Summary
No differences in blood transcriptome, virome, and B-cell/T-cell receptor patterns were found between chronic fatigue syndrome or alternatively diagnosed chronic Lyme syndrome patients and controls. Neither disease was found to be associated with transcriptionally mediated immune dysfunction or active viral infection.
Keywords: chronic fatigue syndrome, RNA-seq, transcriptome, viral infection, B-cell/T-cell receptors,
Abstract
Background.
Chronic fatigue syndrome (CFS) remains poorly understood. Although infections are speculated to trigger the syndrome, a specific infectious agent and underlying pathophysiological mechanism remain elusive. In a previous study, we described similar clinical phenotypes in CFS patients and alternatively diagnosed chronic Lyme syndrome (ADCLS) patients—individuals diagnosed with Lyme disease by testing from private Lyme specialty laboratories but who test negative by reference 2-tiered serologic analysis.
Methods.
Here, we performed blinded RNA-seq analysis of whole blood collected from 25 adults diagnosed with CFS and 13 ADCLS patients, comparing these cases to 25 matched controls and 11 patients with well-controlled systemic lupus erythematosus (SLE). Samples were collected at patient enrollment and not during acute symptom flares. RNA-seq data were used to study host gene expression, B-cell/T-cell receptor profiles (BCR/TCR), and potential viral infections.
Results.
No differentially expressed genes (DEGs) were found to be significant when CFS or ADCLS cases were compared to controls. Forty-two DEGs were found when SLE cases were compared to controls, consistent with activation of interferon signaling pathways associated with SLE disease. BCR/TCR repertoire analysis did not show significant differences between CFS and controls or ADCLS and controls. Finally, viral sequences corresponding to anelloviruses, human pegivirus 1, herpesviruses, and papillomaviruses were detected in RNA-seq data, but proportions were similar (P = .73) across all genus-level taxonomic categories.
Conclusions.
Our observations do not support a theory of transcriptionally mediated immune cell dysregulation in CFS and ADCLS, at least outside of periods of acute symptom flares.
Myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) is a debilitating syndrome of unknown etiology, characterized by profound fatigue exacerbated by physical or mental activity, impaired sleep, cognitive complaints, pain, gastrointestinal symptoms, and/or tender lymph nodes. We previously reported a clinical phenotype that is similar to ME/CFS in patients described as having alternatively diagnosed chronic Lyme syndrome (ADCLS) [1]. These individuals were diagnosed with Lyme disease based on private laboratory testing but were Lyme-negative by reference 2-tiered serology according to Centers for Disease Control and Prevention (CDC) criteria. Systemic lupus erythematosus (SLE) is an autoimmune disease with symptoms that overlap those reported in CFS and ADCLS. However, unlike CFS or ADCLS, some pathophysiologic mechanisms associated with SLE are known and include chronic inflammation and defects in apoptosis clearance [2]. Viral and bacterial infections are known to trigger SLE disease flares by sensitizing B-lymphocytes, thus resulting in the production of autoantibodies [2]. A similar autoimmune hypothesis has been put forward to explain CFS, making SLE cases a useful comparator in studies of CFS and/or ADCLS cases.
The prevailing hypothesis that CFS pathogenesis is immune mediated [3] has been supported by observation of aberrant cytokine expression in early illness and in cerebrospinal fluid [4, 5], natural killer cell dysfunction [6], and an encouraging response to B-cell depletion in clinical trials [7]. Notably, antibiotic-refractory arthritis in Lyme disease patients has been associated with autoimmune T- and B-cell responses against at least 4 human proteins [8]. This naturally leads to the question of whether immune dysregulation or other differential patterns of gene expression in CFS and ADCLS patients can be identified through transcriptional analysis.
