Abstract
Thermotolerant Campylobacter spp. are a major cause of foodborne gastrointestinal infections worldwide. The linkage of human campylobacteriosis and poultry has been widely described. In this study we aimed to investigate the prevalence, antimicrobial resistance and genetic diversity of C. coli and C. jejuni in broilers from Ecuador. Caecal content from 379 randomly selected broiler batches originating from 115 farms were collected from 6 slaughterhouses located in the province of Pichincha during 1 year. Microbiological isolation was performed by direct plating on mCCDA agar. Identification of Campylobacter species was done by PCR. Minimum inhibitory concentration (MIC) values for gentamicin, ciprofloxacin, nalidixic acid, tetracycline, streptomycin, and erythromycin were obtained. Genetic variation was assessed by RFLP-flaA typing and Multilocus Sequence Typing (MLST) of selected isolates. Prevalence at batch level was 64.1%. Of the positive batches 68.7% were positive for C. coli, 18.9% for C. jejuni, and 12.4% for C. coli and C. jejuni. Resistance rates above 67% were shown for tetracycline, ciprofloxacin, and nalidixic acid. The resistance pattern tetracycline, ciprofloxin, and nalidixic acid was the dominant one in both Campylobacter species. RFLP-flaA typing analysis showed that C. coli and C. jejuni strains belonged to 38 and 26 profiles respectively. On the other hand MLST typing revealed that C. coli except one strain belonged to CC-828, while C. jejuni except 2 strains belonged to 12 assigned clonal complexes (CCs). Furthermore 4 new sequence types (STs) for both species were described, whereby 2 new STs for C. coli were based on new allele sequences. Further research is necessary to estimate the impact of the slaughter of Campylobacter positive broiler batches on the contamination level of carcasses in slaughterhouses and at retail in Ecuador.
Keywords: Campylobacter, Ecuador, genetic types, antimicrobial resistance, broilers
INTRODUCTION
Thermotolerant Campylobacter spp. are a major cause of foodborne gastrointestinal infections worldwide. Human campylobacteriosis in its acute phase is characterized by diarrhea, fever, abdominal cramps, and vomiting and has been linked to the development of Guillain-Barré syndrome, reactive arthritis, and irritable bowel syndrome as complications after the acute phase of the disease (Loshaj-Shala et al., 2015). The WHO (2015) estimated that Campylobacter caused 37.600 deaths per year worldwide. For 2014, 237,642 campylobacteriosis cases were registered in the European Union (EFSA and ECDC, 2015). However it has been estimated that the real number of cases occurring yearly may be 9 million (Havelaar et al., 2009). Diarrheal illness caused by these pathogens are especially important in developing countries where the infection in children under the age of two years is frequent and may lead to death (WHO, 2011). Campylobacter has been associated to 11.3 to 21% of diarrhea episodes in children from low-income countries (Platts-Mills and Kosek, 2014). However, the lack of studies on the epidemiology of Campylobacter in developing countries could lead to the underestimation of the burden of Campylobacter infections in these regions (Platts-Mills and Kosek, 2014). In Ecuador data about Campylobacter infections in humans is very limited. Campylobacter has been reported in Ecuadorian low income communities as a possible cause of diarrhea in humans (Vasco et al., 2014). Furthermore, it has been estimated that 50 to 80% of campylobacteriosis cases may be attributed to the chicken reservoir as a whole being poultry the main source of Campylobacter transmission within the European Union (Skarp et al., 2015).
In general, Campylobacter infections do not require antibiotic treatment, however the use of erythromycin, tetracycline, and quinolones is recommended in severe cases (WHO, 2011).
Worldwide the use of antibiotics in husbandry practices is a major concern since this may promote the development of resistant and even multidrug-resistant bacteria. Antibiotics in poultry production systems are widely used to prevent, control, and treat bacterial infections as well as growth promoters in a large number of countries (Seiffert et al., 2013). These facts are of special relevance in developing countries where misuse of antibiotics and the lack of control over their usage is a problem to be addressed (Reardon, 2014). In Latin-American countries, increased rates of antimicrobial resistant Campylobacter have been reported (Pollett et al., 2012; Sierra–Arguello et al., 2016).
In Ecuador chicken meat is frequently consumed and its demand increased over the years (CONAVE, 2014). Although Ecuadorian poultry industry only provides chicken meat for local consumption up to now, it is expected that in the future it can have access to international markets once sanitary conditions are better understood and controlled. Despite of the importance of Campylobacter as a foodborne pathogen, little is known about its epidemiology in poultry farms, slaughterhouses, and retail stores in the main centers of production and consumption of poultry products in Ecuador. This information may help to establish surveillance programs and intervention measures regarding to the presence and antimicrobial resistance of Campylobacter in Ecuadorian poultry.
The aim of this study was to investigate the prevalence, antimicrobial resistance and genetic profiles of Campylobacter in broilers slaughtered in industrial facilities in the province of Pichincha in Ecuador.
MATERIALS AND METHODS
Study Design and Sampling
Pichincha, the province where Quito, the capital city of Ecuador, is located, was selected as the area for the collection of samples since in this province and the surrounding ones 36% of the total Ecuadorian broiler production is located (CGSIN and MAGAP, 2015).
Eight large slaughterhouses are located in Pichincha (CGSIN and MAGAP, 2015). All of them were contacted and asked for their willingness to cooperate in the study. Based on these results sampling was performed in 6 slaughterhouses. From June 2013 to July 2014, a total of 379 batches (birds coming from one house and slaughtered on the same day) were sampled. All sampled batches from a same farm originated from different houses or birds reared on different periods in the same house. In Ecuador commercial broiler management at the farm includes total depopulation of houses, removal of the litter after every reared batch, cleaning and disinfection of the house followed by a down period of 8 to 15 days. All sampled batches were commercially reared and slaughtered at the age of 6 to 7 weeks. From each batch, one caecum from 25 randomly selected chickens was collected, and transported in an ice box within 1 hour to the laboratory for bacteriological analysis.
Isolation and Identification of Campylobacter spp.
