Abstract
STUDY QUESTION
Is the phosphoinositol 1,3-kinase/protein kinase B (PI3K/AKT) pathway expression profile in cumulus cells (CCs) a potential marker of oocyte competence and predictive of pregnancy outcome?
SUMMARY ANSWER
Eleven genes (AKT1, ARHGEF7, BCL2L1, CCND1, E2F1, HRAS, KCNH2, PIK3C2A, SHC1, SOS1 and SPP1) in the PI3K/AKT pathway were significantly down-regulated in CCs from oocytes that went on to produce a pregnancy compared to CCs associated with a negative outcome.
WHAT IS KNOWN ALREADY
The PI3K/AKT pathway plays a pivotal role in the interdependence and continuous feedback between the oocyte and CCs.
STUDY DESIGN SIZE, DURATION
The expression analysis of 92 transcripts in the PI3K/AKT pathway in CCs from patients with negative or positive pregnancy outcome, after single embryo transfer, was performed. Mouse CCs target gene expression was conducted to associate the expression profile of PI3K/AKT pathway to oocyte developmental profile.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Fifty-five good prognosis IVF patients who had been referred to IVF or intracytoplasmic sperm injection treatment for male-factor infertility or tubal disease were enroled. CCs from single cumulus-oocyte complexes (COCs) from 16 patients who underwent a single embryo transfer were analyzed. Twenty-five CD-1 mice were used to assess gene expression in CCs associated with oocytes with different competence in relation to hCG priming. A total 220 human COCs were collected. The RNA extracted from CCs of 16 selected patients was used to analyze PI3K/AKT pathway gene expression employing a 96-well custom TaqMan Array. Expression data of CCs associated to positive IVF outcome were compared to data from negative outcome samples. Mice were sacrificed after 9, 12, 15, 21 and 24 h post-hCG administration to obtain CCs from MII oocytes with different developmental competence. Akt1, Bcl2l2 and Shc1 expression were tested in the collected mouse CCs. In addition, the expression of upstream regulator ESR1, the gene encoding for the oestrogen receptor ERβ, and the downstream effectors of the pathway FOXO1, FOXO3 and FOXO4 was evaluated in human and mouse samples.
MAIN RESULTS AND THE ROLE OF CHANCE
Transcripts involved in the PI3K Signaling Pathway were selectively modulated according to the IVF/ICSI outcome of the oocyte. Eleven transcripts in this pathway were significantly down-regulated in all samples of CCs from oocytes with positive when compared those with a negative outcome. These outcomes were confirmed in mouse CCs associated with oocytes at different maturation stages. Expression data revealed that the down-regulation of ESR1 could be related to oocyte competence and is likely to be the driver of expression changes highlighted in the PI3K/AKT pathway.
LIMITATIONS REASONS FOR CAUTION
Small sample size and retrospective design.
WIDER IMPLICATIONS OF THE FINDINGS
The CCs expression profile of PI3K/AKT signaling genes, disclosed a specific CCs gene signature related to oocyte competence. It could be speculated that CCs associated with competent oocytes have completed their role in sustaining oocyte development and are influencing their fate in response to metabolic and hormonal changes by de-activating anti-apoptotic signals.
STUDY FUNDING/COMPETING INTEREST(S)
Supported by Merck Serono an affiliate of Merck KGaA, Darmstadt, Germany (research grant for the laboratory session; Merck KGaA reviewed the manuscript for medical accuracy only before journal submission. The authors are fully responsible for the content of this manuscript, and the views and opinions described in the publication reflect solely those of the authors). The authors declare no conflict of interest.
Keywords: Oocyte competence, cumulus cells, gene expression, PI3K/AKT, pregnancy outcome, embryo
Introduction
ART as an effective treatment option for infertility and its use is increasing significantly with up to 4% of infants globally conceived in this way (Ozgur et al., 2016). Although ART protocols have improved markedly, still overall only 30% of ART cycles result in the live birth of a baby (Gunby et al., 2009). The absence of an objective, precise, and non-invasive oocyte selection strategy remains one of the major challenges in ART. The gold standard criteria for the assessment of oocyte quality are based on morphological benchmarks such as oocyte cytoplasm, polar body shape, zona pellucida thickness and meiotic spindle position (Coticchio et al., 2004). However, these methods have drawbacks in terms of accuracy, objectivity and consistency (Assidi et al., 2015).