Previous whole transcriptome studies of CFS cases compared to controls included variable sample sizes ranging from 15 to 163 samples per study and showed inconsistent results, with studies reporting a small number of differentially expressed genes (DEGs; 0–88) with little to no overlap [9–14]. However, all gene expression studies of CFS cases to date have been performed using microarrays. The newer RNA-seq approach offers several potential advantages relative to microarrays, including detection of low-abundance and novel transcripts, broader dynamic range, and elimination of background noise, saturation, and probe redundancy [15]. Our previous RNA-seq study of acute Lyme disease patients, diagnosed by clinical criteria and reference serological testing, revealed a large number of DEGs (>1200) at the time of diagnosis, most of which were related to immune cell activation and inflammation pathways in response to acute bacterial infection [16]. In addition to host gene profiling, RNA-seq can provide metagenomic insights into viable infectious organisms in a given host by capturing microbial gene expression [17] and also permits analysis of B-cell and T-cell receptors (BCRs, TCRs) that mediate downstream immune responses [18]. Thus, given the suspected involvement of a pathogen trigger and/or immune dysfunction in CFS and ADCLS, querying an RNA-seq dataset for both the presence of pathogens and BCR/TCR profiles may provide clues as to the mechanisms that underlie these syndromes.
Previously, we demonstrated that CFS, ADCLS, and SLE patients showed significant disability based on physical examination, symptoms, and functional scale scores compared to matched unaffected controls [1]. No differences in baseline clinical data and functional scales were observed between CFS and ADCLS patients. Immune cell counts were significantly lower (and cytokine profiling significantly different) in SLE cases compared to controls. These differences were not seen in CFS and ADCLS cases relative to controls (Supplementary Table 1). Here, we further investigate this patient cohort by deep sequencing whole blood RNA to look for gene expression signatures, chronic viral infections, and BCR/TCR sequence patterns that might differentiate cases from controls.
METHODS
Study Cohort
Our case-control study design has been described previously [1]. Briefly, we enrolled 25 CFS patients who met the Canadian case definition [19], 13 ADCLS cases diagnosed on clinical grounds and supported only by nonreference testing, and 11 SLE cases meeting American College of Rheumatology criteria [20]. As opposed to the CDC Fukuda criteria [21] for CFS, the Canadian case definition is thought to select individuals with less psychiatric comorbidity and more symptoms and functional impairment [22]. Controls were matched by sex and 5-year age strata to CFS participants.
Key differences between the subject groups were as follows (Supplementary Table 1). ADCLS cases showed significant differences in median age when compared to controls or CFS cases (P = .02). CFS cases showed significant differences in ethnicity compared to controls (P = .04). ADCLS, CFS, and SLE cases all showed significant differences (P < .03) in most core symptoms compared to healthy controls. SLE cases showed significant differences in absolute CD4+ (P = .004) and CD57+ (P = .04) cell counts compared to healthy controls, as well as in antinuclear antibody reactivity (P = .01).
Library Prep and Sequencing
Blood samples were drawn into Paxgene blood RNA tubes (Preanalytix) for immediate RNA stabilization of intracellular RNA at collection. Total RNA was extracted using the Paxgene blood RNA kit (Preanalytix) and lyophilized in RNAstable reagent (Biomatrica) for shipment at room temperature and long-term storage. The Ovation human blood RNA-seq kit (Nugen) was used to generate strand-specific RNA-Seq libraries depleted for reads derived from ribosomal RNA (rRNA) and globin genes according to the manufacturer’s protocol. Libraries were sequenced as 100 base pair paired-end runs on a HiSeq 2500 (Illumina).
Whole Transcriptome Analysis
Paired-end reads were mapped to the human genome (hg19), annotated to exons, and normalized to FPKM (fragments per kilobase of exon per million fragments mapped) values for all 25278 human RNA reference sequences in the National Center for Biotechnology Information (NCBI) RefSeq database using version 2 of the Tophat/Cufflinks pipeline [23]. Differential expression of genes was calculated using the voom transformation, which applies precision weights to the matrix count, followed by linear modeling using the Limma package [24]. Genes were considered to be differentially expressed when their fold change was greater than ±1.5, P value < .05, and adjusted P value (or false discovery rate) < .1%. Pathway and network analyses of the transcriptome data were performed using Ingenuity Pathway Analysis software (Qiagen).
