Abstract
Transposable element (TE)-derived sequences make up approximately half of most mammalian genomes, and many TEs have been co-opted into gene regulatory elements. However, we lack a comprehensive tissue- and genome-wide understanding of how and when TEs gain regulatory activity in their hosts. We evaluated the prevalence of TE-derived DNA in enhancers and promoters across hundreds of human and mouse cell lines and primary tissues. Promoters are significantly depleted of TEs in all tissues compared with their overall prevalence in the genome (P < 0.001); enhancers are also depleted of TEs, though not as strongly as promoters. The degree of enhancer depletion also varies across contexts (1.5–3×), with reproductive and immune cells showing the highest levels of TE regulatory activity in humans. Overall, in spite of the regulatory potential of many TE sequences, they are significantly less active in gene regulation than expected from their prevalence. TE age is predictive of the likelihood of enhancer activity; TEs originating before the divergence of amniotes are 9.2 times more likely to have enhancer activity than TEs that integrated in great apes. Context-specific enhancers are more likely to be TE-derived than enhancers active in multiple tissues, and young TEs are more likely to overlap context-specific enhancers than old TEs (86% vs. 47%). Once TEs obtain enhancer activity in the host, they have similar functional dynamics to one another and non-TE-derived enhancers, likely driven by pleiotropic constraints. However, a few TE families, most notably endogenous retroviruses, have greater regulatory potential. Our observations suggest a model of regulatory co-option in which TE-derived sequences are initially repressed, after which a small fraction obtains context-specific enhancer activity, with further gains subject to pleiotropic constraints.
Keywords: transposable elements, gene regulation, evolution, pleiotropy
Introduction
Gene regulation is fundamental to life; it underlies the temporal and spatial heterogeneity of gene expression required for development and differentiation of complex organisms. In addition to regulatory molecules, such as transcription factors (TFs) and RNAs, gene regulation requires noncoding regulatory elements, such as promoters and enhancers, that interact with regulatory molecules to control when and where genes are expressed. Co-option of DNA sequence introduced during invasions of transposable elements (TEs) can create new regulatory elements, including alternate gene promoters (Faulkner etal. 2009; Emera etal. 2012; Kapusta etal. 2013), enhancers (Huda etal. 2010, 2011; Jacques etal. 2013; Xie etal. 2013; del Rosario etal. 2014; Glinsky 2015; Lynch etal. 2015; Notwell etal. 2015), and even insulators (Wang etal. 2015), substantially faster than single mutations, and can influence general chromatin accessibility as well (Gomez etal. 2016). TEs commonly contribute to gene regulatory elements, with up to 40% of genome-wide binding sites for some TFs located in TE-derived regions (Sundaram etal. 2014) and 20% of conserved noncoding elements TE-derived (Lowe etal. 2012). Co-option of the ready-made regulatory elements in TEs may facilitate substantial shifts in gene regulation over short timescales by simultaneously influencing the expression of multiple genes in specific contexts (Rebollo etal. 2011).
Alterations to gene regulation underlie the evolution of many physical traits (Averof and Patel 1997; Cohn and Tickle 1999), but often these changes must be restricted to particular tissues and cell types in order to maintain the integrity of other tissues. Studies have examined the contribution of TEs to enhancers defined through a variety of methods: via histone marks (Huda etal. 2010, 2011; Lynch etal. 2015), DNA methylation (Xie etal. 2013; Glinsky 2015), the binding of proteins associated with enhancer activity (e.g., p300) (Sundaram etal. 2014; Notwell etal. 2015), and sequence conservation (Jacques etal. 2013; del Rosario etal. 2014). Several of these studies reported enrichment of TEs in tissue-specific enhancers; however, until recently, identifying enhancers across many tissues using any of these methods has been both time- and cost-prohibitive, making the true breadth of activity of a regulatory element difficult to determine. Thus, comprehensive examination of how TEs contribute to the specificity of gene regulation across tissues has not been possible.
In order to quantify the role of TEs in regulatory elements active across tissues and their evolutionary dynamics, we analyzed TE contributions to promoters and enhancers defined through cap analysis of gene expression (CAGE) across hundreds of human and mouse cell lines and primary tissues from the FANTOM5 consortium (Andersson etal. 2014). We found that though many TEs have regulatory activity, both enhancers and promoters are depleted of TEs compared with the expectation from their genome-wide prevalence, with promoters significantly more depleted than enhancers. Thus, in spite of their regulatory potential, TEs are significantly less active than expected if they were randomly distributed across the genome. This is consistent with the strong pressure on genomes to repress the activity of TEs due to their mutagenic potential; however, many factors influence this result including the specific sequences integrated and biases in genomic integration sites between families (Sultana etal. 2017). Nevertheless, in the context of this overall depletion, the evolutionary age of a TE correlates positively with its likelihood of contributing to a regulatory element, and tissue-specific enhancers are significantly more likely to be derived from TEs than broadly active enhancers. Overall, with the exception of endogenous retroviruses (ERVs), we observe striking similarity in the likelihood of having regulatory function in the host across TE families. We also observe similar patterns in the relationship of mouse TEs to promoter and enhancer activity. Based on our results, we propose a model of how sequences derived from diverse TE families obtain regulatory functions in host tissues. TE-derived sequences are initially repressed upon integration into host genomes, after which a small fraction obtain context-specific enhancer activity, and over time further gains in regulatory activity are likely, but subject to pleiotropic constraints.
Results
Regulatory Regions Are Depleted of TEs
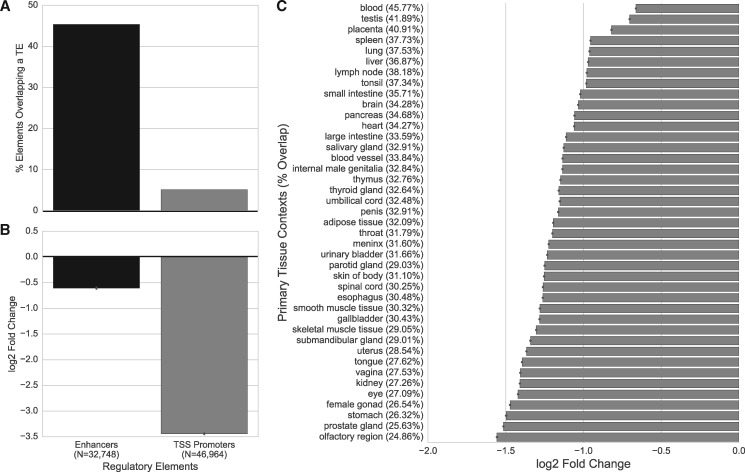
Detectable TE-derived sequences comprise ∼48.5% of the human genome (Smit 1999), so the expectation, even under a null hypothesis of a random distribution across the genome and no enrichment for regulatory function, is that a large proportion of regulatory elements overlap these sequences. We intersected all known TE-derived sequences in the human genome with 32,748 enhancers and 46,964 protein-coding gene promoters defined by CAGE across 112 human cell lines and primary tissues. For simplicity, we will refer to these cell lines and tissues as “contexts.” Overall, 45.4% of enhancers overlapped a TE (fig. 1A). However, the fraction of enhancers overlapping TEs varied considerably across cellular contexts. For example, 24.9% of olfactory region enhancers overlapped a TE while 45.8% of blood enhancers overlapped a TE, with a median of 32.1% across all contexts (fig. 1C andsupplementary fig. S1, Supplementary Material online). Promoters were significantly less likely to overlap a TE than enhancers; only 5.1% of promoters overlapped a TE (fig. 1A;P ≈ 0, one-sided binomial test).
Fig. 1.
Enhancers and promoters are depleted of TEs. (A) The proportion of enhancers and promoters proximal to protein-coding TSSs (“TSS Promoters”) that overlap a TE. (B) The log2-fold difference in observed TE overlap compared with random expectation (median over 10,000 permuted sets). (C) The log2-fold difference from the expected TE overlap with enhancers active in primary tissue contexts. The percent of enhancers from each context overlapping a TE is given in parentheses. Contexts are sorted by enrichment. See supplementary figure S1, Supplementary Material online, for results on cell lines.
