Abstract
STUDY QUESTION
Does selection for mtDNA mutations occur in human oocytes?
SUMMARY ANSWER
We provide statistical evidence in favor of the existence of purifying selection for mtDNA mutations in human oocytes acting between the expulsion of the first and second polar bodies (PBs).
WHAT IS KNOWN ALREADY
Several lines of evidence in Metazoa, including humans, indicate that variation within the germline of mitochondrial genomes is under purifying selection. The presence of this internal selection filter in the germline has important consequences for the evolutionary trajectory of mtDNA. However, the nature and localization of this internal filter are still unclear while several hypotheses are proposed in the literature.
STUDY DESIGN, SIZE, DURATION
In this study, 60 mitochondrial genomes were sequenced from 17 sets of oocytes, first and second PBs, and peripheral blood taken from nine women between 38 and 43 years of age.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Whole genome amplification was performed only on the single cell samples and Sanger sequencing was performed on amplicons. The comparison of variant profiles between first and second PB sequences showed no difference in substitution rates but displayed instead a sharp difference in pathogenicity scores of protein-coding sequences using three different metrics (MutPred, Polyphen and SNPs&GO).
MAIN RESULTS AND THE ROLE OF CHANCE
Unlike the first, second PBs showed no significant differences in pathogenic scores with blood and oocyte sequences. This suggests that a filtering mechanism for disadvantageous variants operates during oocyte development between the expulsion of the first and second PB.
LARGE SCALE DATA
N/A.
LIMITATIONS, REASONS FOR CAUTION
The sample size is small and further studies are needed before this approach can be used in clinical practice. Studies on a model organism would allow the sample size to be increased.
WIDER IMPLICATIONS OF THE FINDINGS
This work opens the way to the study of the correlation between mtDNA mutations, mitochondrial capacity and viability of oocytes.
STUDY FUNDING/COMPETING INTEREST(S)
This work was supported by a SISMER grant. Laboratory facilities and skills were freely provided by SISMER, and by the Alma Mater Studiorum, University of Bologna. The authors have no conflict of interest to disclose.
Keywords: purifying selection, mtDNA variants, human oocytes, first and second polar body, heteroplasmy
Introduction
Whole Genome Sequencing of the polar bodies (PBs) originating from a single human oocyte makes it possible to reconstruct the entire nuclear haploid maternal genome and therefore to detect maternal Mendelian disease-associated alleles in fertilized eggs (Hou et al., 2013). By contrast, previous attempts to detect mitochondrial disease-associated variants in oocytes from sequences extracted from first PBs were not successful (Gigarel et al., 2011). This discrepancy is due to the polyploidy of the mitochondrial genome, which is highly variable in the female germline, containing around 200 copies per cell in primordial germ cells and up to 200 000 copies in mature oocytes (White et al., 2008). Such large variation in the number of mtDNA copies generates a bottleneck, which together with the high substitution rate typical of mtDNA makes it possible that all the phenomena observed in population genetics (founder effect, drift, selection, etc.) may occur during the maturation of oocytes and later during embryo development. The mitochondrion is the crucial source of energy in all cells, which is particularly relevant during oogenesis and embryogenesis. In mice, mature oocytes can still be fertilized when they contain only 4000 copies of the mitochondrial genome per cell, but successful embryo implantation into the uterine wall requires at least 40 000 copies of mtDNA per cell (Steuerwald et al., 2000; Wai et al., 2010). The high mtDNA copy number in mature oocytes guarantees distribution of mitochondrial genomes to all cell progeny during preimplantation embryo development when mtDNA replication is stopped.
Heteroplasmy is the coexistence of two or more variants in the multi-copy mitochondrial population of a single cell or of a tissue. Homoplasmy, where all mtDNAs are identical, was previously considered to be the biological norm. However, Next Generation Sequencing approaches have challenged this dogma because heteroplasmy in humans appears to be more common than previously thought (Payne et al., 2013).
It is controversial whether the mitochondrial genome is able to preserve correctly genetic information over evolutionary time. Several factors oppose the idea that mtDNA is an efficient archive of genetic information, such as the smaller effective population size than the nucleus at the individual level, the greater mutation load documented by higher rate of substitutions in protein-coding synonymous positions (Pesole et al., 1999) and the lack of recombination (Hagström et al., 2014). Given this background, intra-individual selective pressure may play an important role in enabling the mitochondrial genome to preserve information. For example, this seems to occur during the shift from heteroplasmy to homoplasmy of inherited and somatic mtDNA mutations affecting the function of respiratory complex I in oncocytic tumors (Gasparre et al., 2009, 2013). On the other hand, the existence of an ovarian selection system that would eliminate deleterious mtDNA mutations during oocyte maturation is still debated (Wallace, 2013). In Drosophila melanogaster, there is consistent evidence of purifying selection of mtDNA mutations (Ma et al., 2014) and of positive selection of healthy mitochondria (Hill et al., 2014). Other lines of evidence indicate that mtDNA is a place where new adaptive mutations appear (Castellana et al., 2011). Direct proof of the existence of a selective mechanism in mammals was found in the mouse female lineage of a model containing a proofreading-deficient catalytic subunit of mitochondrial DNA polymerase (encoded by nuclear DNA and causing multiple mtDNA mutations) that was repeatedly crossed with wild-type males (Stewart et al., 2008). Starting with the second crossing, the offspring purged the mtDNA mutations from mitochondria and recovered a stable level of variation in the sixth generation. This fast recovery in an experimental setting that showed no differential reproductive capacity can only be explained by internal selection. On the other hand, there are repeated reports of mitochondrial pathogenic variants increasing in frequency in humans (McFarland et al., 2002).