Recently, several studies, aimed at discovering non-invasive and objective tools to evaluate indirectly oocyte quality (Uyar et al., 2013; Fragouli et al., 2014), have focused on cumulus cells (CCs) as a mirror of oocyte characteristics. Inter-communication between the oocyte and the surrounding CCs is essential for the formation of a competent gamete for fertilization, and affects the resultant embryos’ development and the chance of successful pregnancies (Fang et al., 2016). CCs regulate oocyte maturation strictly by producing regulatory signals and paracrine factors. In addition, CCs drive meiotic resumption and modulate changes in the oocyte cytoskeleton (Coticchio et al., 2015). Conversely, alterations in CCs have been linked to oocyte maturation arrest and the formation of non-competent gametes (Ruvolo et al., 2013). In this context, much effort has been spent to develop molecular strategies aimed at selecting the most competent oocyte. Transcriptomic profiling of follicular cells allowed the identifications of several genes expressed in CCs that could function as oocyte regulators (Labrecque and Sirard, 2014). Moreover, CC gene expression profile has been assessed in different species to understand the relationship between CCs and oocytes (Assou et al., 2006; Assidi et al., 2011; Regassa et al., 2011; Sirard, 2011; Xu et al., 2011; Borgbo et al., 2013), maturation and fertilization rate (McKenzie et al., 2004), and pregnancy outcome (Feuerstein et al., 2007; Hamel et al., 2008; Assou et al., 2010; Yerushalmi et al., 2014). Global expression profiling studies on CCs revealed a plethora of putative biomarkers of oocyte competence. However, the general lack of uniformity renders their utility in the clinical setting controversial. Recently, Hammond and colleagues (Hammond et al., 2015) suggested that a combination of time-lapse and CC gene expression assessment might predict embryo quality. In order to acquire this knowledge, we previously performed, using a whole DNA microarray approach, a CC transcriptome study in patients undergoing different ovarian stimulation treatments (Gatta et al., 2013). This work demonstrated that distinct gonadotropin preparations differentially modulate CC transcripts involved in the trafficking of retinoic acid and ovarian steroidogenesis (RXRB, TTR, ALDH8A1), and in the follicular development (IL11 and AKT3) (Gatta et al., 2013).
The present research focused on a specific target-pathway expression study utilizing quantitative real time–polymerase chain reaction (qRT-PCR), to correlate gene expression in CCs to the reproductive outcome. The aim of the study is to disclose a specific CC gene expression signature linked to oocyte competence. We focused on the analysis of 92 transcripts in the PI3K/AKT signalling pathway in CCs associated with oocytes that gave different reproductive outcomes. The PI3K/AKT pathway, already evidenced in our previous study (Gatta et al., 2013), plays a pivotal role in the interdependence and continuous feedback between the oocyte and CCs. It is activated in CCs to fulfil their role in oocyte development via transduction of pro-survival signals (Huang and Wells, 2010).
To investigate further the hypothesis that specific CC genes in the PI3K/AKT pathway could be the signal that the oocyte has become competent for fertilization, a mouse CC target gene expression study was conducted. Different levels of competence based on mice post-ovulatory age were related to gene expression. These experiments were based on the knowledge that oocyte maturation in most mammalian species is completed within 12–24 h, and during this brief period significant qualitative molecular changes take place. These changes initiate and regulate resumption of meiosis, prevent polyspermic fertilization, and promote the exit from the MII arrest stage. In the mouse, oocytes are ovulated in the MII stage around 12 h after the LH surge and are expected to be fertilized within 6 h after ovulation (Ducibella, 1998). Outside this window, post-ovulatory oocyte aging sets in (Miao et al., 2009).
Starting out from this assumption, the expression levels of three genes included in our significant data set (Akt1, Bcl2l1 and Shc1) were analysed in mouse CCs associated to oocytes at different times from hCG priming (9, 12, 15, 21 and 24 h post-hCG administration). The gene expression analysis disclosed similar results in both humans and animal models indicating that CCs associated to the positive outcome showed a reduction in the expression of specific genes involved in cell proliferation.
This study reveal, for the first time, that specific CC genes in the PI3K/AKT signalling are strictly related to oocyte competence and could represent potential non-invasive biomarkers predictive of IVF outcome.