To evaluate whether TE-derived sequences were more likely to have regulatory activity than expected from their prevalence in the genome, we compared the observed overlaps between TEs and regulatory regions to the overlap between TEs and random sets of genomic regions matched to the length and genomic distribution of the enhancers and promoters (see Materials and Methods). We found significantly less overlap between TEs and regulatory elements than expected if they were distributed randomly across the genome. Enhancers overlapped only 44% of the number of TEs expected if they were randomly distributed (P < 0.0001, randomization test; fig. 1A). Promoters were even more depleted; they overlapped less than one-tenth of the expected number of TEs (9.2%; P < 0.001; fig. 1B).
Performing these analyses on mouse promoters and enhancers identified by the FANTOM5 consortium revealed similar levels of depletion as in human. For mouse promoters, 4.6% overlapped a TE, compared with an expected overlap of 9.6% (P < 0.001; supplementary fig. S2, Supplementary Material online); 37.4% of mouse enhancers overlapped a TE, whereas 63.4% was expected under a random distribution (P < 0.001; supplementary fig. S2, Supplementary Material online).
Furthermore, as expected from the overlap results, the degree of depletion among regulatory elements varied widely across tissues. Examining human enhancers on a context-by-context basis revealed relative depletions between 0.34 times for olfactory regions and 0.63 times for blood (fig. 1C and supplementary fig. S1, Supplementary Material online). In general, reproductive and immune contexts, such as blood, testis, placenta, and spleen, were the least depleted of enhancer activity (fig. 1C andsupplementary fig. S1, Supplementary Material online).
Enhancer TEs Are Enriched for Ancient Origins
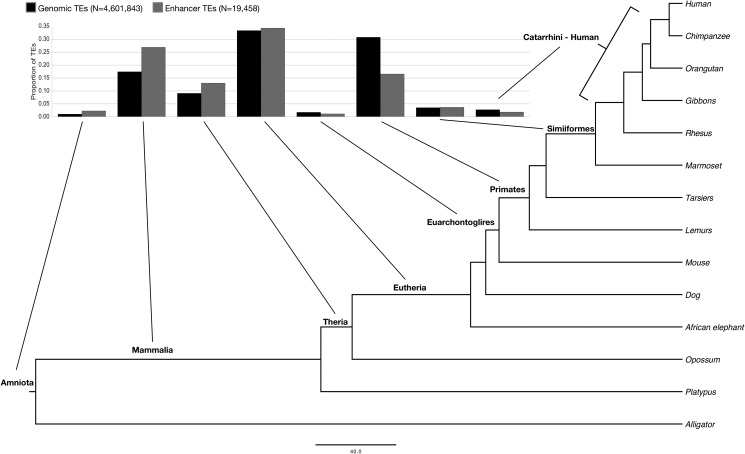
Recent studies in uterine and liver tissue have suggested that enhancers often evolve from ancient TE sequences (Lynch etal. 2015; Villar etal. 2015). To explore the evolutionary dynamics of the contribution of TEs to regulatory activity, we integrated the age of each TE, as inferred from the presence of TEs across taxa, into our analyses of regulatory activity. TEs present in the human genome have diverse evolutionary origins. For example, 17% of human TEs date to the common ancestors of Mammalia, 9% to Theria, 34% to Eutheria, and 31% to primates (fig. 2). TEs that overlap human enhancers (“enhancer TEs”) have qualitatively similar origin patterns to TEs overall (fig. 2B); however, enhancer TEs are significantly older (average 128.7 vs. 111.7 Ma; P < 5E-324, Mann–Whitney U test). Mouse enhancer TEs are also significantly older than mouse TEs overall; the enhancer TE average age is 73.5 Ma versus the genomic TE average of 58.2 Ma (P < 5E-324, Mann–Whitney U test; supplementary fig. S3, Supplementary Material online). In humans, there is a particularly strong depletion for primate-originating TEs (odds ratio [OR] = 0.44, P < 5E-324, two-sided Fisher’s exact test) and enrichment for Mammalia-originating TEs (OR = 1.75, P = 1.97E–237) and ancient TEs (OR = 2.23, P = 2.26E–49). Hereafter, we refer to TEs originating in the most recent common ancestor (MRCA) of amniotes or before as “ancient.” Promoter TEs are also significantly older than TEs overall (average 121.7 vs. 111.7 Ma; P = 2.85E–12, Mann–Whitney U test), but are younger than enhancer TEs (P = 1.5E–14). These results suggest that TEs originating before the divergence of mammals are overrepresented in both enhancers and promoters. Several potential causes for this pattern will be discussed in later sections.
Fig. 2.
Enhancer TEs are enriched for ancient origins. The phylogenetic tree indicates the ancestral branches to which TE origins were mapped. Transposable elements integrated into the human genome at different times. The proportion of all known TEs originating in each lineage is plotted. Regulatory TEs are significantly older than TEs overall; this is true for both enhancer TEs (average 128.7 vs. 111.7 Ma; P < 5E-324, Mann–Whitney U test) and TSS promoter TEs (not plotted; average 121.7 vs. 111.7 Ma; P = 2.85E–12).
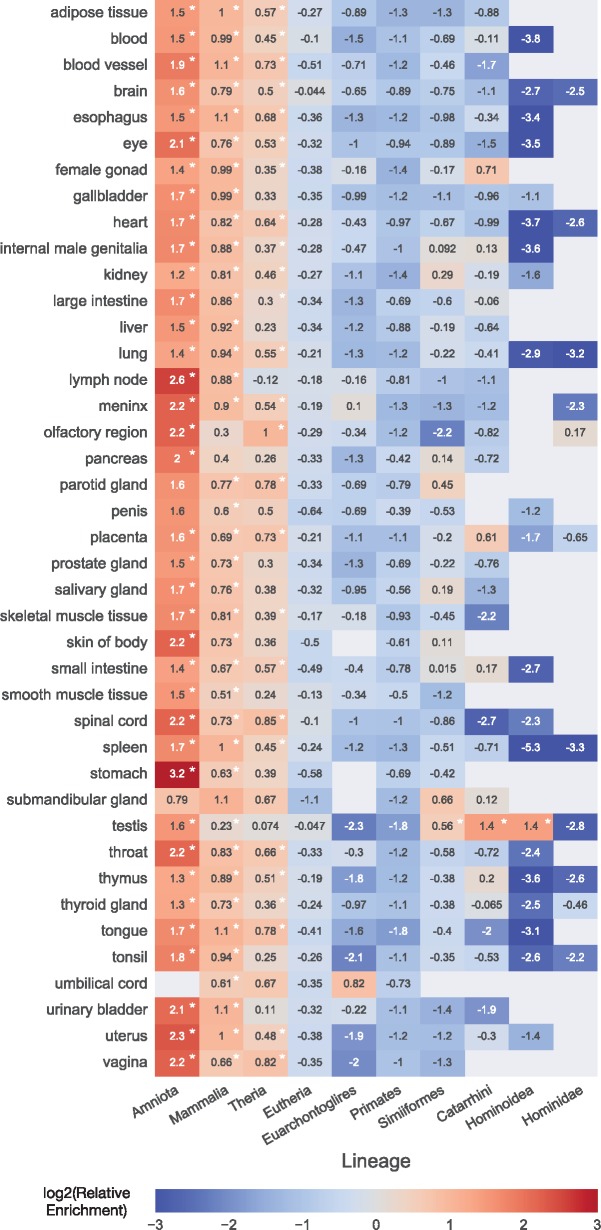
To investigate the relationship between TE age and patterns of enhancer activity across contexts, we tested for enrichment of TEs originating on each lineage among enhancers active in each context. Given the overall depletion of TEs, we investigated whether the proportion of TEs originating on a given lineage was different from the proportion expected if TE age had no effect (see Materials and Methods). We observed significant enrichment for ancient TEs and depletion of young TEs among enhancers from most tissues and cell lines (fig. 3 and supplementary fig. S4, Supplementary Material online). Enhancers from 107 out of the 111 contexts were significantly enriched for ancient TEs (fig. 3 and supplementary fig. S4, Supplementary Material online; q < 0.05, randomization tests with FDR correction). Enhancers from 110 out of the total 112 contexts were similarly enriched for TEs originating in the MRCA of all mammals (q < 0.05). Finally, enhancers from 91 of the 112 contexts were additionally enriched for TEs originating in the MRCA of marsupials and placental mammals (Theria; q < 0.05). Thus, TEs originating before the MRCA of placental mammals (Eutheria) contribute a larger number of enhancers than expected across most contexts.
