Abstract
The substitution rates of transitions are higher than expected by chance relative to those of transversions. Many have argued that selection disfavors transversions, as nonsynonymous transversions are less likely to conserve biochemical properties of the original amino acid. Only recently has it become feasible to directly test this selective hypothesis by comparing the fitness effects of a large number of transition and transversion mutations. For example, a recent study of six viruses and one beta-lactamase gene did not find evidence supporting the selective hypothesis. Here, we analyze the relative fitness effects of transition and transversion mutations from our recently published genome-wide study of mutational fitness effects in influenza virus. In contrast to prior work, we find that transversions are significantly more detrimental than transitions. Using what we believe to be an improved statistical framework, we also identify a similar trend in two HIV data sets. We further demonstrate a fitness difference in transition and transversion mutations using four deep mutational scanning data sets of influenza virus and HIV, which provided adequate statistical power. We find that three of the most commonly cited radical/conservative amino acid categories are predictive of fitness, supporting their utility in studies of positive selection and codon usage bias. We conclude that selection is a major contributor to the transition:transversion substitution bias in viruses and that this effect is only partially explained by the greater likelihood of transversion mutations to cause radical as opposed to conservative amino acid changes.
Keywords: mutation, transition, transversion, fitness, virus
Introduction
Fifty years ago, Walter Fitch noted that the nucleotide substitution pattern in cytochrome c is nonrandom (Fitch 1967). If random, transversions (purine–pyrimidine changes) should be observed twice as often as transitions (purine to purine or pyrimidine to pyrimidine changes) solely due to the accessible mutations. However, Fitch observed that transitions are more common than transversions. In fact, this transition–transversion (Ts:Tv) substitution bias has been noted across many proteins and phyla, and phylogenetic inferences account for this bias by weighting transversions more than transitions (Gojobori et al. 1982; Kumar 1996; Wakeley 1996; Petrov and Hartl 1999; Rosenberg et al. 2003; Lynch 2010; Duchêne et al. 2015).
The underlying reasons for this widespread Ts:Tv substitution bias are largely unknown. Two main hypotheses, which are not mutually exclusive, have emerged to explain this phenomenon: the mutational hypothesis and the selective hypothesis. The mutational hypothesis holds that the transition mutation rates of polymerases are higher than the transversion rates. This hypothesis is supported by the observation of a transitional bias in both coding and noncoding regions (Zhang and Gerstein 2003; Jiang and Zhao 2006) as well as mutation rate analyses showing higher transition mutation rates (Denver et al. 2004; Pauly et al. 2017). The selective hypothesis posits that natural selection disfavors transversions. This hypothesis is based on the observation that, depending on codon usage, nonsynonymous transitions are more likely to conserve important biochemical properties of the original amino acid (Vogel and Kopun 1977; Miyata et al. 1979; Zhang 2000). For example, a mutation that changes the charge of an amino acid is a “radical” change, whereas one that does not is a “conservative” change. However, this provides only indirect evidence for the selective hypothesis and the extent to which radical/conservative distinctions are predictive of fitness is unclear. Radical changes do occur less often than conservative ones during protein evolution. Arguments based on this observation can be circular (Dagan et al. 2002; Yampolsky and Stoltzfus 2005). If the transition mutation rate is higher and transitions are more likely to be conservative, then conservative changes will occur more often simply due to the transitional mutation bias. Furthermore, the radical/conservative amino acid distinctions may be overly broad and arbitrary. For example, the hydrophobicity of amino acids may be more constrained for some proteins, whereas their size may be more constrained for other proteins. One could arbitrarily choose a biochemical distinction that would suggest transversions are more likely to be conservative.
Only recently has it become experimentally tractable to test directly the selective hypothesis by comparing the fitness effects of a large number of transition and transversion mutations. A recent study (Stoltzfus and Norris 2016) compared the fitness effects of missense transitions and transversions reported in eight studies of mutational fitness effects. This meta-analysis included: a beta-lactamase gene (TEM1), two HIV genes (integrase and capsid), and five genome-wide studies of viruses (Sanjuán et al. 2004; Carrasco, Daròs, et al. 2007; Carrasco, de la Iglesia, et al. 2007; Domingo-Calap et al. 2009; Peris et al. 2010; Jacquier et al. 2013; Rihn et al. 2013, 2015). Stoltzfus and Norris did not identify a statistically significant difference in the fitness effects of transitions and transversions (Ts–Tv) in any of the viral data sets (2016). They did find a statistically significant difference after combining the data, which was deemed to be of questionable biological significance.
Here, we revisit this question using our recently published library of randomly distributed point mutations in influenza A virus (Visher et al. 2016). In contrast to other viral data sets, we find that transitions are significantly less detrimental than transversions. We apply what we believe is an improved statistical framework and identify a similar trend in the HIV integrase and capsid data sets (Rihn et al. 2013, 2015). We expand our analysis to include deep mutational scanning studies of one HIV gene and two genes from two different strains of influenza (Bloom 2014; Doud et al. 2015; Doud and Bloom 2016; Haddox et al. 2016). The distribution of fitness effects of transversions is shifted toward more detrimental effects compared with transitions at some points along the fitness distribution in each gene, and transitions are never more detrimental. Three of the most commonly cited radical/conservative distinctions are predictive of mutational fitness effects. However, transversions are more detrimental than transitions even when controlling for their greater likelihood to be radical.
Results
Transitions Are Less Detrimental in Influenza A Virus
We recently published a library of 128 point mutants in influenza A virus (Visher et al. 2016). As in other studies of viral mutational fitness effects, the substitution types were chosen at random and the fitness values were assessed individually. Our library contains 95 mutants distributed across the eight genomic RNA in proportion to the size of each segment and an additional 33 random mutations in the segments encoding the surface proteins hemagglutinin (HA) and neuraminidase (NA). Thus, a total of 57 mutations occur in HA and NA and 71 in the other six segments. For the present study, we excluded 27 synonymous mutations and 6 beneficial missense mutations and considered the remaining 95 missense transitions and transversions that had fitness values ≤1. We performed our analyses on the total library (N = 95), the genes encoding the internal proteins only (N = 53), and the genes encoding the surface proteins only (N = 42) (supple-mentary table S1, Supplementary Material online; Total, Internal, and Surface data sets, respectively). Each of these data subsets is larger than the analogous genome-wide data sets of other viruses.