Viral Metagenomics
Sequencing data from whole transcriptome libraries were analyzed for the presence of RNA sequences corresponding to known human viral pathogens using the sequence-based ultrarapid pathogen identification (SURPI) computational pipeline in comprehensive mode [25]. After computationally subtracting human reads, remaining reads were aligned against all reference microbial sequences in the NCBI GenBank database. The SNAP aligner [26] was used at moderate stringency (edit distance = 12) to align reads to the NCBI nucleotide nt database, allowing for detection of reads with >90% nucleotide identity to known viruses. RAPSearch [27] was used to detect divergent reads from potential novel viruses by translated nucleotide alignment to the NCBI protein nr database. A rapid taxonomic classification algorithm based on the lowest common ancestor was incorporated into SURPI, as previously described [28], and used to assign viral next-generation sequencing reads to the species, genus, or family level. Samples were reported as positive if viral reads were mapped to at least 2 unique regions in the genome. Reads corresponding to potential microbial pathogens other than viruses (ie, bacteria, fungi, and parasites) were not considered because libraries were RNA and were generated from whole blood samples with high human host background, reducing sensitivity, and because a separate, more comprehensive analysis of metagenomic and 16S DNA/RNA sequencing of low-background plasma samples from the same patients had already been performed [29].
BCR/TCR Sequence Analysis
RNA-seq data were analyzed with MiTCR v.1.0.3 [30] using the parameters specified by Brown et al [19] to extract CDR3 TCR data from the transcriptome library data and with MiXCR v1.6 [31] in RNA-seq mode to extract BCR sequences. Within a given case or control sample, if 2 or more distinct nucleotide sequences gave rise to the same peptide BCR/TCR sequence, these were merged into a single record. We looked for both 100% identical BCR/TCR sequences present in samples from at least 10 individuals, as well as clusters of highly similar sequences present in multiple samples. To generate these clusters, all BCR/TCR sequences of length 11–16 amino acids (aa) were inputted into FastTree 2.1.3 [32] for phylogenetic tree-building. The resulting trees were passed to Patristic [33] to calculate all pairwise patristic distances between BCR/TCR sequences. The Gengraph package in adegenet [34] was used to extract clusters of motifs within a specified patristic distance threshold (0.15). We then tallied the number of case and control samples in which clustered motifs were found. A χ2 test in R was used to evaluate the over- and underrepresentation of specific BCR/TCR motifs or motif clusters between subject groups.
RESULTS
RNA-seq Library Sequencing and Analysis
Seventy-four whole blood RNA libraries were sequenced over 10 lanes on an Illumina HiSeq 2500 instrument. On average, 56.1 (±14.5) million reads were obtained per sample. Tophat/Cufflinks detected an average of 77.3% (±5.5%) of all RefSeq isoforms in each sample (Supplementary Figure 1). Principal component analysis of whole transcriptome data from the 74 study participants did not show clusters indicative of technical bias (Supplementary Figure 2). The SURPI metagenomics pipeline subtracted, on average, 99.1% of raw reads by alignment to the human genome. Alignment of the remaining reads to NCBI GenBank identified an average of 150 viral reads per sample (range, 0–2549 reads).
MiTCR returned 18627 TCR sequences; MiXCR returned 22319 BCR sequences. After merging discrete nucleotide sequences encoding identical peptides, 39788 records remained across the samples, representing 28799 unique BCR/TCR motifs of average length 15aa (range, 6–41aa). Clustering the 27876 motifs of length 11–16aa yielded 22707 clusters, 20840 (91.7%) of which contained only a single motif. We focused our analysis on the 35 clusters that contained 20 or more motifs (Supplementary Table 2).