Fig. 3.
Enhancers in most contexts are enriched for ancient TEs and depleted of young TEs. For each primary tissue context, older TEs are more likely to have enhancer activity and younger TEs are less likely to have enhancer activity than expected from their genome-wide prevalence (based on 10,000 permuted enhancer sets). The log2 of the relative difference between observed and expected is given for each comparison. Gray indicates context–lineage pairs with insufficient data. Lineages with fewer than ten contexts with sufficient enhancers were excluded from the figure. Asterisks indicate significant enrichment after controlling the FDR to account for multiple testing (q < 0.05). Results for cell lines were similar (supplementary fig. S4, Supplementary Material online).
There was also strong depletion for enhancer activity among more recent TEs (fig. 3 and supplementary fig. S4, Supplementary Material online). However, a few contexts were enriched for younger TEs. Of the 34 contexts with sufficient data to test, enhancers from 4 were significantly enriched for TEs originating in the MRCA of Haplorrhini (tarsiers, monkeys, and apes; q < 0.05). Enhancers from 3 of the 111 contexts with sufficient data were enriched for TEs originating in the MRCA of Simiiformes (monkeys and apes; q < 0.05). Enhancers from 2 of 103 contexts with sufficient data were enriched for TEs originating in the MRCA of Catarrhini (Old World monkeys and apes; q < 0.05). Only enhancers active in testis (out of 76 contexts with sufficient data) were enriched for TEs originating in the stem lineage of Hominoidea (MRCA of human and gibbons; q = 0.002). Thus, young TEs are not broadly enriched for enhancer function in the contexts analyzed here.
Enhancers overlapping young TEs are enriched for different TF binding motifs than ancient TEs. For example, ancient TEs with enhancer activity are enriched for binding motifs for Jun, Fos, RFX family, and several other TFs, whereas enhancers overlapping young TEs are enriched for NFY, SP, and KLF family TF motifs (supplementary file 1, Supplementary Material online). There was no difference in the age distribution of the TFs with motifs enriched among enhancers overlapping young versus ancient TEs (P = 0.37; Mann–Whitney U test). These results suggest that TF motif analyses have the potential to provide insights into differences in how TEs of different ages influence gene regulation.
Diverse TE Families Exhibit Increases in Enhancer Activity with Age
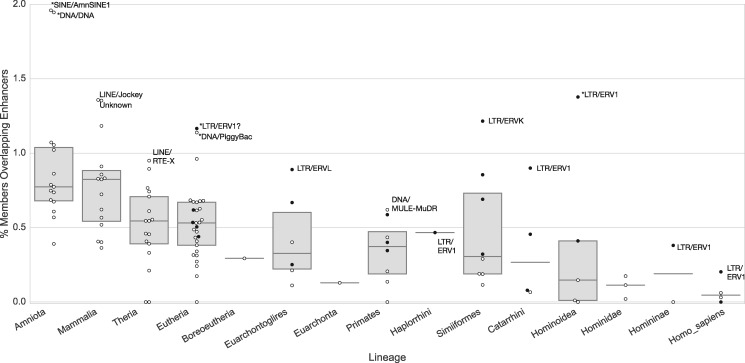
We next evaluated whether the trend of increased enhancer overlap with increased age was universal across TE families. RepeatMasker classifies TEs into classes, families, and subfamilies; for example, an element of the MIRc subfamily is of the family MIR and class SINE. Using this classification, we calculated the proportion of members of each TE family that overlapped an enhancer active in any context. We observed variation across families of similar age; however, the proportion of enhancer TEs increased with the age of the family (fig. 4). Only 0.10% of the members of very young TE families (originating in MRCA of Hominidae or later) overlapped enhancers, whereas the ancient (originating in MRCA of Amniota or earlier) TEs were 9 times more likely to be enhancer TEs on an average (0.92%). This pattern was also observed for TEs overlapped by mouse enhancers (supplementary fig. S5, Supplementary Material online). This trend is likely the product of two forces: the increasing opportunity for co-option the longer a TE-derived sequence spends in the genome and the divergence of older nonfunctional elements to the point that they can no longer be recognized as TE-derived sequences. Results for all families and subfamilies are given in supplementary file 1, Supplementary Material online.
Fig. 4.
TE families of similar ages vary in their likelihood of enhancer activity, but older TEs are more likely to overlap enhancers than young TEs. Each dot represents a TE family. There is substantial variation in the percent of members of a TE family that overlap enhancers among families with similar temporal origins. However, there is a consistent increase in the fraction of members of each family that overlap enhancers with family age. The black dots represent ERV families; ERVs consistently have higher proportions of enhancer activity than other TE families with similar ages. If fewer than five families appeared on a lineage, only the median was plotted. Asterisks indicate outlier families that fall outside 1.5 times the interquartile range.
Whereas the proportion of enhancer TEs increases with age in most TE families, a few TE families were more likely to have enhancer activity than expected based on their age. Several ancient SINE and DNA families overlap TEs beyond the expectation from other families of similar age, but the most consistent group is the ERVs. ERVs consistently had a higher fraction of enhancer overlap than expected in nearly every lineage in which they were present (fig. 4). The ERVs are divided into four main families: ERV1, ERVK, ERVL, and ERVL-MaLR. The oldest extant ERVs appeared in the MRCA of eutherians, and additional subfamilies have appeared on almost every subsequent lineage. When compared with other TE families appearing on the same lineage, one of these four families typically had the highest observed enhancer TE proportion. For example, members of the ERV1 family overlap the largest proportion of enhancers in the stem lineages of Catarrhini, Hominoidea, Homininae, and humans.
We also found that several different tissues are influenced by young ERVs. For example, the testis is strongly enriched for the Hominoidea-originating ERV1 subfamilies LTR12C and LTR12D (log2-fold enrichment > 2, q < 0.01; supplementary file 1, Supplementary Material online). This enrichment is particularly pronounced due to most of the enhancers overlapping these ERV1 subfamilies being testis-specific. Mast cells and monocytes are enriched for the Simiiformes-originating ERV1 subfamily LTR8B (log2-fold enrichment > 2, q < 0.01). In addition to these subfamily analyses, we pooled enhancers active in any primary tissue together and enhancers active in any cell line together into two sets. Enhancers active in both of these sets showed enrichment for HERVFH21 (log2-fold enrichment > 3, q < 0.01), a Catarrhini-originating ERV1 subfamily. Interestingly, no individual context showed enrichment for this subfamily, suggesting that some ERVs contribute strongly to one context, whereas others have a subtle, but consistent influence across contexts.
We then examined whether differences in the length or number of ERVs compared with other TE families could explain their increased overlap with enhancers compared with other families. The ERVs were not substantially different from other TE families appearing at the same time in either dimension (supplementary fig. S6, Supplementary Material online). This suggests that differences in activity are likely due to functionally relevant attributes of these TEs, such as their sequence content, preferred insertion locations, or interactions with the host defense machinery.