The content from the 25 collected caeca was aseptically pooled. Therefore, all caeca were immersed in ethanol, and after evaporation of the ethanol approximately 1 g content/cecum was collected in a sterile plastic bag. The pooled sample was homogenized by hand during 1 min. after the addition of 225 mL buffered peptone water (218103, Difco, BD, Sparks, MD) and a loopful (10 μL) from each homogenate was directly streaked onto a modified Charcoal Cefoperazone Deoxycholate Agar (mCCDA) (Campylobacter blood free selective medium CM0739 plus selective supplement SR0155H [Oxoid, Cheshire, UK]). Plates were incubated under microaerobic conditions at 41.5°C for 48 h. Three presumptive Campylobacter colonies were confirmed by Gram staining and microscopic observation. Colonies containing bacteria with a typical shape were subcultured on mCCDA. After incubation under microaerobic conditions at 41.5°C for 48 h the DNA of one colony per plate was extracted by boiling for 10 minutes in 100 μL of DNA free water. The rest of the culture was transferred into sheep blood and stored at −80°C. Multiplex PCR described by Vandamme et al. (1997) was performed for identification of Campylobacter species. PCR results indicating the presence of both C. jejuni and C. coli were retested after sub-culturing of one colony on mCCDA until only one species was detected.
From each positive batch one isolate was randomly selected for further characterization.
Antimicrobial Resistance
Antimicrobial resistance was evaluated in one isolate per sample. The minimum inhibitory concentration (MIC) was determined using the EUCAMP2 plates (Thermo Scientific, West Palm Beach, FL). The tests were performed according to the manufacturer instructions. The following antibiotics were evaluated: gentamicin, ciprofloxacin, nalidixic acid, tetracycline, streptomycin, and erythromycin. Campylobacter jejuni ATCC 33560 was used as the quality control strain. Epidemiological breakpoint values from the European Committee on Antimicrobial Susceptibility Testing were considered to determine bacterial antibiotic resistance (EUCAST, 2015).
Restriction Fragment Length Polymorphism of the flaA gene (flaA-RFLP)
One Campylobacter isolate per positive batch was tested. For the PCR the consensus pair of primers for the flaA gene described by Wassenaar and Newell (Wassenaar and Newell, 2000) and the reagents and conditions described by Nachamkin et al. (Nachamkin et al., 1993) were applied. For restriction fragment length polymorphism (RFLP) analysis flaA PCR amplicons were treated with restriction enzyme DdeI (Thermo Scientific, West Palm Beach, FL). PCR amplicons (7 μL) were digested according to the manufacturer's instructions and then separated by electrophoresis for 1:40 hours at 120 V. The gels were stained and photographed. The relatedness among the flaA-RFLP profiles was analyzed with GelCompar II software v. 6.6 (Applied Maths, Sint-Martems-Latem, Belgium). Bands representing fragments between 200 bp and 1,100 bp in size were included in the analysis. A similarity dendrogram was constructed by the unweighted pair group method using arithmetic averages algorithm (UPGMA). DICE similarity coefficient with a tolerance position of 1% was calculated. A flaA-RFLP genotype was assigned on the basis of the difference in the presence of at least one band in the Ddel fingerprint.
Multilocus Sequence Typing
Multilocus Sequence Typing (MLST) was carried out on all C. jejuni isolates that still could be subcultured (40 isolates). For C. coli, 40 randomly selected isolates representing 40 farms were typed by MLST.
MLST was performed by the protocol previously described (PubMLST.org, 2016). Sequence types (STs) and clonal complexes (CCs) were assigned by submitting DNA sequences to the Campylobacter MLST database website (http://pubmlst.org/campylobacter). Novel alleles and STs were submitted to the MLST database for the assignation of new numbers.
Statistical Analysis
Statistical analysis was carried out with STATA/IC 11.0 (StataCorp LP, College Station, TX). The survey design corrected prevalence estimates of Campylobacter at batch level were obtained using the linearized Taylor series method. Farms was identified as first-stage cluster. To determine the prevalence of Campylobacter at farm level, a farm was considered positive when at least one of the sampled batches was positive. Farms were assumed to be independent.
Differences of antibiotic resistances between C. coli and C. jejuni were calculated by the chi-square test. Proportions were considered statistical different when the P value was below 0.05.
RESULTS
Prevalence of Campylobacter spp
The 379 sampled batches originated from 115 farms (1 to 9 batches per farm). From all tested batches 243 (64.1%; CI95%: 58.7% to 69.6%) were Campylobacter positive and originated from 97 farms (84.4%; Confidence Interval (CI)95%: 77.6% to 91.1%). From 84 farms, more than one batch was sampled. The number of times that those farms had Campylobacter positive batches ranged from 1 to 6 (Table 1). Initial PCR speciation demonstrated that 167 batches (68.7%; CI95%: 62.9% to 74.6%) were positive for C. coli, 46 (18.9%; CI95%: 14.0% to 23.9%) for C. jejuni and 30 (12.4%; CI95%: 8.2% to 16.5%) for C. coli/C. jejuni. Subculturing of the mixed cultures yielded 22 C. coli and 8 C. jejuni isolates.
Table 1.
Campylobacter positive batches in relation to the number of tested batches per farm.