Two possible mechanisms have been proposed in the past for the intra-individual selection of mtDNA molecules: selective follicular atresia (Krakauer and Mira, 1999; Chu et al., 2014) and selective aggregation in the Balbiani bodies (i.e. the mitochondrial cloud in Zhou et al., 2010). Both mechanisms are associated with the previously cited mitochondrial bottleneck phenomenon (Rand, 2008; Zhou et al., 2010).
In the present work, which follows our preliminary validation of the sequencing method (Gianaroli et al., 2014), we report the distribution of mutations and the in silico evaluation of their pathogenic effect within 17 human oocytes, their corresponding PBs and blood samples from nine donors. We provide evidence in favor of the existence of purifying selection for mtDNA mutations in human oocytes acting between the expulsion of the first and second PBs, thus further defining the initial observation of Gigarel et al. (2011).
Materials and Methods
Ethics statement
The study was approved by Società Italiana Studi di Medicina della Riproduzione (S.I.S.Me.R.) internal Institutional Review Board and by the regional competent Ethical Committee (Comitato Etico Indipendente AUSL di Bologna, E.R. Region Prot. N. 752/CE, Cod. CE: 11033). All patients signed a specific informed consent that had been approved by the Ethical Committee.
DNA extraction and sequencing
The DNA was extracted from 17 triads, that is oocyte, polar body 1 (PB1) and polar body 2 (PB2) derived from oocytes generated by nine different women with age ranging from 38 to 43 years. Only ICSI cycles with ejaculated spermatozoa were included in this study. There were no restrictions with respect to maternal age, the couple's reproductive history or the number of retrieved or fertilized oocytes.
Mitogenomes were amplified by whole genome amplification (WGA) as previously described (Gianaroli et al., 2015). Blood DNA from every donor woman was extracted from 2 ml of whole blood using five PRIME ArchivePure DNA Blood Kit (Eppendorf s.r.l., Gaithersburg, MD, USA). In total, 60 DNA samples were collected and analyzed. WGA DNA from PBs and oocytes was diluted 1:10 and 1 µl was used as template for each reaction to amplify whole mtDNA with the MitoAll kit (Applied Biosystems, Foster City, CA, USA) (Gasparre et al., 2007; Gianaroli et al., 2015). PCR reaction analysis, purification and sequencing were performed as previously described (Gianaroli et al., 2015). For mtDNA regions that failed to amplify or to produce base call of good quality, PCR and sequencing reactions were repeated twice. Electropherograms were visually inspected and aligned to the human mitochondrial reference sequence (NC_012920) using Sequencher 4.10.4 (Genecodes, Ann Arbor, MI, USA).
Preliminary work carried out on the sequences of mtDNA obtained from blood, oocytes, PB1 and PB2 of three donors using the Sanger method showed this to be reliable and free of possible artifacts introduced at random by the amplification and/or sequencing procedures. These random changes would make the mtDNA sequences incompatible with the haplogroups of the donors, as already described (Gianaroli et al., 2015).
Mapping and pathogenicity evaluation
The mtDNA was analyzed by applying the mt-classifier.py module within the MToolBox pipeline (Calabrese et al., 2014) to assign the mitochondrial haplogroup and to report the functional annotation of the variants detected.
The procedure was based on the steps described below:
Haplogroup assignment of the completely assembled or fragmented genome relying on the Reconstructed Sapiens Reference Sequence (RSRS)-based Phylotree resource (van Oven and Kayser, 2009; Behar et al., 2012). The output files here obtained were the reconstructed contig sequence(s) (Contigs.fa) and the list of the best predicted haplogroups. In more fragmented genomes, the number of best predicted haplogroups increases due to missing haplogroup predicting sites.
Functional annotation of the recognized variants. This analysis was carried out by aligning each sample-specific reconstructed contig against the related Macro-Haplogroup-specific Consensus Sequence (MHCS), see Supplementary Information in Calabrese et al. (2014) to recognize private variants via a prioritization process. All variant nucleotide positions and related loci as well as their corresponding codon positions, possible amino acid changes, stop-gains and stop-losses in protein-coding genes were reported in the final annotation file. The process to prioritize the most important candidate variants was mainly based on the recognition of alleles that were not shared with MHCS and sorted by increasing nucleotide variability calculated on multi-aligned complete mitochondrial genomes from 14 144 healthy individuals annotated in Human Mitochondrial DataBase (Rubino et al., 2012). Output values ranged from 0 to 1, where low variability values suggest a private variant. Moreover, we selected three methods of pathogenicity predictions for non-synonymous variants over the seven proposed by MToolbox in order to minimize information redundancy: MutPred (Li et al., 2009), which is based on predicted gain and loss of structural and functional protein property/sites, respectively; PolyPhen-2 HumVar (Adzhubei et al., 2010), which represents human population variations, and SNPs&GO (Calabrese et al., 2009), representing protein profile weighted with Gene Ontology associations. In addition, base changes were compared to MITOMAP (Ruiz-Pesini et al., 2007) annotations related to disease-associated mutations, occurring in coding and control regions and somatic mutations, together with their state of homoplasmy/heteroplasmy previously reported. Furthermore, links to OMIM (http://omim.org) and Mamit-tRNA (Helm et al., 2000) annotations were added to each somatic mutation, when applicable.
Comparison of scores across samples
We analyzed genome variability both in a multi-alignment and in a pairwise fashion. Although multi-alignment is biologically more sound and statistically more powerful, it becomes problematic when some samples (in our case PB genomes) systematically showed more missing data than others.