Context-Specific Enhancers Are Enriched for TEs Compared with Enhancers Active in Multiple Contexts
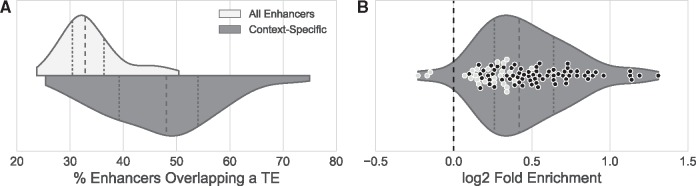
Previous studies have observed an abundance of TE-derived sequence in tissue-specific enhancers (Wilson etal. 2008; Huda etal. 2011; Xie etal. 2013; Notwell etal. 2015). We tested the generality of this pattern across all the contexts in our data set. First, we identified all human enhancers specific to each cell line or primary tissue (“context-specific”). We then tested these for TE enrichment compared with all enhancers active in the context. For 74% (70 out of 95) of the cell lines and primary tissues with sufficient data, context-specific enhancers were significantly enriched for TEs (fig. 5; q < 0.01, FDR-corrected hypergeometric test). Of the 25 contexts that were not significantly enriched, the context-specific enhancers of all but three overlapped more TEs than other enhancers active in, but not specific to the context. These results suggest broad overrepresentation of TEs among context-specific enhancers.
Fig. 5.
Context-specific enhancers are enriched for TEs compared with all active enhancers. (A) Histograms comparing the percent of all enhancers and context-specific enhancers overlapping a TE for each of the 95 contexts (out of 112) containing at least 10 context-specific enhancers. (B) The relative enrichment of context-specific enhancers for TE overlap compared with all enhancers active in each context. The violin plot represents all values for all contexts. Points are plotted for the 70 contexts in which context-specific enhancers are significantly enriched for TEs (black; q < 0.01), and the 25 contexts not significantly enriched (light gray).
Enhancers Overlapping Young TEs Are More Likely to Be Context-Specific than Enhancers Overlapping Old TEs
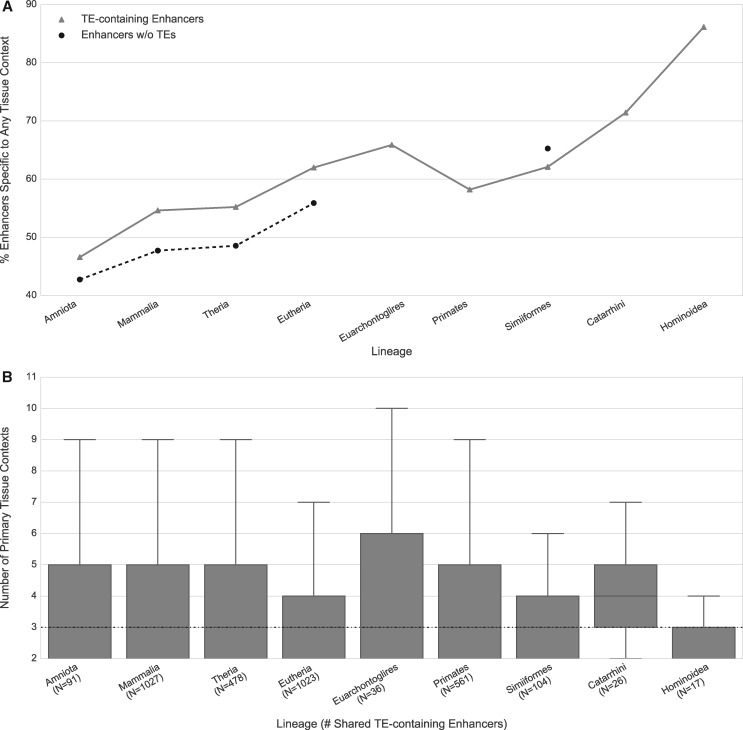
As old and young TEs differ in their likelihood of having enhancer activity (fig. 4), we examined whether the age of a TE is also related to the number of contexts in which it was likely to have enhancer activity. Analyzing the breadth of activity of TE-containing enhancers stratified by TE age, we found that enhancers overlapping young TEs are more likely to be active in a single context compared with enhancers overlapping older elements (fig. 6A and supplementary fig. S7, Supplementary Material online; P = 2.89E-10, Fisher’s exact test). For example, nearly 90% of enhancers containing TEs originating in Hominoidea are tissue-specific, whereas <50% of the ancient TE enhancers are tissue-specific. In other words, the likelihood that a TE-overlapping enhancer is context-specific decreases with the age of the TE.
Fig. 6.
Tissue-specificity decreases with age for both TE and non-TE-containing enhancers, but breadth of activity does not significantly change. (A) The proportion of enhancers that are specific to a single primary tissue context decreases with age. The dashed line represents enhancers that do not overlap a TE. Lineages with fewer than 20 enhancers overlapping a TE were excluded. Non-TE-containing enhancers could only be assigned ages on the Amniota, Mammalia, Theria, Eutheria, and Simiiformes lineages (see Materials and Methods). (B) The breadth of activity in primary tissue contexts for TE-containing enhancers active in at least two contexts is not influenced by age (P = 0.96, Kruskal–Wallis test). Lineages with no enhancers were excluded. The black dotted line indicates the median breadth of activity (three contexts) of non-TE-containing enhancers active in primary tissue contexts. Outliers are not plotted.
However, among enhancers active in more than one context, enhancers overlapping ancient TEs are not active in significantly more contexts than young TEs (fig. 6B and supplementary fig. S7, Supplementary Material online; P = 0.96, Kruskal–Wallis test). The median activity of these TE-containing enhancers is between two and four primary tissue contexts, regardless of age. For each TE subfamily, we also evaluated whether age associates with the breadth of enhancer activity, defined as the size of the union of all contexts of activity across enhancers overlapping a TE from that subfamily. The breadth of activity of pleiotropic enhancers was also not age dependent (supplementary fig. S8, Supplementary Material online). Thus, once a TE-containing enhancer becomes active in more than one context, TE age is not informative about its breadth of activity.
To determine whether these patterns were specific to TE-derived enhancers, we assigned evolutionary origins to enhancers lacking TEs using a recently developed dating strategy based on the pattern of presence or absence of the sequence across species in genome-wide multiple sequence alignments (Emera etal. 2016). We found that the trend of decreasing specificity over time holds in enhancers that do not overlap TEs, though these non-TE-derived enhancers appear to be less likely to be tissue-specific than TE-containing enhancers of the same age. The enhancers that do not contain TEs also exhibited similar median breadth of activity (∼3 contexts; fig. 6B). These results suggest that once TE-derived sequences obtain host enhancer activity, the patterns and dynamics of their activity are similar to those of non-TE-derived enhancers, and thus, that they are constrained by similar pressures.
Histone-Mark-Defined Enhancers Show Similar TE Overlap Patterns as CAGE-Defined Enhancers
The CAGE-defined enhancers from the FANTOM5 consortium provided a high-resolution set of enhancers across an extensive set of human and mouse cells and tissues for our analyses. In order to test whether our results were robust to differences in enhancer identification methodology, we examined eight sets of putative human enhancers defined via the presence of H3K27ac marks from Roadmap Epigenomics: placenta, trophoblast stem cells (TSCs), monocytes, B cells, natural killer (NK) cells, temporal lobe, neuronal stem cells, and lung (Bernstein etal. 2010; Kundaje etal. 2015), and two sets from independent studies of liver (Villar etal. 2015) and decidualized endometrial stromal cells (dESCs) (Lynch etal. 2015). Most of the considered enhancer sets were from immune or reproductive contexts, as these have garnered a great deal of attention with respect to regulatory co-option of TEs, and these contexts showed some of the highest TE regulatory activity in the FANTOM5 enhancers.
