. N Engl J Med. 2006;355(13):1345–1356. doi: 10.1056/NEJMra055323. [DOI] [PubMed] [Google Scholar]
- 3.Astrinidis A, Senapedis W, Henske EP. Hamartin, the tuberous sclerosis complex 1 gene product, interacts with polo-like kinase 1 in a phosphorylation-dependent manner. Hum Mol Genet. 2006;15(2):287–297. doi: 10.1093/hmg/ddi444. [DOI] [PubMed] [Google Scholar]
- 4.Matthew EM, Hart LS, Astrinidis A, Navaraj A, Dolloff NG, Dicker DT. The p53 target Plk2 interacts with TSC proteins impacting mTOR signaling, tumor growth and chemosensitivity under hypoxic conditions. Cell Cycle. 2009;8(24):4168–4175. doi: 10.4161/cc.8.24.10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burns TF, Fei P, Scata KA, Dicker DT, El-Deiry WS. Silencing of the novel p53 target gene Snk/Plk2 leads to mitotic catastrophe in paclitaxel (taxol)-exposed cells. Mol Cell Biol. 2003;23(16):5556–5571. doi: 10.1128/MCB.23.16.5556-5571.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beuvink I, Boulay A, Fumagalli S, Zilbermann F, Ruetz S, O'Reilly T. The mTOR inhibitor RAD001 sensitizes tumor cells to DNA-damaged induced apoptosis through inhibition of p21 translation. Cell. 2005;120(6):747–759. doi: 10.1016/j.cell.2004.12.040. [DOI] [PubMed] [Google Scholar]
- 7.Feng Z, Hu W, de Stanchina E, Teresky AK, Jin S, Lowe S. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 2007;67(7):3043–3053. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- 8.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134(3):451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zitouni S, Nabais C, Jana SC, Guerrero A, Bettencourt-Dias M. Polo-like kinases: structural variations lead to multiple functions. Nat Rev Mol Cell Biol. 2014;15(7):433–452. doi: 10.1038/nrm3819. [DOI] [PubMed] [Google Scholar]
- 10.van Vugt MA, Medema RH. Getting in and out of mitosis with Polo-like kinase-1. Oncogene. 2005;24(17):2844–2859. doi: 10.1038/sj.onc.1208617. [DOI] [PubMed] [Google Scholar]
- 11.Warnke S, Kemmler S, Hames RS, Tsai HL, Hoffmann-Rohrer U, Fry AM. Polo-like kinase-2 is required for centriole duplication in mammalian cells. Curr Biol. 2004;14(13):1200–1207. doi: 10.1016/j.cub.2004.06.059. [DOI] [PubMed] [Google Scholar]
- 12.Matthew EM, Yen TJ, Dicker DT, Dorsey JF, Yang W, Navaraj A. Replication stress, defective S-phase checkpoint and increased death in Plk2-deficient human cancer cells. Cell Cycle. 2007;6(20):2571–2578. doi: 10.4161/cc.6.20.5079. [DOI] [PubMed] [Google Scholar]
- 13.Chase D, Feng Y, Hanshew B, Winkles JA, Longo DL, Ferris DK. Expression and phosphorylation of fibroblast-growth-factor-inducible kinase (Fnk) during cell-cycle progression. Biochem J. 1998;333(Pt 3):655–660. doi: 10.1042/bj3330655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donohue PJ, Alberts GF, Guo Y, Winkles JA. Identification by targeted differential display of an immediate early gene encoding a putative serine/threonine kinase. J Biol Chem. 1995;270(17):10351–10357. doi: 10.1074/jbc.270.17.10351. [DOI] [PubMed] [Google Scholar]
- 15.Syed N, Smith P, Sullivan A, Spender LC, Dyer M, Karran L. Transcriptional silencing of Polo-like kinase 2 (SNK/PLK2) is a frequent event in B-cell malignancies. Blood. 2006;107(1):250–256. doi: 10.1182/blood-2005-03-1194. [DOI] [PubMed] [Google Scholar]
