Ten Eleven Translocation (TET) proteins are a family of dioxygenases (TET1, TET2, and TET3) that catalyze the oxidation of 5-methyl-cytosine (5mC) to 5-hydroxymehylcytosine (5hmC), 5-formlycytosine (5fC), and 5-carboxylcytosine (5caC)1. Mutations involving TET2 (4q24) have widely been reported in the context of age-related clonal hematopoiesis (~10% >80 years of age)2, and hematological malignancies such as myelodysplastic syndromes (MDS 5–20%), myeloproliferative neoplasms (MPN~15%), chronic myelomonocytic leukemia (CMML ~60%), acute myeloid leukemia (AML 8–30%), and T and B cell lymphoproliferative disorders3–5. In CMML, thus far, clonal TET2 mutations in the absence of clonal ASXL1 mutations (ASXL1wt/TET2mt) have been associated with favorable outcomes6. Conversely, mutations in TET1 (10q21.3) and TET3 (2p13.1) are extremely infrequent with a large study of 408 MPN, CMML, and AML patients demonstrating no identifiable mutations in these genes3. In a recent study, whole exome sequencing was performed in 49 CMML patients resulting in the detection of two loss-of-function, subclonal, TET3 mutations (R148H and S1708fs), both in patients with co-existing TET2 mutations7. ASXL2 (additional sex combs-like; 2p23.3) mutations were recently described in adult and pediatric patients with t(8;21)/core binding factor AML (RUNX1–RUNX1T1) (~20%) and were associated with a higher cumulative incidence of relapse8. In MDS/MPN overlap syndromes including CMML, thus far, the frequency and prognostic impact of ASXL2 mutations remain unknown. We carried out this study to estimate the frequency and clinical correlates of TET1, TET3, and ASXL2 mutations in patients with MDS/MPN overlap syndromes.
Eighty three patients meeting the 2016 World Health Organization (WHO) criteria for CMML (n = 30) and MDS/MPN-Unclassifiable (MDS/MPN-U, n = 47) were included in the study9. The median age was 73 years (range, 18–89 years) and 66% were male. All patients had bone marrow (BM) biopsies and cytogenetic studies performed at diagnosis. Target capture-based next generation sequencing (NGS) was carried out on diagnostic BM DNA from all 83 patients for the complete coding regions of the following 42 genes: TET1, TET2,TET3, DNMT3A, IDH1, IDH2, ASXL1, ASXL2, ATM, EED, EZH2, JARID2, SUZ12, BCOR, BCORL1, STAG2, GATA2, TERC, TERT, SRSF2, SF3B1, ZRSR2, U2AF1, PTPN11,PHF6, Tp53, SH2B3, RUNX1, CBL, NRAS, KRAS, JAK2, CSF3R, FLT3, KIT, CALR, MPL, NPM1, CEBPA, IKZF1, ETNK1, and SETBP1 by previously described methods6. Paired-end indexed libraries were prepared from individual patient DNA using the NEBNext Ultra Library prep protocol on the Agilent Bravo liquid handler. Capture libraries were assembled according to Nimblegen standard library protocol. Base-calling was performed using Illumina’s RTA version 1.17.21.3. Genome_GPS v4.0.1 (formerly named as TREAT) was employed to analyze the data10. Specific variants were included if they were cited by the Catalog of Somatic Mutations in Cancer database (COSMIC, http://cancer.sanger.ac.uk) and/or if they were found at less than 0.1% by the Exome Aggregation Consortium (ExAC, Broad Institute, Cambridge, MA) and not associated with a COSMIC identifier. Previously annotated single-nucleotide polymorphisms (http//www.hapmap.org) in these genes were excluded. For ASXL1, only frameshift and nonsense mutations were considered pathogenic11.