Material and Methods
Patients
Cumulus-oocyte complexes (COCs) were collected at the IVF laboratories of the University of Pisa and the Cannizzaro Hospital of Catania from good prognosis IVF patients (healthy patients) undergoing standard assisted reproduction protocols between December 2013 and June 2015. Fifty-five healthy women aged <35 years with normal BMI (18.5–24.9 kg/m2), FSH < 8 pg/ml, antral follicle count >7, and regular cycles who had been referred to IVF or intracytoplasmic sperm injection (ICSI) treatment for male-factor infertility or tubal disease participated in the study after providing written informed consent. The study was approved by the local Ethical Committee of Pisa (protocol number 2007-005397-31) and performed in compliance with the Declaration of Helsinki. The study was also carried out under the supervision of the Italian Society of Embryology and Reproduction.
Patients were down-regulated with GnRH agonist (Enantone; Takeda Pharmaceutical Co.) and received 225 IU hp-hMG daily (Meropur; Ferring Pharmaceuticals), or 150 IU r-hFSH + 75 IU r-hLH daily (Pergoveris; Merck Serono). All patients received gonadotropin treatment for a maximum of 12 days and ovulation was induced by a single administration of human chorionic gonadotropin (hCG) (10000 IU; Gonasi®, IBSA, Lugano, Switzerland) on the day after the last hMG-HP or r-hFSH/r-hLH injection when the leading follicle had reached a mean diameter of ≥17 mm. Aspiration of the oocytes was performed transvaginally by ultrasound guidance 36 hours after hCG administration. The embryos were transferred after 5 days of culture. All patients were tested for pregnancy 12 days after embryo transfer by Beta hCG test. In case of a positive pregnancy test, the patients were followed up to the onset of clinical pregnancy (i.e. gestational sac containing an embryo with a heartbeat).
Human CCs collection
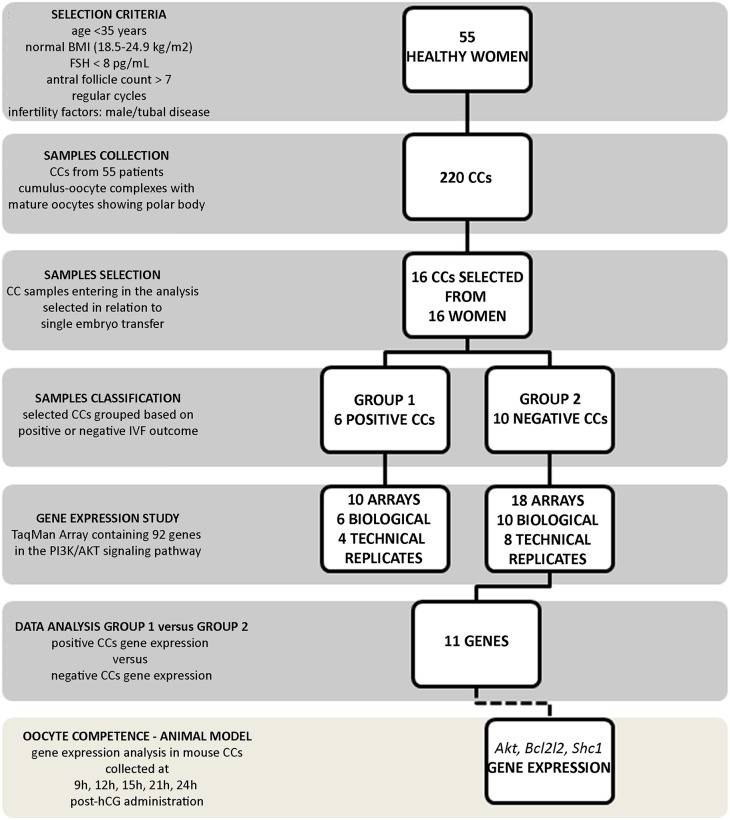
Following follicular aspiration, collected COCs were immediately washed in HEPES-buffered medium (Cook, Brisbane, Australia). CCs were dissected using a sterile scalpel and transferred immediately into a sterile tube containing 50 μl of RNAprotect Cell Reagent (Qiagen Ltd, Crawley, UK) and stored at −80°C until further analysis. A total of 220 CC samples were retrieved from the 55 patients. All were derived from COCs’ containing mature oocytes with a polar body. All COCs included in the analysis were fertilized successfully and embryos were selected and transferred at the blastocyst stage. A single embryo was transferred for 16 patients and transfers were performed only when the level of progesterone on hCG-day and endometrium were adequate. We analysed only the first ET per patient. Among the 16 embryos transferred, 6 achieved a clinical pregnancy (Positive) and 10 did not (Negative) (Fig. 1). The two groups were balanced for the different gonadotropin doses received.
Figure 1.
