Abstract
New genes can arise through duplication of a pre-existing gene or de novo from non-coding DNA, providing raw material for evolution of new functions in response to a changing environment. A prime example is the independent evolution of antifreeze glycoprotein genes (afgps) in the Arctic codfishes and Antarctic notothenioids to prevent freezing. However, the highly repetitive nature of these genes complicates studies of their organization. In notothenioids, afgps evolved from an extant gene, yet the evolutionary origin of afgps in codfishes is unknown. Here, we demonstrate that afgps in codfishes have evolved de novo from non-coding DNA 13–18 Ma, coinciding with the cooling of the Northern Hemisphere. Using whole-genome sequence data from several codfishes and notothenioids, we find higher copy number of afgp in species exposed to more severe freezing suggesting a gene dosage effect. Notably, antifreeze function is lost in one lineage of codfishes analogous to the afgp losses in non-Antarctic notothenioids. This indicates that selection can eliminate the antifreeze function when freezing is no longer imminent. In addition, we show that evolution of afgp-assisting antifreeze potentiating protein genes (afpps) in notothenioids coincides with origin and lineage-specific losses of afgp. The origin of afgps in codfishes is one of the first examples of an essential gene born from non-coding DNA in a non-model species. Our study underlines the power of comparative genomics to uncover past molecular signatures of genome evolution, and further highlights the impact of de novo gene origin in response to a changing selection regime.
Keywords: orphan genes, teleost fishes, molecular adaptation
Introduction
Genomes recurrently acquire new genes, often to take on novel functions in response to a changing selection regime. One notable driver of evolutionary innovation is paleoclimatic changes such as the global cooling and polar icecap formation 10–30 Ma (Kennett 1977; Eastman 1997). This spurred the evolution of the antifreeze proteins (AFPs), which have evolved independently in bacteria, plants (≥ four times), fungi, insects (≥ two times), and teleost fish (≥ seven times) (Cheng 1998; Ewart et al. 1999; Harding et al. 2003; Bildanova et al. 2013; Gupta and Deswal 2014). The most classic way of acquiring new genes is through gene duplications, and many of the AFPs have arisen through neofunctionalization of such duplicates (Liu et al. 2007; Graham et al. 2013). Alternatively, new genes can evolve de novo from non-coding DNA, either by transcripts acquiring an open reading frame (ORF) or consistently transcribed regions of the genome acquiring an ORF (McLysaght and Guerzoni 2015; Schlötterer 2015). De novo gene origin has recently become more widely recognized as a regular source of new genes (Tautz and Domazet-Lošo 2011; Wu et al. 2011; McLysaght and Guerzoni 2015; Schlötterer 2015; McLysaght and Hurst 2016), which often encode novel functions representing lineage specific adaptations to the environment (Khalturin et al. 2009; Tautz and Domazet-Lošo 2011). In notothenioid fishes antifreeze glycoproteins (AFGPs) evolved from neofunctionalization of a duplicate of trypsinogen-like protease (TLP) through a unique recruitment of intronic sequence to form coding DNA (Chen et al. 1997a). In some species within the group of distantly related codfishes (Gadidae) such as Atlantic cod (Gadus morhua), similar AFGP genes (afgps) are the result of convergent evolution (Chen et al. 1997 b). Codfish afgps are suggested to be orphans, i.e. not homologous to any other gene, and likely to have originated de novo from non-genic DNA (Zhuang 2014). Although the exact genesis of cod afgps remains unknown, they are most likely relatively young genes. Such tracing their evolutionary history should be possible, making cod afgps good candidates for studying the role of new genes associated with key innovations such as antifreeze properties.
The evolutionary convergence of AFGPs in notothenioids and codfishes is intriguing as AFGPs in both lineages consist of nearly identical repeats of Thr-Ala(/Pro)-Ala (in codfishes Thr is occasionally substituted with Arg) (Chen et al. 1997 b). These repeats are strung together in large polyproteins that are cleaved after translation yielding isoforms of multiple sizes (Chen et al. 1997a, 1997b), with the shared ability to depress the freezing point of body fluids through thermal hysteresis by binding to ice crystals and preventing them from growing (Kristiansen and Zachariassen 2005). The similar selection pressures imposed by the onset of freezing temperatures in the Arctic and Antarctic have remarkably produced the same function carried out by nearly identical proteins. The independent origin of afgps is evident as the genetic organization and codon usage of afgp are distinctive in the two lineages (Chen et al. 1997b). Codfish afgp has three exons encoding a signal peptide (exon 1, exon 2, and the beginning of exon 3) and the afgp repetitive region (the remainder of exon 3). Notothenioid afgp has two exons, one encoding the signal peptide and another encoding the afgp repeat (Chen et al. 1997b). In addition, notothenioids possess a second AFP known as antifreeze potentiating protein (AFPP) that only exhibits moderate antifreeze activity by itself, but facilitates the function of AFGP (Yang et al. 2013). Especially in species exposed to freezing temperatures year-round, AFPP contributes significantly to a substantial proportion of the total antifreeze activity (Fields and Devries 2015). The evolutionary history of AFPP and how it relates to the appearance of AFGP are not known to date.
There are genetic studies on afgps in a few species in each lineage (reviewed in Cheng 1998; Harding et al. 2003), chiefly on polar cod (Boreogadus saida) in codfishes (Chen et al. 1997 b), and the notothenioid Antarctic toothfish (Dissostichus mawsoni) (Chen et al. 1997a; Cheng 2003; Nicodemus-Johnson et al. 2011; Near et al. 2012). However, none of these studies have looked at afgps in a genomic context, except one attempt to assemble the afgp locus in G. morhua (Zhuang et al. 2012). The afgp locus, including its flanking genes, is still not completely characterized in neither codfishes nor notothenioids, probably due to the challenges of assembling repetitive regions such as those in afgp. Whole genome sequencing (WGS) may provide more accurate estimates of copy number variation (CNV), inference of gene losses, pseudogenization, and resolution of genetic organization and synteny (e.g. Goodwin et al. 2016). Moreover, for comparative studies WGS data have the advantage that all homologous sequences can in most cases be detected by BLAST (Albà and Castresana 2007) to give a complete genomic picture of a gene family.
Here, we use a comparative genomics approach to determine when afgps originated in codfishes, as well as resolving the CNV and genomic organization of afgps in both notothenioids and codfishes. Furthermore, for the less studied AFPP genes (afpps) we have addressed their genomic origin and when they arose in the evolutionary history of notothenioids. To achieve this, we sequenced the genomes of eight notothenioid species; Pleuragramma antarctica, Trematomus newnesi, Harpagifer kerguelensis, Artedidraco skottsbergi, Gymnodraco acuticeps, and Chaenocephalus aceratus as they represent the main notothenioid lineages that have afgp; the non-Antarctic species Eleginops maclovinus that never had afgp, and Patagonothen guntheri, that secondarily left the Antarctic and lost afgp (Near et al. 2012; Miya et al. 2016). In addition, we included the published N. coriiceps genome in the comparison of the notothenioids (Shin et al. 2014). For codfishes we took advantage of the already generated genome assemblies for G. morhua (Tørresen et al. 2017b), haddock (Melanogrammus aeglefinus) (Tørresen et al. 2017a) and 25 additional published codfish genomes (Malmstrøm et al. 2016, 2017). In both lineages, we coupled the presence/absence and copy number of genes in combination with time-calibrated phylogenetic trees (Colombo et al. 2015; Malmstrøm et al. 2016). Our approach reveals that codfish afgp most likely arose de novo from non-genic DNA around 13–18 Ma, which coincides with the onset of freezing temperatures in the Northern Hemisphere (Eastman 1997). Moreover, afgp has been subsequently lost in one lineage of codfishes, analogous to the loss of afgp in non-Antarctic notothenioids. In notothenioids, afpp coevolved with afgp. In both codfishes and notothenioids there is considerable CNV associated with species living in waters with more severe freezing displaying a higher number of afgps. We here demonstrate the importance of WGS data for comparative genomic studies of molecular evolution, by revealing the complex evolution of afgp involving de novo origin in codfishes, coevolution of afgp and afpp in notothenioids as well as extensive CNV, gene losses and pseudogenizations in both lineages.