In the first approach for each of the 17 alignments of individual mitogenomes (blood, oocyte and two PBs), we selected those variable sites that showed no missing data or ambiguous data which would prevent the assignment of the variant to a given branch leading to a cell type. More specifically, we selected sites that had all bases identical but one. We did not try to estimate a tree of mtDNA that could be different from the tree of cell lineage in topology and branch lengths, due to too few variable sites available to assess topology support. We obtained the counts of variants shown in Supplementary data, Table SI. The table is a typical case of sparse matrix with several zero counts, which will not allow to perform a test based on Chi-square distribution (McDonald, 2014). Within a Frequentist approach, the most obvious solution would be a Fisher exact test. But this is a non-optimal solution because the experimental design did not fix the total number of variants observed in each cell type (sum of the columns), as the permutation procedure of the Fisher exact test assumes. The Bayesian approach in this case offers an easier solution thanks to the fact that Dirichlet distribution is the conjugate prior of the multinomial distribution and allows us to calculate posterior distribution in an analytical way. Further, it is possible to combine the results from several tests in a straightforward way with Bayesian factors (BFs), similarly to the G-test, by multiplying the BF scores across the experiment. We therefore performed the test in two ways: summing variant counts per cell type across individuals (the inter-individual approach) and multiplying the BF of the test done in each single cell lineage (intra-individual approach). This allowed to check if the trend within individuals matched the trend across individuals. In greater detail, we estimated the BF between a hypothesis of equal substitution rate versus a hypothesis that assumes different substitution rates between all samples. The test was performed by using an uninformative Dirichelt distribution with all parameters set to 1 as a priori distribution for the test. It is possible to estimate analytically the posterior distribution, simply imposing to the parameters of a Dirichlet distribution the observed counts plus 1. This simple practice, as said before, is possible because Dirichlet distribution is the conjugate prior for the multinomial distribution (Gelman et al., 2003). From the posterior distribution, the BFs were estimated using a Savage–Dickey ratio between the probability of the equal rate hypothesis estimated on posterior and prior of the generic all different rates hypothesis (Verdinelli and Wasserman, 1995). Estimation was done with a random sample from the posterior distribution using R software and the package MCMCpack.
In the pairwise approach, we evaluated the variants resulting in amino acid changes using the pathogenic score discussed above. Each sample was compared with the data sets of any other sample from the same donor with the aim to recognize the cell type-specific variants, disregarding variations across donors. For each comparison, we detected with a python script all sites that were sequenced in both samples and for which a different base was detected. For each of these sites, we calculated the difference in pathogenic score between the two samples with all three methods. The series of differences were tested against the null hypothesis whose values were taken from a population with mean zero, using a single sample T test and a single sample Wilcoxon rank test, both implemented in the R software package stat. With the aim to summarize the results across metrics, the Fisher's method was applied to combine independent P-values (Sokal and Rohlf, 1995) to one metric per family of methods (MutPred, SNPs&GO and PolyPhen-2 HumVar).
To explore the nature of the substitution found in PBs, we mapped them in the 37 different mitochondrial genes (13 of which protein-coding) plus the D-Loop region divided in the three hyper-variable regions and a fourth group that included all remaining sites of the region. The coding region changes were further split in synonymous and non-synonymous sites, accounting for sites where only a subset of variants is non-synonymous. This distribution in PBs was compared with a G-test to three different expectations. The first expectation was the size of each annotation in order to evaluate if the variants were randomly distributed in the genome. The second and the third expectations were the mutation distributions obtained by comparing the different blood samples both in terms of absolute and relative counts, respectively.
Results
Sequencing and haplogroup assignment
Sanger sequencing was performed on a total of 60 mitochondrial genomes consisting of 17 triads each made of the oocyte and the corresponding PB1 and PB2, as well as DNA extracted from blood (Data set SI. Complete FASTA format mitochondrial sequences analyzed in this study. Data set SII. MToolBox pipeline results.). The donors were nine women with ages ranging from 38 to 43. The sequencing coverage were 100% for all the blood samples. Sequencing in oocytes and PBs was partially complete due to the lack of product in some of the amplifications. In particular, the coverage ranged from 80 to 100% for oocytes and from 9 to 95% in the PB samples. Clear double peaks on single sites were manually annotated as heteroplasmic sites following a previously reported protocol which detects heteroplasmic levels higher than 15% (Kurelac et al., 2012). Blood and oocyte samples had no more than three detectable heteroplasmic sites with an average of approximately one, while several samples had none. The PB samples had at least two heteroplasmic sites with an average of approximately five per genome. Details on aneuploidies per chromosome, sequence coverage, haplogroups, number of heteroplasmic sites and number of variants are given in Supplementary data, Table SII.
MToolBox analysis (Calabrese et al., 2014) was performed to determine the mtDNA haplogroups of the sequenced mitogenomes and to annotate functional variants. For incomplete genomes, the haplogroup analysis may give a set of possible haplogroup assignments. In three cases of PBs (W1-PB024, W1-PB026 and W6-PB142), the set did not include the unique assignment of the corresponding blood sample. These three cases can be explained by a lack of diagnostic sites due to incomplete sequencing and by the presence of somatic variants.
Overall, the small sample of nine donors had a good representation of human mtDNA variability having representatives from four out of the eight macro-haplogroups as defined by phylotree (Behar et al., 2012) (see Supplementary data, Tables SI and SIII).
All the detected haplogroups descended from the ancestral macro-haplogroup N including 7 from the macro-haplogroup R (H, J and T—sister haplogroups belonging to the ancestor R*—and K) and two derived from N* (N1b and X). Of the 32 diagnosed PBs, six were euploid (three PB1s and three PB2s) and 26 aneuploid (13 PB1s and 13 PB2s). The incidence of total anomalies (number of aneuploidies per chromosome in all diagnosed PBs) did not differ significantly among the haplogroups (70.8% in haplogroup H and 11.5% in haplogroups J and T). However, the trend was similar to that already reported for a higher number of oocytes and PBs analysed for the D-loop only (Gianaroli et al., 2015) suggesting that the lack of statistically significant difference was due to the limited number of cases analysed in the present study.