| Number of farms with 0 to 6 positive batches | ||||||||
|---|---|---|---|---|---|---|---|---|
| Number of sampled batches/farm | Number of farms | 0 | 1 | 2 | 3 | 4 | 5 | 6 |
| 1 | 31 | 15 | 16 | |||||
| 2 | 19 | 2 | 7 | 10 | ||||
| 3 | 12 | 1 | 2 | 8 | 1 | |||
| 4 | 18 | 1 | 3 | 9 | 5 | |||
| 5 | 15 | 2 | 3 | 1 | 5 | 4 | ||
| 6 | 16 | 4 | 4 | 4 | 3 | 1 | ||
| 7 | 2 | 1 | 1 | |||||
| 8 | 1 | 1 | ||||||
| 9 | 1 | 1 | ||||||
| Total | 115 | 18 | 29 | 28 | 15 | 15 | 7 | 3 |
Antimicrobial Resistance
Twenty-five isolates (19 C. coli and 6 C. jejuni) could not be sub-cultured from −80°C for MIC test; hence 218 isolates were tested (170 C. coli and 48 C. jejuni). The MIC distributions for the different antibiotics of C. coli and C. jejuni are shown in Tables 2 and 3 respectively. C. coli and C. jejuni showed very low resistance rates for gentamicin and the resistance rate was not statistically different between both species (P = 0.752). For streptomycin the resistance rates were 11.2% and 8.3% for C. coli and C. jejuni respectively (P = 0.199). Resistance rate for erythromycin was statistically higher for C. coli (25.9%) compared to C. jejuni (4.2%) (P = 0.024). In contrast the resistance rates for tetracycline was statistically higher for C. jejuni (83.3%) than for C. coli (67.6%) (P = 0.016). Resistance rates of C. coli for ciprofloxacin and nalidixic acid were 100% and 99.4% respectively (P = 0.086). Similarly, C. jejuni presented resistance rates of 97.9% and 100% for ciprofloxacin and nalidixic acid respectively (P = 0.558).
Table 2.
Distribution of the minimal inhibitory concentration values for 170 C. coli isolates collected from broiler batches.
| Number of C. coli isolates with minimal inhibitory concentrations (μg/μL)1 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Antibiotic | 0,12 | 0,25 | 0,5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 |
| Gentamicin | 106 | 49 | 8 | 5 | 1 | 1 | |||||
| Streptomicyn | 1 | 120 | 30 | 19 | |||||||
| Erythromycin | 38 | 33 | 33 | 22 | 6 | 31 | 7 | ||||
| Tetracycline | 34 | 15 | 6 | 1 | 1 | 89 | 24 | ||||
| Ciprofloxacin | 1 | 2 | 134 | 5 | 28 | ||||||
| Nalidixic acid | 1 | 1 | 11 | 157 | |||||||
1Full vertical lines indicate epidemiological break points for resistance described by European Committee on Antimicrobial Susceptibility Testing (EUCAST, 2015).
Table 3.
Distribution of the minimal inhibitory concentration values for 48 C. jejuni isolates collected from broiler batches.
| Number of C. jejuni isolates with minimal inhibitory concentrations (μg/μL)1 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Antibiotic | 0,12 | 0,25 | 0,5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 |
| Gentamicin | 42 | 3 | 2 | 1 | |||||||
| Streptomicyn | 2 | 5 | 35 | 2 | 2 | 2 | |||||
| Erythromycin | 23 | 21 | 2 | 1 | 1 | ||||||
| Tetracycline | 6 | 2 | 1 | 1 | 28 | 10 | |||||
| Ciprofloxacin | 1 | 2 | 33 | 3 | 9 | ||||||
| Nalidixic acid | 1 | 1 | 46 | ||||||||
1Full vertical lines indicate epidemiological break points for resistance described by European Committee on Antimicrobial Susceptibility Testing (EUCAST, 2015).
C. coli and C. jejuni isolates showed 8 and 6 different resistance patterns respectively. C. coli presented resistance against 1 up to 6 antibiotics, whereas for C. jejuni resistance against 2 up to 6 antibiotics were involved. The resistance pattern 5 (C. coli: 42.9%; C. jejuni: 72.9%) was the most frequent one for both species (Table 4). Pattern 3 and, patterns 1, 4, and 6 were presented exclusively for C. jejuni and C. coli respectively.
Table 4.
Antibiotic resistance patterns of C. coli and C. jejuni isolates.
| Pattern | Resistance pattern1 | C. coli (%) | C. jejuni (%) |
|---|---|---|---|
| 1 | C | 1(0.6) | 0 |
| 2 | CN | 49(28.8) | 8(16.7) |
| 3 | TN | 0 | 1(2.1) |
| 4 | CEN | 5(2.9) | 0 |
| 5 | CTN | 73(42.9) | 35(72.9) |
| 6 | CTEN | 23(13.5) | 0 |
| 7 | SCTN | 3(1.8) | 2(4.2) |
| 8 | SCTEN | 14(8.2) | 1(2.1) |
| 9 | GSCTEN | 2(1.2) | 1(2.1) |
| Total | 170(100) | 48(100) |
1C, Ciprofloxacin; E, erythromycin; G, Gentamicin; N, Nalidixic; S, Streptomycin; T, Tetracycline.
RFLP-flaA Typing
For RFLP-flaA typing 38 isolates (26 C. coli and 12 C. jejuni) could not be sub-cultured from −80°C; hence 163 C. coli and 47 C. jejuni isolates were tested. From all tested isolates 1 C. coli and 7 C. jejuni did not present bands in RFLP-flaA typing. For C. coli 38 profiles were obtained, from which 19 profiles contained more than one strain. Each of the later profiles contained 2 up to 25 strains. For C. jejuni 26 profiles were obtained, from which 7 profiles contained 2 to 7 strains. Most of the strains within a RFLP-flaA profiles originated from different farms. However, for profile 5, 9, 18, 19, 21, 22 (C. coli), and 20 (C. jejuni) two strains were found in a single farm, and for profile 16 (C. coli) two farms yielded 2 and 3 strains respectively (Table 5).
Table 5.
Campylobacter spp. RFLP-flaA profiles with more than one isolate.