Results
Presence, Copy Number, and Organization of afgps in Codfishes
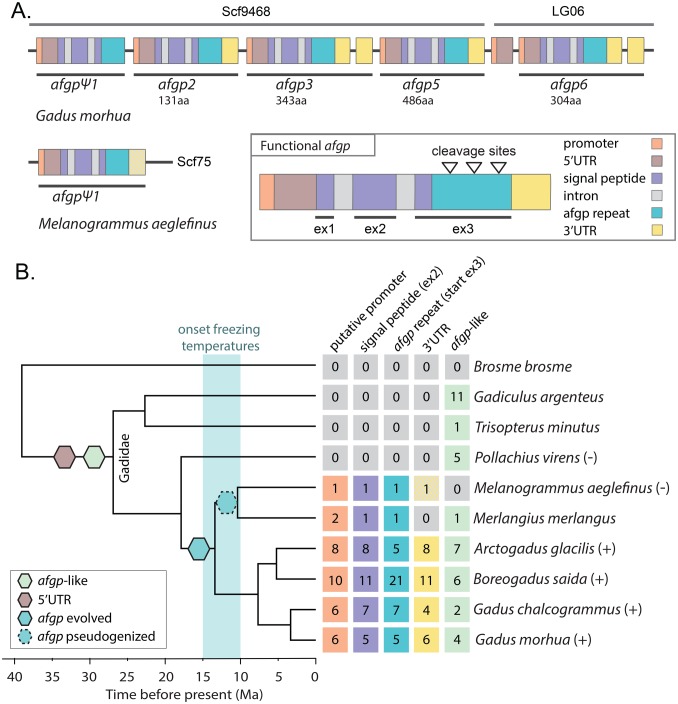
To characterize the genomic organization and micro-synteny of afgps we used the high-quality genome assemblies of G. morhua (Tørresen et al. 2017b) and M. aeglefinus (Tørresen et al. 2017a). Using BLAST we identified four complete copies of afgps on linkage group 6 (LG06) and one copy on scaffold 9468 (Scf9468) in G. morhua (fig. 1). Based on synteny (see further down) Scf9468 was placed within LG06. We defined a full-length afgp gene to contain a promoter, 5′UTR, three exons (abbreviated ‘ex’) that contain both the signal peptide sequence (ex1, ex2, and first part of ex3) and afgp tripeptide repeats (ex3), and a 3′UTR (fig. 1 and supplementary table S1, Supplementary Material online). The afgps were named based on partial afgp sequences from Zhuang et al. (2012). In G. morhua afgp2, afgp3, afgp5, and afgp6 are likely functional genes. afgpψ1 has previously been reported by Zhuang et al. (2012) as a putative pseudogene. We detected a 114 nt insertion in the 5′UTR, a missing 3′UTR and frame-shifting indels rendering the ex3 repeat without the characteristic strings of TAA. Thus, even if this is a functional protein it may not have an antifreeze function.
Fig. 1.
afgps in codfishes. (A) Gene organization of afgps in G. morhua and M. aeglefinus. The afgp genes have been divided up in promoter, 5′UTR, signal peptide, intron, afgp repeat and 3′UTR and colored according to legend. The hatched yellow indicates a truncated 3′UTR. The sequences are labeled with species name, a scaffold (scf) or linkage group (LG) identifier, name of afgps with Ψ signifying a pseudogene, and the length of each gene given as number of amino acids (aa). The organization of a complete, functional afgp gene is shown with triangles indicating cleavage sites of the polyprotein peptide. (B) Presence of afgp in a selection of codfishes in a phylogenetic context, showing copy numbers of different parts of afgps mapped on a time-calibrated species tree modified from (Malmstrøm et al. 2016) with time given in millions of years (Ma). The time period when freezing temperatures appeared in the Northern Hemisphere is shaded in blue (Eastman 1997). Species shown to have functional AFGP and thermal hysteresis are denoted with (+): A. glacilis, B. saida (Praebel 2005), G. chalcogrammus (Tsuda and Miura 2005), and G. morhua (Hew et al. 1981). Species shown not to have functional AFGPs or thermal hysteresis are denoted with (–): M. aeglefinus (Ewart et al. 2000) and P. virens (Denstad et al. 1987). The numbers of putative promoters, ex2, and beginning of ex3 (containing afgp repeat) and 3′UTR are given in the colored boxes. The branches where afgps originated and pseudogenized according to the most parsimonious explanation are indicated in the tree according to the legend along with the first appearance of an afgp 5′UTR sequence.
In M. aeglefinus we identified only one afgp copy on Scf75 (fig. 1). This copy is characterized by a truncated 3′UTR as well as frame-shifting indels and three stop codons in the ex3 repeat; this is a likely pseudogene and homologous to afgpψ1 in G. morhua based on sequence similarity as well as a shared 5′UTR insertion. The absence of functional afgps in M. aeglefinus is in concordance with experimental evidence of no thermal hysteresis and no presence of AFGPs in this species (Ewart et al. 2000). In G. morhua afgp3 and afgp6 have two 3′UTRs and there are a promoter and a 5′UTR that no ORF follows before afgp6 (fig. 1). As there are no afgp-repeat-like sequences upstream of these 3′UTRs and no signal-peptide-like sequences downstream of the 5′UTR, we believe these are not the result of pseudogenization, but rather represent incomplete duplications, or complete duplications followed by deletions of the majority of the gene.
In both G. morhua and M. aeglefinus we found only a single genomic region containing afgp (fig. 1A), which is in accordance with previous findings (Zhuang et al. 2012). However, in G. morhua we found four additional sequences with high similarity to afgp in other genomic regions on LG16, LG23, LG19, and Scf4199. These sequences, denoted as afgp-like, include a promoter-like sequence and a signal peptide-like sequence (ex1, ex2, and beginning of ex3). Although similar, these sequences do not contain the characteristic afgp TAA amino acid repeat or an ORF. The afgp-like ex2 sequences have a sequence identity of 85–91% to our query signal peptide ex2 whereas true signal peptide ex2 are 94–98% identical to each other (supplementary table S2, Supplementary Material online). In addition, we found putative 5′UTR-like sequences of varying length at 376 genomic positions outside the afgp region in G. morhua and 290 genomic positions outside the afgp region in M. aeglefinus with BLAST e-values from 4×10−6 to 5×10−75. 3′UTR-like sequences were not detected outside the region containing the true afgps.
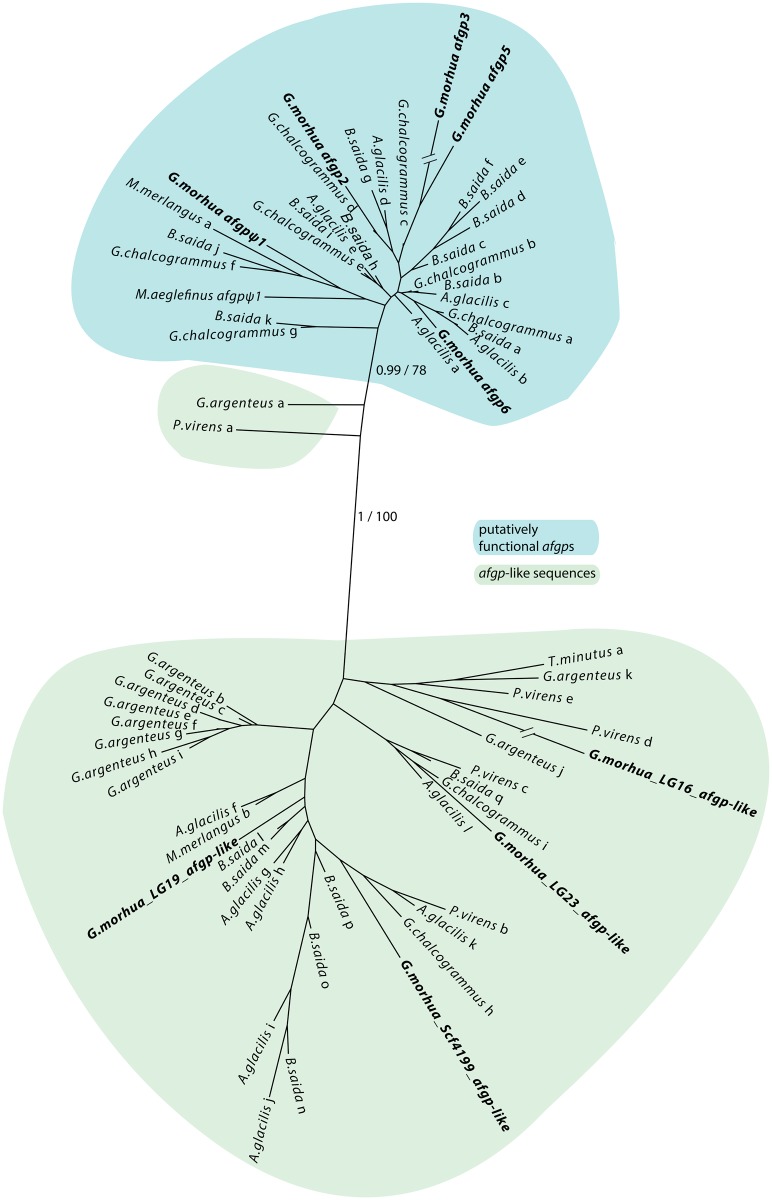
The finding of afgp-like sequences outside the afgp locus in G. morhua complicated the estimation of presence/absence and copy number of afgps in the other Gadiformes genomes, particularly where we could not reconstruct synteny. We located all putative afgp sequences using liberal BLAST searches and then used phylogenetics to determine which sequences were true afgps. All the different components of afgp were found in three codfishes in addition to G. morhua: Gadus chalcogrammus, B. saida, and Arctogadus glacilis (fig. 1B), all of which have been shown to have antifreeze activity in their blood (Hew et al. 1981; Praebel 2005; Tsuda and Miura 2005). Furthermore, we found some segments of afgp in Merlangius merlangus, but just like in its sister species M. aeglefinus a 3′UTR sequence was not detected, suggesting these two species only possess an afgp pseudogene (afgpψ1). There are no afgp-like sequences in Brosme brosme or the 20 codfish genomes investigated outside B. brosme in the phylogeny (see supplementary table S2, Supplementary Material online). However, we did detect some afgp-like sequences in the species more closely related to Atlantic cod: Pollachius virens, Trisopterus minutus, and Gadiculus argenteus. To determine whether the sequences with some similarity to afgp are homologous to the true afgps or the four afgp-like sequences detected in G. morhua we carried out phylogenetic analyses. We included sequences where ex1, ex2, and parts of ex3 were located on one unitig (utg) denoted by a letter (for full utg identifier see supplementary table S3, Supplementary Material online). The un-rooted phylogenetic tree in figure 2 shows that putatively functional afgps together with putative afgp-pseudogenes form a well-supported cluster (posterior probability = 0.99, bootstrap support = 78%). Most of the afgp-like sequences form a single cluster, except G. argenteus_a and P. virens_a; however, these are still outside the cluster of true afgps. Only sequences from G. morhua, G. chalcogrammus, B. saida, A. glacilis, M. merlangus, and M. aeglefinus cluster with true afgps, whereas all sequences from P. virens, T. minutus, and G. argenteus appear to be afgp-like (fig. 2). Furthermore, the afgp-like sequences are located in the G. morhua assembly at LG16 (between calm and kalrn), LG23 (between alox12b and arhgap21), LG19 (between nlpr12 and vldlr), and Scf4199, which contains no genes nor ORFs. In Gasterosteus aculeatus, these genes are not linked, i.e. synteny is not conserved in these regions. The distance between the afgp-like sequence and the closest gene is quite large, ranging from 40 to 160 kb, on average 80 kb. Taken together with the absence of afgp-like sequences outside Gadidae, the evidence for the gadid-specificity of these afgp-like sequences is quite strong, especially considering that the Gadiformes genome assemblies are not repeat-masked, so we would detect these sequences if present.