Quality and quantity of mtDNA variants across cell types
In order to study the variability of the sequenced genomes and to reveal potential signals for purifying selection, the mitogenomes were analyzed by multi-alignment and pairwise alignment approaches. The results of the multi-alignment approach are shown in Table I (with raw data provided in Supplementary data, Table SI). The BF, which is the ratio of the posterior probability of the two hypotheses under examination, supports the hypothesis that the four cell types accumulate a different amount of type-specific variants (BF = 8.10e−07, 1.15e−07 for inter- and intra-individual, respectively). As expected, between oocyte and blood cell types it was approximately seven times more likely to find unequal accumulation of variants, (2 versus 11 across individuals, respectively; see last row of Supplementary data, Table SI). The observed level of accumulation of variants present in blood should not be seen as a baseline level, caused by an absence of purifying selection. As a matter of fact, the low number of variants detected in blood samples could be due to selection that acts during life thanks to the rapid turnover of this cell type as already reported in previously published works (Larsson et al., 1990; Ciafaloni et al., 1991; Rajasimha et al., 2008; Pallotti et al., 2014; Kauppila et al., 2016).
Table I.
Results of the Bayesian factor (BF) analysis for the multi-alignment approach.
| Comparisons | Inter-individuals | Intra-individuals | ||||
|---|---|---|---|---|---|---|
| BF | Best HP | Support | BF | Best HP | Support | |
| Ovo, Blood | 0.133 | Not equal | Positive | 0.158 | Not equal | Positive |
| PB1, PB2 | 5.03 | Equal | Positive | 9.2e−04 | Not equal | Strong |
| All | 8.10e−07 | Not equal | Very strong | 1.15e−07 | Not equal | Very strong |
Each row represents a comparison in the number of cell type-specific variants. Ovo, Blood, PB1 and PB2 stand for oocyte, blood, first and second polar body (PB), respectively. Each comparison was done in two ways: Inter-individual, where counts for each cell type were added and then tested; Intra-individual, where for each cell lineage the test was performed and then the products of the BF was reported. Only the two PBs comparisons support different best hypothesis across the two approaches. Significance values are reported in Supplementary data, Table SIV.
Interestingly, between PBs, the intra-individual approach indicated unequal accumulation as one thousand times more likely (BF = 9.2e−04), but when looking across individuals the result was inverted, and equal accumulation was five times more likely (BF = 5.03). In other words, while in each individual one of the two PBs accumulates significantly more variants than the other, the slightly higher accumulation of variants in PB2 versus PB1 in the total sample (32 private variants versus 27) is likely due to chance.
The pairwise approach analyzes the type of non-synonymous differences found and results are reported in Table II. This approach, although less powerful, can use also the sites that were sequenced in only two genomes of each triad (blood sequence had full sites coverage). The summary P-values obtained using the Fisher procedure suggest that PB1 have a significantly higher pathogenicity score than the three other cell types, either taking into account the face value of the metrics (t test) or only the relative ranking of differences in scores (Wilcoxon rank-sum test). None of the other differences were significant. Inspecting the contribution of the three metrics does not give a result with an opposite sign. The ‘PolyPhen-2 HumVar’ metric that measures pathogenicity based on segregating frequency of the alleles in healthy human populations, coupled with the Wilcoxon rank did not support any conclusion within a risk threshold of 0.05. This is due to the fact that not all of the non-synonymous changes could be scored with this metric, making it difficult to have a significant difference also in the ranked approach, with generally less power than the t test.
Table II.
Results of the pairwise alignment approach on pathogenicity scores.
| Method | Statistics | Ovo-Blood | PB1-Blood | PB2-Blood | PB1-Ovo | PB2-Ovo | PB2–PB1 |
|---|---|---|---|---|---|---|---|
| Fisher's summary | pvalueT | 0.334 | 6.43e−4 | 0.63 | 9.0e-6 | 0.22 | 1.238e−03 |
| pvalueW | 0.156 | 4.734e−3 | 0.73 | 5.8e-5 | 0.19 | 4.955e−03 | |
| MutPred | Mean | −0.83 | 5.506 | 0.68 | 6.336 | 1.51 | −5.25 |
| pvalueW | 0.644 | 0.022 | 0.711 | 0.0058 | 0.346 | 0.032 | |
| pvalueT | 0.421 | 0.0198 | 0.730 | 0.0029 | 0.394 | 0.0196 | |
| PolyPhen-2 HumVar | Mean | −1.00 | 5.558 | 2.103 | 6.558 | 3.103 | −5.544 |
| pvalueW | 1.00 | 0.122 | 0.475 | 0.0138 | 0.155 | 0.089 | |
| pvalueT | 0.323 | 0.038 | 0.422 | 0.0073 | 0.199 | 0.038 | |
| SNPs&GO | Mean | −0.517 | 4.255 | 1.347 | 4.758 | 1.822 | −3.694 |
| pvalueW | 0.0148 | 0.0329 | 0.484 | 0.006 | 0.230 | 0.032 | |
| pvalueT | 0.238 | 0.0105 | 0.376 | 0.0025 | 0.211 | 0.023 | |
| AA changes | Counts | 10 | 27 | 32 | 17 | 24 | 30 |
The columns indicate the six possible comparisons between the four cell types (Ovo, oocyte; PB, polar body). The Fisher summary reports the Fisher procedure to combine results of the T test (pvalueT) and Wilcoxon rank test (pvalueW) across the three selected metrics, respectively. The mean differences between the two cell type scores have a black or gray background if negative or positive, respectively. P values lower than 0.05 are in italics. On the bottom row, the amino acid changes found in the comparison across cell types on which pathogenetic scores difference were calculated.