Transcriptomics
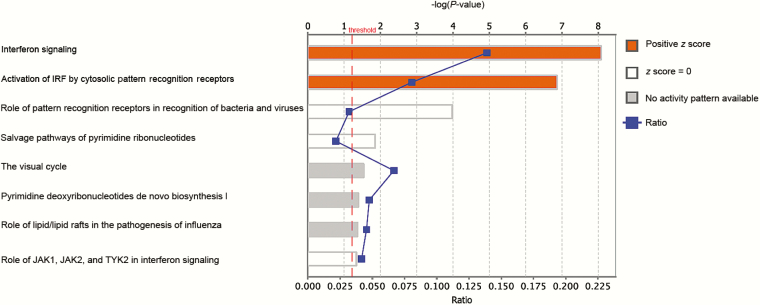
First, we determined whether there were any DEGs between cases and controls in whole blood samples collected during enrollment, at which time ADCLS, CFS, and SLE cases all showed marked disability [1]. A comparison of CFS and/or ADCLS cases with controls yielded no DEGs, nor did a comparison of CFS cases with ADCLS cases. In contrast, 42 DEGs were identified when SLE cases were compared against controls (Supplementary Table 3). Pathway analyses for SLE cases suggested that 10 of 42 (23.8%) DEGs were involved in the activation of interferon signaling pathways mediated by cytosolic pattern recognition receptors (Figure 1).
Figure 1.
Canonical pathways predicted to be involved in systemic lupus erythematosus by RNA-seq analysis. Pathways are ranked by the negative log of the P value of the enrichment score. The color scheme is based on z score, with activation in orange, z score = 0 in white, and undetermined directionality in gray. Also plotted is the ratio of identified differentially expressed genes to the total number of genes involved in each pathway (“ratio”). The yellow line represents the designated significance threshold (P < .05). Abbreviation: IRF, interferon regulating factor
Viral Metagenomics
We detected sequences from a small number of viruses in the RNA-seq data, including anelloviruses/torque teno viruses (TTVs), human pegivirus 1 (formerly designated GB virus C), human papillomaviruses (HPVs), and human herpesviruses (HHVs) (Table 1). The overall virome composition in CFS, ADCLS, and SLE cases did not differ significantly from controls (χ2 test, P = .73). Six of 25 (24%) CFS samples were positive for HPVs, which was a higher detection rate than the 7.6%–9.1% prevalence in other patient groups but was not significant (χ2 test, P = .31). No novel and/or divergent viruses were detected by translated nucleotide alignment, an approach that has previously proven useful for pathogen discovery [35].
Table 1.
Number of Patients with More Than 1 Unique Read to Human Viruses by Metagenomic RNA-seq
| Disease | Human Pegivirus 1 | Human Herpesvirus 4 | Human Herpesvirus 6A | Human Papillomaviruses | Torque Teno Viruses/ Anelloviruses | Total |
|---|---|---|---|---|---|---|
| Alternatively diagnosed chronic Lyme syndrome (n = 13) | 0 | 0 | 0 | 1 | 1 | 2 |
| Chronic fatigue syndrome (n = 25) | 1 | 0 | 1 | 6 | 0 | 8 |
| Systemic lupus erythematosus (n = 11) | 0 | 1 | 0 | 1 | 1 | 3 |
| Controls (n = 25) | 1 | 0 | 0 | 2 | 3 | 6 |
| Total | 2 | 1 | 1 | 10 | 5 | 19 |
BCR/TCR Sequence Analysis
Finally, we examined the BCR/TCR repertoires of each case or control sample. Immune cell counts in CFS and ADCLS patients were no different from those in controls (Supplementary Table 1), thus reducing potential BCR/TCR bias. Of the 28799 unique motifs, we first looked at motifs found in at least 10 study participants. Of the 50 motifs we found, 2 showed unusual distributions between subject groups: motif_2026, underrepresented in SLE cases, and motif_11947, overrepresented in CFS cases (Table 2). Recognizing that receptor–antigen binding is degenerate, we also looked at clusters of related BCR/TCR motifs and their distribution across subject groups. By χ2 testing, we found the following 3 significant clusters of motifs with unusual distributions (Table 2): motifs belonging to cluster 11_285 were overrepresented in control samples—interestingly, this cluster contained motif_2026, which was previously observed to be underrepresented in SLE cases; motifs belonging to cluster 13_2782 were underrepresented in ADCLS cases; and motifs belonging to cluster 14_522 were underrepresented in SLE cases.
Table 2.