Fig. 2.
Phylogeny of afgps and afgp-like sequences in codfishes. Sequences from the genomes of G. morhua, M. aeglefinus, G. chalcogrammus, B. saida, A. glacilis, M. merlangius, P. virens, T. minutus, and G. argenteus are included. The sequences from G. morhua and M. aeglefinus have a scaffold (scf) or linkage group (LG) identifier and sequence annotation (either afgp or afgp-like). Ψ is signifying a pseudogene. The remaining sequences have an assigned letter following the species name (details regarding content and genomic position of each sequence is given in supplementary table S3, Supplementary Material online). The tree topology was constructed with MrBayes. Posterior probabilities are shown for the main branching patterns in addition to bootstrap support for a maximum likelihood topology (using MEGA 7). Putatively functional afgps and afgp-like sequences are highlighted in blue and green, respectively, according to legend.
We estimated copy numbers for the different components of afgp independently, as we did not have complete full-length afgp sequences for many of the species. For signal peptide ex2-like sequences we reconstructed a phylogeny revealing that copies of true afgp ex2 ranges from one in M. aeglefinus and M. merlangus, and 11 in B. saida (supplementary fig. S1, Supplementary Material online). For the other components of afgp it was not feasible to construct phylogenies so we inspected the sequences manually and counted the number of copies. The number of different afgp segments varies somewhat within each species (fig. 1), which is not unexpected given the finding that the number of different segments is not equal to the number of complete genes in G. morhua and M. aeglefinus (fig. 1B). Based on the number of signal peptide ex2 sequences there is still a clear pattern in the number of copies, with a higher number in the lineage with functional copies of afgp, ranging from five in G. morhua to B. saida’s estimated copy number of 11 (fig. 1B). Furthermore, we find no afgp genes in P. virens, T. minutus, and G. argenteus (fig. 1B) based on the absence of afgp repeats and 3′UTR sequences in these species, and that ex2-like sequences found in P. virens, T. minutus, and G. argenteus are not homologous with afgp ex2 (fig. 2 and supplementary fig. S1, Supplementary Material online). The absence of afgp in P. virens is concordant with the finding of no thermal hysteresis in the blood plasma (Denstad et al. 1987).
We were not able to count and map the number of afgp 5′UTRs as we could not determine which 5′UTR BLAST hit belonged to afgp or not. This was due to the many copies of 5′UTR-like sequences in the G. morhua genome, which were indistinguishable from the afgp 5′UTR (see supplementary notes, Supplementary Material online for more information). We did not locate 5′UTR-like sequences in species outside Gadidae, suggesting this is a repeat-specific trait for this family (figs. 1 and 2, and supplementary table S2, Supplementary Material online). Moreover, we did not identify any matches to the signal peptide ex2, 5′UTR, 3′UTR, or the antifreeze repeat sequence in ex3 in our searches in RepBase (Bao et al. 2015), indicating that these sequences are unique to the Gadidae family (fig. 1).
For G. morhua and M. aeglefinus we also have lower coverage draft assemblies constructed in the same way as the codfish genomes (G. chalcogrammus, B. saida, A. glacilis, M. merlangus, P. virens, T. minutus, G. argenteus, and B. brosme) (Malmstrøm et al. 2016). We therefore ran the annotation pipeline on these genome assemblies as a proof of principle on our copy number estimation criteria. The estimated numbers were in concordance with the copy-number estimates from the high-quality genome assemblies for both G. morhua and M. aeglefinus, respectively (supplementary table S2, Supplementary Material online).
Genomic Organization and Synteny of Codfish afgps and Flanking Regions
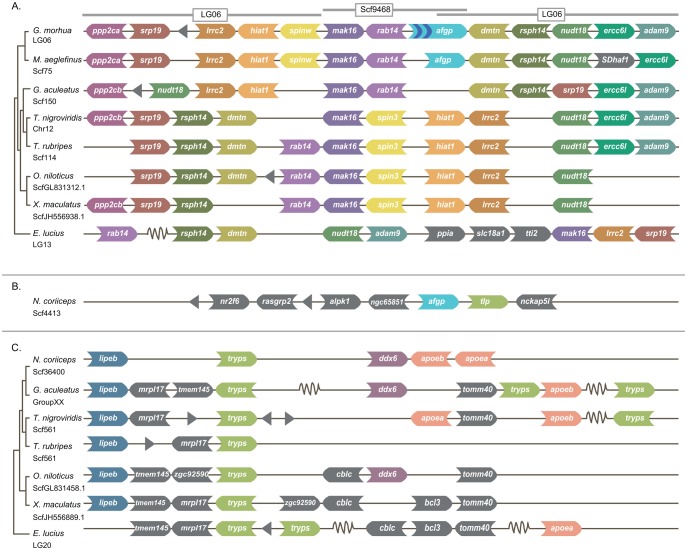
The synteny surrounding the afgp locus is relatively conserved across a selection of teleost fish genomes (fig. 3). This is particularly evident between the phylogenetically close relatives G. morhua and M. aeglefinus; T. nigroviridis, and T. rubripes; and O. niloticus and X. maculatus (fig. 3A). The G. morhua afgp genes are found on LG6 and Scf9468. However, previous studies indicate that there is only one contiguous afgp locus in this species (Zhuang et al. 2012). On LG6, between the genes spinw and dmtn (position LG6: 1941982–1942081), there is a gap—of unknown size—originating from the ordering and orientation of scaffolds into linkage groups (Tørresen et al. 2017 b). Based on the conserved synteny between M. aeglefinus and G. morhua we were able to place Scf9468 in this gap, resulting in the complete, contiguous afgp locus in G. morhua (figs. 1A and 3A). Vista plotting was used to detect short stretches of similar sequences likely to be undetected by BLAST for the region between mak16 and rsph14 in G. morhua, M. aeglefinus, G. aculeatus, O. niloticus, and T. rubripes (supplementary fig. S3, Supplementary Material online). Only the regions containing the flanking genes around afgp (i.e. mak16, rab14, dmtn, and rsph14) are conserved between codfishes and G. aculeatus, O. niloticus, and T. rubripes. There are no regions similar to the afgps in the species outside codfishes, or any conserved non-coding elements. The similarity between G. morhua and M. aeglefinus is high, especially at the flanking genes and the shared afgpΨ1. Furthermore, M. aeglefinus differs from G. morhua at non-coding regions as well as in sequences encoding afgp2, afgp3, afgp5, and afgp6 in G. morhua.
Fig. 3.
Synteny of genes flanking afgp and tryps in codfishes and notothenioids. Scaffold (scf), linkage group (LG), or chromosome (chr) is specified under each species, orientation of genes is indicated as arrows, grey triangles denote unidentified ORFs and truncated lines indicate regions containing genes not shown for practical purposes. (A) Synteny of the afgp genomic region in G. morhua (gadMor2) and M. aeglefinus (melAeg) compared with the following teleosts lacking afgps: G. aculeatus, T. nigroviridis, T. rubripes, O. niloticus, X. maculates, and E. lucius. Scf9468 in G. morhua has been placed in a gap in LG06 based on syntenic context and overlap at the afgp locus. (B) Genomic organization of the afgp locus in N. coriiceps. (C) Synteny of the trypsinogen locus in N. coriiceps and G. aculeatus, T. nigroviridis, T. rubripes, O. niloticus, and E. lucius.
De Novo Origin of Codfish afgp
afgps either evolved from non-coding DNA or pre-existing genes encoding proteins. We did not get any BLAST hits against any part of afgp in genes or ORFs in the high-quality G. morhua and M. aeglefinus genomes, or in the other codfish draft genomes, even with an E-value of 0.1. Furthermore, BLAST got no hits to afgp in Uniprot, the Ensembl genomes or Genbank (except other afgp sequences).