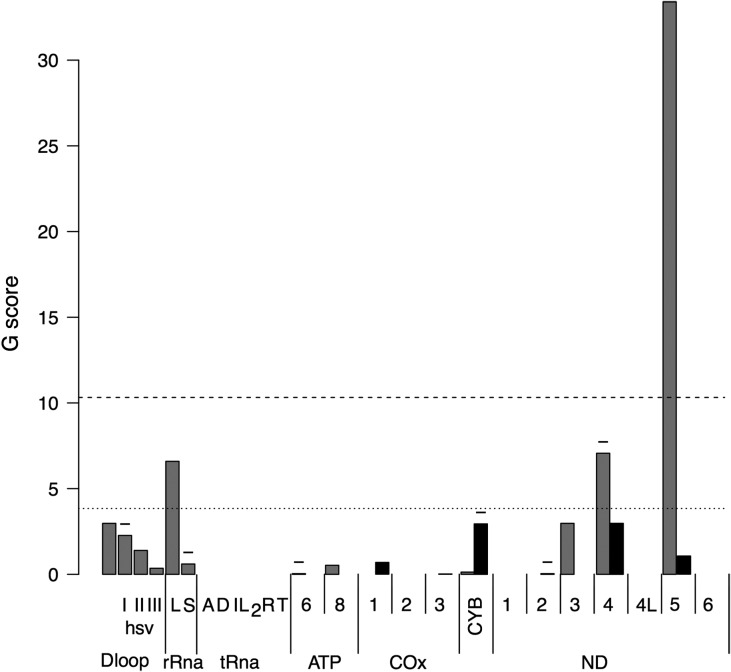
The difference in substitutions found between PBs does not follow the expectation based on annotation size, showing that variants do not seem to occur randomly (Supplementary data, Figs S1 and S2). When the distribution was compared with the absolute rate of substitutions of the blood, all genes for which more than a few counts were observed had a significant accumulation compared to expectation (data not shown). This can be explained with a higher average substitution rate in PBs than in blood. If the relative rates in blood (Fig. 1) are used as reference, only synonymous sites of MT-ND5 have a significant accumulation of substitutions. For the remaining observations, the pattern of substitutions in PBs is comparable with that of blood samples, although with a higher average rate. The MT-ND5 deviation is caused by a compact cluster of recurrent synonymous variants (12684A: 10 times, 12705T: 6 times,12822G: 4 times) that do not appear all at the same time, nor in the same women, nor in the same women at the same time. In fact, 12684A appears in five out of nine women, recurring five times in donor W3 (one of them together with 12705T, the rest alone) and two times in donor W8 accompanied once with 12705T, 12977A, 12612A and the other time only with 12705T and 12612A, even though both PBs have the polymorphic position 12 977 sequenced in this cell lineage. Although these four sites are sequenced on the same amplicon, the fact that no fixed haplotype pattern can be seen in the data and that no non-synonymous sites are involved make the possibility of nuclear pseudogene (called NUMT) (Lopez et al., 1994) amplification unlikely. Moreover, a control experiment showed that this amplicon does not amplify on mtDNA free Rho0 cells using the same PCR conditions, thus excluding non-specific ligation of primers to a nuclear pseudogene region. The lack of a consistent haplotype in the same woman leads to the hypothesis that this region is a mutational hotspot. This high rate of synonymous MT-ND5 variants are not totally a surprise given the high rate of heteroplasmic alleles in MT-ND5 also found in blood samples (Just et al., 2015) and in oncocytic tumors (Gasparre et al., 2007, 2013).
Figure 1.
Polar bodies (PBs) have the same relative variant profile as blood with the exception of ND5. G-test statistics for each mitochondrial gene of PB changes using as expectation the relative change distribution among blood samples. Dashed line indicates 5% risk level with a Bonferroni correction, while dotted line indicates an uncorrected 5% risk level. Negative signs on top of bars indicate that observed values were lower than expected. Gray bar indicates noncoding or synonymous changes, while black bars indicate non-synonymous changes. The name of the genomic region follows the names used on accession NC_012920.1, with the only exception of HSV-I, HSV-II, HSV-III and D-Loop that split the original region annotated as D-Loop in the three hyper-variable regions and the remnant of the D-Loop, respectively.
Discussion
Previous studies have shown that the quality of sequences of nuclear genomes in PBs is comparable to that of oocytes (Hou et al., 2013; Wang et al., 2014; Gianaroli et al., 2015). We can assume that if no degradation pathway acts on nuclear genomes, this may also be true for mtDNA. Consistent with the hypothesis that no preferential degradation is acting on second PBs, we did not observe significant differences in quality (pathogenicity scores) between second PBs and blood or oocyte sequences. But, as shown in Table I, more variants accumulate in PBs than in oocytes or blood. This pattern might be explained by the different amount of molecules found in the different cell types. Oocyte and blood sequences originate from tens of thousands of molecules, while the sequences obtained from PBs originate from a smaller number of molecules (probably a few hundred as in mice as reported in Wang et al., 2014). In mature oocyte sequences, most variants that are present during oogenesis are undetectable because they are present at too low frequencies, while in PBs the small number of total molecules allows the detection of variants present within a few mtDNA molecules as heteroplasmic sites. In fact, heteroplasmic sites are much more abundant in PBs with respect to oocytes and blood (see for details Supplementary data, Table SII).
