Abstract
The fitness effects of mutations can depend on the genetic backgrounds in which they occur and thereby influence future opportunities for evolving populations. In particular, mutations that fix in a population might change the selective benefit of subsequent mutations, giving rise to historical contingency. We examine these effects by focusing on mutations in a key metabolic gene, pykF, that arose independently early in the history of 12 Escherichia coli populations during a long-term evolution experiment. Eight different evolved nonsynonymous mutations conferred similar fitness benefits of ∼10% when transferred into the ancestor, and these benefits were greater than the one conferred by a deletion mutation. In contrast, the same mutations had highly variable fitness effects, ranging from ∼0% to 25%, in evolved clones isolated from the populations at 20,000 generations. Two mutations that were moved into these evolved clones conferred similar fitness effects in a given clone, but different effects between the clones, indicating epistatic interactions between the evolved pykF alleles and the other mutations that had accumulated in each evolved clone. We also measured the fitness effects of six evolved pykF alleles in the same populations in which they had fixed, but at seven time points between 0 and 50,000 generations. Variation in fitness effects was high at intermediate time points, and declined to a low level at 50,000 generations, when the mean fitness effect was lowest. Our results demonstrate the importance of genetic context in determining the fitness effects of different beneficial mutations even within the same gene.
Keywords: epistasis, experimental evolution, historical contingency, pykF, adaptation
Introduction
Early mutational steps along an evolutionary trajectory can interact with subsequent mutations to influence their effects on phenotypes including fitness. For example, the beneficial effect of a late-occurring mutation might depend on the presence of one or more earlier mutations, which thereby change the distribution of potential evolutionary outcomes (Ortlund et al. 2007; Woods et al. 2011; Phillips et al. 2016). For example, one set of studies has shown the importance of “potentiating” mutations for the subsequent evolution of the capacity of Escherichia coli to grow on citrate (Blount et al. 2008, 2012; Quandt et al. 2014, 2015). Here, we examine how the fitness effects of mutations in one E. coli gene, pykF, depend on the genetic context in which they occur, including across replicate evolving populations as well as over time within a population.
Recent theoretical work predicts that the benefits of selected mutations will often depend on the genetic background in which they occur, such that they may provide a smaller benefit, or even be deleterious, when added to earlier genotypes along a particular adaptive trajectory (Draghi and Plotkin 2013; Greene and Crona 2014; Blanquart et al. 2014). A corollary of this prediction is that earlier mutations may increase the benefits of immediately subsequent mutations if the latter depend on the previous ones for their benefits. In that case, early mutations are expected to become selectively “entrenched”, conferring larger benefits and thereby becoming less likely to revert over time (Shah et al. 2015). One study that examined changes over time in the effect of a deleterious mutation introduced into the ancestor of a set of replicate populations showed that reversion of the introduced mutation was, as expected, beneficial at the start of the experiment, but later on became deleterious, indicating the accumulation of mutations that depended on it for their own beneficial effects (Zee et al. 2014). The same mechanism may entrench and thereby stabilize drug-resistance mutations that become beneficial in combination with associated compensatory mutations (Lenski 1998; Maisnier-Patin et al. 2002; Flynn et al. 2017).
In contrast to the theoretical expectation that early beneficial mutations should become more beneficial over time, some experiments have shown a tendency for beneficial mutations to exhibit diminishing-returns epistasis, such that the beneficial effect of a mutation tends to be smaller in higher-fitness genotypes (Chou et al. 2011; Khan et al. 2011; Kryazhimskiy et al. 2014). Most of the mutation combinations considered in these studies, however, were not directly subject to selection. For example, in a population that fixes five mutations, 32 (= 25) combinations can be constructed, but only five are on the mutational path that the evolving population followed. If epistasis is shaped by selection, then combinations of mutations that did not experience selection may interact differently from those combinations that were coselected. As a consequence, the patterns of interaction inferred from the broader set of interactions may not be representative of those that were important during a population’s evolution (Draghi and Plotkin 2013; Greene and Crona 2014; Blanquart et al. 2014).
Here, we study the effects of genetic context, including the importance of coselection, on the fitness effects of mutations that arose in the long-term evolution experiment, or LTEE, with E. coli (Lenski et al. 1991). Genetic and genomic analyses have revealed substantial parallelism in the genetic basis of adaptation of the 12 populations in that experiment (Woods et al. 2006; Tenaillon et al. 2016). One of the genes that has evolved in all of the populations is pykF, which encodes pyruvate kinase I, a key glycolytic enzyme (fig. 1). Pyruvate kinase 1 catalyzes the transfer of a phosphate group from phosphoenolpyruvate (PEP) to ADP, generating ATP and pyruvate (Siddiquee et al. 2004). Pyruvate kinase 1 is also a crucial component of a metabolic sensor that responds to glycolytic flux (Kochanowski et al. 2013). Mutations in pykF occurred early in the evolution of all populations (Woods et al. 2006; Tenaillon et al. 2016), and one of the evolved alleles has been shown directly to be beneficial (Khan et al. 2011).
Fig. 1.
Distribution of evolved mutations across the pyruvate kinase I enzyme. See supplementary table S1, Supplementary Material online for details of each mutation. (A) X and S indicate positions of nonsynonymous and synonymous mutations, respectively; FS indicates a –1 frame-shift mutation; and IS150 indicates the site of an IS150 insertion. (B) Nonsynonymous evolved mutations mapped onto the monomeric structure of pyruvate kinase 1. (C) Nonsynonymous evolved mutations mapped onto the functional unit of pyruvate kinase 1, a homotetramer (Zhu et al. 2010). Colors are consistent across the panels and correspond to functional domains: Lid (gold), catalytic (aqua), and allosteric (purple).
