Abstract
The Leadership and Operations Center (LOC) is responsible for facilitating, coordinating, and implementing the Antibacterial Resistance Leadership Group (ARLG) scientific agenda by engaging thought leaders; soliciting research proposals; and developing the processes, tools, and infrastructure required to operationalize studies and create and sustain the ARLG network. These efforts are ongoing as new projects are developed and the network expands and grows to address the ever-changing priorities in antibacterial resistance. This article describes the innovations, accomplishments, and opportunities of the LOC since the inception of the ARLG in 2013.
Keywords: antibacterial resistance, clinical trial network, infectious disease, mentoring.
The mission of the Antibacterial Resistance Leadership Group (ARLG) is to prioritize, design, and execute clinical research that will reduce the public health threat of antibacterial resistance. The Leadership and Operations Center (LOC) is responsible for facilitating, coordinating, and implementing the ARLG scientific agenda [1] by engaging thought leaders; soliciting research proposals; and developing the processes, tools, and infrastructure required to operationalize studies and create and sustain the ARLG network. The scientific agenda of the ARLG was developed by >50 key leaders in the field who constitute its committee membership. These experts have outlined the areas of greatest unmet need, established a scientific agenda, and formulated and evaluated potential clinical studies to address those needs. This internal expertise is supplemented by soliciting ideas from academia, government, and industry via a proposal portal on the ARLG website (http://arlg.org/how-to-apply).
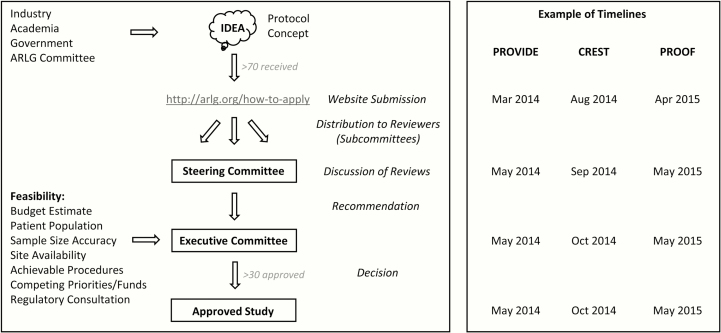
The LOC is physically located within the Duke Clinical Research Institute (DCRI) in Durham, North Carolina. From this position, the LOC’s coordination of ARLG committees, creation and implementation of the proposal portal, and handling of the corresponding application and review system allows for rapid decision making and implementation of proposed studies (Figure 1) [2, 3]. The review process for proprietary proposals is further expedited by the creation of a master Confidentiality and Disclosure Agreement (CDA) system covering committee members for discussion of all proposals. To date, >130 CDAs have been executed to enable ARLG discussions, >70 study proposals have been received and reviewed, and >30 proposals have been approved and implemented as ARLG studies.
Figure 1.
Antibacterial Resistance Leadership Group (ARLG) protocol concept receipt, review, and approval. Example timelines are shown for PROVIDE (Prospective Validation of the Vancomycin Exposure Profile Associated With Optimal Outcomes Among Patients With MRSA Bloodstream Infections) [2], CREST (Carbapenem-Resistant Enterobacteriaceae in Solid Organ Transplant Patients), and PROOF (Pharmacokinetics, Pharmacodynamics and Safety/ Tolerability of Two Dosing Regimens of Oral Fosfomycin Tromethamine in Healthy Adult Participants) [3]. PROVIDE and PROOF concepts were generated by ARLG committees and submitted directly to the ARLG Steering Committee in response to a call for proposals. CREST was submitted via the ARLG website by the CREST investigators.