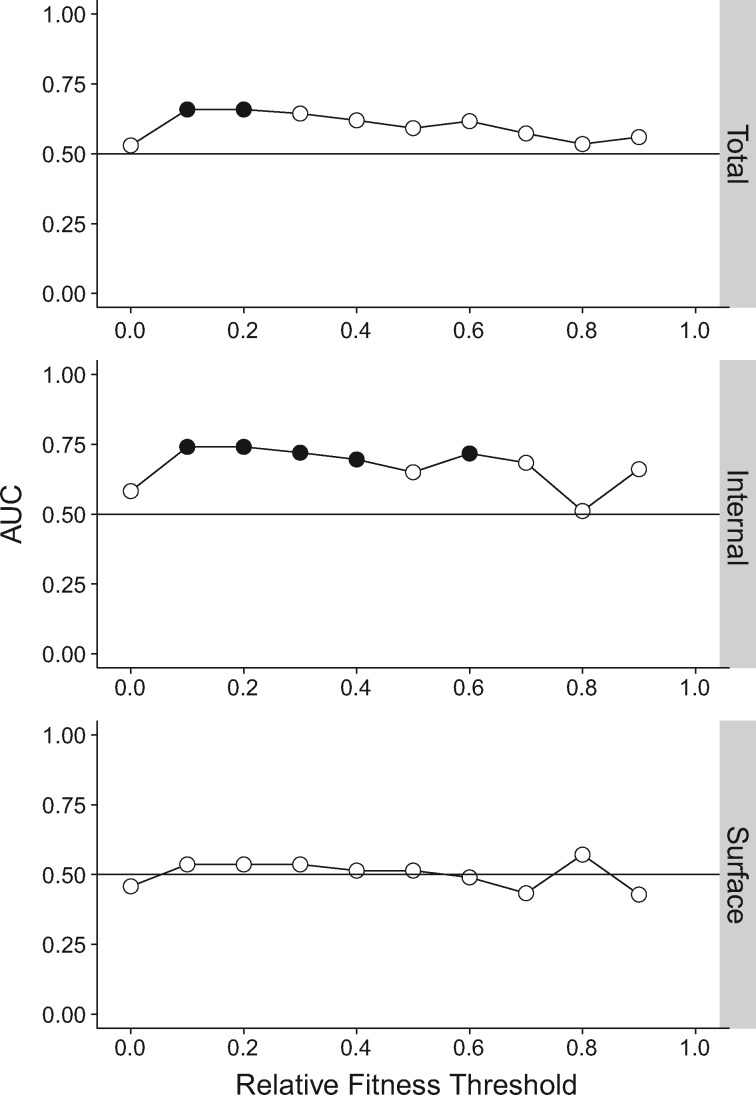
As described by Stoltzfus and Norris (2016), we identified differences in the fitness effects of Ts and Tv by calculating the area under the curve (AUC) of a receiver operating characteristic (ROC) curve. An ROC curve plots the true positive rate against the false positive rate of a binary classifier system as the discrimination threshold varies. Consider a hypothetical example using a binary classifier system to predict whether a mutation is a transition or a transversion without prior knowledge of its identity. If the fitness of the mutation is above a fitness level threshold, it is categorized as a transition, and if it is below the threshold, it is categorized as a transversion. The AUC is equivalent to the probability that a randomly chosen transition is more fit than a randomly chosen transversion. The AUC can be calculated from the Mann–Whitney U test statistic (see Materials and Methods) (Hanley and McNeil 1982; Mason and Graham 2002). An AUC of 1 would indicate that all transitions are more fit than all transversions, and an AUC of 0 would indicate that all transversions are more fit than all transitions. The null expectation is an AUC of 0.50. To identify the points along the fitness distribution at which there is a Ts–Tv difference, we calculated the AUC among mutations at or above 10 successively higher fitness thresholds, starting at 0 and increasing each threshold by 0.10.
Using the AUC criterion, we did not find a statistically significant difference between the fitness effects of transition and transversion mutations across the total data set, the internal data set, or the surface (HA/NA) data set (fig. 1, relative fitness threshold of 0). However, we detected a significant difference when we examined only nonlethal mutations (fig. 1, relative fitness threshold of 0.1; there were no mutations with a fitness value between 0 and 0.1). Among the viable fraction, transitions are significantly more fit than transversions (AUC = 0.65, P = 0.03). This fitness difference is more pronounced in the internal data set (AUC = 0.74, P = 0.02) but is not present among the surface data set. As the thresholds approach 1, the Ts–Tv difference approaches 0.5 and loses significance (perhaps due to decreasing sample size) in both the genome-wide and internal data sets. In contrast, we never found transversions to be significantly more fit than transitions. Thus, viable transitions are more fit than viable transversions, and this fitness difference varies between the internal and surface data sets.
Fig. 1.
Differences in Ts and Tv fitness effects as measured by AUC. Transition–transversion (Ts–Tv) fitness differences as measured by area under the curve of an ROC curve (AUC) for the total, internal, and surface influenza data sets. To identify the points along the fitness distribution at which there is a Ts–Tv difference, the AUC was calculated among mutations at or above 10 successively higher fitness thresholds, starting at 0 and increasing by 0.10. Filled circles denote P<0.05 for a one-sided Mann–Whitney U test where the alternative is transitions are more fit at that threshold. Lines shown to clarify trends only. Plotted data and raw P values can be found at https://github.com/lauringlab/tstv_paper, last accessed September 26, 2017.
Our findings in influenza differ from those in other viruses (HIV integrase, HIV capsid, TEV, F1, VSV, Qβ, φX174) (Stoltzfus and Norris 2016). Although selective constraints could potentially differ among these viruses, another factor could be the greater statistical power of our influenza data set due to our larger sample size as compared with the other genome-wide viral data sets. Our influenza virus data sets were smaller, however, than those in the HIV integrase and capsid studies.
An Alternative Statistical Approach Better Captures Ts–Tv Fitness Differences
The fact that we could only identify a significant difference in influenza by excluding lethal mutations suggests an inherent bias in the AUC threshold analysis and led us to reexamine our statistical framework. If transitions are more likely to be lethal in influenza, this would offset their advantage among viable mutations. More generally, as the fitness threshold increases, the AUC reflects only the Ts–Tv differences at the higher end of the fitness distribution. In fact, when we applied decreasing, as opposed to increasing, thresholds, many of our conclusions were opposite from those obtained with increasing thresholds (supplementary fig. S1, Supplementary Material online). In this case, the influenza total data set is weighted by strongly detrimental transitions and no fitness differences between Ts and Tv were detected at any threshold. In the larger HIV capsid and combined integrase and capsid data sets, we observed a previously unrecognized, and statistically significant, Ts–Tv difference at decreasing thresholds driven by the inclusion of strongly detrimental transversions (Stoltzfus and Norris 2016).
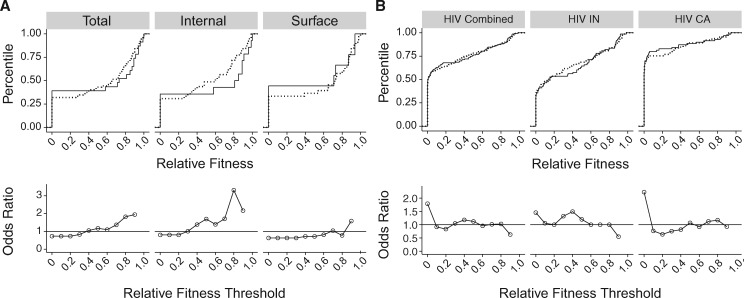
To capture differences in the distribution of fitness effects between transitions and transversions more completely, we compared the empirical cumulative distribution functions (CDF) of each mutation type. The CDF reveal only subtle differences between transitions and transversions in the total influenza data set, with more transitions at low fitness levels and more transversions at intermediate fitness levels (fig. 2A, top). The difference between transitions and transversions is greater for the internal data set, where 75% of transversions have a lower fitness compared with only 50% of transitions at a fitness threshold of 0.8. However, transitions are proportionally overrepresented below a fitness of 0.3. In contrast, the AUC threshold analysis for the internal influenza data set seems to suggest a Ts–Tv fitness difference starting at 0.1 and decreasing to no difference at 0.8 (fig. 1), precisely the opposite of the differences in the distributions (fig. 2A).