We tested the effects of the changing genetic context that occurs during evolution on the fitness benefits conferred by pykF mutations. To do so, we competed constructed strains with and without various pykF mutations in the ancestral genetic background and in clones isolated from the LTEE populations at various generations (fig. 2). Eight tested nonsynonymous mutations all conferred similar fitness benefits in the ancestor. These benefits were greater than that conferred by a loss-of-function deletion allele, indicating that the evolved enzymes retain at least some functionality. The effects of the evolved pykF mutations were much more variable when assessed in clones isolated after 20,000 generations of the LTEE. This variation implies heterogeneous interactions between the pykF mutations and other mutations in the evolved clones. When we tested for interactions in a subset of populations across multiple time points, we further found that interactions were dynamic within a lineage. By 50,000 generations, the evolved pykF alleles, including even a deletion mutation, conferred similar fitness benefits that were, for the most part, somewhat smaller than they had been in the ancestral background. Taken together, these results show that changing genetic context—both over time and across populations—can exert a strong influence on the fitness effects of different beneficial mutations even within the same gene.
Fig. 2.
Schematic overview of pykF mutation transfers. Five sets of strains were constructed. (i) Evolved pykF mutations from 11 of the 12 populations were individually transferred to the ancestor; the Ara+4 population, which had a frameshift allele, was omitted from all experiments. A deletion mutation was used as a proxy for the IS150 insertion mutation in Ara–1. (ii) Evolved pykF alleles were reverted to the ancestral allele in one clone isolated from each population at 20,000 generations. (iii) A deletion allele was transferred to each 20,000-generation clone, replacing its evolved allele. (iv) The A301S allele was transferred to each 20,000-generation clone, replacing its evolved allele. (v) In populations Ara–1, Ara–6, Ara+1, Ara+2, Ara+3, and Ara+5, the evolved pykF alleles, if present, were reverted to the ancestral allele in clones isolated at 2,000, 5,000, 10,000, 20,000, 30,000, and 50,000 generations (see Materials and Methods for details).
Results
Different Evolved pykF Alleles Confer Similar Fitness Benefits in the Ancestral Background
To determine the fitness effects of the nine distinct mutations in pykF that were isolated from 11 independently evolved populations (including A301S mutations in three populations, and the deletion mutation that serves as the proxy for the IS150 insertion in another), we moved each of them into the ancestral chromosome. The mean benefit conferred by these mutations was 10.7% (fig. 3A). The different alleles conferred significantly heterogeneous effects (table 1), though the differences were usually small, with an average pairwise difference of only 2.7%. However, the pykF deletion allele conferred a smaller benefit of 5.7%, which was significantly less than that of all but one of the point mutations (Dunnett's test: P < 0.05 except compared with the D127N mutation). This difference between the nonsynonymous substitutions and the deletion mutation implies that the former are not simply loss-of-function mutations. The heterogeneity in the effect on fitness remains significant even if the deletion mutation is excluded from the analysis (supplementary table S1, Supplementary Material online).
Fig. 3.

Fitness effects of evolved pykF mutations in the ancestor and a 20,000-generation clone isolated from the population in which they occurred. (A) Fitness effects of pykF mutations in the ancestor. (B) Fitness effects of pykF mutations in evolved clones. The A301S mutation evolved independently in the Ara–5, Ara+1, and Ara+5 populations, and thus its effects were measured in three evolved strains. Error bars indicate 95% confidence intervals (with n = 4 in each evolved background and n = 8 in the ancestral background). Asterisks indicate the significance of genetic background on fitness (two-tailed t test): ***P < 0.001, ** 0.001 < P < 0.01, * 0.01 < P < 0.05.
Table 1.
ANOVA Testing for Heterogeneity in Fitness Effects of Mutations in pykF When Moved into the Ancestral Chromosome.
| df | SS | MS | F | P | |
|---|---|---|---|---|---|
| Mutation | 8 | 0.0361 | 0.0045 | 8.591 | <0.001 |
| Block | 1 | 0.0045 | 0.0045 | 8.521 | 0.005 |
| Error | 62 | 0.0326 | 0.0005 |
Note.—The fitness effects of nine evolved mutations (including a deletion that serves as a proxy for an IS150 insertion) were estimated with 8-fold replication performed over two blocks.
The nonsynonymous mutations in pykF occur in gene regions corresponding to all three of the enzyme’s functional domains: Lid, catalytic, and allosteric (Donovan et al. 2016; fig. 1). To test whether the domain in which a mutation occurs contributed to the variation in fitness effects, we performed an ANOVA with the different point mutations nested within the functional domains. There was no significant effect of the functional domain, but the effect of different mutations within the domains remains significant (supplementary table S2, Supplementary Material online).
Fitness Effects of pykF Alleles Are Highly Variable in Evolved Backgrounds
Mutations in pykF arose early in the LTEE populations (Tenaillon et al. 2016), and so there is the potential for subsequent beneficial mutations to depend on a pykF mutation for their benefits. If that were the case, then replacing an evolved pykF allele with the ancestral allele would impose a greater fitness cost in the evolved strain than expected based on its effect in the ancestor. Alternatively, later mutations might reduce the fitness effects of the early pykF mutations, either by specifically making them redundant or as a consequence of a general tendency toward diminishing-returns epistasis, which causes a focal beneficial mutation to confer a smaller benefit in more-fit genetic backgrounds. In that case, reverting a pykF mutation in an evolved strain would produce a smaller fitness cost than expected based on its effect in the ancestor. A combination of these interactions should lead to pykF mutations having more variable fitness effects in the evolved populations than they do in the ancestor.