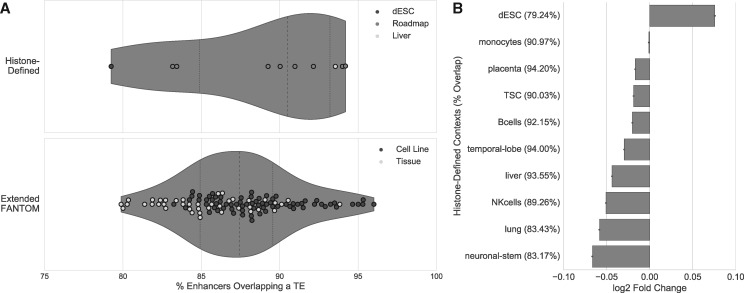
Overall, a much larger proportion of histone-mark-defined enhancers overlap TEs than CAGE-defined enhancers (median of 91% vs. 32.1% across contexts), but this is primarily due to the much lower resolution of the histone-mark-defined enhancers; they are ∼8 times the length of the CAGE-defined enhancers (median length of ∼2.4 kb vs. ∼300 bp). When we extended the CAGE-defined enhancers to the median histone-mark-defined enhancer length, a comparable percentage of these extended enhancers overlapped a TE (median 91% vs. 87.4%; fig. 7A).
Fig. 7.
Histone-mark-defined enhancers overlap more TEs than CAGE-defined enhancers, but are still depleted of TEs. (A) The proportion of enhancers overlapping a TE for histone-mark-defined enhancers and CAGE-defined enhancers that have been extended to the median length of histone-defined enhancers (FANTOM enhancers extended from ∼300 bp to 2.4 kb). A similar percentage of the histone-mark-defined and extended CAGE-defined enhancers overlapped a TE (median 91% vs. 87.4%). (B) Eight of ten histone-mark-defined enhancer sets were significantly depleted of TEs (P < 0.001, randomization test) compared with the genome-wide expectation (median of 1,000 permuted sets), and one set (from dESCs) was enriched. However, the magnitude of the effects for these enhancer sets was much smaller than for the CAGE-defined enhancers (fig. 1B). The percent of each enhancer set overlapping a TE is given in parentheses.
Despite the increased overlap of TEs, eight of the ten sets of histone-mark-defined enhancers are significantly depleted for TEs (P < 0.001, randomization test) compared with the genome-wide expectation (fig. 7B). Unsurprisingly, given their high enhancer overlap, the depletion is weaker than for transcribed enhancers (minimum log2-fold difference of −0.067 vs. −1.61; supplementary fig. S1, Supplementary Material online and fig. 7B). However, enhancers active in dESCs—a cell line not present in the CAGE data—are enriched (P < 0.001, randomization test) for TEs overall, and monocytes are neither enriched nor depleted (P = 0.618, randomization test). As we observed for the CAGE-defined enhancers, each set of histone-mark-defined enhancers is enriched for ancient TEs and depleted of young TEs (q < 0.05; supplementary fig. S9, Supplementary Material online). Together, these results argue that TEs are consistently depleted of enhancer activity and that ancient TEs originating before the MRCA of eutherians are overrepresented in enhancers, regardless of the enhancer identification methodology used.
Discussion
Regulatory Elements Are Depleted of TEs Compared with Their Genomic Prevalence
In this paper, we demonstrate that, even though TEs play important roles in remodeling regulatory networks and enabling functional innovation (Lynch etal. 2011, 2015; Chuong 2013; Chuong etal. 2016a; Trizzino etal. 2016), they are depleted for regulatory activity compared with their prevalence across the human and mouse genomes. This should not be surprising; the integration of TEs often disrupts functional elements with deleterious effects (Chuong etal. 2016b), which leads to strong pressure against TE insertion in functional regions and repression of their activity. In spite of the important contributions of TE-derived sequences to regulatory networks, the vast majority of TE instances do not have consistent regulatory activity in their host genomes. Furthermore, we find that, with the exception of ERVs, the contribution of TEs of different origins to regulatory elements is qualitatively similar across biological contexts. This suggests that consistent forces drive this process across TEs and contexts.
However, we note that the depletion of TEs for regulatory function is contingent on the choice of null model used in the tests. TEs vary with respect to their initial regulatory potential and integration site preferences (Sultana etal. 2017), and taking these factors into account could provide different results. However, these attributes are not well characterized for many TEs. We used a random null because we desired to evaluate the entire process by which TEs gain host regulatory activity—from integration to regulatory function. Furthermore, this approach is agnostic to differences between tissues and cell lines, so it enables comparison between contexts.
We focused our analyses on CAGE-defined enhancers and promoters for two main reasons. First, they are defined for a large number of tissues using consistent protocols and analytical pipelines. This enabled evaluation and comparison of enhancer–TE relationships across diverse tissues. Second, they have higher resolution than most putative enhancers defined by the presence of enhancer-associated histone modifications (e.g., via ChIP-seq for H3K27ac). Nevertheless, we also confirmed our main conclusions in complementary analyses of histone-mark-defined enhancers (fig. 7).
TE Age Has a Strong Effect on Enhancer Overlap
Across contexts and TE families analyzed, ancient TEs are significantly more likely to overlap an enhancer than young TEs. Previous studies have stressed the importance of ancient TEs in gene regulation, but these have focused on the relationship between when regulatory TEs appeared in the genome and specific evolutionary innovations (Chuong etal. 2013; Lynch etal. 2015) or on TE contribution to deeply conserved elements (Lindblad-Toh etal. 2011). Our comprehensive examination of the contribution of TEs across timescales and diverse tissues to enhancer activity demonstrates this pattern is extremely general.
Several factors likely contribute to this pattern. First, opportunities to act as host regulatory elements are correlated with the amount of time spent in the genome; older TEs have had more opportunities to obtain host regulatory functions. Second, TEs with regulatory function, even if it is weak or episodic, are likely under stronger sequence constraint than their nonregulatory family members. Over time, the increased divergence of the sequence of nonfunctional TEs from the original active sequence could make them more difficult to identify as TE-derived, and thus increase the relative fraction of detectable older TEs that are functional. To explore this possibility, we compared the divergence from the consensus sequence for enhancer and nonenhancer TEs from the same subfamilies and found no significant difference (supplementary figs. S10 and S11, Supplementary Material online). More focused analyses that account for TFBS and other functional motifs could reveal more subtle differences between regulatory and nonregulatory TEs. For example, we find that enhancers overlapping ancient TEs are enriched for different TF motifs when compared with enhancers overlapping young TEs (supplementary file 1, Supplementary Material online). Also, it is worth noting that this is heavily contingent on the accuracy of the consensus sequence, which is likely more difficult to determine with very ancient TEs. Nevertheless, lack of difference suggests that sequence divergence is not a major driver of this pattern. Third, it is possible that the regulatory potential of TEs has not been consistent over time. For example, TEs that were active hundreds of millions of years ago might have had greater regulatory potential or integrated into more permissive cellular contexts. However, we would expect these effects to be somewhat family and/or context-specific, so the fact that the increase in activity with age holds generally across families and contexts argues that time spent in the genome is likely the major driver of increased TE regulatory activity with age. Finally, this effect could also be influenced by the greater difficulty of accurately aligning short reads to younger TE sequences given their similarity. However, this is not a major driver of our results as mappability is only weakly correlated with enrichment for regulatory activity (Pearson’s correlation = 0.12, P = 2.9E-05; supplementary fig. S12, Supplementary Material online), and this correlation mainly comes from families lacking significant enrichment for activity.
