Abstract
Neuroblastoma is a commonly occurring extracranial pediatric solid tumor without defined etiology. Polymorphisms in pre-miRNAs have been demonstrated to associate with the risk of several cancers. So far, no such polymorphism has been investigated in neuroblastoma. With this in mind, we performed a two-center case-control study to assess the association of genetic variants in pre-miRNAs and neuroblastoma susceptibility in Chinese children, including 393 cases and 812 controls. We found that miR-34b/c rs4938723 T > C polymorphism was significantly associated with decreased neuroblastoma risk (TC versus TT: adjusted odds ratio [OR] = 0.51, 95% confidence interval [CI] = 0.39–0.67; TC/CC versus TT: adjusted OR = 0.62, 95% CI = 0.48–0.79). We also observed the significant association between the miR-218 rs11134527 A > G polymorphism and decreased neuroblastoma risk (AG versus AA: adjusted OR = 0.73, 95% CI = 0.56–0.96). Stratified analysis further demonstrated that the protective effect of the rs4938723 T > C polymorphism remained prominent in the subgroups, regardless of age, gender, and clinical stages. In term of sites of origin, this polymorphism significantly reduced the risk of tumors originating from the adrenal gland. We further validated the significant results using false-positive report probability analyses. Overall, the miR-34b/c rs4938723 T > C and miR-218 rs11134527 A > G polymorphisms displayed a protective role from neuroblastoma. These findings need further validation.
Keywords: pre-microRNA, miR-34b/c, polymorphism, neuroblastoma, susceptibility
Introduction
Neuroblastoma is one of the most commonly occurring extracranial pediatric solid tumors, which accounts for approximately 8%–10% of all childhood cancers and 15% of pediatric malignancy deaths.1 The 10-year survival rate in patients with low-risk neuroblastoma is around 90%, whereas the long-term survival of high-risk neuroblastoma remains less than 40%, despite great advances achieved in the treatment of cancers.2, 3 Environment risk factors for developing neuroblastoma remain undefined.4, 5 Numerous studies have indicated that genetic factors may play a critical role in the occurrence of neuroblastoma, such as ALK gene mutations6, 7, 8 and genome-wide-association-study-identified susceptibility loci in the CASC15, BARD1, DUSP12, DDX4, IL31RA, HSD17B12, LMO1, HACE1, LIN28B, MLF1, and CPZ genes,9, 10, 11, 12, 13, 14 as well as polymorphisms in the FAS and FASL,15 XPG16 genes.
MicroRNAs (miRNAs) are non-coding single-stranded RNAs of approximately 17–22 nt in length, which is one of the largest classes of gene regulators.17 They can bind to 3′ UTR of mRNA to induce the degradation or translational inhibition of the corresponding mRNAs, consequently silencing target genes.18 In the nucleus, primary miRNA (pri-miRNA) transcripts with lengths from several hundred nucleotides to several kilobases can be cleaved to generate a precursor miRNA (pre-miRNA) of about 70 nt, which can fold to form a stem-loop intermediate.19, 20 Next, the intermediate is further processed to produce a mature miRNA.19 Polymorphisms or mutations in the promoter or in the miRNAs sequence may lead to altered structure or expression of miRNA, thereby influencing the expression of hundreds of target genes.21 Polymorphisms in miRNAs may modify cancer susceptibility and prognosis.22, 23, 24, 25 The association between genetic variants in pre-miRNAs and cancer susceptibility has been investigated in various types of cancer,26 but not in neuroblastoma. Therefore, we performed a two-center case-control study to assess the association of genetic variants in pre-miRNAs and neuroblastoma susceptibility in Chinese children.
Results
Characteristics of the Participants
The demographic and clinical characteristics data of neuroblastoma cases and cancer-free controls are summarized in Table S1. No significant differences were observed between cases and controls for the Southern Chinese children regarding age (p = 0.229) and gender (p = 0.510) and the Northern Chinese children regarding age (p = 0.484) and gender (p = 0.196).