To examine these potential interactions, we measured the fitness effects of replacing the evolved pykF mutations (including the A301S mutation in three cases and the IS150 insertion in another) with the ancestral allele in clones sampled from 11 populations after 20,000 generations (fig. 3B). The different evolved mutations had highly variable effects in their own evolved backgrounds (table 2), ranging from nearly neutral to a benefit of >25%. In all but two cases (the P70T and I264F mutations from populations Ara+6 and Ara–2, respectively), the mutations remained beneficial. In six cases the effect of the evolved allele was more beneficial in its own evolved background than in the ancestor (P70Q, D127N, T462I, the loss-of-function indel, and the A301S mutation in populations Ara+1 and Ara+5), whereas the opposite relationship held in three cases (P70T, I264F, and the A301S mutation in population Ara–5).
Table 2.
ANOVA Testing for Heterogeneity in Fitness Effects of Replacing Evolved pykF Mutations with the Ancestral Allele in Clones Sampled From LTEE Populations at 20,000 Generations.
| df | SS | MS | F | P | |
|---|---|---|---|---|---|
| Population | 10 | 0.2899 | 0.0290 | 25.74 | <0.001 |
| Error | 33 | 0.0372 | 0.0011 |
Note.—The fitness effects of replacements in 11 populations were estimated with 4-fold replication in a single block.
Once again, the heterogeneity in the mutational effects is significant even if the deletion mutation is excluded (supplementary table S4, Supplementary Material online). Moreover, there is significant heterogeneity even in the fitness effects of the three identical A301S substitutions across their independently evolved backgrounds (supplementary table S5, Supplementary Material online), providing a clear demonstration of epistasis between the focal beneficial mutation and its coevolved genetic context. As seen in the ancestral background, there is no significant effect of functional domain, whereas the effect of different mutations within the domains remains significant (supplementary table S6, Supplementary Material online). Four of the 11 LTEE populations used in our study had become mutators by 20,000 generations (Tenaillon et al. 2016). Clones from these mutator populations had between 639 and 1,238 mutations in total, compared with only 33–62 in clones from nonmutator populations (supplementary table S3, Supplementary Material online). To test whether interactions were influenced by the mutator status of the evolved clones, we compared the effects of replacing the pykF mutations between these two groups (supplementary table S7, Supplementary Material online). We found no difference, suggesting that changes in the fitness effects of the pykF mutations are dominated by interactions with other beneficial mutations, which comprise the majority of mutations in the nonmutator clones at 20,000 generations (Tenaillon et al. 2016).
Variation in Evolved Backgrounds Drives Changes in Fitness Effects of pykF Mutations
To confirm the variation in the fitness effects of pykF mutations in the evolved backgrounds, and to explore the causes of that variation, we examined the fitness effects of two particular pykF mutations in the 11 independently evolved 20,000-generation clones. The two mutations were the A301S mutation that arose independently in three LTEE populations and the deletion mutation that is the proxy for the insertion mutation in another population. As an internal control, we also replicated our previous finding that both the A301S and deletion mutations are beneficial when placed in the ancestral chromosome, with the former substantially more so than the latter (fig. 4, far left). An analysis of variance shows that the fitness effects were largely determined by the population’s genetic background, with the specific mutation having only a marginally significant effect (table 3). In fact, the latter effect shows that the pykF deletion mutation tends to be slightly more favorable than the A301S mutation in most of the evolved backgrounds, whereas the opposite tendency held in the ancestral background (fig. 4).
Fig. 4.
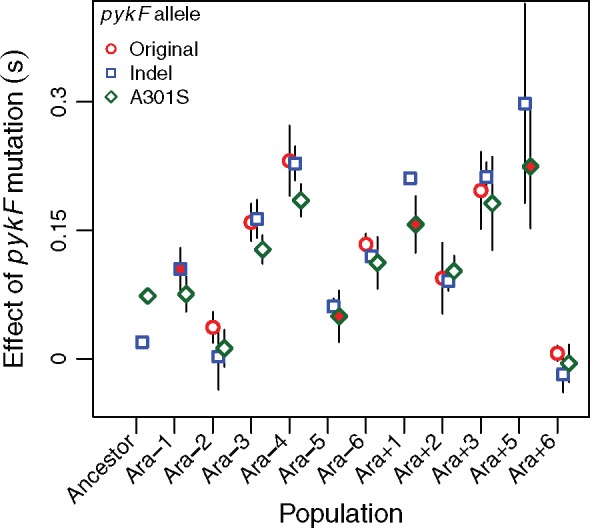
Fitness effects of the deletion and A301S alleles measured in 20,000-generation clones isolated from each population. If the evolved allele present in a clone was different from either of the transferred alleles, then its effect is also shown as the “original” allele. If an evolved clone already had one of the transferred alleles, then this fact is indicated by red fill in the corresponding symbol for the deletion (for Ara–1) or A301S (for Ara–5, Ara+1, and Ara+5) allele. Error bars indicate 95% confidence intervals (n = 3).
Table 3.
Mixed-model ANOVA Testing for the Fitness Effects of Two pykF Mutations in Genetic Backgrounds Sampled from Independently Evolved Populations.
| df | SS | MS | F | P | |
|---|---|---|---|---|---|
| Population | 10 | 0.4424 | 0.0442 | 48.54 | <0.001 |
| Mutation | 1 | 0.0086 | 0.0086 | 7.071 | 0.024 |
| Population × mutation | 10 | 0.0122 | 0.0012 | 1.337 | 0.241 |
| Error | 44 | 0.0401 | 0.0009 |
Note.—The fitness effects of two mutations in genetic backgrounds from 11 populations were estimated with 3-fold replication in a single block. Population and mutation are treated as random and fixed factors, respectively.
These data therefore also indicate that the selective advantage associated with pykF mutations changed as a consequence of the changing genetic context in which they exist. To demonstrate this inference even more directly, we examined the effect of replacing evolved pykF mutations with the ancestral allele over time across several of the LTEE populations, as explained in the next section.