B-Cell/T-Cell Receptor Motifs or Groups of Motifs Over- or Underrepresented Among Sample Classes
| Motif/Cluster ID | Sequence/Consensus | P Value (corrected) | Sample Class Proportion (n) | |||
|---|---|---|---|---|---|---|
| Controls (n = 25) | CFS (n = 25) | ADCLS (n = 13) | SLE (n = 11) | |||
| motif_11947 | CQSYDSSLSGYVF (BCR) | .006 (.006) | 0.24 (6) | 0.52 (13) | 0.23 (3) | 0.18 (2) |
| cluster13_2782 | cssytssstlyvf (BCR) | .014 (.013) | 0.76 (19) | 0.84 (21) | 0.54 (7) | 0.82 (9) |
| cluster11_285 | CmIWHssawvf (BCR) | .031 (.032) | 0.56 (14) | 0.36 (9) | 0.31 (4) | 0.36 (4) |
| motif_2026 | CMIWHSSAWVF (BCR) | .033 (.034) | 0.32 (8) | 0.28 (7) | 0.23 (3) | 0 (0) |
| cluster14_522 | CASslggsTDTQYF (TCR) | .036 (.035) | 0.4 (10) | 0.4 (10) | 0.38 (5) | 0.09 (1) |
| Motif/Cluster | P Value (corrected) | Controls (n = 25) | CFS + ACDLS (n = 38) | |||
| cluster13_2702 | CsSYAGsnnX (BCR) | .015 (0.015) | 0.24 (6) | 0.5 (19) | ||
| cluster12_656 | CcsyagsstwvF (BCR) | .02 (0.021) | 0.64 (16) | 0.87 (33) | ||
| cluster11_1320 | CASssXeTQYF (BCR) | .027 (0.027) | 0.2 (5) | 0.42 (16) | ||
All BCR Motifs Identified were associated with the light chain; the TCR Motif Identified was associated with the beta chain. The boldface text denotes the sample class containing the signfiicantly overrepresented or underrepresented motif.
Abbreviations: ADCLS, alternatively diagnosed chronic Lyme syndrome; BCR, B-cell receptor; CFS, chronic fatigue syndrome; SLE, systemic lupus erythematosus; TCR, T-cell receptor.
Given that our previous work showed similar phenotypes between CFS and ADCLS cases [1], we performed a second comparison of specific motifs and clusters in which CFS and ADCLS samples were grouped and compared to matched controls. Three clusters of motifs not previously seen as significant emerged, all of which were found more frequently in CFS and ADCLS cases than controls: cluster 11_1320, cluster 12_656, and cluster 13_2702. In the earlier 4-group analyses, all of these clusters tended toward underrepresentation in controls but not to a statistically significant degree.
DISCUSSION
Here, we queried RNA-Seq data from a case-control study of patients with complex chronic diseases for various signatures that might offer insight into the pathophysiology of CFS or ADCLS. Whole blood samples were chosen for analysis instead of peripheral blood mononuclear cells, as this enables detection of circulating viral pathogens by metagenomic analysis and as a more clinically accessible sample type for biomarker development. No gene expression signature could be identified when CFS patients were compared to age-, sex-, and geography-matched controls, which is consistent with an earlier study of 44 pairs of twins discordant for CFS in which no differences were observed by microarray [9]. Blood samples in the current study were collected at patient enrollment when patients were ambulatory and thus able to attend a screening appointment, in contrast to during symptom flares when they might otherwise be bed-bound. Thus, we cannot exclude the possibility that there are differences between CFS cases and controls confined to periods of symptom flares. We are presently investigating this hypothesis in a follow-up study involving RNA-seq data collected before and after flares induced by cardiopulmonary exercise stress testing.
Similarly, no differentially expressed genes were identified when ADCLS cases were compared to controls. We previously reported that individuals with documented acute Lyme disease showed differential expression of a large number of transcripts related to inflammation and host responses to infection [17]. There are at least 2 plausible explanations for our failure to observe this signature in the current analysis. First, as suggested by negative reference CDC 2-tiered serological testing, the ADCLS cases in this cohort may not have been infected by Borrelia burgdorferi. Second, the Lyme signature identified in the previous study reflects acute or subacute infection, whereas the 13 ADCLS cases were all chronic, having been sampled, on average, 10.7 years following their self-reported symptom onset.