Association of Selected Polymorphisms with Neuroblastoma Risk
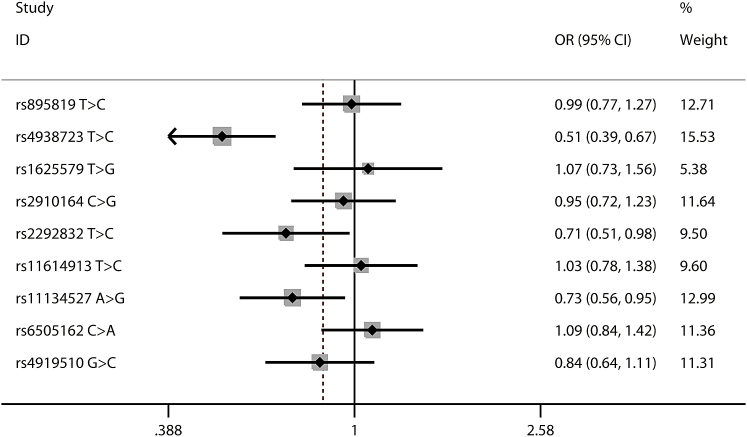
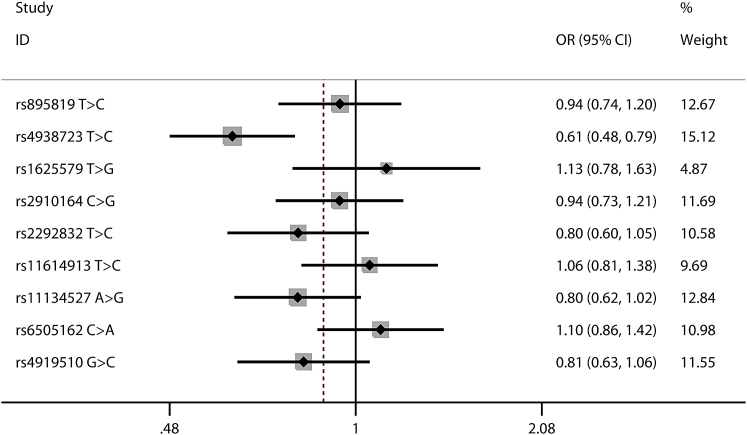
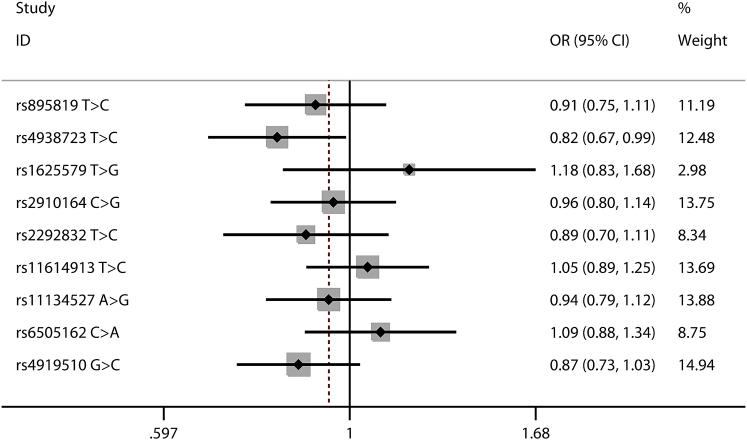
As shown in Tables 1 and S2, all of the nine selected polymorphisms (Table S3) were in accordance with Hardy-Weinberg equilibrium (HWE) in the controls, and the HWE p values ranged from 0.290 to 0.948, except for the miR-149 rs2292832 T > C. As a result, this polymorphism was excluded from further analyses. Of the remaining eight polymorphisms, we found that the miR-34b/c rs4938723 T > C polymorphism was significantly associated with decreased neuroblastoma susceptibility (TC versus TT: adjusted odds ratio [OR] = 0.51, 95% confidence interval [CI] = 0.39–0.67; TC/CC versus TT: adjusted OR = 0.62, 95% CI = 0.48–0.79; and C versus T: adjusted OR = 0.82, 95% CI = 0.68–0.99) (Figures 1, 2, and 3). The miR-218 rs11134527 A > G polymorphism was also shown to significantly decrease neuroblastoma susceptibility (AG versus AA: adjusted OR = 0.73, 95% CI = 0.56–0.96) (Table S4).
Table 1.
Logistic Regression Analyses on Associations between Selected Polymorphisms and Neuroblastoma Risk in Chinese Children
| miRNA | SNP | Allele |
Case (n = 393) |
Control (n = 812) |
Heterozygous (AB versus AA) |
Dominant (AB/BB versus AA) |
Recessive (BB versus AB/AA) |
HWE | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | AA | AB | BB | AA | AB | BB | AOR (95% CI)a | p Valuea | AOR (95% CI)a | p Valuea | AOR (95% CI)a | p Valuea | |||
| miR-27a | rs895819 | T | C | 220 | 153 | 20 | 442 | 312 | 58 | 0.99 (0.77–1.27) | 0.913 | 0.94 (0.74–1.20) | 0.623 | 0.70 (0.42–1.18) | 0.184 | 0.772 |
| miR-34b/c | rs4938723 | T | C | 221 | 107 | 49 | 377 | 358 | 75 | 0.51 (0.39–0.67)b | <0.0001b | 0.62 (0.48–0.79)b | 0.0001b | 1.46 (1.00–2.15) | 0.052 | 0.448 |
| miR-137 | rs1625579 | T | G | 343 | 46 | 3 | 719 | 90 | 1 | 1.07 (0.74–1.57) | 0.717 | 1.13 (0.78–1.64) | 0.519 | 6.23 (0.65–60.00) | 0.114 | 0.290 |
| miR-146a | rs2910164 | C | G | 142 | 189 | 60 | 282 | 397 | 130 | 0.94 (0.72–1.23) | 0.671 | 0.94 (0.73–1.20) | 0.607 | 0.94 (0.68–1.32) | 0.728 | 0.621 |
| miR-149 | rs2292832 | T | C | 286 | 62 | 32 | 560 | 172 | 59 | 0.70 (0.51–0.97)b | 0.032b | 0.79 (0.60–1.05) | 0.106 | 1.14 (0.73–1.79) | 0.559 | <0.001 |
| miR-196a2 | rs11614913 | T | C | 107 | 192 | 94 | 230 | 399 | 183 | 1.04 (0.78–1.39) | 0.793 | 1.06 (0.81–1.39) | 0.673 | 1.08 (0.81–1.43) | 0.607 | 0.691 |
| miR-218 | rs11134527 | A | G | 154 | 164 | 73 | 276 | 403 | 131 | 0.73 (0.56–0.96)b | 0.022b | 0.80 (0.62–1.02) | 0.073 | 1.19 (0.86–1.63) | 0.290 | 0.425 |
| miR-423 | rs6505162 | C | A | 244 | 132 | 17 | 522 | 258 | 31 | 1.09 (0.84–1.42) | 0.497 | 1.10 (0.86–1.41) | 0.448 | 1.13 (0.62–2.07) | 0.694 | 0.900 |
| miR-608 | rs4919510 | G | C | 127 | 190 | 76 | 227 | 405 | 179 | 0.84 (0.64–1.11) | 0.217 | 0.81 (0.63–1.06) | 0.122 | 0.84 (0.62–1.14) | 0.263 | 0.948 |
AOR, adjusted odds ratio; CI, confidence interval; HWE, Hardy-Weinberg equilibrium.