Changes in Fitness Effects of pykF Mutations over Evolutionary Time
The pykF mutations generally arose early in the evolution of the LTEE, but we know the timing and genetic background in only one population (Khan et al. 2011). Thus, we do not know whether the differences in the fitness effects of the pykF mutations in the ancestor and in the evolved backgrounds reflect interactions between the pykF alleles and mutations occurring before or after them, or some combination thereof. Nevertheless, we can assess whether there is a trend or other pattern with respect to the fitness effects of pykF mutations as other mutations arise in the same lineage. To that end, we measured the fitness effects of the evolved pykF mutations by replacing them with the ancestral allele in clones isolated from six populations at 2,000, 5,000, 10,000, 20,000, 30,000, and 50,000 generations (except in four cases as noted in Materials and Methods). For comparison, we also measured the effect of the evolved mutations in the ancestral background (i.e., generation 0) in two of the four experimental blocks. An ANOVA indicates a highly significant interaction between the population and generation factors (table 4, fig. 5A), and the interaction remains highly significant if generation 0 is excluded from the analysis (supplementary table S8, Supplementary Material online). This interaction demonstrates that other mutations accumulating in the LTEE lineages are interacting with the mutations in pykF to change their marginal benefits, sometimes increasing and other times reducing those benefits. The among-population variation in the beneficial effects of the pykF mutations was highest at 10,000 generations and then declined to be undetectable (i.e., the estimated variance component was 0) at 50,000 generations (fig. 5B).
Table 4.
Mixed-model ANOVA Testing for the Effects of Population and Generation on the Fitness Benefits Associated with pykF Mutations in Six Populations of the LTEE.
| df | SS | MS | F | P | |
|---|---|---|---|---|---|
| Population | 5 | 0.1243 | 0.0249 | 21.63 | <0.001 |
| Generation | 6 | 0.1870 | 0.0312 | 1.573 | 0.195 |
| Population × generation | 26 | 0.5151 | 0.0198 | 17.23 | <0.001 |
| Block | 3 | 0.0779 | 0.0260 | 22.59 | <0.001 |
| Error | 97 | 0.1115 | 0.0011 |
Note.—The fitness marginal effects of the evolved pykF mutations were tested in backgrounds from six populations at seven generations in four blocks, with missing data as follows: Two blocks did not include generation 0 for any population; four other genotypes (i.e., combinations of population and generation) were missing from all four blocks; and two other genotypes had one missing value each. Population and generation are treated as random and fixed factors, respectively.
Fig. 5.

Change in effect of pykF mutations over time in six populations. (A) Symbol colors indicate different populations in which evolved pykF mutations in clones isolated at 2,000, 5,000, 10,000, 20,000, 30,000, and 50,000 generations (except the 2,000-generation clone from Ara+5) were reverted to the ancestral allele, and the resulting fitness effects were estimated. The same mutations were added to the ancestor, and their fitness effects were measured to give a reference at 0 generations. Error bars indicate 95% confidence intervals (n = 4 except at the initial time point where n = 2). (B) Estimates of the among-population standard deviation (i.e., the square root of the variance component) in the fitness effects of the pykF mutations at each time point.
Discussion
Interactions between beneficial mutations are important because they influence the topology of the fitness landscape and, therefore, evolutionary outcomes (De Visser and Krug 2014; Hartl 2014). The ability to identify and manipulate beneficial mutations has allowed these interactions to be studied in detail, albeit only for small regions of landscapes (Weinreich et al. 2006; Khan et al. 2011; Chou et al. 2011; Ogbunugafor et al. 2016; Palmer et al. 2017). In this study, we have determined how the fitness effects of mutations in the pykF gene of E. coli changed over the course of a long-term experiment, as other mutations arose in the evolving lineages. Our main findings can be summarized as follows. 1) Eight different nonsynonymous mutations arose that impact all three functional domains of the PykF enzyme. They all confer similar beneficial fitness effects when measured in the ancestral strain. These benefits are greater than for a deletion allele, indicating that the evolved enzymes retain some functionality. 2) The fitness effects of these mutations are much more variable when measured in the evolved strains. Most of this variation results from divergent interactions with the changing genetic backgrounds, rather than from the different pykF alleles. 3) The variation in the fitness effects of the different pykF alleles peaked at intermediate time points and declined to an undetectable level at 50,000 generations. We discuss each of these findings below.
An important question in evolutionary biology is the extent to which different mutations in the same gene can be considered to be equivalent in terms of adaptation (Schluter et al. 2004; Lenormand et al. 2016). The parallel evolution of mutations in the same gene in independent populations provides a signature that those mutations are adaptive (Wichman et al. 1999; Woods et al. 2006; Barrick et al. 2009), but even parallel mutations can have different immediate and long-term consequences (Woods et al. 2011; Vogwill et al. 2014, 2016; Gifford et al. 2016). We found that nonsynonymous mutations in pykF have much more variable fitness effects in 20,000-generation clones than they do in the ancestor. In principle, this variation could reflect the unmasking of different functional changes in the evolved PykF enzymes, divergent interactions between the evolved PykF and other mutations in the different lineages, or some combination thereof. The finding that two distinct pykF alleles had similar effects to one another in many backgrounds, whereas having much more variable effects across the backgrounds, indicates the importance of epistatic interactions between the pykF mutations and the genetic background. In fact, the exact same A301S mutation, which arose independently in three populations, had significantly different effects in clones from those populations. We do not yet have a mechanistic understanding of the effects of the pykF mutations, and so it remains unknown both how mutations in the different functional domains confer similar fitness benefits in a given background and how changing backgrounds modify those benefits.