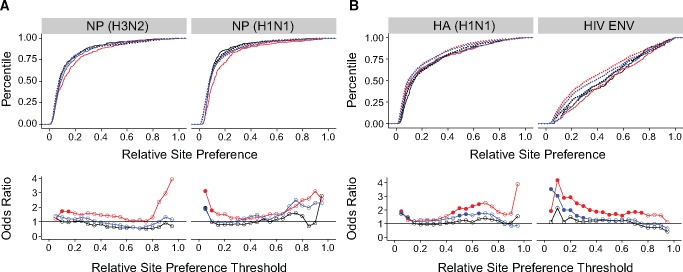
Fig. 2.
Comparisons of the distribution of Ts and Tv fitness effects. Empirical cumulative distribution functions of transitions (solid line) and transversions (dotted line) in our influenza data sets (A, top) and the HIV combined, integrase (IN), and capsid (CA) data sets (B, top). Odds ratios indicate the odds of a transversion versus a transition to be at or below each of 10 relative fitness thresholds as estimated by a Fisher test for our influenza data sets (A, bottom) and HIV data sets (B, bottom). Filled circles denote P<0.05 for a two-sided test with Holm–Bonferroni correction. Lines shown to clarify trends only.
Recognizing these issues, we implemented a different statistical approach that is an explicit comparison of the Ts and Tv CDF and is therefore better able to resolve differences in the distributions of fitness effects. We used 10 fitness thresholds from 0 to 0.9 with a step of 0.1. At each fitness level, we performed a Fisher test to compare the proportion of transversions and transitions at or below the threshold. The estimated effect size is the odds ratio for transversions to be at or below the threshold compared with transitions. As this approach never excludes data, it is less biased by the fitness values at the tails of the distributions. Unlike the AUC (see fig. 1), the odds ratios closely follow the divergence in the corresponding CDF (compare top and bottom panels in fig. 2).
We used this approach to reanalyze data from our influenza data sets as well as those for HIV integrase (IN) and capsid (CA). Using a conservative Holm–Bonferroni correction for multiple comparisons, we did not find a statistically significant difference between transition and transversion mutations in influenza at any fitness level (fig. 2A, bottom). The CDF suggests that the impact of transitions relative to transversions at low fitness levels (0–0.3) is indeed offset by their impact at higher ones (0.6–1). There are no significant Ts–Tv differences in either of the two HIV data sets or in the combined data set. However, there is a trend across the HIV data sets suggesting that transversions are more likely to be lethal as compared with transitions (fig. 2B, bottom, first threshold), an effect missed by the AUC analysis. We note that while the original HIV studies considered a fitness <0.02 to be lethal (Rihn et al. 2013, 2015); we applied a strict and consistent criterion for lethality across the data sets and considered a fitness of 0 to be lethal.
Transversions Are More Detrimental in Larger Influenza and HIV Data Sets
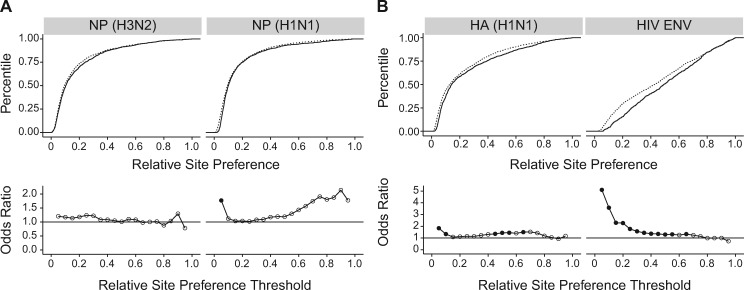
Although the influenza and HIV data sets are relatively large for studies of viral mutational fitness effects, they sample only a small fraction of the total number of possible point mutations. The influenza library contains a median of 11 missense mutations per gene and the HIV IN and CA data sets have 156 and 135 missense mutations, respectively. To increase our power, we analyzed available data from deep mutational scanning (DMS) studies of four viral proteins: the nucleoprotein (NP) of influenza A/Puerto Rico/8/1934 H1N1 and influenza A/Aichi/2/1968 H3N2, the HA protein from the same strain as our mutants (influenza A/WSN/1933 H1N1), and the HIV envelope (ENV) protein (Bloom 2014; Doud et al. 2015; Doud and Bloom 2016; Haddox et al. 2016). Importantly, these four studies all used the same approach and were performed in the same laboratory.
DMS uses high-throughput mutagenesis to introduce every single amino acid substitution in a given gene followed by deep sequencing to measure the change in frequency of each mutation after passage or selection. The effect of each mutation is often reported as a site preference, which represents the expected proportion of an amino acid at a site if all amino acids at that site were present at equal proportions prior to passaging. We derived a relative site preference from these data by dividing the site preference for each mutant by the site preference for the “wild-type” amino acid. We found that relative site preference is a reasonable surrogate for relative fitness, as they are well correlated for mutations in the WSN33 HA gene (Spearman correlation 0.71, P = 2.5×10−5, table 1). Our fitness values for WSN33 NP also exhibit a statistically significant correlation with the DMS data from the closely related PR8 H1N1 strain, but not with the more distant H3N2 strain.
Table 1.
Spearman Correlation between Relative Fitness and Relative Site Preference.
| Gene | Number of Shared Mutations | ρ | P Value |
|---|---|---|---|
| All | 52 | 0.64 | 3.80E-07 |
| HA (H1N1) | 28 | 0.71 | 2.50E-05 |
| NP (H1N1) | 12 | 0.80 | 0.002 |
| NP (H3N2) | 12 | 0.53 | 0.073 |
The DMS studies report changes at the amino acid level. Therefore, for each codon in the nucleotide sequence, we asked which amino acid substitutions could only be made by a single transition (Ts-only) and which could only be made by a single transversion (Tv-only). We excluded the amino acid substitutions that were accessible by both transitions and transversions as well as those that required more than one mutation per codon. We then compared the relative site preferences of Ts-only amino acid changes to those of Tv-only using the Fisher threshold strategy. We used an initial threshold of 0.05 rather than 0, since there are no site preferences of 0 in these data sets, and DMS studies are known to undersample the lethal fraction. The large sample sizes of the DMS studies (supplementary table S1, Supplementary Material online) allowed us to use more thresholds (increasing each by 0.05 instead of 0.01), thereby identifying Ts–Tv differences in the CDF with greater precision.
We found transversions to be significantly more detrimental than transitions at a subset of relative site preference levels in three of the four DMS data sets (fig. 3). In NP (H3N2), transversions tend to be more detrimental than transitions across most of the fitness distribution, but no threshold achieved statistical significance using a Holm–Bonferroni correction (fig. 3A). In NP (H1N1), transversions are significantly more likely to be highly detrimental than transitions (first threshold), and there is a trend for transversions to be more detrimental at higher relative site preferences as well (fig. 3A). Contrary to the trend in our smaller influenza study, the Ts–Tv fitness differences are larger and more broadly distributed in genes coding for the two surface proteins, HA (H1N1) and HIV ENV (fig. 3B), as compared with the genes coding for the internal influenza NP proteins. In HA (H1N1), transversions are significantly more detrimental than transitions across most of the fitness distribution. The Ts–Tv difference is especially pronounced in HIV ENV, for which the odds of a transversion being highly detrimental (thresholds from 0.05 to 0.20) is 2–5 times greater than those of a transition. Across the four data sets, we never found transitions to be significantly more detrimental than transversions, and the odds ratio is rarely <1 for any of the data sets. Thus, with an improved statistical approach and greater power, we found transitions to be less damaging than transversions in proteins from two viruses.