| Campylobacter spp. | ID of RFLP-flaA profiles | Number of isolates within each profile | Number of origin farms |
|---|---|---|---|
| C. coli | 22 | 2 | 1 |
| 7, 17, 25, 31 | 2 | 2 | |
| 1, 11, 32 | 3 | 3 | |
| 21 | 5 | 4 | |
| 5 | 6 | 5 | |
| 3, 30 | 7 | 7 | |
| 6 | 9 | 9 | |
| 8 | 10 | 10 | |
| 18 | 10 | 9 | |
| 9 | 12 | 11 | |
| 29 | 12 | 12 | |
| 16 | 22 | 19 | |
| 19 | 25 | 24 | |
| C. jejuni | 7, 8, 9, 16, 19 | 2 | 2 |
| 14 | 4 | 4 | |
| 20 | 7 | 6 |
MLST Typing
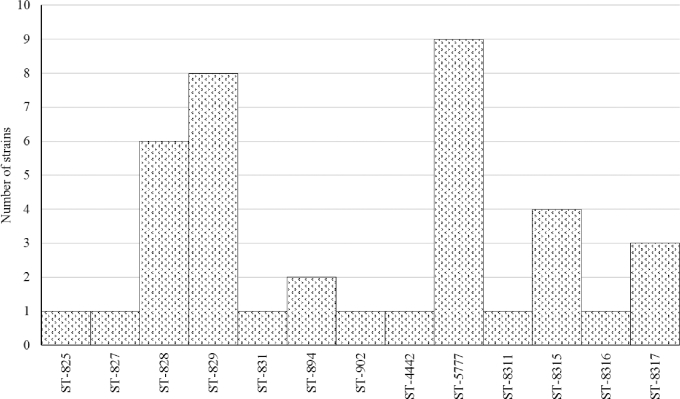
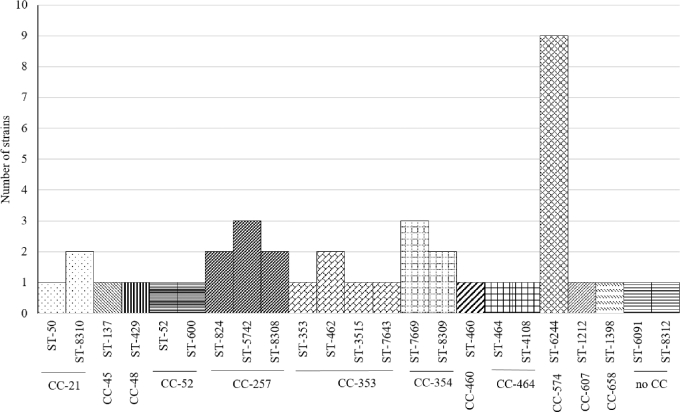
From the 40 C. coli isolates selected for MLST 39 belonged to CC-828 and 1 did not have an assigned CC (ST-1581). The most frequent STs were ST-5777 (9 isolates), followed by ST-829 (8 isolates) and ST-828 (6 isolates) (Figure 1). From the 40 C. jejuni isolates selected for MLST the most common CCs were CC-574 (9 isolates), CC-257 (7 isolates), CC-353 (5 isolates), and CC-354 (5 isolates) (Figure 2). Two C. jejuni isolates did not correspond to an assigned CC. The most ST-diverse CC was CC-353 (4 STs) followed by CC-257 (3 STs), CC-52 (2 STs), CC-354 (2 STs), CC-464 (2 STs), and CC-21 (2 STs).
Figure 1.
Distribution of STs among the 39 C. coli strains belonging to clonal complex 828.
Figure 2.
Distribution of STs and clonal complexes among the 40 C. jejuni strains.
In total, 9 C. coli and 7 C. jejuni strains belonged to STs which were not reported previously. Sequence data from those strains were submitted to the Campylobacter MSLT database (PubMLST.org, 2016) leading to the assignation of 8 novel ST numbers (4 STs for each species) (Table 6). Two novel STs within C. coli (ID PubMLST 48107 and 48108) resulted from novel allele sequences: 5 strains had a novel allele sequence for aspA, of which one strain had also a novel allele sequence for tkt.
Table 6.
MLST profiles of novel STs identified in Campylobacter strains.
| MLST allelic profilea | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Campylobacter species | No. of isolates | Clonal complex | Sequence type | aspA | glnA | gltA | glyA | pgm | tkt | uncA | ID on the PubMLST |
| C. jejuni | 2 | 257 | 8308 | 9 | 2 | 4 | 62 | 606 | 5 | 6 | 48096 |
| C. jejuni | 2 | 354 | 8309 | 8 | 10 | 95 | 2 | 10 | 12 | 6 | 48097 |
| C. jejuni | 2 | 21 | 8310 | 2 | 1 | 5 | 672 | 11 | 1 | 5 | 48113 |
| C. jejuni | 1 | NAb | 8312 | 2 | 84 | 5 | 10 | 11 | 3 | 6 | 48099 |
| C. coli | 1 | 828 | 8311 | 33 | 39 | 30 | 82 | 373 | 56 | 17 | 48106 |
| C. coli | 4 | 828 | 8315 | 441 | 39 | 30 | 82 | 373 | 47 | 17 | 48107 |
| C. coli | 1 | 828 | 8316 | 441 | 39 | 30 | 82 | 113 | 641 | 17 | 48108 |
| C. coli | 3 | 828 | 8317 | 33 | 39 | 30 | 82 | 373 | 47 | 17 | 48110 |
aNew allele sequences are given in bold.
bNA, not assigned.
Comparison of RFLP-flaA profiles and MLST Data
When comparing MLST data with RFLP-flaA profiles, C. coli STs 8315, 8317, 828, 5777, and 829 included 2, 2, 3, 4, and 5 RFLP-flaA profiles respectively, while RFLP-flaA profiles 18, 21, 16, and 19 included 2, 2, 4, and 4 different ST types. For C. jejuni 4 STs (6244, 8308, 8309, and 8310) had two RFLP-flaA profiles and only the RFLP-flaA profile 14 included 2 ST types. No association of RFLP-flaA profiles within STs was found regarding the origin of the isolates.
DISCUSSION
Our findings demonstrated that the prevalence of Campylobacter in broiler batches at slaughter age was 64.1%. Studies from other Latin American countries showed different prevalences. From Brazil and Costa Rica, 100.0% respectively 80.0% of the flocks were reported to be positive for Campylobacter when ceca samples were studied (Giombelli and Gloria, 2014; Zumbaco-Gutiérrez et al., 2014). On the other hand, in Argentina and Chile Campylobacter was found in 33.3% of samples (Rivera et al., 2011; Zbrun et al., 2013a) while in Peru Tresierra-Ayala et al. (1995) reported a prevalence of 35%. Other tropical countries such as Vietnam and South Africa have reported a prevalence of 31.9 and 14.2% respectively (Jonker and Picard, 2010; Carrique-Mas et al., 2014). Although different prevalences are shown in developing countries, it should be keep in mind that differences in methodologies can make direct comparison of results difficult. Moreover, obtained data indicated that at least 84.3% of farms delivered Campylobacter positive batches. For farms delivering only Campylobacter negative batches only a maximum of 3 batches were tested. For those farms it can be hypothesized that when more batches would be sampled also these farms would deliver Campylobacter positive batches for slaughter. On the other hand, the number of positive batches per farm variated considerably which is in concordance with the observations described by McDowell et al. (2008). This variation may be attributed to different risk factors for the introduction of Campylobacter in broilers (Adkin et al., 2006; Torralbo et al., 2014; Sandberg et al., 2015).