The lack of DEGs between CFS and ADCLS cases and controls does not reflect a technical issue, as the SLE cases—a positive control representing a disease with known immune dysfunction—yielded 42 DEGs. This is a low number but could be attributed to disease control, as patients were sampled outside of flares. The differential genes identified in association with SLE patients belonged largely to interferon signaling pathways mediated by pattern recognition receptors; notably, a type I interferon signature has been reported as a potential biomarker for personalized medicine in SLE [36].
Viral pathogens are frequently suggested to be triggers for CFS [37]. We detected a small number of viruses in blood across all cohorts, corresponding to nonpathogenic flora (eg, anelloviruses such as TTV and HPgV-1), likely skin contamination (eg, human papillomaviruses) [38], or latent infections (eg, herpesviruses), none of which were significantly enriched in any subject group. Importantly, no sequence reads corresponding to novel or unexpected viral pathogens were detected. Our failure to detect significant viral infections correlated with subject group is not surprising—assaying pathogens through RNA-seq will only capture active infections and not a prior “hit-and-run” infection often posited to explain CFS onset [39]. The viral metagenomic results presented here are consistent with a separate study by our group using metagenomic and 16S DNA/RNA sequencing of plasma samples from the same patients for pathogen detection, which also failed to identify any significant differences between subject groups with regard to infections [29].
If immune dysfunction indeed plays a critical role in CFS and/or ADCLS, one would expect to observe differences in the adaptive immune responses of cases vs matched controls. To explore this possibility, we looked for BCR and TCR sequences overrepresented or underrepresented in different subject groups. By RNA analysis of whole blood samples, we obtained a high yield of BCR/TCR sequences. Significant TCR motifs across the different sample cohorts were not seen, perhaps because clonal expansion of different TCRs recognizing the same antigen is host major histocompatibility complex–dependent. In contrast, 2 significant BCR motifs were detected. One, motif_11947, was enriched in CFS cases, though we did not observe a parallel enrichment by cluster analysis of related motifs containing motif_11947. Similarly, BCR motif_2026 was significantly underrepresented among lupus cases when examined alone. However, when the larger cluster of related motifs containing motif_2026 was examined, the underrepresentation was no longer apparent—in fact, the larger cluster registered as overrepresented among controls. We also observed 3 BCR motif clusters overrepresented in cases when we combined ADCLS and CFS cases into a single group, which was then compared to controls.
Given the large size of the dataset (we began with nearly 40000 unique motifs and, during clustering, performed more than 142 million pairwise comparisons), it is difficult to say whether these enriched motifs constitute meaningful biological reality or whether they represent an artifact of analyzing data on such a large scale. A larger cohort of ADCLS and CFS cases and controls with longitudinal sampling is needed to explore whether the observed associations between motifs and disease phenotype remain significant and whether these TCR/BCR motifs constitute useful disease biomarkers, as previously shown for cancer [40].
In conclusion, a multipronged RNA-seq–based investigation of patients with complex chronic diseases and matched controls did not yield any signatures that might act as host biomarkers of CFS or ADCLS or suggest an underlying biological mechanism. Thus, these chronic syndromes do not appear to be associated with transcriptionally mediated immune cell dysregulation nor with active viral infection in blood. Other approaches for analysis of these RNA-seq data are feasible, such as a genome-wide association study of single nucleotide polymorphisms, and continued exploration may lend new insights into these debilitating diseases.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Supplementary Material
Notes
Acknowledgments: In addition to the named authors, members of the Complex Chronic Disease Study Team include A. Mattman, S. Sirrs, W. D. Reid, P. Phillips, J. Reynolds, H. Wong, A. Bested, I. Hyams, R. Arseneau, B. Ng, (the late) G. Blaney, J. Spinelli, J. Isaac-Renton, L. Hoang, and M. Krajden.
Financial support. This work was supported by a grant from the British Columbia Centre for Disease Control Foundation for Population and Public Health. J. B. is supported by a grant from the Bay Area Lyme Foundation. This work is also supported by awards to C. Y. C. from the National Institutes of Health (R01-HL105704), Bay Area Lyme Foundation, and the Swartz Foundation. J. L. G. is a Canada Research Chair and a Michael Smith Foundation for Health Research Scholar.