However, the pathogenicity scores of substitutions are higher in first PBs than in other analyzed cell types. The fact that the number of variants are similar in the two PBs makes it unlikely that the higher pathogenicity of such mutations within PB1 is due to a differential degeneration process. Instead, the increase in quality (or decreased pathogenicity level of substitutions) of mtDNA between first and second PBs and the similar quality between second PB and mature oocyte can be explained by a process of selection (see Fig. 2). This selection process would operate between the expulsions of the first and second PB. Because this process is intracellular, it does not fit the selective follicular atresia hypothesis. The process of aggregation within the mitochondrial cloud seems to be active throughout oogenesis making it compatible with our observations from the perspective of time frame and location. Yet, to achieve an increase in mtDNA quality across PBs, defective molecules need to be actively destroyed or taken out from the cytoplasm. The mitochondrial cloud hypothesis, however, would guarantee that only the best molecules will be used for the next generation germline. In that respect, our postulated selective process does not identify with the mitochondrial cloud either. Finally, it is interesting to note that neither mechanism is in line with the results described by Stewart et al. (2008) in mice, where effects are visible in the soma already in the second generation and therefore need to be active on the germline since the first generation.
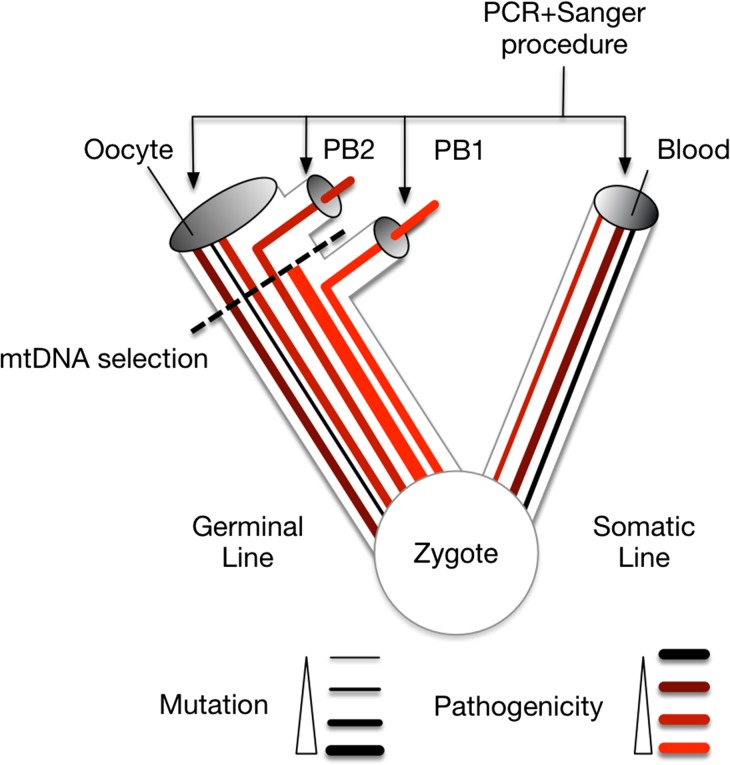
Figure 2.
Schematic interpretation of results. The different cell lines that lead to Oocyte (Ovo) and Blood have both, since their start, consistent amount of heteroplasmy. The different mitochondrial molecules have different number of substitutions (different line thickness) and level of functionality (high pathogenic scores darker lines). Molecules with high pathogenic scores are not allowed to reach mature oocyte by some selection mechanism (dashed line). Sequencing hide low heteroplasmy level per site in tissues with high molecular counts (gray disk).
Two possible scenarios can be suggested to explain the proposed selective mechanism: (i) defective mtDNA molecules are actively accumulated within the first PB; (ii) defective mtDNA molecules are destroyed within the cytoplasm (Gilkerson et al., 2012); or alternatively, more efficient mitochondria duplicate preferentially. It is unlikely that first PBs would represent the final destination of defective molecules given their low mtDNA content. Instead, it is more likely that both PBs represent a random sample of the cytoplasm periphery of the oocyte.
The difference in pathogenicity scores between the two PBs could then be explained as the difference between two random samples taken at two different times from the same population of mtDNA under selection, as assumed in the second scenario. A hypothetical experiment that could analyze a temporal series of cytoplasm samples corresponding in time to the expulsion of first and second PBs in a maturing oocyte would then result in a gradual decrease of the pathogenetic score of the sampled molecules (Stewart et al., 2008). This hypothesis suggests a gradual purifying mechanism, instead of a single step event, as in the first proposed scenario. Yet, the nature of the mechanism underlying this selection process remains to be elucidated and represents the crucial question in the evolution of mtDNA in our species (Wallace, 2015).
These results have clear consequences on the practice of preimplantation diagnostics. In fact, while first and second PBs are both good predictors of the nuclear genetic makeup of the oocyte, for mitochondrial genomes only PB2 seems to be predictive of the genetic status of the oocyte. This distinction might clarify the reason of the discrepancy in results reported by Gigarel et al. (2011), who analyzed PB1 as well as oocytes, blastomeres or whole embryos, and those of Sato et al. (2005) on PB2 in mice. These latter differences might not be caused by species differences but by cell type differences.
Supplementary Material
Acknowledgements
We would like to thank Dr Marcella Attimonelli who put S.V. and D.S. in contact with the rest of the authors and participated in the initial discussion to frame the data analysis strategy of this paper. We thank the NIH Fellows Editorial Board for editorial assistance.
Supplementary data
Supplementary data are available at Human Reproduction online.
Authors’ roles
G.R. contributed to study conception and design. S.D.F., S.V., D.S., M.L. and D.L. performed the experiments, analyzed and interpreted the data. L.G. and C.M. provided samples, funding and contributed in the interpretation of data.
All authors contributed to the drafting of the manuscript, provided critical discussions and approved the final manuscript.