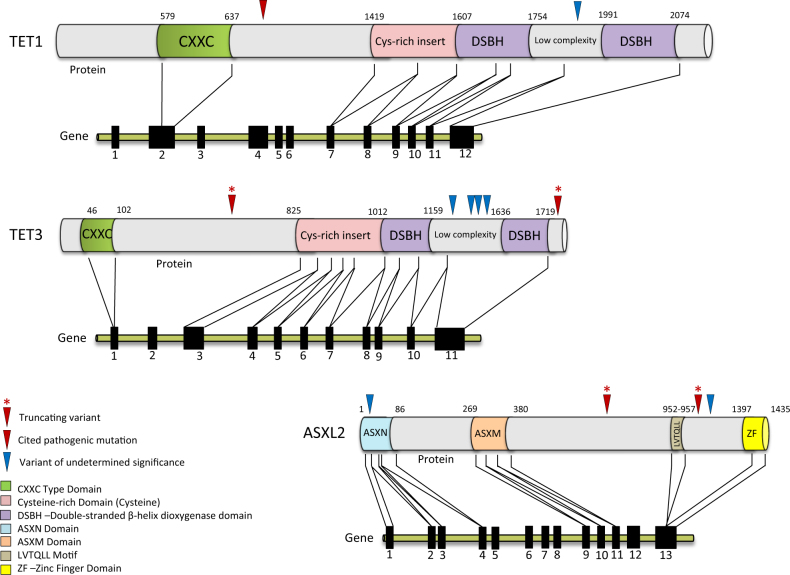
Overall, we observed seven patients (5 CMML [17%] and 2 MDS/MPN-U [4%]) with mutations and/or VUS involving TET1 and TET3 (Table 1 and Fig. 1). All cases were without concurrent TET2 mutations. Of these, loss-of-function TET1 and TET3 mutations were identified in two patients, both with a morphological diagnosis of CMML. Patient one is a 66-year-old male with CMML-0 and normal cytogenetics (Mayo Molecular Model/MMM—intermediate-1 risk) who had presented with monocytosis and thrombocytopenia11. BM NGS analysis revealed a TET3K1760del (48%—variant allele frequency), with an additional PHF6F172Lfs*46 (88%) mutation. He is being treated with 5-azacitidine and at last follow up (15.5 months) remains in a morphological complete remission (CR) after 10 cycles of therapy. Patient two was an 80-year-old female with CMML-0 and normal cytogenetics (MMM—high risk), who had presented with monocytosis and circulating immature myeloid cells. BM NGS at diagnosis identified TET3Y473* (43%) with a coexisting TET1Q683 (46%) mutation (previously cited as pathogenic shown in Table 1), and two additional mutations: ASXL1G646Wfs*12 (26%) and PTPN11N308D (14%). She received supportive care and died within a month of diagnosis without evidence for leukemic transformation.
Table 1.
Spectrum of TET1, TET3 and ASXL2 mutations and variants of unclear significance in patients with MDS/MPN overlap syndromes
| Gene | Chr | Position | Nucleotide nomenclature | Protein consequence | Disease type | Alt Frac | ExAC | dbSNP | Cosmic # | Cited as somatic | Cosmic Annotated Disease Type | Exon | Phenotype Prediction | Concurrent Mutations |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TET1 | 10 | 70404533 | c.2047C > G | Q683E | CMMLb | 46% | 0.08400% | rs139785845 | COSM327333 | yes | ALL1, Sezary Syndrome2 | 4 | MODERATE | TET3Y473a (43%) ASXL1G646Wfsa12 (26%) PTPN11N308D (14%) |
| 70450700 | c.5540G > T | G1847V | MDS/MPNu | 52% | n/a | n/a | n/a | 12 | MODERATE | SRSF2P95R (53%) NRASG13D (41%) | ||||
| TET3 | 2 | 74274463 | c.1419C > A | Y473 a | CMMLb | 43% | n/a | n/a | n/a | 3 | HIGH | TET1Q683E (46%) ASXL1G646Wfsa12 (26%) PTPN11N308D (14%) | ||
| 74329187 | c.5278_5280del | K1760del | CMML | 48% | 0.02900% | rs564392898 | n/a | 11 | MODERATE | PHF6F172Lfsa46 (88%) | ||||
| 74327798 | c.3883G > A | V1295I | CMML | 51% | 0.04400% | rs199849765 | n/a | 11 | MODERATE | CEBPAH195_P196dup (57%) ATML1111P (51%) ASXL1L775a (49%) JARID2R767K (46%) | ||||
| 74327893 | c.3980_3981insACTGAG | N1326_S1327insRL | CMMLa | 41% | 0.00860% | rs768310475 | n/a | 11 | MODERATE | SRSF2P95T (45%) TET3L1328P (42%) | ||||
| 74327898 | c.3983T > C | L1328P | CMMLa | 42% | 0.00860% | rs767538752 | n/a | 11 | MODERATE | SRSF2P95T (45%) TET3N1326_S1327insRL (41%) | ||||
| 74328177 | c.4262C > G | P1421R | CMML | 51% | 0.00940% | rs745953793 | n/a | 11 | MODERATE | SH2B3R140H (58%) JARID2P1229L (52%) NRASQ61K (47%) RUNX1G199W (21%) | ||||
| 74329152 | c.5237G > T | W1746L | MDS/MPNu | 49% | 0.06500% | rs190925009 | n/a | 11 | MODERATE | SRSF2P95L (49%) ASXL1P808H (49%) JAK2V617F (49%) | ||||
| ASXL2 | 2 | 25966302 | c.2902_2903dupCT | P969Cfs a 10 | MDS/MPNu | 22% | n/a | . | n/a | 13 | HIGH | SRSF2P95_R102del (15%) RUNX1R237K (38%) | ||
| 25967305 | c.1901C > A_p.Ser634X | S634 a | MDS/MPNu | 20% | n/a | . | n/a | 13 | HIGH | SRSF2R94dup (39%) | ||||
| 25965934 | c.3272 C>T_p.Ala1091Val | A1091V | CMML | 51% | 0.01300% | rs781151810 | n/a | 13 | MODERATE | ZRSR2 c.400-2A > G (92%) MPLV368L (49%) ASXL1G646Wfsa12 (44%) SETBP1D868N (43%) RUNX1T246Hfsa15 (35%) | ||||
| 26101079 | c.13G > A_p.Gly5Arg | G5R | MDS/MPNu | 51% | 0.01200% | rs371056638 | n/a | 1 | MODERATE | EZH2 c.1411-1G > A (91%) ASXL1R417a (45%) JARID2R326C (48%) SUZ12N263H (47%) |
Fig. 1.