CREATING AND MAINTAINING AN ARLG SITE NETWORK
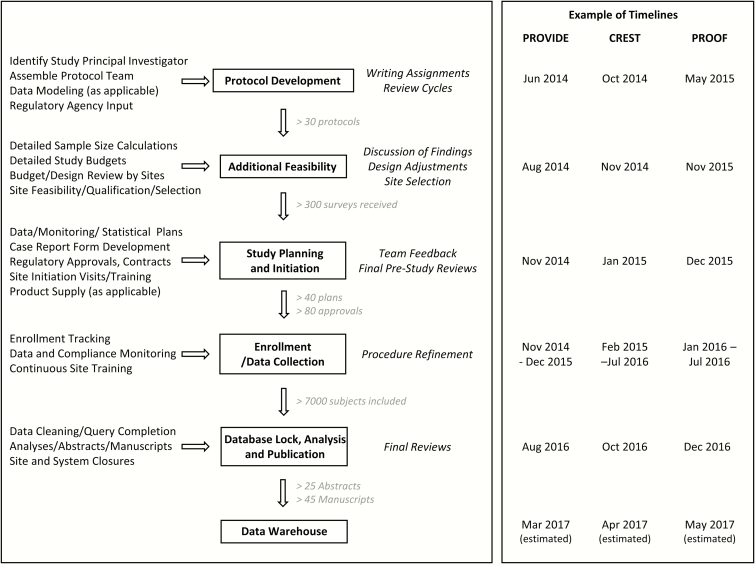
An essential component for any study is identifying clinical sites with the appropriate patient population and resources to complete enrollment on time and within budget. This is particularly important in clinical trials targeting antibacterial resistance, where the utility of an individual site can be influenced by geographic variation in the prevalence of drug-resistant bacteria, national reimbursement trends that discourage the acknowledgement of specific types of nosocomial infections (eg, ventilator-associated bacterial pneumonia) [4], and local expertise that is often syndrome-specific. Accordingly, one of the first priorities of the LOC upon start-up was to create and develop a robust network of clinical sites. The ARLG site network was developed initially from the >37000 sites that previously participated in DCRI studies, through the National Institute of Allergy and Infectious Diseases Vaccine and Treatment Evaluation Unit infrastructure, through site lists donated by industry partners from clinical trials, and through identification of new sites via the study feasibility process. More than 300 detailed site questionnaires have been received by the ARLG thus far, including estimated numbers of cases of key infections and details of typical standards of care, and the site network continues to grow and expand as new sites are identified and approached. The work required to create and maintain a large clinical trial network (Table 1) [5] and to operationalize ARLG studies (Figure 2) [2, 3] is extensive: >100 DCRI staff have played a role in building the ARLG network and operationalizing ARLG studies to date.
Table 1.
Components Required to Support the Antibacterial Resistance Leadership Group Network
| Department and Component | Purpose | No. to Date |
|---|---|---|
| DCRI Contracts Department | ||
| Subawards/agreements | Contract with committee chairs, center directors, vendors, sites | >160 |
| Confidentiality and disclosure agreements | Receive investigational product data from companies and share with committee members for assessment of study proposal priority and viability | >130 |
| Data use agreements | Enable data mining and modeling | 8 |
| Material transfer agreements | Facilitate disbursement of bacterial strains from the ARLG Virtual Biorepository [4] | 10 |
| DCRI Business Development and Duke Office of Research Administration | ||
| Subpacket collection | Preparation for subawards | >100 |
| Ballpark budgets | Pricing estimates to assess feasibility of proposed studies | >26 |
| Full study budgets | Detailed budgets allow correct allocation and efficient use and tracking of funds | >28 |
| Progress reports and noncompeting renewals | Annual compliance with NIH grants policy | 4 y × ~200 pages |
| DCRI Sponsored Projects Administration and Duke Office of Sponsored Programs | ||
| Project-specific and overall grant financial reports | Monitor grant expenditures and compliance | >$35M total funds managed |
| Invoice processing | Facilitate payment of chairs, directors, vendors, sites | >600 |
Abbreviations: DCRI, Duke Clinical Research Institute; NIH, National Institutes of Health.
Figure 2.
Antibacterial Resistance Leadership Group (ARLG) protocol development and study execution. Example timelines are shown for PROVIDE [2], CREST [3], and PROOF [2]. PROVIDE (n = 305) and CREST (n = 170) are noninterventional studies. PROOF is an interventional phase 1 study (n = 19). See Figure 1 legend for study descriptions.
Efficient coordination of such a large network has been facilitated by a number of innovations. First, a database was created to allow querying of feasibility data for each specific ARLG study as it arises. Next, operational innovations such as the DCRI Rapid Start Network (RSN) provide significant time savings. For RSN sites, an initial master agreement outlining general terms is executed far in advance of a specific project need. A study-specific addendum is then added for each study with the individual protocol and study budget included. On average, this reduces site agreement negotiation by 2 months. To date, the DCRI has put in place >140 National Institutes of Health (NIH) grant–specific master RSN agreements.
CREATING AND CONNECTING INFECTIOUS DISEASE CLINICAL TRIAL NETWORKS
Clinical trials in antibacterial resistance, or for new antibiotics, have failed to keep pace with need [6]. This, coupled with rapid emergence of resistant pathogens in a wide variety of disease states with often low prevalence [7], is a significant challenge for operationalizing infectious disease clinical trials. A large network of highly experienced clinical trial sites with sufficient patient populations to address all resistance pathogens in all disease states in both inpatient and outpatient settings did not exist when the ARLG was formed. As such, the ARLG has sought to cultivate networks that can be harnessed for antibacterial research.