Considering Campylobacter species, C. coli was the dominant species in positive batches. This contrasts with other studies from Latin America where C. jejuni has been demonstrated to be the most prevalent species in broilers (Tresierra-Ayala et al., 1995; Rivera et al., 2011; Zbrun et al., 2013b; Giombelli and Gloria, 2014; Zumbaco-Gutiérrez et al., 2014). C. jejuni has also been demonstrated as the most common Campylobacter species from broilers at slaughter age in China and South Africa (Jonker and Picard, 2010; Ma et al., 2014). Meanwhile, the European baseline study on Campylobacter in broilers indicated that the proportion of C. coli/C. jejuni varied considerable between countries and this proportion was generally higher in southern countries than in northern countries (EFSA, 2010).
In this study, C. coli and C. jejuni presented high resistance rates to ciprofloxacin, nalidixic acid and tetracycline while erythromycin, gentamicin, and streptomycin showed lower resistance rates. This is in accordance with a study in Brazil where high resistance rates to ciprofloxacin, nalidixic acid, and tetracycline, and low resistance rates to erythromycin and gentamicin were reported (Ferro et al., 2015). Besides, a similar low resistance rate for erythromycin, a low resistance rate for ciprofloxacin (11,8%) was reported from Chile (Rivera et al., 2011).
In contrast with the data reported in European Union, this study showed that C. jejuni presented higher resistance rates for tetracycline than C. coli (EFSA, 2015). On the other hand, a higher resistance rate to erythromycin was shown for C. coli, which is consistent with data from China and South Africa that showed higher erythromycin resistance rates for C. coli (92.0% and 72.7% respectively) than for C. jejuni (18.8% and 20% respectively) (Jonker and Picard, 2010; Ma et al., 2014).
High resistance rates for (fluoro)quinolones and tetracycline found in the present study may be explained by the common use of these antibiotics as therapeutics in Ecuadorian poultry farms. However it is not clear why the resistance rate to tetracycline was higher for C. jejuni than for C. coli in Ecuador. The low antimicrobial resistance rates to aminoglycosides and macrolides for C. jejuni found in this study indicates that gentamicin and erythromycin can still be used for the treatment of human campylobacteriosis when necessary (WHO, 2011). However, changes in resistance rates presented in this research have to be monitored by the implementation of antimicrobial resistance surveillance on Campylobacter in Ecuador.
Campylobacter typing by RFLP-flaA has been used based on the highly conserved character of this gene. It has also been shown to be a cost-effective alternative to more costly methodologies (Djordjevic et al., 2007). The use of RFLP-flaA as the only typing method is questioned due to intra- and intergenomic recombination within the flagellin genes (Eberle and Kiess, 2012) which can make the comparison of isolates over time difficult. In contrast MLST typing is a more reliable method since it is based on changes in allele sequences of determined housekeeping genes and a library of MLST types is available to compare results from all over the world (PubMLST.org, 2016). Our results showed that the combination of RFLP-flaA and MLST typing led to a further differentiation of a number of isolates. This is in concordance with the results of Duarte et al. (2016) who demonstrated that the combination of both RFLP-flaA and MLST had a higher discriminatory power than both methods separately.
Based on one isolate per batch, our results indicated that a large variation of genetic types were present in Ecuadorian broiler batches. Some genetic types seemed to be more widespread than other ones. Additionally, RFLP-flaA data suggested that over time the persistence of specific genetic types on farms is limited. Analyses of the variable region in the flaA locus (flaA-SVR) have demonstrated that more than one Campylobacter genotype may be present in the same farm (Jorgensen et al., 2011; O’Mahony et al., 2011; Prachantasena et al., 2016). Moreover, some batches were simultaneously colonized with C. coli and C. jejuni in the present study.
To the best of our knowledge, this study is the first report that showed Campylobacter MSLT types from commercial broiler batches in Andes region of Latin America. In this study 39 out of 40 C. coli isolates belonged to CC-828. Predominant distribution of C. coli within CC-828 has also been reported in Europe (Levesque et al., 2013; Piccirillo et al., 2014). It is suggested that the low diversity of CCs in C. coli can be attributed to the presence of a 3-clade C. coli population structure. In this genetic structure, horizontal gene transfer within each clade would be more common than among members of different clades (Sheppard and Maiden, 2015), resulting in a limited number of CCs.
Interestingly, the new reported ST-8315 was present in 4 (10%) C. coli isolates. The implication of this ST in the epidemiology of Campylobacter needs further research. From the 40 C. jejuni isolates tested, the majority belonged to CC-574 (9 isolates), CC-257 (7 isolates), CC-353 (4 isolates), CC-354 (5 isolates), and CC-21 (3 isolates). In Great Britain, an important number of C. jejuni strains were grouped in CC-257, CC-353, and CC-574 (Jorgensen et al., 2011). Meanwhile, CC-354 has been found in commercial poultry in Thailand (Prachantasena et al., 2016). This is in accordance with our results where these CCs were found in 72.5% of the tested samples. Additionally, a Canadian study reported CC-353 in C. jejuni isolates from chickens originated in Peru, Bolivia and Argentina (Lévesque et al., 2008). Other less common CCs found in this study (CC-45, CC-48, CC-52, CC-460, CC-658,CC-464, and CC-607) have also been reported in poultry from Europe, Africa, Asia, and North America (Lévesque et al., 2008; Griekspoor et al., 2010; O’Mahony et al., 2011; Kittl et al., 2013; Ngulukun et al., 2016; Zeng et al., 2016).