De novo genes are more likely to arise in GC-rich genomic regions as these regions are more transcriptionally active and these areas are more likely to obtain an ORF because stop codons are AT-rich (McLysaght and Hurst 2016). Consistent with this we find that the nucleotide composition is indeed skewed towards a high GC-content in the functional afgp copies in G. morhua (table 1). In fact, the GC-content in the afgp copies was as high as 71% vs. 56% on average for all annotated genes in the G. morhua genome assembly (Tørresen et al. 2017 b), implicating that the high alanine content strongly influences the GC-content of afgps (supplementary fig. S4, Supplementary Material online). In addition, by calculating the relative synonymous codon usage (RSCU) we found a significant codon usage bias (RSCU significantly <1 or >1) for the amino acids in the repeats (Thr, Pro, and Ala) across all the afgps in G. morhua and M. aeglefinus, which is consistent across the genes (supplementary table S4, Supplementary Material online). This finding, together with the occurrence of all afgps on a single linkage group and the well conserved synteny (figs. 1 and 3) between G. morhua and M. aeglefinus strongly suggests a common origin of codfish afgps, with subsequent gene duplications.
Table 1.
Nucleotide Composition of afgps in G. morhua.
| Sequence | Percentage of Nucleotide |
Total Number of Nucleotides | ||||
|---|---|---|---|---|---|---|
| T | C | A | G | G + C | ||
| afgp2 | 14.1 | 39.6 | 22.5 | 23.7 | 63.3 | 396 |
| afgp3 | 5.4 | 49.8 | 19 | 25.8 | 75.6 | 1031 |
| afgp5 | 6.2 | 47.1 | 21.3 | 25.4 | 72.5 | 1459 |
| afgp6 | 10.3 | 43.3 | 26.7 | 19.8 | 63.1 | 915 |
| afgp2—ex 3 repeat | 7 | 47.5 | 23.1 | 22.3 | 69.8 | 242 |
| afgp3—ex 3 repeat | 2.1 | 53.4 | 18.8 | 25.8 | 79.2 | 877 |
| afgp5—ex 3 repeat | 3.9 | 49.2 | 21.5 | 25.4 | 74.6 | 1305 |
| afgp6—ex 3 repeat | 7.5 | 46 | 27.9 | 18.7 | 64.7 | 761 |
Note.—For each complete, putatively functional afgp, the percentage of each nucleotide for the complete cds and for only the repeat in ex3 is given, as well as the total number of nucleotides.
One feature that distinguishes natural proteins from a hypothetical protein product of translated non-coding DNA is that the latter is intrinsically more disordered (Romero et al. 1998). We therefore calculated degree of intrinsic structural disorder (ISD) using IUPred (Dosztanyi et al. 2005) for all four putatively functional afgps in G. morhua, including all three potential ORFs for the complete coding sequence and the repetitive region separately. To account for the variation in ISD over the entire ORF we calculated both mean and median ISD for each gene. For the primary ORF, mean ISD ranged from 0.37 to 0.89 with an average of 0.68 and median ISD ranged from 0.40 to 0.98 with an average of 0.75 (supplementary table S5, Supplementary Material online). The level of ISD is consistent across the length of the protein for each afgp gene (supplementary fig. S5, Supplementary Material online), except for afgp2 (supplementary fig. S5A, Supplementary Material online). These values were even higher when only looking at the repetitive region, and the values were consistent even considering alternative ORFs (supplementary table S5, Supplementary Material online). For comparisons, we also calculated ISD for all annotated genes in the G. morhua genome assembly (Tørresen et al. 2017b), where the average mean ISD was 0.36, and the average median ISD was 0.35. These values are clearly much lower than the average mean and median found in afgp (0.68 and 0.75, respectively) (fig. 4 and supplementary fig. S6, Supplementary Material online).
Fig. 4.

ISD for all genes in the G. morhua genome assembly. A histograms of the distribution of mean ISD for all annotated genes in G. morhua with the average mean ISD for afgp is shown. Average mean ISD for all genes in the G. morhua genome assembly was 0.36. ISD was calculated using IUPred for each amino acid position in each annotated gene in G. morhua.
Copy Numbers of afgp in Notothenioids
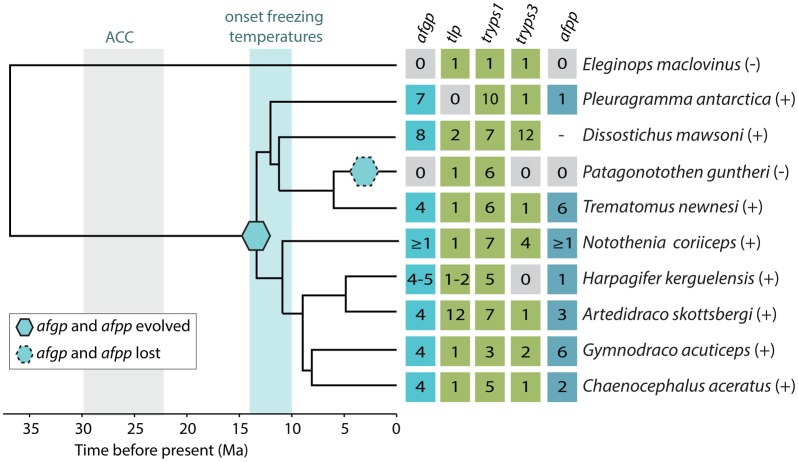
In notothenioid species known to harbor AFGPs (Near et al. 2012; Miya et al. 2016), we identified the afgp repeat in the genomes of P. antarctica, T. newnesi, N. coriiceps, H. kerguelensis, A. skottsbergi, G. acuticeps, and C. aceratus. The presence of afgp, together with data on presence/absence of functional AFGP and thermal hysteresis obtained from the literature, was mapped onto a time-calibrated phylogenetic tree from (Colombo et al. 2015) (fig. 5). As expected from previous studies, we did not detect afgp in the non-Antarctic species E. maclovinus (Cheng and Chen 1999) and P. guntheri (Yang et al. 2013). Based on the number of signal peptide sequences we estimated the copy number of afgp, its evolutionary precursor trypsinogen-like protease gene (tlp) (Chen et al. 1997a), trypsinogen-1 (tryps1), and trypsinogen-3 (tryps3) (fig. 5). In N. coriiceps, the afgp region is poorly assembled, and estimation of copy numbers was unmanageable, but at least one copy is present. In T. newnesi and the lineage containing H. kerguelensis, A. skottsbergi, G. acuticeps, and C. aceratus there are four copies of afgp. The highest copy numbers are seven in P. antarctica and eight in D. mawsoni. The numbers of trypsinogen genes vary even more, between 1 and 12 (fig. 5).
Fig. 5.
afgp, afpp, tlp, tryps1, and tryps3 in notothenioids. Copy numbers of the different genes mapped on a phylogeny modified from (Colombo et al. 2015) and reprinted with permission. Time is given in millions of years (Ma). Species shown to have functional AFGP and thermal hysteresis are signified by (+): P. antarctica (Wöhrmann 1995), D. mawsoni, G. acuticeps (Cheng et al. 2006), T. newnesi (Fields and Devries 2015), N. coriiceps, C. aceratus (DeVries 1971), Harpagifer spp., A. skottsbergi (Miya et al. 2016) and species shown not to have functional AFGPs or thermal hysteresis are signified by (–): E. maclovinus (Cheng 2003), P. guntheri (Miya et al. 2016). The branches with origin and losses of afgp and afpp, that gives the most parsimonious explanation of the occurrence of the events, are indicated as shown in legend, together with the onset of the Antarctic circumpolar current (ACC) and freezing temperatures in the Antarctic (Eastman 1997). Presence of afpp in D. mawsoni is unknown. Copy numbers in D. mawsoni are taken from Nicodemus-Johnson et al. (2011) and N. coriiceps genome assembly was generated by Shin et al. (2014). H. kerguelensis is inserted in the place of its sister species Harpagifer antarcticus (Derome et al. 2002).
Copy Numbers and Evolutionary Origin of afpp in Notothenioids
The AFPP protein sequence available from (Yang et al. 2013) was found to be highly similar to a c1q-like gene (a3ffr1 in D. mawsoni) based on a BLAST search against Uniprot (E-value: 5×10−48, alignment shown in supplementary fig. S7, Supplementary Material online). Based on BLAST alignments with notothenioid genomes there appear to be two exons encoding from 1 to 40 aa and from 40 to 130 aa, which have a high sequence identity with a c1q-like gene (supplementary fig. S7, Supplementary Material online). The first exon seems to be more conserved, yet sufficiently different to distinguish afpp from c1q-like genes (supplementary fig. S7, Supplementary Material online). Therefore, the first exon was used for the detection of afpp presence in notothenioids and for afpp copy number estimation. Echoing the pattern of afgp, afpp is present in the genomes of P. antarctica, T. newnesi, H. kerguelensis, A. skottsbergi, G. acuticeps, and C. aceratus and not present in E. maclovinus and P. guntheri (fig. 5, supplementary table S7, Supplementary Material online). We detected afpp in N. coriiceps, but only in the raw reads and not in the assembly. Thus, we could only confirm presence in N. coriiceps, unable to estimate copy numbers, reconstruct synteny and compare this with other teleosts. However, there are six copies of c1q-like genes in the G. aculeatus genome and in N. coriiceps we found 12 c1q-like sequences (results not shown). This indicates that c1q-genes have undergone extensive gene duplications before one copy was neofunctionalized to form afpp. Notably, we did not get any reliable BLAST hits against afpp in codfishes (supplementary data S2, Supplementary Material online; BLAST alignment for G. morhua).