Adjusted for age and gender.
For these values, the 95% CI excluded 1 or p < 0.05.
Figure 1.
Forest Plot for Association between Selected Polymorphisms and Neuroblastoma Susceptibility by a Heterozygous Model: AB versus AA
For each polymorphism, the estimates of odds ratio and its 95% confidence interval are plotted with a box and a horizontal line.
Figure 2.
Forest Plot for Association between Selected Polymorphisms and Neuroblastoma Susceptibility by a Dominant Model: AB/BB versus AA
For each polymorphism, the estimates of odds ratio and its 95% confidence interval are plotted with a box and a horizontal line.
Figure 3.
Forest Plot for Association between Selected Polymorphisms and Neuroblastoma Susceptibility by a Dominant Model: B versus A
For each polymorphism, the estimates of odds ratio and its 95% confidence interval are plotted with a box and a horizontal line.
Stratified Analysis
We further explored the association of miR-34b/c rs4938723 T > C and miR-218 rs11134527 A > G polymorphisms with neuroblastoma susceptibility by stratified analysis (Table 2). We found that the protective effect of the miR-34b/c rs4938723 T > C polymorphism was significant in subgroups, regardless of age, gender, and clinical stages. Concerning sites of origin, this polymorphism tended to reduce the risk of tumors originated from the adrenal gland but not of tumors from another site. As to the miR-218 rs11134527 A > G polymorphism, the significant association was only observed in male subjects.
Table 2.
Stratified Analyses on Associations of rs4938723 T > C and rs11134527 A > G Polymorphisms with Neuroblastoma Risk
| Variables | rs4938723 (Cases/Controls) |
Crude OR (95% CI) | p Value | Adjusted OR (95% CI)a | p Valuea | rs11134527 (Cases/Controls) |
Crude OR (95% CI) | p Value | Adjusted OR (95% CI)a | p Valuea | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TT | TC/CC | AA | AG/GG | |||||||||
| Age (months) | ||||||||||||
| ≤18 | 75/146 | 45/159 | 0.55 (0.36–0.85)b | 0.007b | 0.55 (0.36–0.85)b | 0.007b | 52/107 | 74/198 | 0.77 (0.50–1.18) | 0.226 | 0.77 (0.50–1.18) | 0.227 |
| >18 | 146/231 | 111/274 | 0.64 (0.47–0.87)b | 0.004b | 0.64 (0.47–0.87)b | 0.004b | 102/169 | 163/336 | 0.80 (0.59–1.10) | 0.166 | 0.81 (0.59–1.10) | 0.168 |
| Gender | ||||||||||||
| Females | 103/160 | 60/181 | 0.52 (0.35–0.76)b | 0.007b | 0.52 (0.35–0.76)b | 0.007b | 63/123 | 103/218 | 0.92 (0.63–1.35) | 0.680 | 0.92 (0.63–1.36) | 0.687 |
| Males | 118/217 | 96/252 | 0.70 (0.51–0.97)b | 0.032b | 0.70 (0.51–0.97)b | 0.031b | 91/153 | 134/316 | 0.71 (0.51–0.99)b | 0.044b | 0.72 (0.52–0.995)b | 0.046b |
| Sites of Origin | ||||||||||||
| Adrenal gland | 101/377 | 51/433 | 0.44 (0.31–0.63)b | <0.0001b | 0.44 (0.31–0.64)b | <0.0001b | 59/276 | 94/534 | 0.82 (0.58–1.18) | 0.286 | 0.83 (0.58–1.19) | 0.312 |
| Retroperitoneal | 35/377 | 42/433 | 1.05 (0.65–1.67) | 0.855 | 1.05 (0.65–1.68) | 0.849 | 32/276 | 54/534 | 0.87 (0.55–1.38) | 0.561 | 0.86 (0.54–1.37) | 0.530 |
| Mediastinum | 60/377 | 46/433 | 0.67 (0.44–1.00) | 0.052 | 0.67 (0.45–1.02) | 0.059 | 47/276 | 61/534 | 0.67 (0.45–1.01) | 0.055 | 0.68 (0.45–1.02) | 0.059 |
| Other | 21/377 | 14/433 | 0.58 (0.29–1.16) | 0.122 | 0.58 (0.29–1.16) | 0.121 | 14/276 | 22/534 | 0.81 (0.41–1.61) | 0.552 | 0.81 (0.41–1.61) | 0.547 |
| Clinical Stagesc | ||||||||||||
| I + II + 4 s | 92/377 | 66/433 | 0.63 (0.44–0.88)b | 0.008b | 0.63 (0.44–0.89)b | 0.008b | 65/276 | 97/534 | 0.77 (0.55–1.09) | 0.141 | 0.78 (0.55–1.10) | 0.151 |
| III + IV | 119/377 | 83/433 | 0.61 (0.44–0.83)b | 0.002b | 0.60 (0.44–0.83)b | 0.002b | 82/276 | 127/534 | 0.80 (0.59–1.10) | 0.164 | 0.80 (0.59–1.10) | 0.174 |
OR, odds ratio; CI, confidence interval; INSS, International Neuroblastoma Staging System.