Fig. 3.
Distribution of Ts and Tv fitness effects in deep mutational scanning data sets. Empirical cumulative distribution functions of transitions (solid line) and transversions (dotted line) in two nucleoprotein (NP) proteins (A, top) and two antigenic surface proteins influenza hemagglutinin (HA) and HIV envelope (ENV) (B, top). Odds ratio estimated by a Fisher test comparing the odds of a transversion versus a transition to be at or below each of 19 fitness thresholds, beginning at a fitness of 0.05 and increasing by 0.05, for the same data sets (A and B, bottom). Filled circles denote P<0.05 for a two-sided test with Holm–Bonferroni correction. Lines shown to clarify trends only. Plotted data and raw P values can be found at https://github.com/lauringlab/tstv_paper, last accessed September 26, 2017.
Differences in Ts and Tv Fitness Effects within Radical and Conservative Substitution Classes
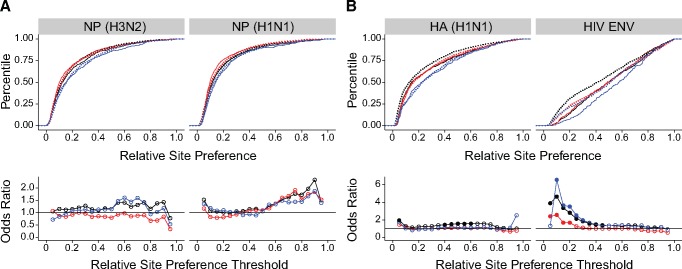
We next asked why transversions are more detrimental than transitions. The genetic code constrains the type of amino acid substitutions accessible by mutation, and it has been proposed that transversions are more detrimental because they are more likely to cause substitutions that radically alter biochemical properties of the original amino acid. We therefore examined whether the observed fitness differences could be explained by the differences in the accessibility of radical versus conservative amino acid changes by transitions and transversions.
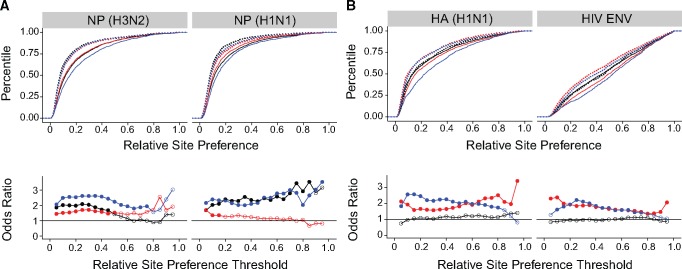
We used the Fisher threshold strategy to test whether radical amino acid changes are more detrimental than conservative changes for three of the most commonly cited biochemical distinctions, which categorize amino acids based on charge, polarity, and polarity and size, (Miyata et al. 1979; Zhang 2000) (see supplementary table S2, Supplementary Material online). Similar categories have been used in other studies of protein evolution (Epstein 1967; Grantham 1974). Radical amino acid substitutions of all three types are more detrimental than conservative changes in the two NP proteins across much of the fitness distribution (fig. 4). Radical changes of polarity (red) and polarity and size (blue) are also more detrimental than conservative changes in the two surface proteins HA (H1N1) and HIV ENV. Radical charge changes (black) have similar effects on fitness as compared with conservative changes in both HA (H1N1) and HIV ENV. Despite this variation in the impact of changes in charge, these simple categories are remarkably predictive of fitness effects across these four proteins.
Fig. 4.
Distribution of radical and conservative amino acid changes. Empirical cumulative distribution functions (CDF) of conservative (solid lines) and radical (dotted lines) amino acid changes in deep mutational scanning data sets of two NP proteins (A, top) and two antigenic surface proteins (B, top). Shown for amino acid changes classified by charge (black), polarity (red), and polarity and size (blue). Odds ratio estimated by a Fisher test comparing the odds of a radical versus a conservative amino acid change to be at or below each of 19 fitness thresholds, beginning at a fitness of 0.05 and increasing in steps of 0.05, for the same data sets (A and B, bottom). Color scheme is the same as in the CDFs. Filled circles denote P<0.05 for a two-sided test with Holm–Bonferroni correction. Lines shown to clarify trends only. Plotted data and raw P values can be found at https://github.com/lauringlab/tstv_paper, last accessed September 26, 2017.
Transversions may be more detrimental than transitions in these four proteins if they are more likely than transitions to cause a radical amino acid change. As above, we considered amino acid substitutions that could only be made by a single transition (Ts-only) or by a single transversion (Tv-only). Using a Fisher test, we compared the odds that a Tv-only amino acid change is radical to the odds that a Ts-only amino acid change is radical as defined by each of the three categories above. We considered all possible Tv-only and Ts-only amino acid changes in these four proteins. For all four proteins, the odds of a transversion causing a radical change of any of these three types is significantly greater than the odds of a transition causing a radical change (table 2). This difference is greatest for amino acid substitutions that affect polarity for all genes.
Table 2.
Tv/Ts Odds Ratio for Causing a Radical Amino Acid Substitution.
| Gene | Charge | Polarity | Polarity and Size |
|---|---|---|---|
| NP (H3N2) | 1.54* | 2.5* | 1.24* |
| NP (H1N1) | 1.53* | 2.46* | 1.22* |
| HA (H1N1) | 1.35* | 2.26* | 1.31* |
| HIV ENV | 1.5* | 2.07* | 1.46* |
Two-sided Fisher test, P < 0.05.
We next examined whether the fact that transversions are more likely to be radical explains all of the observed differences in Ts–Tv fitness effects in the DMS studies. If the fitness differences can be accounted for by this bias, the differences should be eliminated when comparing both radical Ts to radical Tv and conservative Ts to conservative Tv. Alternatively, if this bias does not account for the difference, one would see a difference in fitness between transitions and transversions among radical or among conservative changes of a given category.
We first compared radical transitions to radical transversions. In NP (H3N2), there are no significant Ts–Tv differences among radical charge (black) or radical polarity and size (blue) changes. However, among radical polarity (red) changes, transversions are more detrimental than transitions—an effect not seen in the overall data set (fig. 3A). In NP (H1N1), radical transversions are more detrimental than radical transitions for all three amino acid classifications at the first threshold (fig. 5A). In both NP proteins, the Ts–Tv fitness difference is greater in magnitude among radical polarity changes as compared with the differences among all mutations (the odds ratio is > 3 in fig. 5A but is ≤ 2 in fig. 3A), indicating that controlling for radical transversions can increase rather than eliminate Ts–Tv differences. In HA (H1N1), there are no significant Ts–Tv differences among radical charge changes. However, among radical polarity and polarity and size changes, transversions are more detrimental than transitions. Similarly, in HIV ENV, there are no Ts–Tv differences among radical charge changes (fig. 5B). Transversions are more detrimental than transitions among radical polarity and polarity and size changes, although to a lesser degree as compared with overall in figure 3. Thus, even among radical substitutions of three different amino acid categories, transitions tend to be less detrimental than transversions.
Fig. 5.