Moreover, a study in Ecuador demonstrated that CC-353, CC-354 and CC-607 were present in C. jejuni isolates from backyard poultry and other domestic animals kept in households (Graham et al., 2016). A query in the Campylobacter jejuni/coli PubMSLT database (PubMLST.org, 2016) (Last accessed: 21/07/2016) showed that in Latin America, Brazil, and Uruguay reported Campylobacter MLST profiles from chicken samples. These isolates belonged to CC-257, CC-52 (C. jejuni), and CC-828 (C. coli) in Uruguay, while in Brazil a no determined CC (ST-7370) was reported.
Although there are new STs in some of our strains C. coli isolates, the most of CCs found in this study have been reported in chicken samples (PubMLST.org, 2016).
This study gives insights on the epidemiology of Campylobacter in commercial reared poultry in Ecuador. Since high levels of Campylobacter on carcasses has been linked to an increasing risk of Campylobacter infections in humans (EFSA, 2011), it would be interesting to collect data about the contamination including contamination levels, of broiler meat and related risk factors for contamination at the following stages of the broiler meat chain. Campylobacter types and its antimicrobial resistance have not been studied from humans in Ecuador. Therefore, it is not possible to link human campylobacteriosis to the genotypes found in this study. Therefore further research on Campylobacter isolates from the broiler meat chain and humans may give more insights on the epidemiology of Campylobacter in Ecuador.
Acknowledgments
We acknowledge for the financial suport of this research to Secretaría de Educación Superior Ciencia y Tecnología del Ecuador. Programa de Becas Convocatoria Abierta 2012 Segunda Fase. http://www.educacionsuperior.gob.ec. We acknowledge the slaughterhouses that participated in this study and Andrea Galárraga, Valeria Poma and Jonathan Falcon for sample collection in the slaughterhouses.
REFERENCES
- Adkin A., Hartnett E., Jordan L., Newell D., Davison H.. 2006. Use of a systematic review to assist the development of Campylobacter control strategies in broilers. J. Appl. Microbiol. 100:306–315. [DOI] [PubMed] [Google Scholar]
- Carrique-Mas J. J., Bryant J. E., Cuong N. V, Hoang N. V. M., Campbell J., Hoang N. V, Dung T. T. N., Duy D. T., Hoa N. T., Thompson C., Hien V. V, Phat V. V, Farrar J., Baker S.. 2014. An epidemiological investigation of Campylobacter in pig and poultry farms in the Mekong delta of Vietnam. Epidemiol. Infect. 142:1425–1436. Available at http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4045178&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CGSIN and MAGAP 2015. Registro Nacional de Producción Avícola. Quito. [Google Scholar]
- CONAVE 2014. Estadísticas de Producción Avícola 2013. 1.
- Djordjevic S. P., Unicomb L. E., Adamson P. J., Mickan L., Rios R., Adamson P., Cheung K., Combs B., Dalton C., Doyle R., Ferguson J., Gilbert L., Givney R., Gordon D., Gregory J., Hogg G., Inglis T., Jelfs P., Kirk M., Lalor K., Lanser J., O’Reilly L., Sarna M., Sharma H., Smith H., Valcanis M.. 2007. Clonal complexes of Campylobacter jejuni identified by multilocus sequence typing are reliably predicted by restriction fragment length polymorphism analyses of the flaA gene. J. Clin. Microbiol. 45:102–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte A., Seliwiorstow T., Miller W. G., De Zutter L., Uyttendaele M., Dierick K., Botteldoorn N.. 2016. Discriminative power of Campylobacter phenotypic and genotypic typing methods. J. Microbiol. Methods. 125:33–39. Available at http://linkinghub.elsevier.com/retrieve/pii/S0167701216300380. [DOI] [PubMed] [Google Scholar]
- Eberle K. N., Kiess A. S.. 2012. Phenotypic and genotypic methods for typing Campylobacter jejuni and Campylobacter coli in poultry. Poult. Sci. 91:255–264. Available at http://ps.oxfordjournals.org/content/91/1/255.full. [DOI] [PubMed] [Google Scholar]
- EFSA 2010. Analysis of the baseline survey on the prevalence of Campylobacter n broiler batches and of Campylobacter and Salmonella on broiler carcasses in the EU, 2008, Part A: Campylobacter and Salmonella prevalence estimates. EFSA J. 15:100. [Google Scholar]
- EFSA 2011. Scientific Opinion on Campylobacter in broiler meat production: control options and performance objectives and/or targets at different stages of the food chain. EFSA J. 9:2105 Available at http://doi.wiley.com/10.2903/j.efsa.2011.2105. [Google Scholar]
- EFSA 2015. SCIENTIFIC REPORT OF EFSA AND ECDC. EU Summary Report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2013. EFSA J. 13:1–178. [Google Scholar]
- EFSA, and ECDC 2015. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2014. EFSA J. 13:4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EUCAST 2015. European committee on antimicrobial susceptibility testing. Data from the EUCAST MIC distribution website. Available at http://mic.eucast.org (verified 19 October 2016). [Google Scholar]
- Ferro I. D., Benetti T. M., Oliveira T. C. R. M., Abrahão W. M., Farah S. M. S. S., Luciano F. B., Macedo R. E. F.. 2015. Evaluation of antimicrobial resistance of Campylobacter spp. isolated from broiler carcasses. Br. Poult. Sci. 56:66–71. Available at http://www.ncbi.nlm.nih.gov/pubmed/25567139. [DOI] [PubMed] [Google Scholar]
- Giombelli A., Gloria M. B. A.. 2014. Prevalence of salmonella and campylobacter on broiler chickens from farm to slaughter and efficiency of methods to remove visible fecal contamination. J. Food Prot. 77:1851–1859. Available at http://openurl.ingenta.com/content/xref?genre=article&issn=0362-028X&volume=77&issue=11&spage=1851. [DOI] [PubMed] [Google Scholar]