Experimental design. The flowchart depicts the experimental design of the study including the number of samples enroled, stimulation protocol used and patient inclusion criteria. CCs, cumulus cells.
The CC samples associated to each transferred embryo were then processed for molecular analysis.
As shown in Table I, both groups were comparable for age, infertility duration, basal levels of FSH, number of antral follicles, serum estradiol levels on hCGhCG-day and number of oocytes collected.
Table I.
Patient demographics and baseline characteristics. Data are presented as mean ± SD.
| Group 1 positive | Group 2 negative | P-valuea | |
|---|---|---|---|
| No. of patients | 6 | 10 | |
| Age (y) | 33.1 ± 1.8 | 34.1 ± 1.0 | NS |
| Duration of infertility (y) | 3.1 ± 0.8 | 2.9 ± 1.2 | NS |
| Basal FSH (mU/ml) | 5.2 ± 1.5 | 4.9 ± 1.4 | NS |
| AFC | 16.4 ± 2.1 | 15.5 ± 3.1 | NS |
| Treatment length (d) | 11.5 ± 1.5 | 11.7 ± 1.9 | NS |
| Total gonadotropin dose (IU) | 2565 ± 1000 | 2650 ± 1125 | NS |
| E2 on hCGhCG-day (pg/ml) | 1435.4 ± 480 | 1515.5 ± 540 | NS |
| No. of oocytes collected | 11.3 ± 2.8 | 10.6 ± 2.5 | NS |
AFC, antral follicle count; E2, estradiol.
aStudent t-test. NS, not statistically significant (P > 0.05).
Mice
CD-1 mice were obtained from Charles River Italia s.r.l. (Calco, Italy). Animal care and experiments were carried out in accordance with the Guiding Principles in the Care and Use of Animals approved by the University of L’Aquila (Italy). Mice were housed at 22 ± 2°C, 12–12 h light–dark cycle, with lights on from 8 a.m. to 8 p.m., free access to water and food, four animals per cage). At the age of 4–8 weeks, females were superovulated by intraperitoneal injection of 10 IU of PMSG (Folligon; Intervet-International, Boxmeer, Holland) and 10 IU of hCG (Profasi HP 2000; Serono, Roma, Italy) 48 h apart. Mice were killed by cervical dislocation at different times post-hCG. To collect COCs in the periovulatory phase, ovaries were removed from mice at 9 h post-hCG by puncturing large follicles. To collect ovulated COCs, oviducts were excised after 12, 15, 21 and 24 h post-hCG. Three animals per time point were used.
Mouse CCs collection
Cumulus masses were transferred in M2 medium (Sigma, St. Louis, MO) containing 0.1 mg/ml hyaluronidase (Sigma) for 7 min. MII oocytes were removed and CCs were transferred into a sterile tube containing 50 μl of RNAprotect Cell Reagent (Qiagen Ltd, Crawley, UK) by using a stripping pipette. CCs from at least 30 COCs were used for each experimental condition.
RNA extraction and qRT-PCR
Arcturus PicoPure RNA Isolation Kit (ThermoFisher Scientific, Waltham, MA, USA) was used for total RNA purification from all human CC samples reported in Table I and mouse CC samples.
PI3K/AKT signalling pathway TaqMan Array
Total purified RNA was linearly amplified using the AminoAllylMessageAmp™ II aRNA Amplification Kit (Ambion, Austin, TX, USA) in two amplification rounds following manufacturer's instructions. About 7 μg of the amplified RNA was reverse transcribed using the High Capacity RNA-to-cDNA Kit (ThermoFisher Scientific, Waltham, MA, USA). A 96-well TaqMan Array Human PI3K signalling Pathway (ThermoFisher Scientific, Waltham, MA, USA) was employed for expression analysis. Each array contains 92 assays for genes involved in the pathway and four assays as reference genes (Supplementary Table SI).
cDNA was mixed with DNase-free water and added to 2XTaqMan Universal PCR Master Mix No AmpErase UNG (ThermoFisher Scientific, Waltham, MA, USA) following manufacturer's instructions. A 96-well TaqMan Array Human PI3K Pathway was loaded with 20 μl per well of the mix and run on an Abi 7900HT Sequencing Detection System (ThermoFisher Scientific, Waltham, MA, USA). The amplification conditions were 10 min at 95°C followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Two independent experiments were run for 12 samples, the remaining 4 samples had insufficient RNA for the technical replicate, for a total of 28 plates as reported in Fig. 1.