For example, CRACKLE (Consortium on Resistance Against Carbapenems in Klebsiella pneumoniae and Other Enterobacteriaceae) is an ARLG network developed by van Duin et al [8], which now includes 17 states across the United States as well as sites in Colombia. In addition to findings regarding the epidemiology, outcomes, and impact of carbapenem-resistant Enterobacteriaceae (CRE) molecular characteristics in CRE-infected patients, as summarized by Doi et al [3], CRACKLE and CRACKLE II have enabled assessment of risk factors, frequency, geographic location, and clinical characteristics of CRE-infected patients. These data are being utilized to inform realistic inclusion/exclusion criteria, assist in early consenting procedures, and identify appropriate clinical sites for the ARLG’s planned interventional CRE studies. Additional plans are underway to include data from patients infected with Pseudomonas aeruginosa and Acinetobacter baumannii to inform design of interventional clinical trials addressing these key pathogens.
In addition to creating new networks, the ARLG has leveraged existing networks like the Duke Infection Control Outreach Network (DICON) and Duke Antibiotic Stewardship Outreach Network (DASON) to conduct a first-in-kind study, DICON I [9], of antimicrobial stewardship in a community hospital setting. DICON I is now being expanded to DICON II [9] to introduce research capabilities to additional community hospitals, in which the majority of US patients receive their care but where few research studies are conducted [10].
The ARLG is also closely affiliated with the hospital-acquired bacterial pneumonia (HABP) and ventilator-associated bacterial pneumonia (VABP) risk factor and pilot studies from Duke’s Clinical Trials Transformation Initiative [11]. These projects—collaborations between academia, the US Food and Drug Administration, and industry—aim to develop and utilize novel techniques, such as consenting at-risk patients at the time of intensive care unit admission for enrollment into a trial should HABP or VABP develop, to increase the feasibility of these extremely challenging trials. The risk factor study, which has already enrolled >3000 subjects, also provides data that can inform realistic inclusion/exclusion criteria and identify appropriate clinical sites for the types of interventional pneumonia trials that the ARLG is planning.
The ARLG is also collaborating with Duke’s Pediatric Trials Network (PTN) to assess the pharmacokinetics of a novel aminoglycoside in the pediatric population [3]. The PTN was created in response to the Best Pharmaceuticals for Children Act to provide pediatric clinical trial data on products commonly used in the pediatric population yet not approved specifically for such use [12]. To date, the PTN has completed 11 clinical trials in >4000 patients, including trials of several antibiotics. As such, the network serves as a valuable source of experienced clinical trial sites for ARLG pediatric infectious disease studies.
In addition, the ARLG is in discussions with the Innovative Medicines Initiative–funded Combatting Bacterial Resistance in Europe (COMBACTE) project to collaborate with its pan-European clinical trial network (CLIN-Net) and has actively initiated collaborations with networks in Colombia [13], Chile, Argentina, Brazil, Mexico, Asia, Australia, and New Zealand [14]. Future collaborations with the International Collaboration on Endocarditis and the critical care clinical trial networks in Canada and Australia/New Zealand are also envisioned. Laboratory-based collaborations with China and India are also in discussion, which may also ultimately lead to ARLG clinical trials in those regions.
FOSTERING THE NEXT GENERATION OF INFECTIOUS DISEASE CLINICAL INVESTIGATORS
Another key focus of the ARLG is mentoring. Providing infectious disease fellows and junior faculty with research opportunities and mentoring in clinical trials is crucial to the future of antibacterial resistance research. The ARLG LOC has a Mentoring Committee dedicated to supporting and nurturing new investigators through 3 types of training opportunities: early-stage investigator (ESI) seed grants, the ARLG fellowship, and mentee involvement in ARLG projects.
To date, 5 ESI seed grants have been awarded (Table 2) and there are currently 26 mentees participating in 16 ARLG studies. The ESI seed grants in particular are designed to allow researchers to generate preliminary data leading to additional external funding. Recipients of the 3 initial seed grants have received subsequent grants outside of the ARLG and their ARLG-funded work has been published in high-profile journals [15, 16]. Altogether, 13 ARLG abstracts and 12 ARLG manuscripts have included mentee authors.
Table 2.