In the Introduction, we presented two distinct possibilities for how the effect of an early occurring beneficial mutation might change over time as a lineage continues to evolve and adapt. First, the effect of the early mutation could follow a pattern of diminishing-returns epistasis, such that its marginal benefit declines as the lineage’s fitness increases (Khan et al. 2011; Kryazhimskiy et al. 2014; Chou et al. 2011; Wang et al. 2016; Wünsche et al. 2017). This relationship is “global” in the sense that the marginal fitness effect of adding a particular beneficial mutation depends on the fitness of the recipient rather than its specific genotype (Kryazhimskiy et al. 2014). The dynamics of fitness improvement and mutation accumulation in the LTEE are largely consistent with this global tendency toward diminishing-returns epistasis (Wiser et al. 2013; Tenaillon et al. 2016). Second, selection may favor new mutations that interact positively with the particular genetic background in which they occur. As a consequence, early mutations might become increasingly entrenched as later mutations depend on their presence for their own benefit (Shah et al. 2015). If so, then the marginal fitness effect of removing early beneficial mutations should increase over time (Zee et al. 2014). However, the high variation in the marginal effects of pykF mutations in the evolved clones from 20,000 generations—ranging from nearly neutral to benefits almost twice that seen in the ancestor—does not align with either scenario.
The high variation in the fitness effects of pykF mutations in evolved backgrounds might simply indicate the need for caution when extrapolating from specific cases to general patterns. Indeed, in a previous study, the pykF deletion allele included here was the only one of five mutations that did not show a pattern of diminishing-returns epistasis when tested across 16 genetic backgrounds that differed at one to four other loci (Khan et al. 2011). This background-dependent variation in the fitness effects of pykF mutations could reflect some combination of diminishing-returns and positive epistasis. Previous findings of diminishing-returns epistasis between beneficial mutations compared strains that had evolved for <2,000 generations and usually harboring <10 mutations relative to their ancestors (Khan et al. 2011; Chou et al. 2011; Kryazhimskiy et al. 2014). In this work, by contrast, pykF effects were measured in strains that had evolved for up to 50,000 generations, and which had accumulated at that point between 67 (in Ara+2) and 1,091 (in Ara–1) mutations in addition to the one in pykF (supplementary table S3, Supplementary Material online; Tenaillon et al. 2016). Thus, even if specific interactions between pykF and other mutations are rare, the large number of potential interactions means they might overwhelm any global trend. Supporting this possibility, a study that analyzed interactions among mutations in several experimentally determined fitness landscapes found at least one higher-order epistatic interaction in each landscape, even after controlling for global effects (Sailer and Harms 2017). We note, however, that the average fitness effect of pykF mutations not differ significantly between mutator and nonmutator clones, despite an order-of-magnitude difference in the number of background mutations. This result suggests that the most important interactions with respect to the fitness effects of pykF mutations involve other beneficial mutations. On the other hand, the relatively low mean and variance of the fitness effects of pykF mutations across genetic backgrounds at the final 50,000-generation time point suggests some statistical repeatability in the relevant interactions, even if the mechanistic basis for that convergence remains unknown at present.
In summary, we have shown that different mutations in the pykF gene that were selected in independently evolving E. coli populations had similar effects on fitness when each was moved into their common ancestor’s genome. However, these same mutations had very different effects when they were replaced by the ancestral allele in clones sampled from each population after 20,000 generations. By 50,000 generations, however, their effects on fitness were again similar, with an overall tendency toward negative interactions with later-occurring mutations. Our results thus show that the fitness effects of beneficial mutations in some genes can be highly dynamic in evolving populations, with changing genetic backgrounds sometimes increasing and other times reducing the effect size of the focal mutations.
Materials and Methods
Strains and Mutations
We examined the fitness effects of various pykF mutations in the ancestral strain and evolved clones isolated from a long-term evolution experiment (Lenski et al. 1991). That experiment started with two strains of E. coli B (Jeong et al. 2009) that were isogenic except for a mutation in araA, which impacts the ability to use the sugar arabinose and serves as a neutral marker (Lenski et al. 1991) and a second mutation, also evidently neutral, that arose inadvertently (Tenaillon et al. 2016). Each strain was used to found six populations: Populations Ara–1 to Ara–6 were founded by the Ara– ancestral strain, REL606, and populations Ara+1 to Ara+6 were founded by the Ara+ ancestral strain, REL607. The 12 populations are grown at 37 °C in Davis Mingioli minimal medium supplemented with 25 µg/ml glucose (DM25) as the limiting resource, and they are propagated by daily 1:100 transfers into fresh medium (Lenski et al. 1991). Clones were isolated periodically from each evolving populations and then frozen. Here, we use the “A” clones taken from the populations at various time points, as noted in the text. A mutation in pykF was first found in a clone sampled from population Ara–1 at 20,000 generations, and subsequent work identified other mutations in this gene in the other 11 evolved populations (Schneider et al. 2000; Woods et al. 2006; figs. 1 and2, and supplementary table S9, Supplementary Material online). Here, we consider the effects of the eight unique nonsynonymous point mutations that were identified as well as a loss-of-function deletion mutation; we omit a frameshift mutation that was found in one population. The mutation found in the Ara–1 population involved the transposition of an IS150 element into the pykF reading frame; we use as a proxy a deletion allele because it was technically easier to transfer into the recipient strains (Khan et al. 2011). IS elements can affect fitness not only by disrupting genes but also by influencing the expression of nearby genes (Chou et al. 2009). In the case of the pykF:: IS150 insertion, changes in expression seem unlikely because the genes on either side have independent transcriptional control (Gama-Castro et al. 2016). In any case, the fitness effect of the insertion allele is indistinguishable from that of the deletion allele, indicating that any polar effects are small (Khan et al. 2011).