Potential conflicts of interest. C. Y. C. is the director of the University of California–San Francisco–Abbott Viral Diagnostics and Discovery Center and receives research support from Abbott Laboratories. All other authors report no conflicts of interest. The authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Patrick DM, Miller RR, Gardy JL, et al. ; Complex Chronic Disease Study Group Lyme disease diagnosed by alternative methods: a phenotype similar to that of chronic fatigue syndrome. Clin Infect Dis 2015; 61:1084–91. [DOI] [PubMed] [Google Scholar]
- 2. Lisnevskaia L, Murphy G, Isenberg D. Systemic lupus erythematosus. Lancet 2014; 384(9957):1878–88. [DOI] [PubMed] [Google Scholar]
- 3. Morris G, Berk M, Galecki P, Maes M. The emerging role of autoimmunity in myalgic encephalomyelitis/chronic fatigue syndrome (ME/cfs). Mol Neurobiol 2014; 49:741–56. [DOI] [PubMed] [Google Scholar]
- 4. Hornig M, Montoya JG, Klimas NG, et al. Distinct plasma immune signatures in ME/CFS are present early in the course of illness. Sci Adv 2015; 1 pii: e1400121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hornig M, Gottschalk G, Peterson DL, et al. Cytokine network analysis of cerebrospinal fluid in myalgic encephalomyelitis/chronic fatigue syndrome. Mol Psychiatry 2016; 21:261–9. [DOI] [PubMed] [Google Scholar]
- 6. Bansal AS, Bradley AS, Bishop KN, Kiani-Alikhan S, Ford B. Chronic fatigue syndrome, the immune system and viral infection. Brain Behav Immun 2012; 26:24–31. [DOI] [PubMed] [Google Scholar]
- 7. Fluge Ø, Risa K, Lunde S, et al. B-lymphocyte depletion in myalgic encephalopathy/chronic fatigue syndrome. An open-label phase ii study with rituximab maintenance treatment. PLoS One 2015; 10:e0129898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arvikar SL, Crowley JT, Sulka KB, Steere AC. Autoimmune arthritides, rheumatoid arthritis, psoriatic arthritis, or peripheral spondyloarthropathy, following Lyme disease. Arthritis Rheumatol 2016. doi: 10.1002/art.39866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Byrnes A, Jacks A, Dahlman-Wright K, et al. Gene expression in peripheral blood leukocytes in monozygotic twins discordant for chronic fatigue: no evidence of a biomarker. PLoS One 2009; 4:e5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frampton D, Kerr J, Harrison TJ, Kellam P. Assessment of a 44 gene classifier for the evaluation of chronic fatigue syndrome from peripheral blood mononuclear cell gene expression. PLoS One 2011; 6:e16872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gow JW, Hagan S, Herzyk P, Cannon C, Behan PO, Chaudhuri A. A gene signature for post-infectious chronic fatigue syndrome. BMC Med Genomics 2009; 2:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kaushik N, Fear D, Richards SC, et al. Gene expression in peripheral blood mononuclear cells from patients with chronic fatigue syndrome. J Clin Pathol 2005; 58:826–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kerr JR, Petty R, Burke B, et al. Gene expression subtypes in patients with chronic fatigue syndrome/myalgic encephalomyelitis. J Infect Dis 2008; 197:1171–84. [DOI] [PubMed] [Google Scholar]
- 14. Pihur V, Datta S, Datta S. Meta analysis of chronic fatigue syndrome through integration of clinical, gene expression, SNP and proteomic data. Bioinformation 2011; 6:120–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao S, Fung-Leung WP, Bittner A, Ngo K, Liu X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One 2014; 9:e78644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bouquet J, Soloski MJ, Swei A, et al. Longitudinal transcriptome analysis reveals a sustained differential gene expression signature in patients treated for acute Lyme disease. MBio 2016; 7:e00100–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Razzauti M, Galan M, Bernard M, et al. A comparison between transcriptome sequencing and 16S metagenomics for detection of bacterial pathogens in wildlife. PLoS Negl Trop Dis 2015; 9:e0003929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brown SD, Raeburn LA, Holt RA. Profiling tissue-resident T cell repertoires by RNA sequencing. Genome Med 2015; 7:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carruthers BM, van de Sande MI, De Meirleir KL, et al. Myalgic encephalomyelitis: international consensus criteria. J Intern Med 2011; 270:327–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 1997; 40:1725. [DOI] [PubMed] [Google Scholar]