Distribution of radical Ts and radical Tv. Empirical cumulative distribution functions of radical transitions (solid lines) and radical transversions (dotted lines) in deep mutational scanning data sets of two NP proteins (A, top) and two antigenic surface proteins (B, top). Computed for radical amino acid changes classified by charge (black), polarity (red), and polarity and size (blue). Odds ratio estimated by a Fisher test comparing the odds of a radical transversion versus a radical transition to be at or below each of 19 fitness thresholds, beginning a t a fitness of 0.05 and increasing by 0.05, for the same data sets (A and B, bottom). Color scheme is the same as in the CDFs. Filled circles denote P<0.05 for a two-sided test with Holm–Bonferroni correction. Lines shown to clarify trends only. Plotted data and raw P values can be found at https://github.com/lauringlab/tstv_paper, last accessed September 26, 2017.
We then compared conservative transitions to conservative transversions (fig. 6). In both NP proteins, there are no significant Ts–Tv differences among conservative changes of all three amino acid classes (fig. 6A). In HA (H1N1), there are no significant Ts–Tv differences among conservative polarity or polarity and size changes. However, transversions are more detrimental than transitions among conservative charge changes. In HIV ENV, transversions are more detrimental than transitions among conservative changes of all three types (fig. 6B). Among conservative polarity and size changes, the odds ratio at the second threshold (>6) is higher than the odds ratios when comparing all Ts and Tv mutations (fig. 3B, <4), indicating that controlling for the conservation of transitions can increase rather than eliminate Ts–Tv differences. However, the Ts–Tv differences are reduced among conservative polarity changes as compared with overall (compare the first thresholds—the odds ratio is ∼2 in fig. 6B but ∼5 in fig. 3B). Thus, conservative transversions are more detrimental than conservative transitions for these three categories in some of the data sets.
Fig. 6.
Distribution of conservative Ts and conservative Tv. Empirical cumulative distribution functions of conservative transitions (solid lines) and conservative transversions (dotted lines) in deep mutational scanning data sets of two NP proteins (A, top) and two antigenic surface proteins (B, top). Computed for conservative amino acid changes classified by charge (black), polarity (red), and polarity and size (blue). Odds ratio estimated by a Fisher test comparing the odds of a conservative transversion versus a conservative transition to be at or below each of 19 fitness thresholds, beginning at a fitness of 0.05 and increasing by 0.05, for the same data sets (A and B, bottom). Color scheme is the same as in the CDFs. Filled circles denote P<0.05 for a two-sided test with Holm–Bonferroni correction. Lines shown to clarify trends only. Plotted data and raw P values can be found at https://github.com/lauringlab/tstv_paper, last accessed September 26, 2017.
In sum, transversions are more detrimental than transitions either among radical or among conservative changes of all three amino acid classes in all four proteins (table 3). In some cases, Ts–Tv fitness differences were increased when we constrained the analysis to just radical or conservative changes. For NP (H3N2), the constrained analysis revealed a Ts–Tv fitness difference among radical polarity changes that was not observed overall. In other cases, Ts–Tv fitness differences were eliminated or reduced, mostly when constraining the analysis to conservative changes. Thus, these three amino acid categories at best only partially explain the Ts–Tv fitness differences in these proteins, with conservative transitions and transversions generally being more similar in fitness than overall.
Table 3.
Summary of Results.
| Study |
Ts versus Tv |
Radical Ts versus Radical Tv |
Conservative Ts versus Conservative Tv |
|
|---|---|---|---|---|
| Analysis Type | Increasing AUC | CDF/Fisher | CDF/Fisher | CDF/Fisher |
| Influenza Total | Tv worse* | Tv worse | ||
| HIV ENV | Tv worse* | Tv worse*, except charge | Tv worse*, charge | |
| HA H1N1 | Tv worse* | Tv worse*, except charge | Tv worse*, all types | |
| NP H1N1 | Tv worse* | Tv worse*, polarity | No sig. differences | |
| NP H3N2 | Tv worse | Tv worse*, all types | No sig. differences | |
Statistically significant as described in Results.
Discussion
We addressed a longstanding question in molecular evolution, whether the observed Ts:Tv substitution bias is due to a mutational bias or to selection disfavoring transversions. We found that missense transversions are more detrimental to fitness than transitions in two RNA viruses, influenza and HIV. Our study therefore provides direct support for the selective hypothesis. Furthermore, transversions are more detrimental even when controlling for their greater likelihood of causing a radical amino acid change. These data demonstrate that commonly used classifications of amino acid changes may not adequately capture the varying selective constraints on different proteins.
The fitness differences between transitions and transversions can be measured in multiple ways and are not well described by a single summary statistic. In four analyzed DMS data sets, the distribution of fitness effects of transversions is shifted toward more deleterious effects. However, they differ at the fitness level at which the shift occurs. We suggest that one explanation for finding a null result is the use of AUC as a summary statistic, which can overweight effects at the ends of the fitness distribution and obscure differences in other regions. In contrast, our use of a Fisher test and large DMS data sets allowed for explicit comparisons of Ts and Tv fitness effects along the distributions without sacrificing power.
Several observations support the idea that the small but significant Ts–Tv fitness differences we identify are biologically relevant and can plausibly explain the Ts:Tv substitution bias. First, despite variation in the Ts–Tv fitness differences, transitions are never more detrimental than transversions, and transversions are either similar to transitions or more detrimental (table 3). This consistent trend suggests that we are identifying a biologically important generality in the effects of transitions and transversions and not simply subtle variations in effects in the highly powered DMS data sets. Second, the Ts–Tv fitness differences in many cases are similar to or even greater than those between radical and conservative amino acid changes. For example, at the low-fitness end of the HIV ENV distribution, the Ts–Tv fitness difference (fig. 3) is greater than that between radical and conservative changes of any type at any fitness level for any gene studied (fig. 4). The radical/conservative distinction has widely accepted evolutionary consequences—conservative substitutions occur more often than radical ones in proteins under purifying selection (Epstein 1967; Clarke 1970; Miyata et al. 1979; Zhang 2000; Miller and Kumar 2001; Duda et al. 2002; Eyre-Walker et al. 2002; Popadin et al. 2007). If the fitness differences between radical and conservative changes have consequences for protein evolution, then the similar or greater fitness differences between transversions and transitions are likely to be consequential as well. Finally, the biological relevance of these effects is also supported by our own and other measurements of Ts and Tv mutation rates in several influenza strains (Bloom 2014; Pauly et al. 2017). The Ts:Tv mutational bias is 2–3.6, significantly less than the average observed Ts:Tv substitution ratio of 5.24 in influenza (Duchêne et al. 2015). These measured mutational biases demonstrate that the selective and mutational hypotheses for the Ts:Tv substitution bias are not mutually exclusive. An important area for future work will be to determine the relative impact of the transitional mutational and selective biases on the overall Ts:Tv substitution bias, particularly in varying genomic contexts (e.g., coding vs. noncoding regions).