Genetic Manipulations
We constructed five sets of strains in order to measure the fitness effects of evolved pykF alleles in different genetic contexts. In all cases, the alleles were moved into the recipient chromosome, replacing the recipient strain’s pykF allele. Unless otherwise noted, these manipulations involve the eight nonsynonymous mutations and the deletion allele (fig. 2). The five strain sets are: 1) each pykF mutation separately moved into the ancestral strain, REL606; 2) the various evolved pykF mutations replaced with the ancestral allele in evolved clones isolated from each population at 20,000 generations; 3) the pykF deletion allele added to the evolved clones isolated at 20,000 generations; 4) the pykF A301S allele added to the evolved clones isolated at 20,000 generations; and 5) the evolved pykF alleles replaced by the ancestral allele in clones isolated from six populations at 2,000, 5,000, 10,000, 20,000, 30,000 and 50,000 generations. A schematic of these strain sets is presented in figure 2.
Allele replacement was performed using a suicide-vector protocol (Philippe et al. 2004). Briefly, PCR products of ∼800 bp in length and centered on a pykF mutation were cloned into plasmid pCR2.1 using the TA cloning kit, and the resulting construct was used to transform TOP10F’ cells (ThermoFisher, MA). Inserts containing the pykF mutations were then cut from pCR2.1 and cloned into a suicide plasmid, pDS132, which was used to transform MFDpir cells (Philippe et al. 2004; Ferrieres et al. 2010). MFDpir cells were used to mobilize the constructed pDS132:: pykF plasmids to the relevant recipient strain. Selection for plasmid transfer and subsequent allele exchange were performed as described previously (Cooper et al. 2003). To reduce the possibility that secondary mutations arising during the strain construction might confound the effect of the transferred pykF allele, we isolated several strain pairs from the end of independent transfer experiments, where one strain had the transferred allele and the other retained its original allele. In the absence of secondary mutations, the strain that retained the original allele should be identical to the relevant recipient strain. Only if those two strains were indistinguishable based on fitness assays was the strain with the transferred allele used in subsequent work. All mutation-clone combinations were successfully constructed except for the following: The Ara+5 clone isolated at 2,000 generations, which had not substituted the pykF mutation that later fixed in that population; the Ara+1 clone at 5,000 generations, for which we were unable to introduce a neutral marker; and Ara+3 clones at 5,000 and 50,000 generations, for which no constructs that were free of secondary mutations were isolated.
Most assays of competitive fitness were performed between strains differing only at the transferred pykF allele. To facilitate these competitions, we introduced a neutral marker into one of the competing strains, so that the strains could be distinguished on an agar-based indicator medium. Strains that were initially Ara+ were reverted to Ara– through the addition of the araA allele present in REL606 using the same allelic replacement method outlined above. Strains that were initially Ara– were selected on minimal-arabinose plates and a spontaneous Ara+ mutant isolated. We were unable to isolate a neutral Ara+ derivative of the clone isolated from Ara–4 at 20,000 generations. In this case we used “gene gorging” (Herring et al. 2003) to introduce a point mutation into lacZ that disrupted its function by introducing a stop codon (nucleotide change: G1556A). This mutation caused the constructed strain to produce white colonies on LB+X-gal+IPTG medium, whereas its progenitor formed blue colonies. All markers were tested for neutrality by competing the constructed marked strains against their progenitor, using the fitness protocol described below.
Measurements of Fitness Effects
The fitness effects of pykF mutations were estimated by head-to-head competitions. Competing strains that differed at a neutral marker, either araA or lacZ, were inoculated from freezer stocks into lysogeny broth (LB) and incubated overnight. Cultures were incubated at 37 °C in this and all subsequent steps. The overnight cultures were diluted 104-fold into 50-ml Erlenmeyer flasks containing 10 ml of DM25 and grown with shaking for 24 h. This environment is the same as used in the LTEE in which the pykF mutations evolved (Lenski et al. 1991). Cultures were then diluted 1:100 into fresh DM25 medium and incubated for a second 24-h cycle. After these two days of preconditioning, competitors were diluted 1:200 and mixed at an equal volumetric ratio to start the competition. The initial density of each competitor was estimated by plating an aliquot from the competition mixture onto either tetrazolium arabinose (TA) or LB+IPTG+X-gal indicator plates, for competitions using the araA or lacZ, markers, respectively. After two one-day growth cycles, an aliquot from each competition was plated again onto indicator agar to estimate the competitor’s final cell densities. The relative fitness was calculated as the ratio of each competitor’s realized growth rate (Lenski et al. 1991); the fitness effect of a mutation, s, was calculated simply by subtracting one from the corresponding relative fitness (figs. 3–5).
Statistical Analyses
We performed statistical analyses using R version 3.3.2 (R Core Team 2015). Some ANOVAs were one-way whereas others were mixed models or nested, as described below. The Anova function from the car package was used to compute Type II sums of squares, which are preferable when data are unbalanced owing to missing values. The nlme function and the confint function using the profile method setting, both from the lme4 package, were used to estimate confidence intervals of variance components. In mixed models, the F statistics of the fixed and random main effects are calculated and tested in comparison to the interaction and error mean squares, respectively; in nested models, the F statistics of the nested and top-level factors are calculated and tested in comparison to the error and nested mean squares, respectively (Sokal and Rohlf 1981). Dunnett’s test was carried out using the glht function in the multcomp package. Depending on the specific question addressed, the relevant set of competitions may have been performed simultaneously or in blocks, as noted in the corresponding tables.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Science Foundation (DEB-1253650 to T.F.C.), the US Army Research Office (W911NF-11-1-0481 to R.C.J.D. and T.F.C.), and the Royal Society of New Zealand Marsden Fund (UOC1013 to R.C.J.D.). The long-term evolution experiment is supported by funds from the National Science Foundation (DEB-1451740) and Michigan State University to R.E.L. T.F.C. will make the strains constructed in this study available to qualified recipients following completion of an institutional material transfer agreement. The results of the competition experiments, summary input data, and analysis scripts that pertain to the experiments and analyses reported in this paper have been deposited at http://doi:10.5061/dryad.7ck38.