Adjusted for age and gender, omitting the corresponding stratification factor.
For these values, the 95% CI excluded 1 or p < 0.05.
INSS criteria defined stage 4 s as age <1 year old, with localized primary tumor as delineated in stage I or II, and with dissemination limited to liver, skin, or bone marrow.
False-Positive Report Probability Results
We preset 0.2 as the false-positive report probability (FPRP) threshold. As shown in Table 3, at the prior probability of 0.1, all of the significant findings for the miR-34b/c rs4938723 T > C polymorphism remained noteworthy, except for the results on subjects no more than 18 months old, males, and the allele contrast model. Moreover, the association with the miR-218 rs11134527 A > G polymorphism (AG versus AA) was also noteworthy, with a statistical power of 0.820 and the FPRP value of 0.187.
Table 3.
Results of False-Positive Report Probability Analysis for Significant Findings
| Genotype and Variables | OR (95% CI) | p Valuea | Statistical Powerb | Prior Probability |
||||
|---|---|---|---|---|---|---|---|---|
| 0.25 | 0.1 | 0.01 | 0.001 | 0.0001 | ||||
| rs4938723 T > C | ||||||||
| TC versus TT | 0.51 (0.39–0.67) | <0.0001 | 0.039 | 0.000c | 0.000c | 0.003c | 0.033c | 0.254 |
| TC/CC versus TT | 0.62 (0.48–0.79) | 0.0001 | 0.243 | 0.001c | 0.004c | 0.039c | 0.291 | 0.805 |
| C versus T | 0.82 (0.68–0.99) | 0.039 | 0.976 | 0.108c | 0.266 | 0.799 | 0.976 | 0.998 |
| TC/CC versus TT | ||||||||
| ≤18 months old | 0.55 (0.36–0.85) | 0.007 | 0.194 | 0.096c | 0.242 | 0.779 | 0.973 | 0.997 |
| >18 months old | 0.64 (0.47–0.87) | 0.004 | 0.392 | 0.030c | 0.084c | 0.503 | 0.911 | 0.990 |
| Females | 0.52 (0.35–0.76) | 0.001 | 0.096 | 0.021c | 0.062c | 0.420 | 0.880 | 0.987 |
| Males | 0.70 (0.51–0.97) | 0.032 | 0.607 | 0.136c | 0.320 | 0.838 | 0.981 | 0.998 |
| Adrenal gland as site of origin | 0.44 (0.31–0.63) | <0.0001 | 0.015 | 0.002c | 0.006c | 0.062c | 0.399 | 0.869 |
| I + II + 4 s | 0.63 (0.44–0.88) | 0.008 | 0.351 | 0.060c | 0.161c | 0.679 | 0.955 | 0.995 |
| III + IV | 0.61 (0.44–0.83) | 0.002 | 0.278 | 0.019c | 0.055c | 0.391 | 0.866 | 0.985 |
| rs11134527 A > G | ||||||||
| AG versus AA | 0.73 (0.56–0.95) | 0.021 | 0.820 | 0.071c | 0.187c | 0.717 | 0.962 | 0.996 |
| Males | 0.71 (0.51–0.99) | 0.044 | 0.648 | 0.169c | 0.378 | 0.870 | 0.985 | 0.999 |
OR, odds ratio; CI, confidence interval.
Chi-square test was used to calculate the genotype frequency distributions.
Statistical power was calculated using the number of observations in each subgroup and the corresponding ORs and p values in this table.
The level of false-positive report probability threshold was set at 0.2 and noteworthy findings are presented.
Discussion
In the current two-center case-control study, we investigated the association of nine polymorphisms in pre-miRNAs with neuroblastoma susceptibility in Chinese children. We found that the miR-34b/c rs4938723 T > C and miR-218 rs11134527 A > G polymorphisms were significantly associated with a decreased neuroblastoma risk. The associations were further validated by stratified analyses and FPRP analyses. Our results indicate that the polymorphisms in pre-miRNAs may play critical roles in the etiology of neuroblastoma.
miRNAs can negatively regulate gene expressions at the posttranscriptional level and, thereby, affect cell proliferation, differentiation, apoptosis, metabolism, and carcinogenesis.27 Particularly, miR-34 family members can serve as direct transcriptional targets of TP53. Loss of function of miR-34 impairs TP53-mediated cell death, while overexpression of miR-34 induces apoptosis.28, 29, 30 miR-34b/c has been reported to target TP53 and cooperate to suppress cell proliferation and adhesion-independent growth. Furthermore, TP53 can bind to the promoter region of miR-34b/c to increase the expression of miR-34b/c.31 The rs4938723 C > T polymorphism is located at the promoter region of pri-miR-34b/c (423 bp from the transcription start site), which may alter GATA-X transcription factor binding capacity and, consequently, affect the expression of target genes related to carcinogenesis.32, 33 In 2011, Xu et al.33 first found that carriers of the miR-34b/c rs4938723 T allele had a significantly increased risk of hepatocellular carcinoma. Since then, numerous epidemiology studies have been carried out to assess the role of this polymorphism in various cancers.34 So far, no study investigating the association between miR-34b/c rs4938723 C > T polymorphism and neuroblastoma has been reported.