There is considerable variation in the number of afpp copies, from one copy in P. antarctica and H. kerguelensis to six copies in T. newnesi and G. acuticeps (fig. 5). Together, these results indicate that afpp evolved from a c1q-like gene at the root of the Antarctic notothenioids. This coincides with the cooling of the Southern Ocean and the evolutionary origin of afgps (Chen et al. 1997a) (fig. 5).
Genomic Organization and Synteny of afgps in Notothenioids
In the notothenioid N. coriiceps, we identified only a single large scaffold, Scf4413, containing afgp where synteny could be reconstructed (fig. 3B). The genes flanking afgp were not syntenic with other teleost genome assemblies such as G. aculeatus, T. nigroviridis, T. rubripes, D. rerio, O. niloticus, and X. maculatus (data not shown). Afgp is juxtaposed with tlp, its evolutionary precursor. tlp is a gene encoding an enzyme which belongs to a larger family of trypsins, and we also identified a larger scaffold containing a trypsinogen gene (tryps1), Scf36400 (fig. 3C). This region is quite conserved across teleosts (fig. 3C). However, a higher genomic resolution is needed to pinpoint the genomic origin of the tryps gene ancestral to the afgps.
Discussion
Here, we show that AFGPs evolved around 13–18 Ma in codfishes ancestral to the lineage including by M. aeglefinus to G. morhua in figure 1B, congruent with the cooling of the Northern Hemisphere and first glaciations 10–15 Ma (Eastman 1997). As the oceans cooled in the Northern Hemisphere, organisms faced three possibilities: migrate south to warmer waters, die out, or adapt to the new, freezing conditions. The birth of an entirely new gene, afgp, from previous non-genic DNA allowed the ancestor of afgp-bearing codfishes to survive in freezing conditions and take advantage of the highly productive Arctic waters. The alternative scenario is that afgp evolved from a pre-existing protein encoding gene so diverged from afgp to not be recognized by BLAST. Although the subject of homology detection using BLAST has been heavily debated (Elhaik et al. 2005; Albà and Castresana 2007), the consensus is that for relatively young genes homologous sequences are unlikely to avoid detection by BLAST. For example, genes originating after the split between tetrapods and other vertebrates ∼400 Ma are readily detected using a BLAST cutoff of 10−4 (Elhaik et al. 2005). Furthermore, a gene is not expected to evolve at a high rate along the entire sequence, so there are usually conserved domains (Albà and Castresana 2007). This is exemplified in the notothenioids, where afgps have diverged to a high degree from its evolutionary precursor (tlp), yet the signal peptide and regulatory sequences are still highly similar (Chen et al. 1997a). We also detect the putative pseudogene afgpψ1 in M. aeglefinus (fig. 1), even though pseudogenes accumulate mutations at a much higher rate than functional genes (Echols et al. 2002). Since we have used very relaxed search settings (E-value cutoff = 0.1) and many different BLAST algorithms, it is highly unlikely that we have missed the hypothetical evolutionary precursor protein to AFGP. Beyond sequence similarity, the absence of coding sequence in an orthologous region within an outgroup genome is strong evidence for de novo gene appearance (McLysaght and Guerzoni 2015; McLysaght and Hurst 2016). We found no conserved non-coding elements or afgp-like sequences in the genomic region syntenic to afgp in figure. 3A and supplementary figure S3, Supplementary Material online. Together with the absence of orthologous genes to afgp in species closely related to afgp-bearing codfishes this is strong evidence that afgps are de novo genes (fig. 1B).
For a non-genic region to evolve into a gene, two key evolutionary events must occur: the region must be consistently transcribed and translated into a protein, and it must procure an ORF (reviewed in McLysaght and Guerzoni 2015; Schlötterer 2015; McLysaght and Hurst 2016). Evolution of de novo genes can thus take two alternative routes, transcription first or ORF first (McLysaght and Guerzoni 2015; Schlötterer 2015), and it remains to be seen which one will be more prevalent in de novo gene evolution. Our data do not allow reliable pinpointing which route afgp evolution undertook, although the presence of afgp-like sequences in the genome of G. morhua (fig. 2) opens up the possibility that an afgp ORF evolved first. The afgp-like sequences only occur in the Gadidae family, even in species that do not have the characteristic afgp repeat (P. virens, T. minutus, and G. argenteus, respectively) (figs. 1 and 2). Thus, in the ancestor of Gadidae we propose there were afgp-like sequences that had the potential to evolve into the signal peptide of afgp. In the ancestor of afgp-bearing codfishes, the afgp-like ex2 sequence occurred together with a stretch of the afgp-repeat. In the ancestral state this repeat did not have to be long, as the smallest functional AFGP is only 14 amino acids long in G. morhua (Hew et al. 1981). If this ancestral afgp sequence then became transcribed (either by utilizing another gene’s regulatory sequence or acquiring its own) and translated, it became subject to selection. The small, ancestral AFGP probably had a rudimentary ice-binding activity giving its carrier a fitness benefit, leading to afgp eventually becoming fixed in the population as a new gene. If the afgp-like sequences in P. virens, T. minutus, and G. argenteus are true pseudogenes, meaning afgp first appeared in the ancestor of Gadidae 27 Ma (fig. 1), our de novo origin hypothesis still holds water as there are no afgp homologs outside Gadidae. Furthermore, the evolution of antifreeze function more than 10 Ma before the onset of freezing temperatures in the Northern Hemisphere is a less plausible scenario.
Two hypotheses have been proposed to explain de novo gene birth; either new genes evolve through a series of intermediate stages between non-coding DNA and gene (‘continuum hypothesis’) (Carvunis et al. 2012), or arise from non-coding DNA that happens to be gene-like (‘preadaptation’ hypothesis) (Masel 2006; Wilson and Masel 2011). Preadaptation of de novo genes does not invoke selection on noncoding DNA, but refers to the conditional probability that given that a gene was born it likely originated from a non-coding sequence with more favorable characteristics for gene birth than the average noncoding sequence (Wilson and Masel 2011). Thus, the preadaptation hypothesis of de novo gene appearance predicts that young genes are more disordered (higher ISD) than older genes, and the continuum hypothesis predicts that older genes are more disordered than younger genes. ISD in genes of different ages of origin in vertebrates ranged from 0.35 in pre-vertebrates to 0.55 in rodents, using mouse as a focal species (Wilson et al. 2017). For afgp ISD is very high (on average 0.68, supplementary table S5, Supplementary Material online) compared to the average ISD of 0.36 in the annotated genes in G. morhua (fig. 4), consistent with being young genes. Furthermore, the high GC-content in afgps (63–75%, table 1) increases the chances of obtaining a coding sequence uninterrupted by stop codons, which are TA rich, and increases the chance of transcription. Taken together with the presence of signal-peptide like sequences in Gadidae the sequences ancestral to afgps seem to have been preadapted to become genes.
The genomic processes leading to the genesis of afgp in notothenioids and codfishes seem to have been quite different. The evolution of afgp did not result in any major genomic rearrangements in codfishes as judged from the conserved synteny of the genes flanking afgp across teleosts (fig. 3A). In contrast, the evolution of notothenioid afgp seems to have involved genome rearrangements since the afgp locus is not syntenic with other teleosts (fig. 3B). afgp together with tlp—its evolutionary precursor—is flanked by genes that are not linked in other teleosts examined (fig. 3B). On the other hand, tryps, which is paralogous to tlp and afgp, is in a region with high degree of synteny with other teleosts (fig. 3C). This suggests that in the ancestor of afgp-bearing notothenioids tryps got duplicated to a new genomic location to form at least two copies of tlp, and afgp subsequently evolved from one of the tlp copies (Chen et al. 1997a). The event that resulted in the duplication of tlp may have caused reshuffling of other genes as well, ensuing the lack of synteny in the afgp locus in the notothenioid N. coriiceps.
Our data demonstrate that WGS is essential to get a complete overview of the afgp gene family, especially due to their repetitive nature and the presence of multiple copies (fig. 1), pseudogenes, incomplete duplications (fig. 1), and afgp-like sequences (fig. 2). To get complete afgp sequences suitable for resolving the organization of afgps (fig. 1A) and their syntenic context (fig. 3), long-read sequencing technologies such as PacBio is an advantage because short reads from e.g. Illumina do not span repetitive regions longer than the read length, leading to collapsed repeats and assembly gaps. The assembly challenge associated with such complex repeats may explain why afgps were not properly assembled in the first version of the Atlantic cod (G. morhua) genome (Star et al. 2011). Yet, for detecting the presence/absence and CNV of afgp, we have shown that genomes generated from short-read sequencing are sufficient.