- Graham J. P., Vasco K., Trueba G.. 2016. Hyperendemic Campylobacter jejuni in guinea pigs (Cavia porcellus) raised for food in a semi-rural community of Quito, Ecuador. Environ. Microbiol. Rep. 8:382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griekspoor P., Engvall E. O., Olsen B., Waldenstrom J.. 2010. Multilocus sequence typing of Campylobacter jejuni from broilers. Vet. Microbiol. 140:180–185. [DOI] [PubMed] [Google Scholar]
- Havelaar A. H., van Pelt W., Ang C. W., Wagenaar J. A., van Putten J. P. M., Gross U., Newell D. G.. 2009. Immunity to Campylobacter: its role in risk assessment and epidemiology. Crit. Rev. Microbiol. 35:1–22. Available at http://www.ncbi.nlm.nih.gov/pubmed/19514906. [DOI] [PubMed] [Google Scholar]
- Jonker A., Picard J. A.. 2010. Antimicrobial susceptibility in thermophilic Campylobacter species isolated from pigs and chickens in South Africa. J. S. Afr. Vet. Assoc. 81:228–236. [DOI] [PubMed] [Google Scholar]
- Jorgensen F., Ellis-Iversen J., Rushton S., Bull S. A., Harris S. A., Bryan S. J., Gonzalez A., Humphrey T. J.. 2011. Influence of season and geography on Campylobacter jejuni and C. coli subtypes in housed broiler flocks reared in Great Britain. Appl. Environ. Microbiol. 77:3741–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittl S., Heckel G., Korczak B. M., Kuhnert P.. 2013. Source attribution of human Campylobacter isolates by MLST and Fla-typing and association of genotypes with quinolone resistance. PLoS One. 8:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque S., Fournier E., Carrier N., Frost E., Arbeit R. D., Michaud S.. 2013. Campylobacteriosis in urban versus rural areas: A case-case study integrated with molecular typing to validate risk factors and to attribute sources of infection. PLoS One. 8:17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévesque S., Frost E., Arbeit R. D., Michaud S.. 2008. Multilocus sequence typing of Campylobacter jejuni isolates from humans, chickens, raw milk, and environmental water in Quebec, Canada. J. Clin. Microbiol. 46:3404–3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loshaj-Shala A., Regazzoni L., Daci A., Orioli M., Brezovska K., Panovska A. P., Beretta G., Suturkova L.. 2015. Guillain Barré syndrome (GBS): new insights in the molecular mimicry between C. jejuni and human peripheral nerve (HPN) proteins. J. Neuroimmunol. 289:168–176. Available at http://www.ncbi.nlm.nih.gov/pubmed/26616887. [DOI] [PubMed] [Google Scholar]
- Ma L., Wang Y., Shen J., Zhang Q., Wu C.. 2014. Tracking Campylobacter contamination along a broiler chicken production chain from the farm level to retail in China. Int. J. Food Microbiol. 181:77–84. Available at http://dx.doi.org/10.1016/j.ijfoodmicro.2014.04.023. [DOI] [PubMed] [Google Scholar]
- McDowell S. W. J., Menzies F. D., McBride S. H., Oza A. N., McKenna J. P., Gordon A. W., Neill S. D.. 2008. Campylobacter spp. in conventional broiler flocks in Northern Ireland: Epidemiology and risk factors. Prev. Vet. Med. 84:261–276. [DOI] [PubMed] [Google Scholar]
- Nachamkin I., Bohachick K., Patton C. M.. 1993. Flagellin gene typing of Campylobacter jejuni by restriction fragment length polymorphism analysis. J. Clin. Microbiol. 31:1531–1536. Available at http://www.ncbi.nlm.nih.gov/pubmed/8100241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngulukun S., Oboegbulem S., Klein G.. 2016. Multilocus sequence typing of Campylobacter jejuni and Campylobacter coli isolates from poultry, cattle and humans in Nigeria. J. Appl. Microbiol. Available at http://doi.wiley.com/10.1111/jam.13185. [DOI] [PubMed] [Google Scholar]
- O’Mahony E., Buckley J. F., Bolton D., Whyte P., Fanning S.. 2011. Molecular epidemiology of campylobacter isolates from poultry production units in southern Ireland (T Kimman, Ed.). PLoS One 6:e28490 Available at http://dx.plos.org/10.1371/journal.pone.0028490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo A., Giacomelli M., Salata C., Bettanello S., De Canale E., Palù G.. 2014. Multilocus sequence typing of Campylobacter jejuni and Campylobacter coli from humans and chickens in North-Eastern Italy. 557–562. [PubMed]
- Platts-Mills J. A., Kosek M.. 2014. Update on the burden of Campylobacter in developing countries. Curr. Opin. Infect. Dis. 27:444–450. Available at http://www.ncbi.nlm.nih.gov/pubmed/24655651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollett S., Rocha C., Zerpa R., Patiño L., Valencia A., Camiña M., Guevara J., Lopez M., Chuquiray N., Salazar-Lindo E., Calampa C., Casapia M., Meza R., Bernal M., Tilley D., Gregory M., Maves R., Hall E., Jones F., Arriola C. S., Rosenbaum M., Perez J., Kasper M.. 2012. Campylobacter antimicrobial resistance in Peru: a ten-year observational study. BMC Infect. Dis. 12:193 Available at http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3482591&tool=pmcentrez&rendertype=abstract (verified 16 April 2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prachantasena S., Charununtakorn P., Muangnoicharoen S., Hankla L., Techawal N., Chaveerach P., Tuitemwong P., Chokesajjawatee N., Williams N., Humphrey T., Luangtongkum T.. 2016. Distribution and Genetic Profiles of Campylobacter in Commercial Broiler Production from Breeder to Slaughter in Thailand. PLoS One. 11:e0149585 Available at http://dx.plos.org/10.1371/journal.pone.0149585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PubMLST.org 2016. Campylobacter MLST Home Page. Available at PubMLST.org (verified 11 July 2016).