In the present study, we found that the rs4938723 C > T polymorphism was associated with a significantly decreased neuroblastoma risk. The miR-34b/c rs4938723 C > T polymorphism has been suggested to decrease the risk of intracranial aneurysm,35 colorectal cancer,36 esophageal squamous cell carcinoma,37, 38 gastric cancer,39, 40 and childhood acute lymphoblastic leukemia.41, 42 Moreover, we also found that the miR-218 rs11134527 A > G polymorphism was associated with a decreased neuroblastoma risk. This finding is consistent with those of some previous studies, such as studies conducted in cervical cancer43, 44 and esophageal squamous cell carcinoma.45 Opposite results were also observed. For instance, Han et al. found that the same polymorphism was associated with an increased hepatocellular carcinoma risk.46 Polymorphisms may have diverse genetic effects on cancer susceptibility, depending on different cancer types, regions, and ethnicities. It is possible that the methylation status of miR-34b/c may vary among different types of cancer, which could also have an impact on the risk of cancer.36
This is the first and largest study to investigate the associations between polymorphisms in pre-miRNAs and neuroblastoma susceptibility in Chinese children; however, several limitations should be addressed. First, the sample size is still moderate, even though we pooled together samples from two hospitals, partially due to the low incidence rate of neuroblastoma (approximately 7.7 per million in Chinese children).47 As a result, the statistical power of this study was relatively limited. Second, we only included nine polymorphisms in pre-miRNAs. More polymorphisms should be investigated to fully illuminate the contribution of polymorphisms in pre-miRNAs to neuroblastoma susceptibility. Third, other than polymorphisms, low-frequency coding variants and mutations undetectable by genome-wide association studies (GWASs) may also play important roles in neuroblastoma risk.8 More comprehensive studies are encouraged. Fourth, functional analysis is warranted to prove the biological plausibility of our findings from observational studies, which would reveal the underlying mechanisms by which the significant polymorphisms modify neuroblastoma susceptibility. Additionally, in the current hospital-based case-control study, selection bias may exist. Thus, these findings cannot be directly applied to the general population. Finally, due to the nature of retrospective studies, some demographic, environmental, and clinical characteristics were not available, which limited our ability to conduct gene-environmental interactions analysis.
In conclusion, our study provides evidence that miR-34b/c rs4938723 T > C and miR-218 rs11134527 A > G polymorphisms may play a protective role against neuroblastoma. Further prospective studies with different ethnicities and a large sample size are warranted to confirm our findings. In the near future, functional experiments should be performed to explore the possible mechanisms by which these polymorphisms in pre-miRNAs modulate the development of neuroblastoma.
Materials and Methods
Participants
The current two-center case-control study was composed of two independent retrospective studies. One study enrolled 275 histopathologically confirmed neuroblastoma cases enrolled from the Guangzhou Women and Children’s Medical Center (Guangdong Province, China), mainly between February 2010 and March 2017, and 531 cancer-free controls recruited from the same hospital as we described previously.48, 49, 50, 51 The other study incorporated 118 cases and 281 controls recruited from the First Affiliated Hospital of Zhengzhou University (Henan Province, China) from August 2011 to April 2017.52 Informed written consent was obtained from the guardians of all participants. The study protocol was approved by the institutional review boards of the participating institutions.
Polymorphism Selection and Genotyping
Nine widely investigated polymorphisms (miR-27a rs895819 T > C, miR-34b/c rs4938723 T > C, miR-137 rs1625579 T > G, miR-146a rs291016 C > G, miR-149 rs2292832 T > C, miR-196a2 rs11614913 T > C, miR-218 rs11134527 A > G, miR-423 rs6505162 C > A, and miR-608 rs4919510 G > C) were selected (Table S3). The minor allele frequency for all of the nine polymorphisms was larger than 0.05. Of them, eight were located in the transcription factor binding sites, as predicted by SNPinfo (https://snpinfo.niehs.nih.gov/), and the miR-137 rs1625579 T > G polymorphism was significantly associated with schizophrenia risk.53 Genomic DNA was mainly extracted from EDTA-anticoagulated blood samples by using the TIANamp Blood DNA Kit (TianGen Biotech, Beijing, China).54 Genotyping was performed by TaqMan methodology.55, 56, 57 For quality control, 10% samples were retested, and the genotype concordance was 100%.
Statistical Analysis
The χ2 test was used to compare the differences in the frequency distributions of demographic variables and genotypes between cases and controls. The goodness-of-fit χ2 test was adopted to evaluate departure from HWE for the selected polymorphisms in control subjects. ORs and 95% CIs, calculated by multivariate logistic regression, were used to assess the association between the nine selected polymorphisms and neuroblastoma susceptibility. Additionally, stratified analyses were performed by age, gender, tumor sites, and clinical stages. Moreover, we also performed FPRP analysis to verify significant results from the combined subjects.16, 58 All data were analyzed using SAS software (v9.4; SAS Institute, Cary, NC, USA). The p values less than 0.05 were considered as statistically significant.
Author Contributions
Participated in research design: J.H., and H.X.; Conducted experiments: J.H., X.L., and R.Z.; Collected samples: J.H., Y.Z., J. Zhang, and T.Y.; Performed data analysis: J.H. and J. Zhu. Wrote or contributed to the writing of the manuscript: J.H., J. Zhu, and H.X.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by grants from the Pearl River S&T Nova Programme of Guangzhou (no. 201710010086), the National Natural Science Foundation of China (nos. 81502046 and 81602199), the Guangzhou Science Technology and Innovation Commission (no. 201607010395), the Natural Science Foundation of Guangdong Province (no. 2016A030313496), and the State Clinical Key Specialty Construction Project (Paediatric Surgery) 2013 (no. GJLCZD1301).
Footnotes
Supplemental Information includes four tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.01.003.
Contributor Information
Jing He, Email: hejing198374@gmail.com.
Huimin Xia, Email: xia-huimin@foxmail.com.