The Context-Specific Contribution of TEs
Our results establish that context-specific enhancers are enriched for TEs across nearly all contexts (fig. 5)—a pattern previously observed in a handful of tissues (Huda etal. 2011; Xie etal. 2013). CAGE-based identification of eRNA has been previously demonstrated to be a robust predictor of cell-specific enhancer activity (Andersson etal. 2014), so these context-specific enhancer patterns are unlikely to be due to false positives. Furthermore, our results suggest that context-specific enhancer activity may be connected to the age of sequences that become enhancers, as well as TE origins. Enhancers overlapping young TEs are more likely to be context-specific than those overlapping ancient TEs. This trend also holds for enhancers formed from young DNA not derived from a TE. The increased contribution of TEs to context-specific gene regulation could be driven by the fact that most young DNA is derived from TE sequence (Emera etal. 2016). Thus, if enhancers derived from young DNA—regardless of TE content—are more likely to be context-specific and most young DNA is comprised of TE-derived sequence, TEs should make a strong contribution to context-specific gene regulation. This theory is supported by a recent study of primate liver enhancers, which showed that species-specific liver enhancers were more likely to be context-specific and to overlap recently integrated TEs (Trizzino etal. 2016). Despite this strong contribution, the lack of enrichment for young (i.e., more lineage-specific) TEs in any of the contexts tested suggests that the exaptation of young TEs into regulatory elements is relatively rare compared with the number that have integrated into the genome.
The Contribution of TEs to Gene Regulatory Evolution in Rapidly Evolving Tissues
Because co-option of TEs into enhancers or promoters has the potential to change the expression of many genes at once and thus accelerate regulatory evolution, TEs have garnered particular attention in tissues that are rapidly evolving, such as those involved in reproduction (Lynch etal. 2011, 2015; Chuong 2013; Chuong etal. 2013) and the immune system (Flajnik and Kasahara 2010; Chuong etal. 2016a). Our data set includes multiple immunological cell lines, whole blood, placenta, and several cell lines and primary tissue samples from male and female reproductive organs. This allowed us to examine whether tissues undergoing rapid phenotypic changes substantially differ from others, either in number or origins of enhancers created by TEs.
Testis displays a unique regulatory TE signature, mostly driven by tissue-specific enhancers created from young ERV subfamilies, and both testis and placenta have very strong enrichments for TEs among their context-specific enhancers (q < 0.01, FDR-controlled hypergeometric test). However, other reproductive contexts do not show clear enrichment for specific TE families or ages. Immunological contexts are particularly enriched for mammalian-originating TEs, and several of these are also enriched for young ERV subfamilies. Additionally, enhancers specific to whole blood, which is comprised of many immune cell types, are very strongly enriched for TEs (q < 0.01, FDR-controlled hypergeometric test). These results suggest that some rapidly evolving tissues may be more likely to co-opt TEs into regulatory elements than other tissues; however, these effects are not strong enough to overcome the overall TE depletion in their regulatory regions. It is possible that similar effects were missed in some rapidly evolving tissues due to the fact that the active enhancer sets are not available at all relevant physiological time points (e.g., different stages of pregnancy in female reproductive tissues).
These analyses have shown ERVs to be particularly represented in enhancers. Several features of ERVs contribute to their potential for obtaining regulatory function: they frequently undergo partial deletion that removes the internal genes required for retrotransposition, but leaves the long terminal repeat ends intact (Thompson etal. 2016). These long terminal repeats typically contain many TFBS, which enrich ERVs for combinatorial TF binding as seen in ENCODE cell lines (Teng etal. 2014) and placenta (Chuong etal. 2013). Furthermore, ERVs often escape repression in hypo-methylated tissues, such as embryonic stem cells, placenta, and testis, which can result in transcription or enhancer activity (Chuong etal. 2013; Pavlicev etal. 2015; Gerdes etal. 2016). For example, ERVs have been found to be involved in fundamental organismal processes, such as the response to interferon stimulation in multiple cell types (Chuong etal. 2016a). In our data set, we find ERVs to be enriched for enhancer activity in testis, but not placenta. Additionally, most contexts have at least one enhancer that overlaps an ERV, suggesting that ERV enhancer activity is not restricted to rapidly evolving tissues (supplementary file 1, Supplementary Material online). Our results suggest that ERVs exhibit different dynamics of function acquisition compared with other TEs across many contexts, rather than only in a few particular tissues.
Pleiotropic Constraints on Enhancer Function
We observe that once TE-derived sequences gain enhancer activity—either through accrual of gain-of-function mutations or escaping repression by the host—their breadth of activity does not dramatically increase with age or show differences from non-TE enhancers. The median activity of shared TE-containing enhancers is similar to enhancers that do not contain TEs: ∼3 primary tissue contexts. This number is remarkably consistent with the low degree of pleiotropy among many functional genetic elements that has been independently estimated with multiple approaches (Wagner etal. 2008; reviewed in Stearns 2010; Wagner and Zhang 2011). Thus, general evolutionary constraints likely contribute to the observation that TE-containing enhancers do not commonly function in more than a few contexts or show substantial difference in their breadth of activity compared with enhancers not overlapping TEs. As traits become exposed to conflicting selective pressures, coupled variation may constrain adaptive response and divergence of traits.
Model and Conclusions
We find that human and mouse promoters are strikingly depleted of TEs despite the regulatory potential of many TE-derived sequences. Enhancers from both species show a more modest depletion that is consistent across contexts. However, this depletion is influenced by the time that a TE invaded the genome. Age plays a large role in enhancer tissue-specificity and the proportion of TE families that contribute to enhancers, though some TE families, such as ERVs, appear more primed for enhancer activity than others. Enhancers overlapping sequence derived from TEs are not strikingly different from non-TE enhancers, particularly with respect to breadth of activity and the connection between tissue-specificity and age. This suggests that enhancers, regardless of whether the underlying sequence originated from a TE, are subject to similar constraints both when obtaining initial activity, as well as pleiotropic constraints on the breadth of activity.
Based on these observations, we propose the following model of TE co-option into regulatory function. First, a TE integrates into the genome, replicates for some time, and is eventually repressed by the host genome. After mutations have reduced its mutagenic potential and/or increased its regulatory potential, a TE may be co-opted into an enhancer or alternative promoter, though this is quite rare. This new element is likely to be context-specific, but may gain activity in additional contexts based on the regulatory potential of the TE and as permitted by evolutionary constraints. The likelihood of a TE being co-opted in this fashion increases with its age in the genome, likely due to the increased opportunity for beneficial co-option with time. Our results and model provide a framework for evaluating evolutionary hypotheses about the dynamics of TE co-option into regulatory function. Future collection of comprehensive enhancer data sets from diverse species and tissues will enable more precise modeling of the evolutionary and functional dynamics of TE sequences.
Materials and Methods
Genomic Data
All analyses were carried out in the context of the GRCh37/hg19 build of the human genome. Annotations in reference to other builds of the human genome were mapped to hg19 using liftOver from the UCSC Kent tools with default parameters (http://hgdownload.cse.ucsc.edu/admin/jksrc.zip; last accessed August 23, 2017). All comparisons between genomic region sets were performed using the bedtools suite (Quinlan and Hall 2010).
Transcribed Enhancers
The genomic locations (in hg19 coordinates) of transcribed human enhancers defined by CAGE (Andersson etal. 2014) were downloaded from the FANTOM5 Phase 1 release (http://enhancer.binf.ku.dk/presets/; last accessed August 23, 2017). These consisted of 71 “cell facets,” which were derived from cell lines, and 41 “organ facets,” derived from primary tissue samples. For simplicity, we refer to both as “contexts.” The two sets were considered separately in most analyses, but showed similar patterns. We also analyzed mouse enhancers from the FANTOM5 Phase 2 release. However, these were not separated by “facet” of activity, limiting the analyses that we could perform. When directly comparing human and mouse enhancers, we used the human enhancers defined by FANTOM5 Phase 2.