Antibacterial Resistance Leadership Group Early-Stage Investigator Seed Grants
| Study Name | Description | Status |
|---|---|---|
| BCID | Assessment of clinical and economic outcomes of the rapid FilmArray blood culture identification panel test for gram-positive bloodstream infections | Complete [15] |
| CEF-BP | Ceftriaxone breakpoints—a gram-negative database to establish clinically relevant antibiotic breakpoint interpretive criteria for ceftriaxone | Complete [16] |
| CRKP-LTACH | Study of carbapenem-resistant Klebsiella pneumoniae in long-term acute care hospitals | Analysis phase |
| VENOUS | Cancer patients with vancomycin-resistant Enterococcus faecium bacteremia—prospective evaluation of clinical outcomes | Protocol development |
| MICROFIRE | Study of microbiota colonization in the presence of intestinal fluoroquinolone-resistant Escherichia coli | Protocol development |
COMMUNICATING ARLG OPPORTUNITIES AND PROGRESS
Communication of the ARLG scientific agenda, opportunities, progress, and study findings has been accomplished via a website (http://arlg.org/), at meetings, and in presentations and publications. Formation of the ARLG publication committee and its corresponding process has facilitated publication approval, tracking, and compliance with the NIH Public Access Policy. To date, >45 manuscripts have been published and 25 abstracts have been presented from ARLG work.
CHALLENGES AND FUTURE DIRECTIONS
The ARLG LOC has successfully developed new innovative processes, collaborations, and mentorships to create and sustain a research network and its associated clinical trials. The LOC will continue to innovate in order to address the challenge of operationalizing the more complex multinational studies that are currently being developed. With the inevitable emergence and spread of new resistant pathogens, the LOC will facilitate the ARLG scientific agenda to ensure that the most critical priorities in antibacterial resistance are being addressed.
APPENDIX
ARLG Leadership and Operations Center. Faculty: Henry “Chip” Chambers, MD; Vivian Chu, MD*; Ralph Corey, MD; Sarah Doernberg, MD; Vance Fowler, MD, MHS; Anthony Harris, MD, MPH; Thomas Holland, MD; Christoph Hornik, MD*; Julia Messina, MD*; Joshua Thaden, MD, PhD. Staff: Brigette Adamkiewicz, Peter Anderson, Peggy Arias, Malik Awan, Paula Aycock, Alina Barnes, Keri Baum, Corey Brennan*, Kimberly Brown, Ivra Bunn, Heather Cross, Morgan deBlecourt, Nancie Deckard, Michele Downing*, Weiying Drake*, Beth Evans, Lee Greiner, Peidi Gu, Joseph Gugliotti, Stephanie Harrison, Catherine Hart, Entrane Harvey, Anne Heath, Brian Hoegg, Peter Hoffman, Deborah Hopkins, Ethan Hughes, David Jensen, Patrick Jordan*, Chloe Katz, Wallace Lamb, Brenda Lane, Paula Lanning, Camille Leeds, Darcy Louzao*, James Lynch, John MacNeela, Marsha Marquess, Brenda Mickley, Elizabeth Mocka, Christina Murphy*, Samantha Murray, Norman Mustafa, Anastasia Ngugi, Theresa O’Reilly, Sara Patillo, Brenda Pattison, Elizabeth Petzold, Andy Richardson, Kevin Roddy, Ronald Roddy*, Carolyn Rugloski, Carl Schuler, Jonathan Shepherd*, Anna Suojanen, David Souto, Denise Sturdy, Jodi Tipper*, Rupal Vora, Duncan Wallace, Cathy Wickward, Nancy Wood, Laura Wrightson, Hirra Zahir (*denotes former member).
ARLG Mentoring Committee. John Bartlett, MD; Robert Bonomo, MD; Henry “Chip” Chambers, MD; Ralph Corey, MD; Sara Cosgrove, MD, MS; Heather Cross, DPhil; Robert Daum, MD; Scott Evans, PhD; Vance Fowler, MD, MHS; Anthony Harris, MD, MPH; Ebbing Lautenbach, MD, MPH, MSCE; Loren Miller, MD, MPH; Barbara Murray, MD; Robin Patel, MD; Daniel Sexton, MD* (*denotes former member).
Notes
Acknowledgments. The authors acknowledge Brenda Lane, Brenda Mickley, and Rupal Vora for facilitation of the ARLG network and Nancie Deckard and Norman Mustafa for leadership of the LOC team.
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health (NIH).
Financial support. This article was supported by the National Institute of Allergy and Infectious Diseases of the NIH (award number UM1AI104681).
Supplement sponsorship. This article appears as part of the supplement “Antibacterial Resistance Leadership Group (ARLG): Productivity and Innovation,” sponsored by the Antibacterial Resistance Leadership Group.