Supplemental Information
References
- 1.Maris J.M., Hogarty M.D., Bagatell R., Cohn S.L. Neuroblastoma. Lancet. 2007;369:2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 2.Spix C., Pastore G., Sankila R., Stiller C.A., Steliarova-Foucher E. Neuroblastoma incidence and survival in European children (1978-1997): report from the Automated Childhood Cancer Information System project. Eur. J. Cancer. 2006;42:2081–2091. doi: 10.1016/j.ejca.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein M.L., Leclerc J.M., Bunin G., Brisson L., Robison L., Shuster J., Byrne T., Gregory D., Hill G., Dougherty G. A population-based study of neuroblastoma incidence, survival, and mortality in North America. J. Clin. Oncol. 1992;10:323–329. doi: 10.1200/JCO.1992.10.2.323. [DOI] [PubMed] [Google Scholar]
- 4.De Roos A.J., Olshan A.F., Teschke K., Poole C., Savitz D.A., Blatt J., Bondy M.L., Pollock B.H. Parental occupational exposures to chemicals and incidence of neuroblastoma in offspring. Am. J. Epidemiol. 2001;154:106–114. doi: 10.1093/aje/154.2.106. [DOI] [PubMed] [Google Scholar]
- 5.De Roos A.J., Teschke K., Savitz D.A., Poole C., Grufferman S., Pollock B.H., Olshan A.F. Parental occupational exposures to electromagnetic fields and radiation and the incidence of neuroblastoma in offspring. Epidemiology. 2001;12:508–517. doi: 10.1097/00001648-200109000-00008. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y., Takita J., Choi Y.L., Kato M., Ohira M., Sanada M., Wang L., Soda M., Kikuchi A., Igarashi T. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;455:971–974. doi: 10.1038/nature07399. [DOI] [PubMed] [Google Scholar]
- 7.George R.E., Sanda T., Hanna M., Fröhling S., Luther W., 2nd, Zhang J., Ahn Y., Zhou W., London W.B., McGrady P. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature. 2008;455:975–978. doi: 10.1038/nature07397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janoueix-Lerosey I., Lequin D., Brugières L., Ribeiro A., de Pontual L., Combaret V., Raynal V., Puisieux A., Schleiermacher G., Pierron G. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature. 2008;455:967–970. doi: 10.1038/nature07398. [DOI] [PubMed] [Google Scholar]
- 9.Maris J.M., Mosse Y.P., Bradfield J.P., Hou C., Monni S., Scott R.H., Asgharzadeh S., Attiyeh E.F., Diskin S.J., Laudenslager M. Chromosome 6p22 locus associated with clinically aggressive neuroblastoma. N. Engl. J. Med. 2008;358:2585–2593. doi: 10.1056/NEJMoa0708698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Capasso M., Devoto M., Hou C., Asgharzadeh S., Glessner J.T., Attiyeh E.F., Mosse Y.P., Kim C., Diskin S.J., Cole K.A. Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nat. Genet. 2009;41:718–723. doi: 10.1038/ng.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen B., Diskin S.J., Capasso M., Wang K., Diamond M.A., Glessner J., Kim C., Attiyeh E.F., Mosse Y.P., Cole K. Phenotype restricted genome-wide association study using a gene-centric approach identifies three low-risk neuroblastoma susceptibility Loci. PLoS Genet. 2011;7:e1002026. doi: 10.1371/journal.pgen.1002026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang K., Diskin S.J., Zhang H., Attiyeh E.F., Winter C., Hou C., Schnepp R.W., Diamond M., Bosse K., Mayes P.A. Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature. 2011;469:216–220. doi: 10.1038/nature09609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diskin S.J., Capasso M., Schnepp R.W., Cole K.A., Attiyeh E.F., Hou C., Diamond M., Carpenter E.L., Winter C., Lee H. Common variation at 6q16 within HACE1 and LIN28B influences susceptibility to neuroblastoma. Nat. Genet. 2012;44:1126–1130. doi: 10.1038/ng.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDaniel L.D., Conkrite K.L., Chang X., Capasso M., Vaksman Z., Oldridge D.A., Zachariou A., Horn M., Diamond M., Hou C. Common variants upstream of MLF1 at 3q25 and within CPZ at 4p16 associated with neuroblastoma. PLoS Genet. 2017;13:e1006787. doi: 10.1371/journal.pgen.1006787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han W., Zhou Y., Zhong R., Wu C., Song R., Liu L., Zou L., Qiao Y., Zhai K., Chang J. Functional polymorphisms in FAS/FASL system increase the risk of neuroblastoma in Chinese population. PLoS ONE. 2013;8:e71656. doi: 10.1371/journal.pone.0071656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He J., Wang F., Zhu J., Zhang R., Yang T., Zou Y., Xia H. Association of potentially functional variants in the XPG gene with neuroblastoma risk in a Chinese population. J. Cell. Mol. Med. 2016;20:1481–1490. doi: 10.1111/jcmm.12836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bentwich I., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 18.Carthew R.W., Sontheimer E.J. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schanen B.C., Li X. Transcriptional regulation of mammalian miRNA genes. Genomics. 