Promoters
We downloaded all Phase 1 promoters identified by the FANTOM5 consortium (http://pressto.binf.ku.dk/about.php; last accessed August 23, 2017); however, promoter activity profiles across cell lines and primary tissues as defined for the enhancers above were not available. We merged promoters on the same strand that fell within 100 bp of one another. This reduced the number of human promoters from 182,476 to 113,916. We took gene annotations from Ensembl Genes 82 and GENCODE v23. From the Ensembl set, we considered all possible gene start, transcript start, and transcription start sites (TSS). From GENCODE, we used all transcript starts. We then filtered the promoters to those that were on the same strand and were within 1 kb of an annotated TSS. Results using GENCODE TSSs were similar to Ensembl TSSs, so we report only the Ensembl results here. Although we focus on protein-coding promoters, we also analyzed all promoters identified by CAGE due to the contribution of TEs to lncRNAs demonstrated in a previous study (Kapusta etal. 2013). This larger set of promoters was also depleted of TEs (18.2% overlap; P < 0.001). For the mouse FANTOM5 Phase 2 analyses, mouse promoter data had been previously intersected with Ensembl annotations. We considered all promoters with any of the annotations: “TSSregion500, S,” “TSS, S,” or “upstream1000, S.” We then merged those that were within 100 bp of each other, regardless of annotation.
Transposable Elements
Transposable element genomic locations were retrieved from RepeatMasker v4.0.5 (Smit etal. 2013–2015). The clades in which each TE is present were taken from Dfam v1.4 (Wheeler etal. 2013). In situations where Dfam provided multiple clades, the most recent shared branch was designated as the origin. As there were few identifiable TEs with origins before the last common ancestor of amniotes, we collapsed all TEs originating in the last common ancestor of Amniota or before into one category. For the FANTOM Phase 2 analyses, we used Dfam v2.0.
Histone-Mark-Defined Enhancers
Liver enhancers defined via genome-wide profiling of H3K27ac and H3K4me3 histone marks were downloaded from the supplementary material of Villar etal. (2015). dESC histone marks were collected and processed as described in Lynch etal. (2015). We defined the other eight sets of enhancers based on histone modification data from the Roadmap Epigenomics consortium for: monocytes, neuronal progenitor, temporal lobe, trophoblast stem cell, B cell, lung, natural killer cells, and placenta (Bernstein etal. 2010; Kundaje etal. 2015). We downloaded gapped ChIP-seq peaks for both H3K27ac and H3K4me3 for all contexts. To exclude potential promoters in our enhancer sets, we removed H3K27ac peaks that overlapped H3K4me3 peaks. These “H3K27ac-only” peaks comprised our enhancer sets.
Creation of Random Sets and Significance Testing
We used shuffleBed (Quinlan and Hall 2010) to shuffle enhancer and promoter regions around the genome. We constrained the shuffled regions to the chromosome of the corresponding observed region and did not allow shuffled regions overlap one another, gaps in the genome assembly, or ENCODE blacklist regions (Bernstein etal. 2012). For the transcribed enhancers, we created 10,000 sets of shuffled regions. For the CAGE-defined promoters and histone-defined and mouse enhancers, we created 1,000 sets of shuffled regions for each set. We calculated the permutation-based P value for each subfamily for all TEs by calculating the number of permuted sets that overlapped more or the same number of TEs in a set of interest, for example, a subfamily. Tests were only performed if at least ten enhancers overlapped a TE in the set of interest. Given the overall depletion of TEs, we additionally compared the distribution of the ages of TE-containing enhancers to the randomized sets. These analyses compared the proportion of TEs originating on a given lineage to the average proportion of TEs originating on that lineage by the permuted sets. To account for multiple testing here and in all other relevant analyses, we controlled the FDR and report q values (Storey and Tibshirani 2003; Bass etal. 2015) for all contexts within subfamily, family, and lineage. Corrections were applied to histone-mark-defined enhancer sets and transcribed enhancer contexts separately.
Context-Specific Enhancer TE Enrichment
For the analyses of context-specific enhancers, we identified all enhancers active in a single cell line or primary tissue context. We defined context-specific enhancers separately for cell lines and primary tissues, since many of the cell lines were originally collected from a primary tissue type examined. For example, we would expect enhancers found in the neuron (cell line, CL) context would likely be found in the brain (primary tissue, PT) context and vice versa. We tested the enrichment of TEs within context-specific enhancers with a one-tailed hypergeometric test.
To compare the tendency toward context-specific activity of young and old TEs, we collapsed TEs originating in Catarrhini, Hominoidea, Hominidae, Homininae, and human into the “young TE” category. We performed a two-sided Fisher’s exact test comparing the number of ancient shared (PT: 94, CL: 289), ancient specific (PT: 82, CL: 97), young shared (PT: 43, CL: 124), and young specific (PT: 154, CL: 79). To determine if there was a difference between the breadth of activity of shared enhancers overlapping either ancient or young TEs, we collapsed enhancers overlapping any of the young TEs described earlier and performed a Kruskal–Wallis test comparing the activity of these two groups of enhancers. We performed this separately for enhancers active in primary tissues and cell lines.
Dating the Origins of Non-TE-Derived Sequences
Following a recent approach (Emera etal. 2016), we assigned ages to genomic regions based on the presence/absence of homologous regions across species from the 46-way MultiZ multiple sequence alignment from the UCSC genome browser. Enhancers not containing a TE were required to overlap a genomic segment with an assigned age by at least 10 bp. If an enhancer overlapped multiple segments of different ages, it was assigned the oldest age. For consistency with our Dfam labels, we examined the species they used for aging and categorized their “primate” designation as “Simiiformes” and their “ape” designation as “Hominidae.” We also collapsed their older age designations into the single “Amniota or before.”
TE Sequence Divergence
RepeatMasker v4.0.5 (Smit etal. 2013–2015) quantifies the sequence divergence of each TE instance from the consensus TE model. Briefly, this is the percent of bases that diverge from the consensus for each TE. We compared the average divergence of all nonenhancer TE members to the average divergence of enhancer-TE members over all TE subfamilies (supplementary fig. S10, Supplementary Material online). Only subfamilies that have at least one enhancer TE and five nonenhancer TEs are plotted. We also calculated the log2 of the ratio of the divergence of each enhancer TE to the average divergence of all TE subfamily instances that did not overlap an enhancer. The ratio of divergence for all enhancer TEs is plotted in supplementary figure S11, Supplementary Material online.
Alignability
We downloaded the alignablility of all 24-mers (wgEncodeMapability track) across the hg19 build of the human genome (Derrien etal. 2012) from the UCSC genome browser. We then used the bigWigAverageOverBed tool to calculate the average alignability across all TEs in the human genome as well as all human CAGE-defined enhancers examined in this study. We then correlated the minimum enrichment (−log10(P)) for each TE subfamily over all contexts with alignability (supplementary fig. S12, Supplementary Material online).
Transcription Factor Motif Analysis
We identified occurrences of TF motifs from the JASPAR 2016 vertebrate database (Mathelier etal. 2016) in sequences of interest using FIMO (Grant etal. 2011) with the default settings. We only considered TF motif matches with a q value < 0.1. We then calculated the number of motifs belonging to unique TFs for each enhancer. For enhancers overlapping TEs originating in Amniota and in Catarrhini and younger (see above), we calculated enrichment using a one-sided binomial test (supplementary file 1, Supplementary Material online). After identifying TF motifs that were enriched in either age set, we used ProteinHistorian (Capra etal. 2012) to identify the ages of these TFs and compare the age distributions of those significantly enriched (P < 1E-03; one-sided binomial test) in either ancient or young TEs.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Laura Colbran for her assistance with the transcription factor motif analysis. This work was supported by the National Institutes of Health (T32 EY021453 to C.N.S. and R01 GM115836 to J.A.C.) and the March of Dimes (Ohio Collaborative Innovation Catalyst Award to J.A.C.). M.P. is supported by the March of Dimes Prematurity Research Center Ohio Collaborative (#22-FY14-470).