- Reardon S. 2014. Antibiotic resistance sweeping developing world. Nature. 509:141–142. Available at http://www.ncbi.nlm.nih.gov/pubmed/24805322 (verified 9 June 2014). [DOI] [PubMed] [Google Scholar]
- Rivera N., Bustos R., Montenegro H. S., Sandoval M. M., Castillo N. J., Fernández J. H., Maturana M.R., Delgado R L., Contreras S. Á., Chávez N D., Quevedo L. I.. 2011. Genotipificación y resistencia antibacteriana de cepas de Campylobacter spp aisladas en niños y en aves de corral. Rev. Chil. infectología. 28:555–562. Available at http://www.scielo.cl/scielo.php?script=sci_arttext&pid=S0716-10182011000700008&lng=es&nrm=iso&tlng=es. [PubMed] [Google Scholar]
- Sandberg M., Sørensen L. L., Steenberg B., Chowdhury S., Ersbøll A. K., Alban L.. 2015. Risk factors for Campylobacter colonization in Danish broiler flocks, 2010 to 2011. Poult. Sci. 94:447–453. Available at http://ps.oxfordjournals.org/content/94/3/447.abstract. [DOI] [PubMed] [Google Scholar]
- Seiffert S. N., Hilty M., Perreten V., Endimiani A.. 2013. Extended-spectrum cephalosporin-resistant Gram-negative organisms in livestock: an emerging problem for human health? Drug Resist. Updat. 16:22–45. Available at http://www.ncbi.nlm.nih.gov/pubmed/23395305 (verified 18 December 2014). [DOI] [PubMed] [Google Scholar]
- Sheppard S. K., Maiden M. C. J.. 2015. The Evolution of Campylobacter jejuni and Campylobacter coli. 1–14. [DOI] [PMC free article] [PubMed]
- Sierra–Arguello Y. M., Perdoncini G., Morgan R. B., Salle C. T. P., Moraes H. L. S., Gomes M. J. P., do Nascimento V. P.. 2016. Fluoroquinolone and Macrolide Resistance in Campylobacter Jejuni Isolated from Broiler Slaughterhouses in Southern Brazil. Avian Pathol. 9457:1–22. Available at http://www.tandfonline.com/doi/full/10.1080/03079457.2015.1120272. [DOI] [PubMed] [Google Scholar]
- Skarp C. P. A., Hänninen M.-L., Rautelin H. I. K.. 2015. Campylobacteriosis: the role of poultry-meat. Clin. Microbiol. Infect. 1–7. Available at http://linkinghub.elsevier.com/retrieve/pii/S1198743X15010253. [DOI] [PubMed] [Google Scholar]
- Torralbo A., Borge C., Allepuz A., García-Bocanegra I., Sheppard S. K., Perea A., Carbonero A.. 2014. Prevalence and risk factors of Campylobacter infection in broiler flocks from southern Spain. Prev. Vet. Med. 114:106–113. Available at http://dx.doi.org/10.1016/j.prevetmed.2014.01.019. [DOI] [PubMed] [Google Scholar]
- Tresierra-Ayala A., Fernández H., Bendayán M. E., Pereyra G., Bernuy A.. 1995. Aislamiento de especies termotolerantes de Campylobacter en dos poblaciones de pollos criados con y sin confinamiento. Rev. Saude. Publica. 29:389–392. Available at http://www.scielo.br/scielo.php?script=sci_arttext&pid=S0034-89101995000500008&lng=es&nrm=iso&tlng=es. [DOI] [PubMed] [Google Scholar]
- Vandamme P., Van Doorn L. J., al Rashid S. T., Quint W. G., van der Plas J., Chan V. L., On S. L.. 1997. Campylobacter hyoilei Alderton et al. 1995 and Campylobacter coli Véron and Chatelain 1973 are subjective synonyms. Int. J. Syst. Bacteriol. 47:1055–1060. Available at http://www.ncbi.nlm.nih.gov/pubmed/9336905 (verified 28 July 2013). [DOI] [PubMed] [Google Scholar]
- Vasco G., Trueba G., Atherton R., Calvopiña M., Cevallos W., Andrade T., Eguiguren M., Eisenberg J. N. S.. 2014. Identifying etiological agents causing diarrhea in low income ecuadorian communities. Am. J. Trop. Med. Hyg. 91:563–569. Available at http://www.ncbi.nlm.nih.gov/pubmed/25048373 (verified 25 September 2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenaar T. M., Newell D. G.. 2000. Genotyping of Campylobacter spp. Appl. Environ. Microbiol. 66:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO 2011. Campylobacter. Fact sheet N°255. Available at http://www.who.int/mediacentre/factsheets/fs255/en/ (verified 1 February 2016).
- WHO 2015. WHO Estimates of the Global Burden of Foodborne Diseases. First edit.World Health Organization, Geneva. [Google Scholar]
- Zbrun M. V, Romero-Scharpen A., Olivero C., Rossler E., Soto L. P., Rosmini M. R., Sequeira G. J., Signorini M. L., Frizzo L. S.. 2013a. Occurrence of thermotolerant Campylobacter spp. at different stages of the poultry meat supply chain in Argentina. N. Z. Vet. J. 61:337–343. Available at <Go to ISI>://000325844500005. [DOI] [PubMed] [Google Scholar]
- Zbrun M., Romero-Scharpen A., Olivero C., Rossler E., Soto L., Rosmini M., Sequeira G., Signorini M., Frizzo L.. 2013b. Occurrence of thermotolerant Campylobacter spp. at different stages of the poultry meat supply chain in Argentina. N. Z. Vet. J. Available at http://www.ncbi.nlm.nih.gov/pubmed/23906333 (verified 6 August 2013). [DOI] [PubMed] [Google Scholar]
- Zeng D., Zhang X., Xue F., Wang Y., Jiang L., Jiang Y.. 2016. Phenotypic characters and molecular epidemiology of campylobacter jejuni in East China. J. Food Sci. 81:M106–M113. [DOI] [PubMed] [Google Scholar]
- Zumbaco-Gutiérrez L., Arévalo-Madrigal A., Donado-Godoy M., Romero-Zúniga J.. 2014. DIAGNÓSTICO MOLECULAR DE Campylobacter EN LA Cadena. Agron. Mesoam. 25:357–363. Available at http://www.mag.go.cr/rev_meso/v25n02_357.pdf. [Google Scholar]