Domain architecture of TET1, TET3, and ASXL2 with observed gene mutations and variants of unclear significance
We identified ASXL2 mutations or VUS in four patients (3 MDS/MPN-U [6%] and 1 CMML [3%]). Of these, two patients with MDN/MPN-U harbored loss-of-function ASXL2 mutations. Patient one was a 67-year-old male with trisomy 21 who had presented with transfusion-dependent anemia and thrombocytopenia. BM NGS at diagnosis identified ASXL2S634* (20%) and SRSF2R94dup (39%). He was treated with 5-azacitdine and had no response after four cycles. He died shortly thereafter with no evidence for leukemic transformation. Patient two was a 75-year-old female with MDS/MPN-U and trisomy 8 and trisomy 9, who had presented with transfusion-dependent anemia. BM NGS at diagnosis identified ASXL2P969Cfs*10 (22%), RUNX1R237K (38%), and SRSF2P95_R102del (15%). She was treated with transfusional supportive care and was lost to follow-up.
Our study reveals that although uncommon, loss-of-function TET1, TET3, and ASXL2 mutations can be seen in patients with MDS/MPN overlap syndromes. TET1 and TET3 mutations were seen exclusively in CMML, were found to coexist with each other (TET1 and TET3), and occurred independent of TET2 mutations. ASXL2 mutations were seen in MDS/MPN-U, were associated with numerical chromosomal aberrations, and occurred independent of ASXL1 mutations. The current study was limited by a small number of informative cases to opine on clinical correlates and survival outcomes. Studies exploring the functional redundancy of TET1 and TET3 mutations with TET2 activity, the impact of TET1, TET3, and ASXL2 mutations on global and sequence-specific 5-mC and 5-hmC levels and post-translational histone modifications (H3K27me3), and the impact of these mutations on survival are currently being planned.
Acknowledgements
Current publication is supported in part by grants from the “The Gerstner Family Career Development Award” and the Mayo Clinic Center for Individualized Medicine, Mayo Clinic, Rochester, MN, USA”. This publication was supported by CTSA Grant Number KL2 TR000136 from the National Center for Advancing Translational Science (NCATS). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wu X, Zhang Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat. Rev. Genet. 2017;18:517–34. doi: 10.1038/nrg.2017.33. [DOI] [PubMed] [Google Scholar]
- 2.Jaiswal S, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014;371:2488–98. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdel-Wahab O, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114:144–7. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patnaik MM, et al. Number and type of TET2 mutations in chronic myelomonocytic leukemia and their clinical relevance. Blood Cancer J. 2016;6:e472. doi: 10.1038/bcj.2016.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bejar R, et al. TET2 mutations predict response to hypomethylating agents in myelodysplastic syndrome patients. Blood. 2014;24:2705–12. doi: 10.1182/blood-2014-06-582809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patnaik MM, et al. Prognostic interaction between ASXL1 and TET2 mutations in chronic myelomonocytic leukemia. Cancer J. 2016;6:e385. doi: 10.1038/bcj.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merlevede J, et al. Mutation allele burden remains unchanged in chronic myelomonocytic leukaemia responding to hypomethylating agents. Nat. Commun. 2016;7:10767. doi: 10.1038/ncomms10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Micol JB, et al. Frequent ASXL2 mutations in acute myeloid leukemia patients with t(8;21)/RUNX1-RUNX1T1 chromosomal translocations. Blood. 2014;124:1445–9. doi: 10.1182/blood-2014-04-571018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arber DA, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 10.Asmann YW, et al. TREAT: a bioinformatics tool for variant annotations and visualizations in targeted and exome sequencing data. Bioinformatics. 2012;28:277–8. doi: 10.1093/bioinformatics/btr612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patnaik MM, et al. ASXL1 and SETBP1 mutations and their prognostic contribution in chronic myelomonocytic leukemia: a two-center study of 466 patients. Leukemia. 2014;28:2206–12. doi: 10.1038/leu.2014.125. [DOI] [PubMed] [Google Scholar]
- 12.Kalender Atak Z, et al. High accuracy mutation detection in leukemia on a selected panel of cancer genes. PLoS One. 2012;7:e38463. doi: 10.1371/journal.pone.0038463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiel MJ, et al. Genomic analyses reveal recurrent mutations in epigenetic modifiers and the JAK–STAT pathway in Sézary syndrome. Nat. Commun. 2015;6:8470. doi: 10.1038/ncomms9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palomo L, et al. Targeted deep sequencing improves outcome stratification in chronic myelomonocytic leukemia with low risk cytogenetic features. Oncotarget. 2016;7:57021–57035. doi: 10.18632/oncotarget.10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weissmann S, et al. Landscape of TET2 mutations in acute myeloid leukemia. Leukemia. 2011;26:934. doi: 10.1038/leu.2011.326. [DOI] [PubMed] [Google Scholar]