- 16.Ma S, Charron J, Erikson RL. Role of Plk2 (Snk) in mouse development and cell proliferation. Mol Cell Biol. 2003;23(19):6936–6943. doi: 10.1128/MCB.23.19.6936-6943.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simmons DL, Neel BG, Stevens R, Evett G, Erikson RL. Identification of an early-growth-response gene encoding a novel putative protein kinase. Mol Cell Biol. 1992;12(9):4164–4169. doi: 10.1128/mcb.12.9.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winkles JA, Alberts GF. Differential regulation of polo-like kinase 1, 2, 3, and 4 gene expression in mammalian cells and tissues. Oncogene. 2005;24(2):260–266. doi: 10.1038/sj.onc.1208219. [DOI] [PubMed] [Google Scholar]
- 19.Liby K, Wu H, Ouyang B, Wu S, Chen J, Dai W. Identification of the human homologue of the early-growth response gene Snk, encoding a serum-inducible kinase. DNA Seq. 2001;11(6):527–533. doi: 10.3109/10425170109041337. [DOI] [PubMed] [Google Scholar]
- 20.Ma S, Liu MA, Yuan YL, Erikson RL. The serum-inducible protein kinase Snk is a G1 phase polo-like kinase that is inhibited by the calcium- and integrin-binding protein CIB. Mol Cancer Res. 2003;1(5):376–384. [PubMed] [Google Scholar]
- 21.Yogosawa S, Hatakeyama S, Nakayama KI, Miyoshi H, Kohsaka S, Akazawa C. Ubiquitylation and degradation of serum-inducible kinase by hVPS18, a RING-H2 type ubiquitin ligase. J Biol Chem. 2005;280(50):41619–41627. doi: 10.1074/jbc.M508397200. [DOI] [PubMed] [Google Scholar]
- 22.Weichert W, Denkert C, Schmidt M, Gekeler V, Wolf G, Kobel M. Polo-like kinase isoform expression is a prognostic factor in ovarian carcinoma. Br J Cancer. 2004;90(4):815–821. doi: 10.1038/sj.bjc.6601610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weichert W, Schmidt M, Gekeler V, Denkert C, Stephan C, Jung K. Polo-like kinase 1 is overexpressed in prostate cancer and linked to higher tumor grades. Prostate. 2004;60(3):240–245. doi: 10.1002/pros.20050. [DOI] [PubMed] [Google Scholar]
- 24.Weichert W, Kristiansen G, Winzer KJ, Schmidt M, Gekeler V, Noske A. Polo-like kinase isoforms in breast cancer: expression patterns and prognostic implications. Virchows Arch. 2005;446(4):442–450. doi: 10.1007/s00428-005-1212-8. [DOI] [PubMed] [Google Scholar]
- 25.Knecht R, Elez R, Oechler M, Solbach C, von Ilberg C, Strebhardt K. Prognostic significance of polo-like kinase (PLK) expression in squamous cell carcinomas of the head and neck. Cancer Res. 1999;59(12):2794–2797. [PubMed] [Google Scholar]
- 26.Kanaji S, Saito H, Tsujitani S, Matsumoto S, Tatebe S, Kondo A. Expression of polo-like kinase 1 (PLK1) protein predicts the survival of patients with gastric carcinoma. Oncology. 2006;70(2):126–133. doi: 10.1159/000093003. [DOI] [PubMed] [Google Scholar]
- 27.Eckerdt F, Yuan J, Strebhardt K. Polo-like kinases and oncogenesis. Oncogene. 2005;24(2):267–276. doi: 10.1038/sj.onc.1208273. [DOI] [PubMed] [Google Scholar]
- 28.Andrysik Z, Bernstein WZ, Deng L, Myer DL, Li YQ, Tischfield JA. The novel mouse Polo-like kinase 5 responds to DNA damage and localizes in the nucleolus. Nucleic Acids Res. 2010;38(9):2931–2943. doi: 10.1093/nar/gkq011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301(5895):89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 30.Ushijima T. Detection and interpretation of altered methylation patterns in cancer cells. Nat Rev Cancer. 2005;5(3):223–231. doi: 10.1038/nrc1571. [DOI] [PubMed] [Google Scholar]