Shared features in notothenioids and codfishes—revealed by WGS—are a common origin of afgp and high CNV, resulting from gene duplications and losses leading to unique afgp repertoires in different species (figs. 1 and 5). Since afgps consist mainly of repeats, unequal crossing over events are likely to be the driver in CNV—and further reinforced by selection in the various species (figs. 1 and 5). Furthermore, there have been pseudogenization and loss of functional afgps in both codfishes (fig. 1) and notothenioids (fig. 5) (Miya et al. 2016). In both lineages there seems to be a gene dosage effect related to the harshness of climate different species are exposed to, yet there are still some notable differences. In notothenioids, the evolution of afgp was a matter of survival, as they became isolated in the freezing Antarctic waters at the onset of the circumpolar current. In the Antarctic notothenioids, the number of afgps ranges from at least one to eight (fig. 5). Most notothenioid species are demersal or benthopelagic, including the species in figure 5, except for the two pelagic species P. antarctica and D. mawsoni. These species have about twice as many afgps as the other species (fig. 5), indicating that a larger afgp repertoire is associated with a pelagic distribution. This could be because pelagic species are exposed to more variable abiotic factors requiring more diverse afgps, or a larger diversity of afgps is associated with the physiology of more active, pelagic species. However, the high-Antarctic species T. newnesi and G. acuticeps have the same number of afgps as the sub-Antarctic H. kerguelensis. To fully determine if there is a gene dosage effect, CNV data for more taxa are required. Given that gene dosage effects have been demonstrated in freeze-preventing zona pellucida proteins in notothenioids (Cao et al. 2016), an analogous scenario for afgps seems plausible. The notothenioid P. guntheri has completely lost afgp (Miya et al. 2016), and we cannot find trace of any afgp pseudogene in its genome (fig. 5). This species has colonized waters north of the polar front and is not exposed to freezing temperatures (Miya et al. 2016). In contrast, the Arctic is not an isolated system and it is possible to escape freezing waters by swimming south or down to deeper waters. Furthermore, many teleost species thrive in the Arctic without afgps. The main advantage of afgps might be allowing its bearer to escape both predation and competition in freezing waters unavailable to animals without special adaptations. Consequently, as A. glacilis and B. saida are Arctic specialists occupying latitudes north of 60 and 70°N, respectively, they possess more afgps than G. morhua and G. chalcogrammus, which are not restricted to the high Arctic and have a broader thermal niche (Eschemeyer and Fricke 2017). In the most parsimonious scenario, the loss of AFGP function of M. merlangius and M. aeglefinus was a single event in the ancestor of these species (fig. 1B). Their distribution ranges between latitudes of 72–35°N for M. merlangius and 79–35°N for M. aeglefinus (Eschemeyer and Fricke 2017). Part of these regions can have freezing temperatures, but not throughout the water column. If the ancestor of these species had a more southerly distribution or avoided freezing waters by for instance swimming deeper, relaxed selection on afgp could have led to pseudogenization. There could also have been selection against this gene due to a harmful effect of ice build-up, which has been demonstrated in notothenioids (Cziko et al. 2014), or simply because of energy expenditure associated with production and circulation of AFGP. Alternatively, it could be lost due to genetic drift. However, given the large effective population sizes observed for M. aeglefinus (Tørresen et al. 2017a) we find this a less likely scenario.
In notothenioids, antifreeze activity is enhanced by the presence of AFPP. Although not essential, AFPP can contribute significantly to the total antifreeze activity, especially in species exposed to freezing temperatures year-round (Fields and Devries 2015). Here, we show that afpp most likely evolved from a c1q-like gene concurrently with afgp 13–20 Ma (fig. 5). This coevolution makes the evolution of antifreeze more complex and raises the question whether afpp evolved just after or simultaneously as afgp. In codfishes afpp is not present, according to a PhD thesis abstract (Jin 2003) and given the independent origin of afgp in these lineages it seems unlikely codfish should possess afpp, although codfishes could have a protein with an analogous function to afpp.
Most likely, codfish afgps arose from entirely non-coding DNA making them type I de novo genes, according to the classification in McLysaght and Hurst (2016). Notothenioid afgp is a type II de novo gene, as part of its sequence, the signal peptide, has previously been under selection, but not the region with an acquired new function (i.e. antifreeze). The afgp-repeat of the sequence arose from intronic, non-coding DNA (Chen et al. 1997a). Intriguingly, genes encoding AFP type I (AFPI) in cunner, snailfish, sculpins, and flounder has no homologous gene (Graham et al. 2013) and are therefore also candidates for de novo gene evolution. As antifreeze is a completely new function in these organisms even a protein with rudimentary antifreeze function can become selectively advantageous and set off the process of evolutionary tinkering, finally ending up with the plethora of AFPs detected to date. De novo genes are often involved in response to biotic and abiotic stress, being related to functions that require rapid change to a new selective regime (Schlötterer 2015). This could explain the apparent prevalence of de novo gene evolution in freeze avoidance in teleosts. Interestingly, in animals there is a peak of emergence of new genes around 800 Ma, which precedes the major radiations of animals in the time period Earth underwent a series of freezing cycles (Tautz and Domazet-Lošo 2011).
This is the first study employing WGS data in a phylogenetic context to shed new light on the evolution of antifreeze genes in codfishes and notothenioids. Using different types of genomic data we have been able to fill some of the gaps in the intriguing evolutionary history of afgps in codfishes and notothenioids and compare two paths of de novo gene birth. Even though de novo origin of genes is currently seen as more prevalent than previously thought, examples of new genes being essential for survival such as afgp in codfishes and notothenioids are few. As more high quality genomes are being sequenced we will get a better picture of which functions are underpinned by de novo genes across species.
Materials and Methods
Annotation of afgp in G. morhua and M. aeglefinus
For G. morhua and M. aeglefinus we have annotated genomes assemblies of high contiguity denoted as gadMor2 (Tørresen et al. 2017 b) and melAeg (Tørresen et al. 2017a), respectively. To determine the organization of afgps in these genomes we used BLAST (v. 2.2.26+) (Altschul et al. 1990) with queries from G. morhua and B. saida (Zhuang 2014). Our query sequences contain both non-coding sequences like the putative promoter, 5′UTR, and 3′UTR sequences, as well as protein-coding sequences divided into the signal peptide in ex2, and the ex3 which contains 84 nt of non-repetitive sequence and sequences encoding the tripeptide-repeats in the mature afgps (annotation of gene organization based on gene-prediction in Zhuang 2014). Using these different parts of afgps also allowed us to identify potentially homologous regions in the genomes. We BLASTed using both proteins (tBLASTn) and nucleotides (BLASTn) as queries against nuclear databases for each species. We also used the option BLASTn-short, which is more optimal to detect the short sequences used as queries (Altschul et al. 1990). For all BLAST searches we used an E-value cutoff of 0.1 and otherwise default options, unless explicitly noted.
We analyzed codon usage bias and GC-content of the afgps using MEGA v.7 (Kumar et al. 2016). We also calculated the GC-content of all annotated genes in the G. morhua genome assembly (gadMor2) (Tørresen et al. 2017 b). Codon usage bias was calculated as the observed frequency of a codon divided by the expected one (RSCU values).
The degree of ISD was calculated using IUPred (Dosztanyi et al. 2005) for afgps and all other annotated genes in the gadMor2 assembly. We calculated ISD for sequences translated into proteins using all three ORFs.
Copy Number Estimation and Evolutionary Origin of afgp in Codfishes
We used BLAST to annotate afgps in the genomes of 23 codfishes available (Malmstrøm et al. 2016, 2017). However, as these genomes are assembled from only short reads, afgps with their long repetitive regions are not in contiguous sequences in these species. Hits against afgp repeats were thus only used to detect presence/absence of afgp. To establish afgp copy number we used the signal peptide sequences, as well as the non-coding sequences of the promoter, 5′UTR and 3′UTR. However, gadMor2 5′UTRs gave significantly more hits than 3′UTR and were excluded from the analysis. Annotation was validated by aligning the reported BLAST hit regions and generating phylogenetic trees using maximum likelihood (Tamura-Nei model, partial deletion of missing data, 1,000 bootstraps) in MEGA v.7 (Kumar et al. 2016).
To distinguish between afgps and afgp-like sequences, a phylogenetic tree was constructed with all putative afgp sequences which included ex1, ex2, and the start of ex3 from G. chalcogrammus, B. saida, A. glacilis, M. merlangius, P. virens, T. minutus, and G. argenteus, and afgps and afgp-like sequences from G. morhua and M. aeglefinus. The main topology of the phylogenetic tree was constructed using a Bayesian method in MrBayes 3.2.2 (Ronquist and Huelsenbeck 2003) with standard priors, four chains of simulations for 1×106 generations sampling every 1×103 generation. For each run convergence was considered reached when the likelihood scores leveled off asymptotically. Trees sampled before convergence were discarded and support (posterior probability) was calculated based on a consensus of the remaining 1,502 trees. Bootstrap support values were obtained by constructing a phylogenetic tree with maximum likelihood (Tamura-Nei model, partial deletion of missing data, 1,000 bootstraps) in MEGA v.7 (Kumar et al. 2016).
To determine evolutionary origin the presence of afgp repeat as well as copy number of promoter, signal peptide ex2, beginning of ex3 and 3′UTR were mapped on a time calibrated phylogeny from (Malmstrøm et al. 2016). Furthermore, we searched for the presence of signal peptide ex2, beginning of 5′UTR, 3′UTR, and the antifreeze repeat sequence in ex3 in RepBase (Bao et al. 2015).
Assembly of Notothenioid Genomes
To obtain representatives from most lineages of notothenioids (Colombo et al. 2015), we did WGS for the following eight species; E. maclovinus, P. antarctica, P. guntheri, T. newnesi, H. kerguelensis, A. skottsbergi, G. acuticeps, and C. aceratus. We sequenced paired end libraries with an average insert size of 350 bp (2 × 150 bp reads on Illumina HiSeq 2000) with coverage ranging from 31 to 67× (average coverage 44×). The Celera assembler (Miller et al. 2008) was used to assemble the genomes, with contig N50 ranging from 5 to 9.6 kb with an average of 6.3 kb. CEGMA (Parra et al. 2009) and BUSCO (Simão et al. 2015) were used to evaluate gene completeness; CEGMA gave, on average, complete or partial hits for 69% of the conserved eukaryotic genes included in the CEGMA analysis and BUSCO gave, on average, 68% of the conserved genes belonging to the Actinopterygii lineage in the BUSCO analysis. A list of species with relevant genome statistics is given in supplementary table S6, Supplementary Material online. Sequences are deposited in ENA.