- 21. Fukuda K, Straus SE, Hickie I, Sharpe MC, Dobbins JG, Komaroff A. The chronic fatigue syndrome: a comprehensive approach to its definition and study. International Chronic Fatigue Syndrome Study Group. Ann Intern Med 1994; 121:953–9. [DOI] [PubMed] [Google Scholar]
- 22. Brown AA, Jason LA. Validating a measure of myalgic encephalomyelitis/chronic fatigue syndrome symptomatology. Fatigue 2014; 2:132–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012; 7:562–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Naccache SN, Federman S, Veeraraghavan N, et al. A cloud-compatible bioinformatics pipeline for ultrarapid pathogen identification from next-generation sequencing of clinical samples. Genome Res 2014; 24:1180–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zaharia M, Bolosky WJ, Curtis K, et al. Faster and More Accurate Sequence Alignment with SNAP. ArXiv11115572 Cs Q-Bio 2011; Available at: http://arxiv.org/abs/1111.5572 Accessed 25 July 2016.
- 27. Zhao Y, Tang H, Ye Y. RAPSearch2: a fast and memory-efficient protein similarity search tool for next-generation sequencing data. Bioinformatics 2012; 28:125–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Greninger AL, Naccache SN, Messacar K, et al. A novel outbreak enterovirus D68 strain associated with acute flaccid myelitis cases in the USA (2012–14): a retrospective cohort study. Lancet Infect Dis 2015; 15:671–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miller RR, Uyaguari-Diaz M, McCabe MN, et al. Metagenomic Investigation of Plasma in Individuals with ME/CFS Highlights the Importance of Technical Controls to Elucidate Contamination and Batch Effects. Switzer WM, ed. PLoS ONE. 2016;11(11):e0165691. doi:10.1371/journal.pone.0165691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bolotin DA, Shugay M, Mamedov IZ, et al. MiTCR: software for T-cell receptor sequencing data analysis. Nat Methods 2013; 10:813–4. [DOI] [PubMed] [Google Scholar]
- 31. Bolotin DA, Poslavsky S, Mitrophanov I, et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods 2015; 12:380–1. [DOI] [PubMed] [Google Scholar]
- 32. Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 2010; 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fourment M, Gibbs MJ. PATRISTIC: a program for calculating patristic distances and graphically comparing the components of genetic change. BMC Evol Biol 2006; 6:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jombart T, Ahmed I. Adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 2011; 27:3070–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chiu CY. Viral pathogen discovery. Curr Opin Microbiol 2013; 16:468–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Biesen R, Rose T, Hoyer BF, Alexander T, Hiepe F. Autoantibodies, complement and type I interferon as biomarkers for personalized medicine in SLE. Lupus 2016; 25:823–9. [DOI] [PubMed] [Google Scholar]
- 37. Navaneetharaja N, Griffiths V, Wileman T, Carding SR. A role for the intestinal microbiota and virome in myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS)? J Clin Med 2016; 5 pii: E55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bzhalava D, Mühr LSA, Lagheden C, et al. Deep sequencing extends the diversity of human papillomaviruses in human skin. Sci Rep 2014; 4:5807; Available at: http://www.nature.com/articles/srep05807 Accessed 17 October 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hickie I, Davenport T, Wakefield D, et al. ; Dubbo Infection Outcomes Study Group Post-infective and chronic fatigue syndromes precipitated by viral and non-viral pathogens: prospective cohort study. BMJ 2006; 333:575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Han Y, Li H, Guan Y, Huang J. Immune repertoire: a potential biomarker and therapeutic for hepatocellular carcinoma. Cancer Lett 2016; 379:206–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.