- 31.Blagosklonny MV. p53 from complexity to simplicity: mutant p53 stabilization, gain-of-function, and dominant-negative effect. FASEB J. 2000;14(13):1901–1907. doi: 10.1096/fj.99-1078rev. [DOI] [PubMed] [Google Scholar]
- 32.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pidsley R, CCYW, Volta M, Lunnon K, Mill J, Schalkwyk LC. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013;14:293. doi: 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiang GG, Abraham RT. Phosphorylation of mammalian target of rapamycin (mTOR) at Ser-2448 is mediated by p70S6 kinase. J Biol Chem. 2005;280(27):25485–25490. doi: 10.1074/jbc.M501707200. [DOI] [PubMed] [Google Scholar]
- 36.Greijer AE, Delis-van Diemen PM, Fijneman RJA, Giles RH, Voest EE, van Hinsbergh VWM. Presence of HIF-1 and related genes in normal mucosa, adenomas and carcinomas of the colorectum. Virchows Arch. 2008;452(5):535–544. doi: 10.1007/s00428-008-0578-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benetatos L, Dasoula A, Hatzimichael E, Syed N, Voukelatou M, Dranitsaris G. Polo-like kinase 2 (SNK/PLK2) is a novel epigenetically regulated gene in acute myeloid leukemia and myelodysplastic syndromes: genetic and epigenetic interactions. Ann Hematol. 2011;90(9):1037–1045. doi: 10.1007/s00277-011-1193-4. [DOI] [PubMed] [Google Scholar]
- 38.Stirzaker C, Taberlay PC, Statham AL, Clark SJ. Mining cancer methylomes: prospects and challenges. Trends Genet. 2014;30(2):75–84. doi: 10.1016/j.tig.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 39.Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21(16):3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 40.Aubele DL, Hom RK, Adler M, Galemmo RA, Jr., Bowers S, Truong AP. Selective and brain-permeable polo-like kinase-2 (Plk-2) inhibitors that reduce alpha-synuclein phosphorylation in rat brain. ChemMedChem. 2013;8(8):1295–1313. doi: 10.1002/cmdc.201300166. [DOI] [PubMed] [Google Scholar]
- 41.Shan HM, Wang T, Quan JM. Crystal structure of the polo-box domain of polo-like kinase 2. Biochem Biophys Res Commun. 2015;456(3):780–784. doi: 10.1016/j.bbrc.2014.11.125. [DOI] [PubMed] [Google Scholar]
- 42.Kim JH, Ku B, Lee KS, Kim SJ. Structural analysis of the polo-box domain of human Polo-like kinase 2. Proteins. 2015;83(7):1201–1208. doi: 10.1002/prot.24804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Q, Dunbrack RL., Jr. The protein common interface database (ProtCID)--a comprehensive database of interactions of homologous proteins in multiple crystal forms. Nucleic Acids Res. 2011;39(Database issue):D761–70. doi: 10.1093/nar/gkq1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu J, Shen C, Wang T, Quan J. Structural basis for the inhibition of Polo-like kinase 1. Nat Struct Mol Biol. 2013;20(9):1047–1053. doi: 10.1038/nsmb.2623. [DOI] [PubMed] [Google Scholar]
- 45.Valenti F, Fausti F, Biagioni F, Shay T, Fontemaggi G, Domany E. Mutant p53 oncogenic functions are sustained by Plk2 kinase through an autoregulatory feedback loop. Cell Cycle. 2011;10(24):4330–4340. doi: 10.4161/cc.10.24.18682. [DOI] [PubMed] [Google Scholar]