Funding
Società Italiana Studi di Medicina della Riproduzione (S.I.S.Me.R.) grant; Laboratory facilities and skills were freely provided by SISMER; the Alma Mater Studiorum, University of Bologna.
Conflict of interest
None declared.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar DM, van Oven M, Rosset S, Metspalu M, Loogväli EL, Silva NM, Kivisild T, Torroni A, Villems R. A ‘Copernican’ reassessment of the human mitochondrial DNA tree from its root. Am J Hum Genet 2012;90:675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese C, Simone D, Diroma MA, Santorsola M, Guttà C, Gasparre G, Picardi E, Pesole G, Attimonelli M. MToolBox: a highly automated pipeline for heteroplasmy annotation and prioritization analysis of human mitochondrial variants in high-throughput sequencing. Bioinformatics 2014;30:3115–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese R, Capriotti E, Fariselli P, Martelli PL, Casadio R. Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum Mutat 2009;30:1237–1244. [DOI] [PubMed] [Google Scholar]
- Castellana S, Vicario S, Saccone C. Evolutionary patterns of the mitochondrial genome in Metazoa: exploring the role of mutation and selection in mitochondrial protein coding genes. Genome Biol Evol 2011;3:1067–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu HP, Liao Y, Novak JS, Hu Z, Merkin JJ, Shymkiv Y, Braeckman BP, Dorovkov MV, Nguyen A, Clifford PM et al. . Germline quality control: eEF2K stands guard to eliminate defective oocytes. Dev Cell 2014;28:561–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciafaloni E, Ricci E, Servidei S, Shanske S, Silvestri G, Manfredi G, Schon EA, DiMauro S. Widespread tissue distribution of a tRNALeu(UUR) mutation in the mitochondrial DNA of a patient with MELAS syndrome. Neurology 1991;41:1663–1664. [DOI] [PubMed] [Google Scholar]
- Gasparre G, Iommarini L, Porcelli AM, Lang M, Ferri GG, Kurelac I, Zuntini R, Mariani E, Pennisi LF, Pasquini E et al. . An inherited mitochondrial DNA disruptive mutation shifts to homoplasmy in oncocytic tumor cells. Hum Mutat 2009;30:391–396. [DOI] [PubMed] [Google Scholar]
- Gasparre G, Porcelli AM, Bonora E, Pennisi LF, Toller M, Iommarini L, Ghelli A, Moretti M, Betts CM, Martinelli GN. Disruptive mitochondrial DNA mutations in complex I subunits are markers of oncocytic phenotype in thyroid tumors. Proc Natl Acad Sci USA 2007;104:9001–9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparre G, Porcelli AM, Lenaz G, Romeo G. Relevance of mitochondrial genetics and metabolism in cancer development. Cold Spring Harb Perspect Biol 2013;5:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman A, Carlin JB, Stern HS, Rubin DB. Bayesian Data Analysis, 2nd edn Boca Raton, Florida, USA: CRC Press, 2003. [Google Scholar]
- Gianaroli L, Luiselli D, Crivello AM, Lang M, Ferraretti AP, De Fanti S, Magli MC, Romeo G. Mitogenomes of polar bodies and corresponding oocytes. PLoS One 2014;9:e102182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianaroli L, Luiselli D, Crivello AM, Lang M, Ferraretti AP, De Fanti S, Magli MC, Romeo G. Mitochondrial DNA analysis and numerical chromosome condition in human oocytes and polar bodies. Mol Hum Reprod 2015;21:46–57. [DOI] [PubMed] [Google Scholar]
- Gigarel N, Hesters L, Samuels DC, Monnot S, Burlet P, Kerbrat V, Lamazou F, Benachi A, Frydman R, Feingold J et al. . Poor correlations in the levels of pathogenic mitochondrial DNA mutations in polar bodies versus oocytes and blastomeres in humans. Am J Hum Genet 2011;88:494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkerson RW, De Vries RL, Lebot P, Wikstrom JD, Torgyekes E, Shirihai OS, Przedborski S, Schon EA. Mitochondrial autophagy in cells with mtDNA mutations results from synergistic loss of transmembrane potential and mTORC1 inhibition. Hum Mol Genet 2012;21:978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagström E, Freyer C, Battersby BJ, Stewart JB, Larsson NG. No recombination of mtDNA after heteroplasmy for 50 generations in the mouse maternal germline. Nucleic Acids Res 2014;42:1111–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M, Brulé H, Friede D, Giegé R, Pütz D, Florentz C. Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA 2000;6:1356–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JH, Chen Z, Xu H. Selective propagation of functional mitochondrial DNA during oogenesis restricts the transmission of a deleterious mitochondrial variant. Nat Genet 2014;46:389–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Fan W, Yan L, Li R, Lian Y, Huang J, Li J, Xu L, Tang F, Xie XS et al. . Genome analyses of single human oocytes. Cell 2013;155:1492–1506. [DOI] [PubMed] [Google Scholar]
- Just RS, Irwin JA, Parson W. Mitochondrial DNA heteroplasmy in the emerging field of massively parallel sequencing. Forensic Sci Int Genet 2015;18:131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass RE, Raferty AE. Bayes factors. J Am Stat Assoc 1995;90:773–795. [Google Scholar]
- Kauppila JH, Baines HL, Bratic A, Simard ML, Freyer C, Mourier A, Stamp C, Filograna R, Larsson NG, Greaves LC et al. . A phenotype-driven approach to generate mouse models with pathogenic mtDNA mutations causing mitochondrial disease. Cell Rep 2016;16:2980–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakauer DC, Mira A. Mitochondria and germ-cell death. Nature 1999;400:125–126. [DOI] [PubMed] [Google Scholar]