Copy Number Estimation in Notothenioids
The afgps in notothenioids consist of a signal peptide (ex1) and tripeptide repeats of various lengths (ex2). We used both exons to BLAST against the genomes of E. maclovinus, P. antarctica, P. guntheri, T. newnesi, H. kerguelensis, A. skottsbergi, G. acuticeps, and C. aceratus, as well as the published genome of Notothenia coriiceps (Shin et al. 2014). The hits against ex2 were only used to determine presence/absence of afgp. Because afgp are homologous with tlp, we used the signal peptide from tlp, as well as the related sequences encoding trypsinogen-1 (tryps1) and trypsinogen-3 (tryps3), as query sequences. Query sequences were obtained from D. mawsoni (Nicodemus-Johnson et al. 2011). We then mapped the number of signal peptide copies from these genes on a phylogeny from (Colombo et al. 2015), together with copy number estimates from (Nicodemus-Johnson et al. 2011) for D. mawsoni. According to Nicodemus-Johnson et al. (2011) there are two haplotypes in D. mawsoni with different copy numbers of afgp, tlp, tryps1, and tryps3. We chose copy numbers from haplotype 2 (accession number HQ447060) (fig. 5), as we do not trust the evidence for haplotype 1.
We estimated the number of afpps using BLAST hits against AFPP amino acid sequence from G. acuticeps (Yang et al. 2013). As the different exons were usually on different utgs, we used the putative first exon to count the number of copies of afpps, using BLAST sequence identity to distinguish afpp from related gene sequences. Copy number for each species was then mapped onto the phylogeny in figure 5.
Synteny of afgp and Trypsinogen Locus
We investigated the flanking sequences of afgp in G. morhua (gadMor2), M. aeglefinus (melAeg) and N. coriiceps (Shin et al. 2014); in the N. coriiceps we also investigated sequences flanking tryps1 and tryps3. In codfish we delimited synteny analyses between the genes ppp2ca and adam9; in N. coriiceps we delimited synteny analyses between the genes nr2f6 and nckap5l for afgp and lipeb and apoea for tryps. We then identified homologous regions in release 86 of the Ensembl database (Yates et al. 2016) for the following species; tilapia (Oreochromis niloticus), platyfish (Xiphophorus maculatus), three-spined stickleback (Gasterosteus aculeatus), tetraodon (Tetraodon nigroviridis), and fugu (Takifugu rubribes), as well as the genome of the northern pike (Esox lucius) (Rondeau et al. 2014). In cases where the automatic annotation was incomplete we manually annotated genes by BLASTing against the Uniprot and Ensembl databases.
mVista comparisons between species were carried out using LAGAN alignments (Frazer et al. 2004).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
Sequencing was carried out at the Norwegian Sequencing Centre (NSC), and McGill University and Genome Quebec Innovation Centre.
Author Contributions
S.J. and H.T.B. initially conceived and designed the study, with input from K.S.J. and W.S. Samples for the eight notothenioid genomes were provided by W.S. and R.H. The genome assembly pipeline was set up by O.K.T. and carried out by O.K.T. and H.T.B. Annotation of afgp genes in codfishes was carried out by H.T.B. with assistance from M.H.S. Genome-mining, phylogenetic analyses and construction of all figures and tables were done by H.T.B. Synteny analyses were carried out by H.T.B. and O.K.T. The manuscript was written by H.T.B. together with S.J. and K.S.J. with additional input from all other authors.
Funding
This study was funded by grant awarded to K.S.J. from the Research Council of Norway (RCN grant 222378).
References
- Albà MM, Castresana J.. 2007. On homology searches by protein Blast and the characterization of the age of genes. BMC Evol Biol. 7:53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O.. 2015. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA 6(1):462–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bildanova LL, Salina EA, Shumny VK.. 2013. Main properties and evolutionary features of antifreeze proteins. Russ J Genet Appl Res. 3(1):66–82.http://dx.doi.org/10.1134/S207905971301005X [Google Scholar]
- Cao L, Huang Q, Wu Z, Cao D-D, Ma Z, Xu Q, Hu P, Fu Y, Shen Y, Chan J.. 2016. Neofunctionalization of zona pellucida proteins enhances freeze-prevention in the eggs of Antarctic notothenioids. Nat Comms. 7:12987–12911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvunis A-R, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N, Charloteaux B, Hidalgo CA, Barbette J, Santhanam B, et al. 2012. Proto-genes and de novo gene birth. Nature 487(7407):370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Chen LB.. 1999. Evolution of an antifreeze glycoprotein. Nature 401(6752):443–444.http://dx.doi.org/10.1038/46721 [DOI] [PubMed] [Google Scholar]
- Cheng C-H. 1998. Evolution of the diverse antifreeze proteins. Curr Opin Genet Dev. 8(6):715–720.http://dx.doi.org/10.1016/S0959-437X(98)80042-7 [DOI] [PubMed] [Google Scholar]
- Cheng C-HC. 2003. Functional antifreeze glycoprotein genes in temperate-water New Zealand Nototheniid fish infer an antarctic evolutionary origin. Mol Biol Evol. 20(11):1897–1908.http://dx.doi.org/10.1093/molbev/msg208 [DOI] [PubMed] [Google Scholar]
- Cheng C-HC, Cziko PA, Evans CW.. 2006. Nonhepatic origin of notothenioid antifreeze reveals pancreatic synthesis as common mechanism in polar fish freezing avoidance. Proc Natl Acad Sci U S A. 103(27):10491–10496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, DeVries AL, Cheng C-H.. 1997a. Evolution of antifreeze glycoprotein gene from a trypsinogen gene in Antarctic notothenioid fish. Proc Natl Acad Sci U S A. 94:3811–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, DeVries AL, Cheng C-H.. 1997. Convergent evolution of antifreeze glycoproteins in Antarctic notothenioid fish and Arctic cod. Proc Natl Acad Sci U S A. 94(8):3817–3822.http://dx.doi.org/10.1073/pnas.94.8.3817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M, Damerau M, Hanel R, Salzburger W, Matschiner M.. 2015. Diversity and disparity through time in the adaptive radiation of Antarctic notothenioid fishes. J Evol Biol. 28(2):376–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cziko PA, Devries AL, Evans CW, Cheng C-HC.. 2014. Antifreeze protein-induced superheating of ice inside Antarctic notothenioid fishes inhibits melting during summer warming. Proc Natl Acad Sci U S A. 111(40):14583–14588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denstad J-P, Aunaas T, Börseth JF, Aarset AV, Zachariassen KE.. 1987. Thermal hysteresis antifreeze agents in fishes from Spitsbergen waters. Pol Res. 5(2):1–4. [Google Scholar]
- Derome N, Chen W-J, Dettaï A, Bonillo C, Lecointre G.. 2002. Phylogeny of Antarctic dragonfishes (Bathydraconidae, Notothenioidei, Teleostei) and related families based on their anatomy and two mitochondrial genes. Mol Phyl Evol. 24(1):139–152. [DOI] [PubMed] [Google Scholar]
- DeVries AL. 1971. Glycoproteins as biological antifreeze agents in Antarctic fishes. Science 172(3988):1152–1155.http://dx.doi.org/10.1126/science.172.3988.1152 [DOI] [PubMed] [Google Scholar]
- Dosztanyi Z, Csizmok V, Tompa P, Simon I.. 2005. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21(16):3433–3434.http://dx.doi.org/10.1093/bioinformatics/bti541 [DOI] [PubMed] [Google Scholar]
- Eastman JT. 1997. Comparison of the Antarctic and Arctic fish faunas. Cybium 21:335–352. [Google Scholar]
- Echols N, Harrison P, Balasubramanian S, Luscombe NM, Bertone P, Zhang Z, Gerstein M.. 2002. Comprehensive analysis of amino acid and nucleotide composition in eukaryotic genomes, comparing genes and pseudogenes. Nucleic Acids Res. 30(11):2515–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhaik E, Sabath N, Graur D.. 2005. The “Inverse Relationship Between Evolutionary Rate and Age of Mammalian Genes” is an artifact of increased genetic distance with rate of evolution and time of divergence. Mol Biol Evol. 23(1):1–3. [DOI] [PubMed] [Google Scholar]
- Eschemeyer WN, Fricke R.. 2017. Catalog of fishes. http://research.calacademy.org/researchichthyology/catalog/fishcatmain.asp; last accessed June 15, 2017.