References
- Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, Lenski RE, Kim JF.. 2009. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461(7268):1243–1247. [DOI] [PubMed] [Google Scholar]
- Blanquart F, Achaz G, Bataillon T, Tenaillon O.. 2014. Properties of selected mutations and genotypic landscapes under Fisher's geometric model. Evolution 68(12):3537–3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount ZD, Borland CZ, Lenski RE.. 2008. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc Natl Acad Sci U S A. 105(23):7899–7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount ZD, Barrick JE, Davidson CJ, Lenski RE.. 2012. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature 489(7417):513–518.http://dx.doi.org/10.1038/nature11514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H-H, Berthet J, Marx CJ, Matic I.. 2009. Fast growth increases the selective advantage of a mutation arising recurrently during evolution under metal limitation. PLoS Genet. 5(9):e1000652.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou HH, Chiu HC, Delaney NF, Segre D, Marx CJ.. 2011. Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science 332(6034):1190–1192.http://dx.doi.org/10.1126/science.1203799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TF, Rozen DE, Lenski RE.. 2003. Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc Natl Acad Sci U S A. 100(3):1072–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Visser JAGM, Krug J.. 2014. Empirical fitness landscapes and the predictability of evolution. Nat Rev Gen. 15(7):480–490.http://dx.doi.org/10.1038/nrg3744 [DOI] [PubMed] [Google Scholar]
- Donovan KA, Atkinson SC, Kessans SA, Peng F, Cooper TF, Griffin MDW, Jameson GB, Dobson RCJ.. 2016. Grappling with anisotropic data, pseudo-merohedral twinning and pseudo-translational noncrystallographic symmetry: a case study involving pyruvate kinase. Acta Crystallogr D Struct Biol. 72(4):512–519. [DOI] [PubMed] [Google Scholar]
- Draghi JA, Plotkin JB.. 2013. Selection biases the prevalence and type of epistasis along adaptive trajectories. Evolution 67(11):3120–3131.http://dx.doi.org/10.1111/evo.12192 [DOI] [PubMed] [Google Scholar]
- Ferrieres L, Hemery G, Nham T, Guerout AM, Mazel D, Beloin C, Ghigo JM.. 2010. Silent mischief: bacteriophage Mu insertions contaminate products of Escherichia coli random mutagenesis performed using suicidal transposon delivery plasmids mobilized by broad-host-range RP4 conjugative machinery. J Bacteriol. 192(24):6418–6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn WF, Haldane A, Torbett BE, Levy RM.. 2017. Inference of epistatic effects leading to entrenchment and drug resistance in HIV-1 protease. Mol Biol Evol. 34(6):1291–1306.http://dx.doi.org/10.1093/molbev/msx095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Castro S, Salgado H, Santos-Zavaleta A, Ledezma-Tejeida D, Muñiz-Rascado L, García-Sotelo JS, Alquicira-Hernández K, Martínez-Flores I, Pannier L, Castro-Mondragón JA, et al. 2016. RegulonDB version 9.0: high-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res. 44(D1):D133–D143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford DR, Toll-Riera M, MacLean RC.. 2016. Epistatic interactions between ancestral genotype and beneficial mutations shape evolvability in Pseudomonas aeruginosa. Evolution 70(7):1659–1666.http://dx.doi.org/10.1111/evo.12958 [DOI] [PubMed] [Google Scholar]
- Greene D, Crona K.. 2014. The changing geometry of a fitness landscape along an adaptive walk. PLoS Comp Biol. 10(5):e1003520.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL. 2014. What can we learn from fitness landscapes? Curr Opin Microbiol. 21:51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring CD, Glasner JD, Blattner FR.. 2003. Gene replacement without selection: regulated suppression of amber mutations in Escherichia coli. Gene 311:153–163.http://dx.doi.org/10.1016/S0378-1119(03)00585-7 [DOI] [PubMed] [Google Scholar]
- Jeong H, Barbe V, Lee CH, Vallenet D, Yu DS, Choi S-H, Couloux A, Lee S-W, Yoon SH, Cattolico L, et al. 2009. Genome sequences of Escherichia coli B strains REL606 and BL21(DE3). J Mol Biol. 394(4):644–652. [DOI] [PubMed] [Google Scholar]
- Khan AI, Dinh DM, Schneider D, Lenski RE, Cooper TF.. 2011. Negative epistasis between beneficial mutations in an evolving bacterial population. Science 332(6034):1193–1196. [DOI] [PubMed] [Google Scholar]
- Kochanowski K, Volkmer B, Gerosa L, Haverkorn van Rijsewijk BR, Schmidt A, Heinemann M.. 2013. Functioning of a metabolic flux sensor in Escherichia coli. Proc Natl Acad Sci U S A. 110(3):1130–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryazhimskiy S, Rice DP, Jerison ER, Desai MM.. 2014. Global epistasis makes adaptation predictable despite sequence-level stochasticity. Science 344(6191):1519–1522.http://dx.doi.org/10.1126/science.1250939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand T, Chevin L-M, Bataillon T.. 2016. Parallel evolution: what does it (not) tell us and why is it (still) interesting? In: Pence, Ramsey, editors. Chance in evolution. Chicago: Univ. Chicago Press. [Google Scholar]
- Lenski RE. 1998. Bacterial evolution and the cost of antibiotic resistance. Int Microbiol. 1(4):265–270. [PubMed] [Google Scholar]
- Lenski RE, Rose MR, Simpson SC, Tadler SC.. 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat. 138(6):1315–1341. [Google Scholar]
- Maisnier-Patin S, Berg OG, Liljas L, Andersson DI.. 2002. Compensatory adaptation to the deleterious effect of antibiotic resistance in Salmonella typhimurium. Mol Microbiol. 46(2):355–366. [DOI] [PubMed] [Google Scholar]