Although transversions are more likely to be radical than transitions, this bias only partially accounts for the observed differences in fitness effects. This is perhaps not surprising, as the radical/conservative distinction may not capture the varying constraints on proteins of diverse structure and function. For example, the radical/conservative distinction did not always predict fitness in viral genes. We suggest that the Ts–Tv distinction might be able to better capture these differing functional constraints because transversions are more likely to be radical for a number of different amino acid categories, not just the three analyzed here (Stoltzfus and Norris 2016). Dozens of amino acid categories and many other metrics, such those provided by Polyphen and SIFT, exist for predicting the fitness effects of amino acid substitutions (Kawashima et al. 2007; Kumar et al. 2009; Stoltzfus and Norris 2016). Any one of our simple categories cannot themselves explain the Ts–Tv fitness difference, but their combination, represented in the Ts–Tv distinction, can be quite generally predictive of fitness. Therefore, just as the radical/conservative substitution ratio has been used to detect relaxed selection or positive selection (Hughes et al. 1990; Eyre-Walker et al. 2002; Zhang et al. 2002; Pupko et al. 2003; Zhang and Webb 2004; Tennessen 2005; Shen et al. 2009; Wernegreen 2011), our data support the use of the Ts:Tv ratio as an independent, and perhaps more general, test of selection.
We focused on nonsynonymous point mutations because the available evidence suggests that these have greater fitness impacts than synonymous or noncoding mutations (Cuevas et al. 2012). Significant fitness effects from synonymous substitutions are more often observed with large scale changes rather than individual mutations; for example, a complete change in the codon usage of a gene (Lauring et al. 2012). Other selective pressures on synonymous or noncoding sequences include regulation of replication and translation (Groeneveld et al. 1995; Klovins et al. 1998), targeting by host RNAses (Klovins et al. 1997), and G + C content and thermostability of RNA structures with various functions (Schultes et al. 1997; Smit et al. 2009; Watts et al. 2009). Although it is possible that these selective pressures also contribute to the observed fitness disadvantage of transversions, the main factor is likely the amino acid change.
Our data suggest that the predictive value of the radical/conservative amino acid distinctions may vary due to differing functions of the structural and nonstructural proteins of viruses. Genes encoding for the surface proteins often have a history of intense frequency-dependent selection and may exhibit tolerance to mutations that allow for immune escape while preserving their essential functions of binding and fusion (Stephens and Waelbroeck 1999; Plotkin and Dushoff 2003; Thyagarajan and Bloom 2014; Doud and Bloom 2016; Visher et al. 2016). We therefore expected radical amino acid changes, which may allow for immune escape, to exhibit a less pronounced fitness disadvantage in the surface proteins (HA and ENV) as compared with the internal NP proteins. This is true for charge changes, but not for polarity and polarity and size changes. This observation is also in agreement with two studies of codon usage bias in HA and HIV ENV (Stephens and Waelbroeck 1999; Plotkin and Dushoff 2003). These studies found that, as compared with nonantigenic regions or genes, the antigenic regions exhibit a bias toward codons that tend to mutate nonsynonymously, but not toward codons that tend to mutate to radical polarity and size changes. Charge changes were not evaluated. Thus, these genes may be more tolerant of charge changes that allow for immune escape. If correct, one might expect a bias toward codons that preferentially mutate to radical charge changes and that charge changes, rather than polarity and/or size changes, more often lead to escape from host immune pressure.
The observed Ts–Tv fitness differences suggest an evolutionarily informed approach to improving antiviral strategies. Mutagenic drugs have been used to cause extinction of a variety of viruses in cell culture, a strategy called lethal mutagenesis (Anderson et al. 2004; Bull et al. 2007). There has been little consideration regarding the choice of mutagenic drug, and most commonly employed mutagens cause transitions (Crotty et al. 2001; Ruiz-Jarabo et al. 2003; Graci and Cameron 2008; Dapp et al. 2009). We suggest that the most effective way to achieve lethal mutagenesis may be by using drugs that increase the rate of the more deleterious transversion mutations. In fact, a previous report from our lab showed that the influenza RNA polymerase makes fewer transversions than transitions (Pauly and Lauring 2015). Additionally, 5-azacytidine, a mutagenic drug that causes transversions, is more effective at reducing viral infectivity than two drugs that cause transitions (Pauly and Lauring 2015). Given our results, we speculate that the same may be true for HIV.
Here we find that despite being broad mutational categories, transitions and transversions can capture functional constraints in very different proteins in two viruses. Although the underlying reason for the relative fitness advantage of transitions likely depends on the structure of the genetic code and the accessibility of different types of amino acids, we have shown that the reason is not as simple as the lower likelihood of a transition causing radical changes of certain broad categories. One possibility is that the codon usage in RNA viruses may have evolved in part to buffer a transitional mutation load (Sanjuán 2010; Lauring et al. 2012) due to their high mutation rates and underlying transitional mutation bias (Drake and Holland 1999; Pauly et al. 2017). Identifying the combination of biochemical factors that lead to the fitness advantage of transitions, the relative effects of selection and mutational biases on the overall Ts:Tv substitution bias, and the degree to which these results extend beyond RNA viruses will be important areas of further research.
Materials and Methods
Data
All fitness and site preference data were obtained from sup-plementary material in the published articles or provided by the authors directly. Please see the original papers for details on measurements of fitness and site preference (Rihn et al. 2013, 2015; Thyagarajan and Bloom 2014; Doud et al. 2015; Doud and Bloom 2016; Haddox et al. 2016; Visher et al. 2016). To identify transition-only and transversion-only accessible amino acid substitutions for the mutational scanning data, we obtained the backbone nucleotide sequence of the genes in which the amino acid substitutions were made. These were provided in supplemental files in the published articles for HA (H1N1), NP (H3N2), and HIV ENV. The sequence for NP (H1N1) was obtained from Genbank (Accession number EF467822.1; last accessed September 26, 2017). All sequences can be found online at https://github.com/lauringlab/tstv_paper, last accessed September 26, 2017. For all our analyses, we excluded beneficial mutations, synonymous mutations, and amino acid substitutions accessible by both transitions and transversions or requiring more than one nucleotide mutation.
AUC Analysis
Our AUC analysis was performed exactly as in Stoltzfus and Norris 2016. An ROC curve plots the true positive rate against the false positive rate of a binary classifier system as the discrimination threshold varies. The AUC is the area under this curve. Consider a hypothetical example in which a fitness value between 0 and 1 serves as a discrimination threshold used to predict whether a mutation is a transition or a transversion. A mutation with a fitness value above the threshold level will be classified as a transition and below the threshold level as a transversion. Thus, the true positive rate is the proportion of transitions above the threshold and the false positive rate is the proportion of transversions above the threshold. If transitions and transversions do not differ in their fitness effects, the true positive rate will be equal to the false positive rate at all threshold levels and an ROC curve would show a 1:1 line. The AUC in this null case is half of the total ROC plot area, or 0.5. If transitions generally have a higher fitness than transversions, the true positive rate will be higher than the false positive rate at most threshold levels. The corresponding ROC curve would have a steeper slope than a 1:1 line and have an AUC >0.5. The greater the difference in fitness between transitions and transversions, the greater the difference between the true positive rates and the false positive rates, leading to a steeper ROC curve and a greater AUC. The AUC is mathematically equivalent to the chance that a randomly chosen positive instance of the classifier system is ranked higher than a randomly chosen negative instance (Hanley and McNeil 1982; Mason and Graham 2002). Thus, for our analysis, the AUC is the probability that a randomly chosen transition has a higher fitness value than a randomly chosen transversion. The AUC is calculated from the Mann–Whitney U test (Hanley and McNeil 1982; Mason and Graham 2002): AUC = (pairs−statistic)/pairs, where pairs = number of transitions number of transversions and statistic is the Mann–Whitney U test statistic comparing the fitness values of transitions and transversions. Statistics were calculated using the wilcox.test() function in R. All P values are for a one-sided Mann–Whitney U test where the alternative hypothesis is that transitions are ranked higher than transversions.