- Ewart KV, Blanchard B, Johnson SC, Bailey WL, Martin-Robichaud DJ, Buzeta MI.. 2000. Freeze susceptibility in haddock (Melanogrammus aeglefinus). Aquaculture 188(1–2):91–101. [Google Scholar]
- Ewart KV, Lin Q, Hew CL.. 1999. Structure, function and evolution of antifreeze proteins. Cell Mol Life Sci. 55(2):271–283.http://dx.doi.org/10.1007/s000180050289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields LG, Devries AL.. 2015. Variation in blood serum antifreeze activity of Antarctic Trematomus fishes across habitat temperature and depth. Comp Biochem Physiol, Part A Mol. Integr Physiol. 185:43–50.http://dx.doi.org/10.1016/j.cbpa.2015.03.006 [DOI] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I.. 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32(Web Server issue):W273–W279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin S, McPherson JD, McCombie WR.. 2016. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 17(6):333–351.http://dx.doi.org/10.1038/nrg.2016.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham LA, Hobbs RS, Fletcher GL, Davies PL.. 2013. Helical antifreeze proteins have independently evolved in fishes on four occasions. PLoS ONE 8(12):e81285.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Deswal R.. 2014. Antifreeze proteins enable plants to survive in freezing conditions. J Biosci. 39(5):931–944.http://dx.doi.org/10.1007/s12038-014-9468-2 [DOI] [PubMed] [Google Scholar]
- Harding MM, Anderberg PI, Haymet ADJ.. 2003. “Antifreeze” glycoproteins from polar fish. Eur J Biochem. 270(7):1381–1392. [DOI] [PubMed] [Google Scholar]
- Hew CL, Slaughter D, Fletcher GL, Joshi SB.. 1981. Antifreeze glycoproteins in the plasma of Newfoundland Atlantic cod (Gadus morhua). Can J Zool. 59(11):2186–2192.http://dx.doi.org/10.1139/z81-296 [Google Scholar]
- Jin Y. 2003. Freezing avoidance of antarctic fishes: the role of a novel antifreeze potentiating protein and the antifreeze glycoproteins. PhD thesis University of Illinois, Urbana-Champaign
- Kennett JP. 1977. Cenozoic evolution of antarctic glaciation, circum-antarctic ocean, and their impact on global paleoceanography. J Geophys Res Oceans 82(27):3843–3860.http://dx.doi.org/10.1029/JC082i027p03843 [Google Scholar]
- Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TCG.. 2009. More than just orphans: are taxonomically-restricted genes important in evolution? Trends Genet. 25(9):404–413. [DOI] [PubMed] [Google Scholar]
- Kristiansen E, Zachariassen KE.. 2005. The mechanism by which fish antifreeze proteins cause thermal hysteresis. Cryobiology 51(3):262–280.http://dx.doi.org/10.1016/j.cryobiol.2005.07.007 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 33(7):1870–1874.http://dx.doi.org/10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Li Z, Lin Q, Kosinski J, Seetharaman J, Bujnicki JM, Sivaraman J, Hew C-L.. 2007. Structure and evolutionary origin of Ca2+-dependent herring Type II antifreeze protein. PLoS ONE 2(6):e548–e511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmstrøm M, Matschiner M, Tørresen OK, Jakobsen KS, Jentoft S.. 2017. Whole genome sequencing data and de novo draft assemblies for 66 teleost species. Sci Data 4:1–13.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmstrøm M, Matschiner M, Tørresen OK, Star B, Snipen LG, Hansen TF, Baalsrud HT, Nederbragt AJ, Hanel R, Salzburger W, et al. 2016. Evolution of the immune system influences speciation rates in teleost fishes. Nat Genet. 48(10):1204–1210. [DOI] [PubMed] [Google Scholar]
- Masel J. 2006. Cryptic genetic variation is enriched for potential adaptations. Genetics 172(3):1985–1991.http://dx.doi.org/10.1534/genetics.105.051649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLysaght A, Guerzoni D.. 2015. New genes from non-coding sequence: the role of de novo protein-coding genes in eukaryotic evolutionary innovation. Philos Trans R Soc Lond B Biol Sci. 370(1678):20140332–20140338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLysaght A, Hurst LD.. 2016. Open questions in the study of de novo genes: what, how and why. Nat Rev Genet. 17(9):579–579. [DOI] [PubMed] [Google Scholar]
- Miller JR, Delcher AL, Koren S, Venter E, Walenz BP, Brownley A, Johnson J, Li K, Mobarry C, Sutton G.. 2008. Aggressive assembly of pyrosequencing reads with mates. Bioinformatics 24(24):2818–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya T, Gon O, Mwale M, Cheng CHC.. 2016. Multiple independent reduction or loss of antifreeze trait in low Antarctic and sub-Antarctic notothenioid fishes. Antarct Sci. 28(01):17–28.http://dx.doi.org/10.1017/S0954102015000413 [Google Scholar]
- Near TJ, Dornburg A, Kuhn KL, Eastman JT, Pennington JN, Patarnello T, Zane L, Fernandez DA, Jones CD.. 2012. Ancient climate change, antifreeze, and the evolutionary diversification of Antarctic fishes. Proc Natl Acad Sci U S A. 109(9):3434–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicodemus-Johnson J, Silic S, Ghigliotti L, Pisano E, Cheng CHC.. 2011. Assembly of the antifreeze glycoprotein/trypsinogen-like protease genomic locus in the Antarctic toothfish Dissostichus mawsoni (Norman). Genomics 98(3):194–201. [DOI] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Ning Z, Keane T, Korf I.. 2009. Assessing the gene space in draft genomes. Nucleic Acids Res. 37(1):289–297.http://dx.doi.org/10.1093/nar/gkn916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praebel K. 2005. Antifreeze activity in the gastrointestinal fluids of Arctogadus glacialis (Peters 1874) is dependent on food type. J Evol Biol. 208(13):2609–2613. [DOI] [PubMed] [Google Scholar]
- Romero P, Obradovic Z, Kissinger CR, Villafranca JE, Garner E, Guilliot S, Dunker AK.. 1998. Thousands of proteins likely to have long disordered regions. Pac Symp Biocomput. 3:437–448. [PubMed] [Google Scholar]
- Rondeau EB, Minkley DR, Leong JS, Messmer AM, Jantzen JR, von Schalburg KR, Lemon C, Bird NH, Koop BF, Yin T.. 2014. The genome and linkage map of the northern pike (Esox lucius): conserved synteny revealed between the Salmonid Sister Group and the Neoteleostei. PLoS ONE 9(7):e102089–e102018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP.. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19(12):1572–1574.http://dx.doi.org/10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Schlötterer C. 2015. Genes from scratch—the evolutionary fate of de novo genes. Trends Genet. 31(4):215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SC, Ahn DH, Kim SJ, Pyo CW, Lee H, Kim M-K, Lee J, Lee JE, Detrich HW, Postlethwait JH, et al. 2014. The genome sequence of the Antarctic bullhead notothen reveals evolutionary adaptations to a cold environment. Genome Biol. 15(9):468.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Star B, Nederbragt AJ, Jentoft S, Grimholt U, Malmstrøm M, Gregers TF, Rounge TB, Paulsen J, Solbakken MH, Sharma A, et al. 2011. The genome sequence of Atlantic cod reveals a unique immune system. Nature 477(7363):207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Domazet-Lošo T.. 2011. The evolutionary origin of orphan genes. Nat Rev Genet. 12(10):692–702. [DOI] [PubMed] [Google Scholar]
- Tsuda S, Miura A.. 2005. Antifreeze proteins originating in fishes. United States patent US20050019856 A1
- Tørresen OK, Brieuc MSO, Solbakken MH, Sørhus E, Nederbragt AJ, Jakobsen KS, Meier S, Edvardsen RB, Jentoft S.. 2017a. Genomic architecture of codfishes featured by expansions of innate immune genes and short tandem repeats. bioRxiv: 163949. doi: https://doi.org/10.1101/163949. [DOI] [PMC free article] [PubMed]
- Tørresen OK, Star B, Jentoft S, Reinar WB, Grove H, Miller JR, Walenz BP, Knight J, Ekholm JM, Peluso P.. et al. 2017b. An improved genome assembly uncovers prolific tandem repeats in Atlantic cod. BMC Genomics 18:311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BA, Foy SG, Neme R, Masel J.. 2017. Young genes are highly disordered as predicted by the preadaptation hypothesis of de novo gene birth. Nat Ecol Evol. 1(6):0146–0146..http://dx.doi.org/10.1038/s41559-017-0146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BA, Masel J.. 2011. Putatively noncoding transcripts show extensive association with ribosomes. Genome Biol Evol. 3:1245–1252.http://dx.doi.org/10.1093/gbe/evr099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wöhrmann AP. 1995. Antifreeze glycopeptides of the high-Antarctic silverfish Pleuragramma antarcticum (Notothenioidei). Comp Biochem Physiol C Pharmacol Toxicol Endocrinol. 111(1):121–129. [DOI] [PubMed] [Google Scholar]
- Wu D-D, Irwin DM, Zhang Y-P.. 2011. De novo origin of human protein-coding genes. PLoS Genet 7(11):e1002379–e1002378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S-H, Wojnar JM, Harris PWR, Devries AL, Evans CW, Brimble MA.. 2013. Chemical synthesis of a masked analogue of the fish antifreeze potentiating protein (AFPP). Org Biomol Chem. 11(30):4935–4939. [DOI] [PubMed] [Google Scholar]
- Yates A, Akanni W, Amode MR, Barrell D, Billis K, Carvalho-Silva D, Cummins C, Clapham P, Fitzgerald S, Gil L, et al. 2016. 2016. Ensembl 2016. Nucleic Acids Res. 44(D1):D710–D716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X. 2014. Creating sense from non-sense DNA: de novo genesis and evolutionary history of antifreeze glycoprotein gene in northern codfishes (gadidae). PhD thesis, University of Illinois, Urbana-Champaign.
- Zhuang X, Yang C, Fevolden S-E, Cheng CHC.. 2012. Protein genes in repetitive sequence-antifreeze glycoproteins in Atlantic cod genome. BMC Genomics 13:293.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.