2011;97:1–6. doi: 10.1016/j.ygeno.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee Y., Jeon K., Lee J.T., Kim S., Kim V.N. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu Z., Chen J., Tian T., Zhou X., Gu H., Xu L., Zeng Y., Miao R., Jin G., Ma H. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J. Clin. Invest. 2008;118:2600–2608. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mishra P.J., Humeniuk R., Mishra P.J., Longo-Sorbello G.S., Banerjee D., Bertino J.R. A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. Proc. Natl. Acad. Sci. USA. 2007;104:13513–13518. doi: 10.1073/pnas.0706217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu Z., Liang J., Wang Z., Tian T., Zhou X., Chen J., Miao R., Wang Y., Wang X., Shen H. Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum. Mutat. 2009;30:79–84. doi: 10.1002/humu.20837. [DOI] [PubMed] [Google Scholar]
- 24.He B., Pan Y., Xu Y., Deng Q., Sun H., Gao T., Wang S. Associations of polymorphisms in microRNAs with female breast cancer risk in Chinese population. Tumour Biol. 2015;36:4575–4582. doi: 10.1007/s13277-015-3102-2. [DOI] [PubMed] [Google Scholar]
- 25.He B.S., Pan Y.Q., Lin K., Ying H.Q., Wang F., Deng Q.W., Sun H.L., Gao T.Y., Wang S.K. Evaluation the susceptibility of five polymorphisms in microRNA-binding sites to female breast cancer risk in Chinese population. Gene. 2015;573:160–165. doi: 10.1016/j.gene.2015.07.052. [DOI] [PubMed] [Google Scholar]
- 26.Ryan B.M., Robles A.I., Harris C.C. Genetic variation in microRNA networks: the implications for cancer research. Nat. Rev. Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 28.He L., He X., Lim L.P., de Stanchina E., Xuan Z., Liang Y., Xue W., Zender L., Magnus J., Ridzon D. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raver-Shapira N., Marciano E., Meiri E., Spector Y., Rosenfeld N., Moskovits N., Bentwich Z., Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 30.Chang T.C., Wentzel E.A., Kent O.A., Ramachandran K., Mullendore M., Lee K.H., Feldmann G., Yamakuchi M., Ferlito M., Lowenstein C.J. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corney D.C., Flesken-Nikitin A., Godwin A.K., Wang W., Nikitin A.Y. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 32.Bossard P., Zaret K.S. GATA transcription factors as potentiators of gut endoderm differentiation. Development. 1998;125:4909–4917. doi: 10.1242/dev.125.24.4909. [DOI] [PubMed] [Google Scholar]
- 33.Xu Y., Liu L., Liu J., Zhang Y., Zhu J., Chen J., Liu S., Liu Z., Shi H., Shen H., Hu Z. A potentially functional polymorphism in the promoter region of miR-34b/c is associated with an increased risk for primary hepatocellular carcinoma. Int. J. Cancer. 2011;128:412–417. doi: 10.1002/ijc.25342. [DOI] [PubMed] [Google Scholar]
- 34.Li H., Diao S., Li J., Ma B., Yuan S. An updated meta-analysis of 23 case-control studies on the association between miR-34b/c polymorphism and cancer risk. Oncotarget. 2017;8:28888–28896. doi: 10.18632/oncotarget.16322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li L., Sima X., Bai P., Zhang L., Sun H., Liang W., Liu J., Zhang L., Gao L. Interactions of miR-34b/c and TP53 polymorphisms on the risk of intracranial aneurysm. Clin. Dev. Immunol. 2012;2012:567586. doi: 10.1155/2012/567586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao L.B., Li L.J., Pan X.M., Li Z.H., Liang W.B., Bai P., Zhu Y.H., Zhang L. A genetic variant in the promoter region of miR-34b/c is associated with a reduced risk of colorectal cancer. Biol. Chem. 2013;394:415–420. doi: 10.1515/hsz-2012-0297. [DOI] [PubMed] [Google Scholar]
- 37.Yin J., Wang X., Zheng L., Shi Y., Wang L., Shao A., Tang W., Ding G., Liu C., Liu R. Hsa-miR-34b/c rs4938723 T>C and hsa-miR-423 rs6505162 C>A polymorphisms are associated with the risk of esophageal cancer in a Chinese population. PLoS ONE. 2013;8:e80570. doi: 10.1371/journal.pone.0080570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J., Huang X., Xiao J., Yang Y., Zhou Y., Wang X., Liu Q., Yang J., Wang M., Qiu L. Pri-miR-124 rs531564 and pri-miR-34b/c rs4938723 polymorphisms are associated with decreased risk of esophageal squamous cell carcinoma in Chinese populations. PLoS ONE. 2014;9:e100055. doi: 10.1371/journal.pone.0100055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang C., Ma X., Liu D., Wang Y., Tang R., Zhu Y., Xu Z., Yang L. Promoter polymorphisms of miR-34b/c are associated with risk of gastric cancer in a Chinese population. Tumour Biol. 2014;35:12545–12554. doi: 10.1007/s13277-014-2574-9. [DOI] [PubMed] [Google Scholar]
- 40.Pan X.M., Sun R.F., Li Z.H., Guo X.M., Qin H.J., Gao L.B. Pri-miR-34b/c rs4938723 polymorphism is associated with a decreased risk of gastric cancer. Genet. Test. Mol. Biomarkers. 2015;19:198–202. doi: 10.1089/gtmb.2014.0287. [DOI] [PubMed] [Google Scholar]