- 46.Argyriou P, Papageorgiou SG, Panteleon V, Psyrri A, Bakou V, Pappa V. Hypoxia-inducible factors in mantle cell lymphoma: implication for an activated mTORC1-->HIF-1alpha pathway. Ann Hematol. 2011;90(3):315–322. doi: 10.1007/s00277-010-1070-6. [DOI] [PubMed] [Google Scholar]
- 47.Rosner M, Siegel N, Valli A, Fuchs C, Hengstschlager M. mTOR phosphorylated at S2448 binds to raptor and rictor. Amino Acids. 2010;38(1):223–228. doi: 10.1007/s00726-008-0230-7. [DOI] [PubMed] [Google Scholar]
- 48.Bryne M, Boysen M, Alfsen CG, Abeler VM, Sudbo J, Nesland JM. The invasive front of carcinomas. The most important area for tumour prognosis? Anticancer Res. 1998;18(6B):4757–4764. [PubMed] [Google Scholar]
- 49.Aoki K, Tamai Y, Horiike S, Oshima M, Taketo MM. Colonic polyposis caused by mTOR-mediated chromosomal instability in Apc+/Delta716 Cdx2+/- compound mutant mice. Nat Genet. 2003;35(4):323–330. doi: 10.1038/ng1265. [DOI] [PubMed] [Google Scholar]
- 50.Zhang YJ, Dai Q, Sun DF, Xiong H, Tian XQ, Gao FH. mTOR signaling pathway is a target for the treatment of colorectal cancer. Ann Surg Oncol. 2009;16(9):2617–2628. doi: 10.1245/s10434-009-0555-9. [DOI] [PubMed] [Google Scholar]
- 51.Johnson SM, Gulhati P, Rampy BA, Han Y, Rychahou PG, Doan HQ. Novel expression patterns of PI3K/Akt/mTOR signaling pathway components in colorectal cancer. J Am Coll Surg. 2010;210(5):767–776. doi: 10.1016/j.jamcollsurg.2009.12.008. [76-8] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bergstrom C, Palmqvist R, Denekamp J, Oberg A, Tavelin B, Stenling R. Factors influencing the estimates of proliferative labelling indices in rectal cancer. Radiother Oncol. 1998;46(2):169–177. doi: 10.1016/s0167-8140(97)00190-4. [DOI] [PubMed] [Google Scholar]
- 53.Jung A, Schrauder M, Oswald U, Knoll C, Sellberg P, Palmqvist R. The invasion front of human colorectal adenocarcinomas shows co-localization of nuclear beta-catenin, cyclin D1, and p16INK4A and is a region of low proliferation. Am J Pathol. 2001;159(5):1613–1617. doi: 10.1016/s0002-9440(10)63007-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Palmqvist R, Sellberg P, Oberg A, Tavelin B, Rutegard JN, Stenling R. Low tumour cell proliferation at the invasive margin is associated with a poor prognosis in Dukes' stage B colorectal cancers. Br J Cancer. 1999;79(3-4):577–581. doi: 10.1038/sj.bjc.6690091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramires M, David L, Leitao D, Seixas M, Sansonetty F, Sobrinho-Simoes M. Ki67 labelling index in gastric carcinomas. An immunohistochemical study using double staining for the evaluation of the proliferative activity of diffuse-type carcinomas. J Pathol. 1997;182(1):62–67. doi: 10.1002/(SICI)1096-9896(199705)182:1<62::AID-PATH849>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 56.Chan AO, Broaddus RR, Houlihan PS, Issa JP, Hamilton SR, Rashid A. CpG island methylation in aberrant crypt foci of the colorectum. Am J Pathol. 2002;160(5):1823–1830. doi: 10.1016/S0002-9440(10)61128-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36(4):417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 58.Strebhardt K. Multifaceted polo-like kinases: drug targets and antitargets for cancer therapy. Nat Rev Drug Discov. 2010;9(8):643–660. doi: 10.1038/nrd3184. [DOI] [PubMed] [Google Scholar]