- Ogbunugafor CB, Wylie CS, Diakite I, Weinreich DM, Hartl DL, Bergstrom CT.. 2016. Adaptive landscape by environment interactions dictate evolutionary dynamics in models of drug resistance. PLoS Comp Biol. 12(1):e1004710–e1004720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortlund EA, Bridgham JT, Redinbo MR, Thornton JW.. 2007. Crystal structure of an ancient protein: evolution by conformational epistasis. Science 317(5844):1544–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AC, Toprak E, Baym M, Kim S, Veres A, Bershtein S, Kishony R.. 2017. Delayed commitment to evolutionary fate in antibiotic resistance fitness landscapes. Nat Commun. 6:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe N, Alcaraz J-P, Coursange E, Geiselmann J, Schneider D.. 2004. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51(3):246–255. [DOI] [PubMed] [Google Scholar]
- Phillips KN, Castillo G, Wünsche A, Cooper TF.. 2016. Adaptation of Escherichia coli to glucose promotes evolvability in lactose. Evolution 70(2):465–470. [DOI] [PubMed] [Google Scholar]
- Quandt EM, Deatherage DE, Ellington AD, Georgiou G, Barrick JE.. 2014. Recursive genomewide recombination and sequencing reveals a key refinement step in the evolution of a metabolic innovation in Escherichia coli. Proc Natl Acad Sci U S A. 111(6):2217–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt EM, Gollihar J, Blount ZD, Ellington AD, Georgiou G, Barrick JE.. 2015. Fine-tuning citrate synthase flux potentiates and refines metabolic innovation in the Lenski evolution experiment. eLife 4:e09696.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2015. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Sailer ZR, Harms MJ.. 2017. Detecting high-order epistasis in nonlinear genotype-phenotype maps. Genetics 205(3):1079–1088.http://dx.doi.org/10.1534/genetics.116.195214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter D, Clifford EA, Nemethy M, McKinnon JS.. 2004. Parallel evolution and inheritance of quantitative traits. Am Nat. 163(6):809–822. [DOI] [PubMed] [Google Scholar]
- Schneider D, Duperchy E, Coursange E, Lenski RE, Blot M.. 2000. Long-term experimental evolution in Escherichia coli. IX. Characterization of insertion sequence-mediated mutations and rearrangements. Genetics 156:477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, McCandlish DM, Plotkin JB.. 2015. Contingency and entrenchment in protein evolution under purifying selection. Proc Natl Acad Sci U S A. 112(25):E3226–E3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiquee KAZ, Arauzo-Bravo MJ, Shimizu K.. 2004. Effect of a pyruvate kinase (pykF-gene) knockout mutation on the control of gene expression and metabolic fluxes in Escherichia coli. FEMS Microbiol Lett. 235(1):25–33. [DOI] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ.. 1981. Biometry, second edition New York: W. H. Freeman. [Google Scholar]
- Tenaillon O, Barrick JE, Ribeck N, Deatherage DE, Blanchard JL, Dasgupta A, Wu GC, Wielgoss S, Cruveiller S, Médigue C, et al. 2016. Tempo and mode of genome evolution in a 50,000-generation experiment. Nature 536(7615):165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogwill T, Kojadinovic M, Furió V, MacLean RC.. 2014. Testing the role of genetic background in parallel evolution using the comparative experimental evolution of antibiotic resistance. Mol Biol Evol. 31(12):3314–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogwill T, Kojadinovic M, Furió V, MacLean RC.. 2016. Epistasis between antibiotic resistance mutations and genetic background shape the fitness effect of resistance across species of Pseudomonas. Proc R Soc Lond B. 283(1830):20160151.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Díaz Arenas C, Stoebel DM, Flynn K, Knapp E, Dillon MM, Wünsche A, Hatcher PJ, Moore FB-G, Cooper VS, et al. 2016. Benefit of transferred mutations is better predicted by the fitness of recipients than by their ecological or genetic relatedness. Proc Natl Acad Sci U S A. 113(18):5047–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich DM, Delaney NF, Depristo MA, Hartl DL.. 2006. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312(5770):111–114. [DOI] [PubMed] [Google Scholar]
- Wichman HA, Badgett MR, Scott LA, Boulianne CM, Bull JJ.. 1999. Different trajectories of parallel evolution during viral adaptation. Science 285(5426):422–424. [DOI] [PubMed] [Google Scholar]
- Wiser MJ, Ribeck N, Lenski RE.. 2013. Long-term dynamics of adaptation in asexual populations. Science 342(6164):1364–1367.http://dx.doi.org/10.1126/science.1243357 [DOI] [PubMed] [Google Scholar]
- Woods RJ, Barrick JE, Cooper TF, Shrestha U, Kauth MR, Lenski RE.. 2011. Second-order selection for evolvability in a large Escherichia coli population. Science 331(6023):1433–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods R, Schneider D, Winkworth CL, Riley MA, Lenski RE.. 2006. Tests of parallel molecular evolution in a long-term experiment with Escherichia coli. Proc Natl Acad Sci U S A. 103(24):9107–9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wünsche A, Dinh DM, Satterwhite RS, Diaz Arenas C, Stoebel DM, Cooper TF.. 2017. Diminishing-returns epistasis decreases adaptability along an evolutionary trajectory. Nat Ecol Evol. 1(4):0061.. [DOI] [PubMed] [Google Scholar]
- Zee PC, Mendes-Soares H, Yu Y-TN, Kraemer SA, Keller H, Ossowski S, Schneeberger K, Velicer GJ.. 2014. A shift from magnitude to sign epistasis during adaptive evolution of a bacterial social trait. Evolution 68(9):2701–2708. [DOI] [PubMed] [Google Scholar]
- Zhu T, Bailey MF, Angley LM, Cooper TF, Dobson RCJ.. 2010. The quaternary structure of pyruvate kinase type 1 from Escherichia coli at low nanomolar concentrations. Biochimie 92(1):116–120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