Empirical CDF and Odds Ratios
Empirical CDF were computed using the ggplot2 stat_ecdf function in R. Odds ratios were estimated by Fisher’s exact test using the fisher.test() function in R. When the odds ratio was calculated as infinite (e.g., when transversions fall below a relative fitness threshold but no transitions fall below the threshold), the estimated lower 95% confidence interval of the odds ratio was plotted. All P values are for a two-sided test. Holm–Bonferroni correction was implemented when comparisons were performed at multiple fitness level thresholds for a given data set.
Availability of Computer Code and Data
R version 3.3.2 was used for all data analysis and to create all figures. Scripts and data are available online at https://github.com/lauringlab/tstv_paper, last accessed September 26, 2017. as are all plotted data along with unadjusted P values for all figures.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank George Zhang for helpful guidance and a critical reading of the manuscript. We thank Kayla Peck for helpful comments on the manuscript and review of R scripts. We thank Jesse Bloom for suggestions on appropriate use of the DMS data. We thank Rafael Sanjuan for providing the raw data for the VSV study and Suzannah Rihn and Paul Bieniasz for providing the raw data for the HIV IN and CA studies. This work was supported by a Clinician Scientist Development Award from the Doris Duke Charitable Foundation (CSDA 2013105) and R01 AI118886, both to A.S.L. D.M.L. was supported by University of Michigan Medical Scientist Training Program (T32GM007863).
References
- Anderson JP, Daifuku R, Loeb LA.. 2004. Viral error catastrophe by mutagenic nucleosides. Annu Rev Microbiol. 58:183–205. [DOI] [PubMed] [Google Scholar]
- Bloom JD. 2014. An experimentally determined evolutionary model dramatically improves phylogenetic fit. Mol Biol Evol. 318:1956–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ, Sanjuán R, Wilke CO.. 2007. Theory of lethal mutagenesis for viruses. J Virol. 816:2930–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco P, Daròs JA, Agudelo-Romero P, Elena SF.. 2007. A real-time RT-PCR assay for quantifying the fitness of tobacco etch virus in competition experiments. J Virol Methods 1392:181–188. [DOI] [PubMed] [Google Scholar]
- Carrasco P, de la Iglesia F, Elena SF.. 2007. Distribution of fitness and virulence effects caused by single-nucleotide substitutions in tobacco etch virus. J Virol. 8123:12979–12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke B. 1970. Selective constraints on amino-acid substitutions during the evolution of proteins. Nature 2285267:159–160. [DOI] [PubMed] [Google Scholar]
- Crotty S, Cameron CE, Andino R.. 2001. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc Natl Acad Sci U S A. 9812:6895–6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas JM, Domingo-Calap P, Sanjuán R.. 2012. The fitness effects of synonymous mutations in DNA and RNA viruses. Mol Biol Evol. 291:17–20. [DOI] [PubMed] [Google Scholar]
- Dagan T, Talmor Y, Graur D.. 2002. Ratios of radical to conservative amino acid replacement are affected by mutational and compositional factors and may not be indicative of positive Darwinian selection. Mol Biol Evol. 197:1022–1025. [DOI] [PubMed] [Google Scholar]
- Dapp MJ, Clouser CL, Patterson S, Mansky LM.. 2009. 5-Azacytidine can induce lethal mutagenesis in human immunodeficiency virus type 1. J Virol. 8322:11950–11958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Morris K, Lynch M, Thomas WK.. 2004. High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature 4307000:679–682. [DOI] [PubMed] [Google Scholar]
- Domingo-Calap P, Cuevas JM, Sanjuá NR.. 2009. The fitness effects of random mutations in single-stranded DNA and RNA bacteriophages. PLoS Genet 5(11): e1000742. [DOI] [PMC free article] [PubMed]
- Doud MB, Ashenberg O, Bloom JD.. 2015. Site-specific amino acid preferences are mostly conserved in two closely related protein homologs. Mol Biol Evol. 3211:2944–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doud MB, Bloom JD.. 2016. Accurate measurement of the effects of all amino-acid mutations on influenza hemagglutinin. Viruses 86:155.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JW, Holland JJ.. 1999. Mutation rates among RNA viruses. Proc Natl Acad Sci U S A. 9624:13910–13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchêne S, Ho SY, Holmes EC.. 2015. Declining transition/transversion ratios through time reveal limitations to the accuracy of nucleotide substitution models. BMC Evol Biol. 15:36.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda TF, Vanhoye D, Nicolas P.. 2002. Roles of diversifying selection and coordinated evolution in the evolution of amphibian antimicrobial peptides. Mol Biol Evol. 196:858–864. [DOI] [PubMed] [Google Scholar]
- Epstein CJ. 1967. Non-randomness of ammo-acid changes in the evolution of homologous proteins. Nature 2155099:355–359. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley PD, Smith NGC, Gaffney D.. 2002. Quantifying the slightly deleterious mutation model of molecular evolution. Mol Biol Evol. 1912:2142–2149. [DOI] [PubMed] [Google Scholar]
- Fitch WM. 1967. Evidence suggesting a non-random character to nucleotide replacements in naturally occurring mutations. J Mol Biol. 263:499–507. [DOI] [PubMed] [Google Scholar]
- Gojobori T, Li W-H, Graur D.. 1982. Patterns of nucleotide substitution in pseudogenes and functional genes. J Mol Evol. 185:360–369. [DOI] [PubMed] [Google Scholar]
- Graci JD, Cameron CE.. 2008. Therapeutically targeting RNA viruses via lethal mutagenesis. Future Virol. 36:553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R. 1974. Amino acid difference formula to help explain protein evolution. Science 1854154:862–864. [DOI] [PubMed] [Google Scholar]
- Groeneveld H, Thimon K, van Duin J.. 1995. Translational control of maturation-protein synthesis in phage MS2: a role for the kinetics of RNA folding? RNA 11:79–88. [PMC free article] [PubMed] [Google Scholar]
- Haddox HK, Dingens AS, Bloom JD.. 2016. Experimental estimation of the effects of all amino-acid mutations to HIV’s envelope protein on viral replication in cell culture. PLoS Pathog. 1212:e1006114.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley JA, McNeil BJ.. 1982. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1431:29–36. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Ota T, Nei M.. 1990. Positive Darwinian selection promotes charge profile diversity in the antigen-binding cleft of class I major-histocompatibility-complex molecules. Mol Biol Evol. 7:515–524. [DOI] [PubMed] [Google Scholar]
- Jacquier H, Birgy A, Le Nagard H, Mechulam Y, Schmitt E, Glodt J, Bercot B, Petit E, Poulain J, Barnaud G, et al. 2013. Capturing the mutational landscape of the beta-lactamase TEM-1. Proc Natl Acad Sci U S A. 11032:13067–13072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Zhao Z.. 2006. Mutational spectrum in the recent human genome inferred by single nucleotide polymorphisms. Genomics 885:527–534. [DOI] [PubMed] [Google Scholar]