- 41.Hashemi M., Bahari G., Naderi M., Sadeghi-Bojd S., Taheri M. Pri-miR-34b/c rs4938723 polymorphism is associated with the risk of childhood acute lymphoblastic leukemia. Cancer Genet. 2016;209:493–496. doi: 10.1016/j.cancergen.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 42.Tong N., Chu H., Wang M., Xue Y., Du M., Lu L., Zhang H., Wang F., Fang Y., Li J. Pri-miR-34b/c rs4938723 polymorphism contributes to acute lymphoblastic leukemia susceptibility in Chinese children. Leuk. Lymphoma. 2016;57:1436–1441. doi: 10.3109/10428194.2015.1092528. [DOI] [PubMed] [Google Scholar]
- 43.Zhou X., Chen X., Hu L., Han S., Qiang F., Wu Y., Pan L., Shen H., Li Y., Hu Z. Polymorphisms involved in the miR-218-LAMB3 pathway and susceptibility of cervical cancer, a case-control study in Chinese women. Gynecol. Oncol. 2010;117:287–290. doi: 10.1016/j.ygyno.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 44.Shi T.Y., Chen X.J., Zhu M.L., Wang M.Y., He J., Yu K.D., Shao Z.M., Sun M.H., Zhou X.Y., Cheng X. A pri-miR-218 variant and risk of cervical carcinoma in Chinese women. BMC Cancer. 2013;13:19. doi: 10.1186/1471-2407-13-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang L., Wang C., Sun C., Xu Y., Ding Z., Zhang X., Huang J., Yu H. The impact of pri-miR-218 rs11134527 on the risk and prognosis of patients with esophageal squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 2014;7:6206–6212. [PMC free article] [PubMed] [Google Scholar]
- 46.Han Y., Pu R., Han X., Zhao J., Li W., Yin J., Zhang Y., Shen Q., Xie J., Zhang Q. Association of a potential functional pre-miR-218 polymorphism and its interaction with hepatitis B virus mutations with hepatocellular carcinoma risk. Liver Int. 2014;34:728–736. doi: 10.1111/liv.12313. [DOI] [PubMed] [Google Scholar]
- 47.Bao P.P., Li K., Wu C.X., Huang Z.Z., Wang C.F., Xiang Y.M., Peng P., Gong Y.M., Xiao X.M., Zheng Y. [Recent incidences and trends of childhood malignant solid tumors in Shanghai, 2002-2010] Zhonghua Er Ke Za Zhi. 2013;51:288–294. [PubMed] [Google Scholar]
- 48.He J., Yang T., Zhang R., Zhu J., Wang F., Zou Y., Xia H. Potentially functional polymorphisms in the LIN28B gene contribute to neuroblastoma susceptibility in Chinese children. J. Cell. Mol. Med. 2016;20:1534–1541. doi: 10.1111/jcmm.12846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He J., Zhong W., Zeng J., Zhu J., Zhang R., Wang F., Yang T., Zou Y., Xia H. LMO1 gene polymorphisms contribute to decreased neuroblastoma susceptibility in a Southern Chinese population. Oncotarget. 2016;7:22770–22778. doi: 10.18632/oncotarget.8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He J., Wang F., Zhu J., Zhang Z., Zou Y., Zhang R., Yang T., Xia H. The TP53 gene rs1042522 C>G polymorphism and neuroblastoma risk in Chinese children. Aging (Albany N.Y.) 2017;9:852–859. doi: 10.18632/aging.101196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He J., Zou Y., Wang T., Zhang R., Yang T., Zhu J., Wang F., Xia H. Genetic variations of GWAS-identified genes and neuroblastoma susceptibility: a replication study in Southern Chinese children. Transl. Oncol. 2017;10:936–941. doi: 10.1016/j.tranon.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang J., Lin H., Wang J., He J., Zhang D., Qin P., Yang L., Yan L. LMO1 polymorphisms reduce neuroblastoma risk in Chinese children: a two-center case-control study. Oncotarget. 2017;8:65620–65626. doi: 10.18632/oncotarget.20018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He J., Zhang R., Zou Y., Zhu J., Yang T., Wang F., Xia H. Evaluation of GWAS-identified SNPs at 6p22 with neuroblastoma susceptibility in a Chinese population. Tumour Biol. 2016;37:1635–1639. doi: 10.1007/s13277-015-3936-7. [DOI] [PubMed] [Google Scholar]
- 55.He J., Qiu L.X., Wang M.Y., Hua R.X., Zhang R.X., Yu H.P., Wang Y.N., Sun M.H., Zhou X.Y., Yang Y.J. Polymorphisms in the XPG gene and risk of gastric cancer in Chinese populations. Hum. Genet. 2012;131:1235–1244. doi: 10.1007/s00439-012-1152-8. [DOI] [PubMed] [Google Scholar]
- 56.Li J., Zou L., Zhou Y., Li L., Zhu Y., Yang Y., Gong Y., Lou J., Ke J., Zhang Y. A low-frequency variant in SMAD7 modulates TGF-β signaling and confers risk for colorectal cancer in Chinese population. Mol. Carcinog. 2017;56:1798–1807. doi: 10.1002/mc.22637. [DOI] [PubMed] [Google Scholar]
- 57.Lou J., Gong J., Ke J., Tian J., Zhang Y., Li J., Yang Y., Zhu Y., Gong Y., Li L. A functional polymorphism located at transcription factor binding sites, rs6695837 near LAMC1 gene, confers risk of colorectal cancer in Chinese populations. Carcinogenesis. 2017;38:177–183. doi: 10.1093/carcin/bgw204. [DOI] [PubMed] [Google Scholar]
- 58.Fu W., Zhu J., Xiong S.W., Jia W., Zhao Z., Zhu S.B., Hu J.H., Wang F.H., Xia H., He J., Liu G.C. BARD1 gene polymorphisms confer nephroblastoma susceptibility. EBioMedicine. 2017;16:101–105. doi: 10.1016/j.ebiom.2017.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.