Potential conflicts of interest. H. R. C.’s salary is paid by an ARLG grant via Duke University. H. F. C. has served on advisory boards for Allergan and Genentech and has received grant support from The Medicines Company and Genentech. V. G. F. has received grants from NIH, MedImmune, Cerexa/Forest/Actavis/Allergan, Pfizer, Advanced Liquid Logics, Theravance, Novartis, Cubist/Merck, Medical Biosurfaces, Locus, Affinergy, Contrafect, Karius, and the Centers for Disease Control and Prevention; has received personal fees from Merck, Pfizer, Novartis, Galderma, Novadigm, Durata, Debiopharm, Genentech, Achaogen, Affinium, The Medicines Company, Cerexa, Tetraphase, Trius, MedImmune, Bayer, Theravance, Cubist, Basilea, Affinergy, Janssen, Contrafect, xBiotech, Green Cross, Cubist, and UpToDate; and has a patent pending for sepsis diagnostics. All other authors report no potential conflicts. The authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Chambers HF, Bartlett JG, Bonomo RA, et al. Antibacterial resistance leadership group: open for business. Clin Infect Dis 2014; 58:1571–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Doernberg SB, Lodise TP, Thaden JT, et al. Gram-positive bacterial infections: research priorities, accomplishments, and future directions of the Antibacterial Resistance Leadership Group. Clin Infect Dis 2017; 64(Suppl 1):S24–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Doi Y, Bonomo RA, Hooper DC, et al. Gram-negative bacterial infections: research priorities, accomplishments, and future directions of the Antibacterial Resistance Leadership Group. Clin Infect Dis 2017; 64(Suppl 1):S24–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Talbot TR, Bratzler DW, Carrico RM, et al. ; Healthcare Infection Control Practices Advisory Committee Public reporting of health care-associated surveillance data: recommendations from the Healthcare Infection Control Practices Advisory Committee. Ann Intern Med 2013; 159:631–5. [DOI] [PubMed] [Google Scholar]
- 5. Manca C, Hill C, Hujer AM, et al. Leading antibacterial laboratory research by integrating conventional and innovative approaches: the Laboratory Center of the Antibacterial Resistance Leadership Group. Clin Infect Dis 2017; 64(Suppl 1):S13–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boucher HW, Talbot GH, Bradley JS, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 2009; 48:1–12. [DOI] [PubMed] [Google Scholar]
- 7. Spellberg B, Guidos R, Gilbert D, et al. ; Infectious Diseases Society of America The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin Infect Dis 2008; 46:155–64. [DOI] [PubMed] [Google Scholar]
- 8. van Duin D, Perez F, Rudin SD, et al. Surveillance of carbapenem-resistant Klebsiella pneumoniae: tracking molecular epidemiology and outcomes through a regional network. Antimicrob Agents Chemother 2014; 58:4035–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Anderson DJ, Jenkins T, Evans S, et al. The role of stewardship in antibiotic resistance: the Stewardship and Infection Control Committee of the Antibacterial Resistance Leadership Group. Clin Infect Dis 2017; 64(Suppl 1):S36–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. National Center for Health Statistics. Health, United States, 2011: with special feature on socioeconomic status and health 2012. Available at: http://www.cdc.gov/nchs/data/hus/hus11.pdf. Accessed 1 November 2016.
- 11. Cox E, Cavaleri M, Eichler HG, Woodcock J, Borio L. Facilitating antibacterial drug development in a time of great need. Clin Infect Dis 2016; 63:S27–8. [DOI] [PubMed] [Google Scholar]
- 12. Laughon MM, Benjamin DK, Jr, Capparelli EV, et al. Innovative clinical trial design for pediatric therapeutics. Expert Rev Clin Pharmacol 2011; 4:643–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Correa A, Del Campo R, Perenguez M, et al. Dissemination of high-risk clones of extensively drug-resistant Pseudomonas aeruginosa in Colombia. Antimicrob Agents Chemother 2015; 59:2421–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harris PN, Peleg AY, Iredell J, et al. Meropenem versus piperacillin-tazobactam for definitive treatment of bloodstream infections due to ceftriaxone non-susceptible Escherichia coli and Klebsiella spp (the MERINO trial): study protocol for a randomised controlled trial. Trials 2015; 16:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Banerjee R, Teng CB, Cunningham SA, et al. Randomized trial of rapid multiplex polymerase chain reaction-based blood culture identification and susceptibility testing. Clin Infect Dis 2015; 61:1071–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tamma PD, Han JH, Rock C, et al. ; Antibacterial Resistance Leadership Group Carbapenem therapy is associated with improved survival compared with piperacillin-tazobactam for patients with extended-spectrum β-lactamase bacteremia. Clin Infect Dis 2015; 60:1319–25. [DOI] [PMC free article] [PubMed] [Google Scholar]