- Kawashima S, Pokarowski P, Pokarowska M, Kolinski A, Katayama T, Kanehisa M.. 2007. AAindex: amino acid index database, progress report 2008. Nucleic Acids Res. 36(Database):D202–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klovins J, Berzins V, Duin JV.. 1998. A long-range interaction in Qβ RNA that bridges the thousand nucleotides between the M-site and the 3′ end is required for replication. RNA 48:948–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klovins J, van Duin J, Olsthoorn RCL.. 1997. Rescue of the RNA phage genome from RNase III cleavage. Nucleic Acids Res. 2521:4201–4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC.. 2009. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 47:1073–1081. [DOI] [PubMed] [Google Scholar]
- Kumar S. 1996. Patterns of nucleotide substitution in mitochondrial protein coding genes of vertebrates. Genetics 1431:537–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring AS, Acevedo A, Cooper SB, Andino R.. 2012. Codon usage determines the mutational robustness, evolutionary capacity, and virulence of an RNA virus. Cell Host Microbe 125:623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. 2010. Rate, molecular spectrum, and consequences of human mutation. Proc Natl Acad Sci U S A. 1073:961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason SJ, Graham NE.. 2002. Areas beneath the relative operating characteristics (ROC) and relative operating levels (ROL) curves: statistical significance and interpretation. Q J R Meteorol Soc. 128584:2145–2166. [Google Scholar]
- Miller MP, Kumar S.. 2001. Understanding human disease mutations through the use of interspecific genetic variation. Hum Mol Genet. 1021:2319–2328. [DOI] [PubMed] [Google Scholar]
- Miyata T, Miyazawa S, Yasunaga T.. 1979. Two types of amino acid substitutions in protein evolution. J Mol Evol. 123:219–236. [DOI] [PubMed] [Google Scholar]
- Pauly MD, Lauring AS.. 2015. Effective lethal mutagenesis of influenza virus by three nucleoside analogs. J Virol. 897:3584–3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly MD, Procario MC, Lauring AS.. 2017. A novel twelve class fluctuation test reveals higher than expected mutation rates for influenza A viruses. eLife 6:e26437.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris JB, Davis P, Cuevas JM, Nebot MR, Sanjuán R.. 2010. Distribution of fitness effects caused by single-nucleotide substitutions in bacteriophage f1. Genetics 1852:603–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov DA, Hartl DL.. 1999. Patterns of nucleotide substitution in Drosophila and mammalian genomes. Proc Natl Acad Sci U S A. 964:1475–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin JB, Dushoff J.. 2003. Codon bias and frequency-dependent selection on the hemagglutinin epitopes of influenza A virus. Proc Natl Acad Sci U S A. 10012:7152–7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popadin K, Polishchuk LV, Mamirova L, Knorre D, Gunbin K.. 2007. Accumulation of slightly deleterious mutations in mitochondrial protein-coding genes of large versus small mammals. Proc Natl Acad Sci U S A. 10433:13390–13395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pupko T, Sharan R, Hasegawa M, Shamir R, Graur D.. 2003. Detecting excess radical replacements in phylogenetic trees. Gene 319:127–135. [DOI] [PubMed] [Google Scholar]
- Rihn SJ, Hughes J, Wilson SJ, Bieniasz PD.. 2015. Uneven genetic robustness of HIV-1 integrase. J Virol. 891:552–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rihn SJ, Wilson SJ, Loman NJ, Alim M, Bakker SE, Bhella D, Gifford RJ, Rixon FJ, Bieniasz PD.. 2013. Extreme genetic fragility of the HIV-1 capsid. PLoS Pathog. 96:e1003461.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg MS, Subramanian S, Kumar S.. 2003. Patterns of transitional mutation biases within and among mammalian genomes. Mol Biol Evol. 206:988–993. [DOI] [PubMed] [Google Scholar]
- Ruiz-Jarabo CM, Ly C, Domingo E, de la Torre JC.. 2003. Lethal mutagenesis of the prototypic arenavirus lymphocytic choriomeningitis virus (LCMV). Virology 3081:37–47. [DOI] [PubMed] [Google Scholar]
- Sanjuán R. 2010. Mutational fitness effects in RNA and single-stranded DNA viruses: common patterns revealed by site-directed mutagenesis studies. Philos Trans R Soc Lond B Biol Sci. 3651548:1975–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuán R, Moya A, Elena SF.. 2004. The distribution of fitness effects caused by single-nucleotide substitutions in an RNA virus. Proc Natl Acad Sci U S A. 10122:8396–8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultes E, Hraber PT, LaBean TH.. 1997. Global similarities in nucleotide base composition among disparate functional classes of single-stranded RNA imply adaptive evolutionary convergence. RNA 37:792–806. [PMC free article] [PubMed] [Google Scholar]
- Shen Y-Y, Shi P, Sun Y-B, Zhang Y-P.. 2009. Relaxation of selective constraints on avian mitochondrial DNA following the degeneration of flight ability. Genome Res. 1910:1760–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit S, Knight R, Heringa J.. 2009. RNA structure prediction from evolutionary patterns of nucleotide composition. Nucleic Acids Res. 375:1378–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens CR, Waelbroeck H.. 1999. Codon bias and mutability in HIV sequences. J Mol Evol. 484:390–397. [DOI] [PubMed] [Google Scholar]
- Stoltzfus A, Norris RW.. 2016. On the causes of evolutionary transition: transversion bias. Mol Biol Evol. 333:595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA. 2005. Molecular evolution of animal antimicrobial peptides: widespread moderate positive selection. J Evol Biol. 186:1387–1394. [DOI] [PubMed] [Google Scholar]
- Thyagarajan B, Bloom JD.. 2014. The inherent mutational tolerance and antigenic evolvability of influenza hemagglutinin. eLife 3:e03300.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visher E, Whitefield SE, McCrone JT, Fitzsimmons W, Lauring AS.. 2016. The mutational robustness of influenza A virus. PLoS Pathog. 128:e1005856.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel F, Kopun M.. 1977. Higher frequencies of transitions among point mutations. J Mol Evol. 92:159–180. [DOI] [PubMed] [Google Scholar]
- Wakeley J. 1996. The excess of transitions among nucleotide substitutions: new methods of estimating transition bias underscore its significance. Trends Ecol Evol. 114:158–162. [DOI] [PubMed] [Google Scholar]
- Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Swanstrom R, Burch CL, Weeks KM.. 2009. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 4607256:711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernegreen JJ. 2011. Reduced selective constraint in endosymbionts: elevation in radical amino acid replacements occurs genome-wide. PLoS One 612:e28905.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yampolsky LY, Stoltzfus A.. 2005. The exchangeability of amino acids in proteins. Genetics 1704:1459–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. 2000. Rates of conservative and radical nonsynonymous nucleotide substitutions in mammalian nuclear genes. J Mol Evol. 501:56–68. [DOI] [PubMed] [Google Scholar]
- Zhang J, Webb DM.. 2004. Rapid evolution of primate antiviral enzyme APOBEC3G. Hum Mol Genet. 1316:1785–1791. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang Y, Rosenberg HF.. 2002. Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nat Genet. 304:411–415. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Gerstein M.. 2003. Patterns of nucleotide substitution, insertion and deletion in the human genome inferred from pseudogenes. Nucleic Acids Res. 3118:5338–5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.