- Kurelac I, Lang M, Zuntini R, Calabrese C, Simone D, Vicario S, Santamaria M, Attimonelli M, Romeo G, Gasparre G. Searching for a needle in the haystack: comparing six methods to evaluate heteroplasmy in difficult sequence context. Biotechnol Adv 2012;30:363–371. [DOI] [PubMed] [Google Scholar]
- Larsson NG, Holme E, Kristiansson B, Oldfors A, Tulinius M. Progressive increase of the mutated mitochondrial DNA fraction in Kearns-Sayre syndrome. Pediatr Res 1990;28:131–136. [DOI] [PubMed] [Google Scholar]
- Li B, Krishnan VG, Mort ME, Xin F, Kamati KK, Cooper DN, Mooney SD, Radivojac P. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics 2009;25:2744–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez JV, Yuhki N, Masuda R, Modi W, O'Brien SJ. Numt, a recent transfer and tandem amplification of mitochondrial DNA to the nuclear genome of the domestic cat. J Mol Evol 1994;39:174–190. [DOI] [PubMed] [Google Scholar]
- Ma H, Xu H, O'Farrell PH. Transmission of mitochondrial mutations and action of purifying selection in Drosophila melanogaster. Nat Genet 2014;46:393–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH. Handbook of Biological Statistics, 3rd edn Baltimore, Maryland: Sparky House, 2014. [Google Scholar]
- McFarland R, Clark KM, Morris AA, Taylor RW, Macphail S, Lightowlers RN, Turnbull DM. Multiple neonatal deaths due to a homoplasmic mitochondrial DNA mutation. Nat Genet 2002;30:145–146. [DOI] [PubMed] [Google Scholar]
- Pallotti F, Binelli G, Fabbri R, Valentino ML, Vicenti R, Macciocca M, Cevoli S, Baruzzi A, DiMauro S, Carelli V. A wide range of 3243A>G/tRNALeu(UUR) (MELAS) mutation loads may segregate in offspring through the female germline bottleneck. PLoS One 2014;9:e96663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne BA, Wilson IJ, Yu-Wai-Man P, Coxhead J, Deehan D, Horvath R, Taylor RW, Samuels DC, Santibanez-Koref M, Chinnery PF. Universal heteroplasmy of human mitochondrial DNA. Hum Mol Genet 2013;22:384–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesole G, Gissi C, De Chirico A, Saccone C. Nucleotide substitution rate of mammalian mitochondrial genomes. J Mol Evol 1999;48:427–434. [DOI] [PubMed] [Google Scholar]
- Rajasimha HK, Chinnery PF, Samuels DC. Selection against pathogenic mtDNA mutations in a stem cell population leads to the loss of the 3243A-->G mutation in blood. Am J Hum Genet 2008;82:333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM. Mitigating mutational meltdown in mammalian mitochondria. PLoS Biol 2008;6:e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubino F, Piredda R, Calabrese FM, Simone D, Lang M, Calabrese C, Petruzzella V, Tommaseo-Ponzetta M, Gasparre G, Attimonelli M. HmtDB, a genomic resource for mitochondrion-based human variability studies. Nucleic Acids Res 2012;40:1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Pesini E, Lott MT, Procaccio V, Poole JC, Brandon MC, Mishmar D, Yi C, Kreuziger J, Baldi P, Wallace DC. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res 2007;35:823–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Kono T, Nakada K, Ishikawa K, Inoue S, Yonekawa H, Hayashi J. Gene therapy for progeny of mito-mice carrying pathogenic mtDNA by nuclear transplantation. Proc Natl Acad Sci USA 2005;102:16765–16770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry: The Principles and Practice of Statistics in Biological Research, Vol. 3. San Francisco, California, USA: Freeman, 1995. [Google Scholar]
- Steuerwald N, Barritt JA, Adler R, Malter H, Schimmel T, Cohen J, Brenner CA. Quantification of mtDNA in single oocytes, polar bodies and subcellular components by real-time rapid cycle fluorescence monitored PCR. Zygote 2000;8:209–215. [DOI] [PubMed] [Google Scholar]
- Stewart JB, Freyer C, Elson JL, Wredenberg A, Cansu Z, Trifunovic A, Larsson NG. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol 2008;6:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat 2009;30:386–394. [DOI] [PubMed] [Google Scholar]
- Verdinelli I, Wasserman L. Computing Bayesian Factors Using a Generalization of the Savage-Dickey Density Ratio. J Am Stat Assoc 1995;90:614–618. [Google Scholar]
- Wai T, Ao A, Zhang X, Cyr D, Dufort D, Shoubridge EA. The role of mitochondrial DNA copy number in mammalian fertility. Biol Reprod 2010;83:52–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Bioenergetics in human evolution and disease: implications for the origins of biological complexity and the missing genetic variation of common diseases. Philos Trans R Soc Lond B Biol Sci 2013;368:20120267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial DNA variation in human radiation and disease. Cell 2015;163:33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Sha H, Ji D, Zhang HL, Chen D, Cao Y, Zhu J. Polar body genome transfer for preventing the transmission of inherited mitochondrial diseases. Cell 2014;157:1591–1604. [DOI] [PubMed] [Google Scholar]
- White DJ, Wolff JN, Pierson M, Gemmell NJ. Revealing the hidden complexities of mtDNA inheritance. Mol Ecol 2008;17:4925–4942. [DOI] [PubMed] [Google Scholar]
- Zhou RR, Wang B, Wang J, Schatten H, Zhang YZ. Is the mitochondrial cloud the selection machinery for preferentially transmitting wild-type mtDNA between generations? Rewinding Müller's ratchet efficiently. Curr Genet 2010;56:101–107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.