References
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T.. 2014. An atlas of active enhancers across human cell types and tissues. Nature 5077493: 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averof M, Patel NH.. 1997. Crustacean appendage evolution associated with changes in Hox gene expression. Nature 3886643: 682–686. [DOI] [PubMed] [Google Scholar]
- Bass A, Storey J, Dabney A, Robinson D.. 2015. qvalue: Q-value estimation for false discovery rate control. Available from: http://github.com/jdstorey/qvalue
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M.. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, et al. 2010. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 28: 1045–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra JA, Williams AG, Pollard KS, Prlic A.. 2012. ProteinHistorian: tools for the comparative analysis of eukaryote protein origin. PLoS Comput Biol. 8: e1002567.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB. 2013. Retroviruses facilitate the rapid evolution of the mammalian placenta. BioEssays 3510: 853–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C.. 2016a. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science (80-.) 351: 1083–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C.. 2016b. Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet. 18: 71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Rumi MAK, Soares MJ, Baker JC.. 2013. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat Genet. 45: 325–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn MJ, Tickle C.. 1999. Developmental basis of limblessness and axial patterning in snakes. Nature 3996735: 474–479. [DOI] [PubMed] [Google Scholar]
- del Rosario RCH, Rayan NA, Prabhakar S.. 2014. Noncoding origins of anthropoid traits and a new null model of transposon functionalization. Genome Res. 24: 1469–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Estellé J, Marco Sola S, Knowles DG, Raineri E, Guigó R, Ribeca P, Ouzounis CA.. 2012. Fast computation and applications of genome mappability. PLoS One 71: e30377.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emera D, Casola C, Lynch VJ, Wildman DE, Agnew D, Wagner GP.. 2012. Convergent evolution of endometrial prolactin expression in primates, mice, and elephants through the independent recruitment of transposable elements. Mol Biol Evol. 29: 239–247. [DOI] [PubMed] [Google Scholar]
- Emera D, Yin J, Reilly SK, Gockley J, Noonan JP.. 2016. Origin and evolution of developmental enhancers in the mammalian neocortex. Proc Natl Acad Sci USA. 11319: E2617–E2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al. 2009. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 41: 563–571. [DOI] [PubMed] [Google Scholar]
- Flajnik MF, Kasahara M.. 2010. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet. 11: 47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes P, Richardson SR, Mager DL, Faulkner GJ.. 2016. Transposable elements in the mammalian embryo: pioneers surviving through stealth and service. Genome Biol. 17: 100.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinsky GV. 2015. Transposable elements and DNA methylation create in embryonic stem cells human-specific regulatory sequences associated with distal enhancers and noncoding RNAs. Genome Biol Evol. 7: 1432–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez NC, Hepperla AJ, Dumitru R, Simon JM, Fang F, Davis IJ.. 2016. Widespread chromatin accessibility at repetitive elements links stem cells with human cancer. Cell Rep. 176: 1607–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS.. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 277: 1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huda A, Mariño-Ramírez L, Jordan IK.. 2010. Epigenetic histone modifications of human transposable elements: genome defense versus exaptation. Mob DNA. 11: 2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huda A, Tyagi E, Mariño-Ramírez L, Bowen NJ, Jjingo D, Jordan IK, Dalal Y.. 2011. Prediction of transposable element derived enhancers using chromatin modification profiles. PLoS One 611: e27513.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques P-É, Jeyakani J, Bourque G.. 2013. The majority of primate-specific regulatory sequences are derived from transposable elements. PLoS Genet. 9: e1003504.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay LA, Bourque G, Yandell M, Feschotte C.. 2013. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet. 9: e1003470.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 5187539: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, et al. 2011. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 4787370: 476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe CB, Haussler D, Yu J-KS.. 2012. 29 mammalian genomes reveal novel exaptations of mobile elements for likely regulatory functions in the human genome. PLoS One 78: e43128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch VJ, Leclerc RD, May G, Wagner GP.. 2011. Transposon-mediated rewiring of gene regulatory networks contributed to the evolution of pregnancy in mammals. Nat Genet. 43: 1154–1159. [DOI] [PubMed] [Google Scholar]
- Lynch VJ, Nnamani MC, Kapusta A, Brayer K, Plaza SL, Mazur EC, Emera D, Sheikh SZ, Grützner F, Bauersachs S, et al. 2015. Ancient transposable elements transformed the uterine regulatory landscape and transcriptome during the evolution of mammalian pregnancy. Cell Rep. 104: 551–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelier A, Fornes O, Arenillas DJ, Chen CY, Denay G, Lee J, Shi W, Shyr C, Tan G, Worsley-Hunt R, et al. 2016. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 44(D1): D110–D115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notwell JH, Chung T, Heavner W, Bejerano G.. 2015. A family of transposable elements co-opted into developmental enhancers in the mouse neocortex. Nat Commun. 6: 6644.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlicev M, Hiratsuka K, Swaggart KA, Dunn C, Muglia L.. 2015. Detecting endogenous retrovirus-driven tissue-specific gene transcription. Genome Biol Evol. 7: 1082–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo R, Romanish MT, Mager DL.. 2011. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet. 46: 21–42. [DOI] [PubMed] [Google Scholar]
- Smit AF. 1999. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 9: 657–663. [DOI] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P. RepeatMasker Open-4.0. 2013–2015. Available from: http://www.repeatmasker.org
- Stearns FW. 2010. One hundred years of pleiotropy: a retrospective. Genetics 186: 767–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R.. 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 10016: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana T, Zamborlini A, Cristofari G, Lesage P.. 2017. Integration site selection by retroviruses and transposable elements in eukaryotes. Nat Rev Genet. 18: 292–308. [DOI] [PubMed] [Google Scholar]
- Sundaram V, Cheng Y, Ma Z, Li D, Xing X, Edge P, Snyder MP, Wang T.. 2014. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 2412: 1963–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng L, He B, Gao P, Gao L, Tan K.. 2014. Discover context-specific combinatorial transcription factor interactions by integrating diverse ChIP-Seq data sets. Nucleic Acids Res. 42: e24.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PJ, Macfarlan TS, Lorincz MC.. 2016. Long terminal repeats: from parasitic elements to building blocks of the transcriptional regulatory repertoire. Mol Cell. 625: 766–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trizzino M, Park Y, Holsbach-Beltrame M, Aracena K, Mika K, Caliskan M, Perry GH, Lynch VJ, Brown CD.. 2016. Transposable element exaptation is the primary source of novelty in the primate gene regulatory landscape. bioRxiv 83980. Available from: http://www.biorxiv.org/content/early/2016/10/27/083980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar D, Berthelot C, Aldridge S, Rayner TF, Lukk M, Pignatelli M, Park TJ, Deaville R, Erichsen JT, Jasinska AJ, et al. 2015. Enhancer evolution across 20 mammalian species. Cell 1603: 554–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP, Kenney-Hunt JP, Pavlicev M, Peck JR, Waxman D, Cheverud JM.. 2008. Pleiotropic scaling of gene effects and the “cost of complexity.” Nature 4527186: 470–472. [DOI] [PubMed] [Google Scholar]
- Wagner GP, Zhang J.. 2011. The pleiotropic structure of the genotype-phenotype map: the evolvability of complex organisms. Nat Rev Genet. 12: 204–213. [DOI] [PubMed] [Google Scholar]
- Wang J, Vicente-García C, Seruggia D, Moltó E, Fernandez-Miñán A, Neto A, Lee E, Gómez-Skarmeta JL, Montoliu L, Lunyak VV, et al. 2015. MIR retrotransposon sequences provide insulators to the human genome. Proc Natl Acad Sci USA. 11232: E4428–E4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TJ, Clements J, Eddy SR, Hubley R, Jones TA, Jurka J, Smit AFA, Finn RD.. 2013. Dfam: a database of repetitive DNA based on profile hidden Markov models. Nucleic Acids Res. 41(D1): D70–D82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MD, Barbosa-Morais NL, Schmidt D, Conboy CM, Vanes L, Tybulewicz VLJ, Fisher EMC, Tavaré S, Odom DT.. 2008. Species-specific transcription in mice carrying human chromosome 21. Science 3225900: 434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Hong C, Zhang B, Lowdon RF, Xing X, Li D, Zhou X, Lee HJ, Maire CL, Ligon KL, et al. 2013. DNA hypomethylation within specific transposable element families associates with tissue-specific enhancer landscape. Nat Genet. 45: 836–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.