Target genes qRT-PCR
One μg of total purified RNA from eight patients (four in the positive group, four in the negative group) and from mouse samples was reverse transcribed using the High Capacity RNA-to-cDNA Kit (ThermoFisher Scientific, Waltham, MA, USA). The remaining patients were not analysed due to inadequate amounts of RNA. qRT-PCR was performed in a total volume of 20 μl containing 2XTaqMan Universal PCR Master Mix No AmpErase UNG and 25 ng of cDNA using TaqMan Gene Expression Assays (20X) (mouse assays: Mm01331626_m1 for Akt1, Mm00437783_m1 for Bcl2l1, Mm00468942_g1for Shc, Mm00433149_m1 for Esr1, Mm00490671_m1 for Foxo1, Mm01185722_m1 for Foxo3, and Mm00840140_g1 for Foxo4; human assays: Hs01046816_m1 for ESR1, Hs00231106_m1 for FOXO1, Hs00818121_m1 for FOXO3, Hs00172973_m1 for FOXO4) on an Abi 7900HT Sequencing Detection System. GAPDH (mouse assay Mm99999915_g1; human assay Hs02786624_g1) and GUSB (mouse assay Mm01197698_m1; human assay Hs00939627_m1) were used as housekeeping genes (ThermoFisher Scientific, Waltham, MA, USA). Real time amplification conditions were 10 min at 95°C followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Each sample was run in triplicate.
Data analysis
PI3K/AKT signalling pathway TaqMan Array
Raw data were analysed by DataAssist software (ThermoFisher Scientific, Waltham, MA, USA). Each analysis underwent a global normalization using GAPDH, 18 s, and GUSB as selected internal controls. Only genes showing no outlier replicates and high expression in CCs (Ct < 30) were included in the analysis. The transcripts that showed high degree of variability across samples, and small fold differences between the two classes studied (positive and negative outcome) were excluded. A gene was considered differentially expressed in positive CCs versus negative CCs when showing a fold change >1.4 or <0.7 and a P-value < 0.05 (ANOVA). P-values were adjusted using Benjamini–Hochberg FDR correction test. The fold changes and statistical significance for each gene in the array are reported in Supplementary Table I.
qRT-PCR
The genes’ relative expression was corrected against GAPDH and GUSB genes as internal controls. The ΔΔCt method was employed to compare relative fold changes between samples (12, 15, 21, 24 h post-hCG) and control (9 h post-hCG) for mouse CCs; human CC relative fold changes were calculated in positive group versus negative group. ANOVA was used to assess the P-value, considering data significant when P < 0.05.
IPA-inferred Biological Networks and Upstream Regulators analysis
The 11 significant de-regulated genes were analysed by Ingenuity Pathway Analysis (IPA) software (Qiagen, Hilden, Germany). IPA-inferred network analysis was generated for the 11 transcripts linking their functionality to the function of other genes and a mechanistic network of these genes was highlighted based on their connectivity and enrichment statistics. A Fisher's exact test was used to generate the network score, based on the number and size of eligible genes and the total number of genes that could be included in the network. An upstream effect analysis was run to predict the drivers of the expression modulation evidenced. The predicted effect is based on a value calculated by the IPA z-score algorithm. The z-score predicts the direction of change for the function (negative or positive), being significant when showing a P < 0.005.
Results
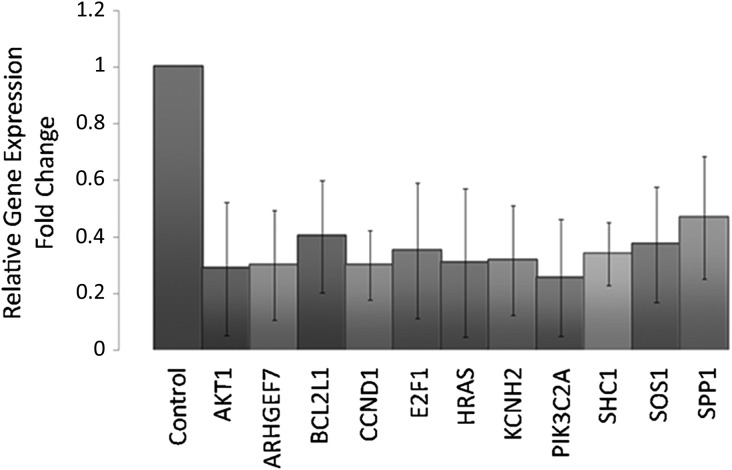
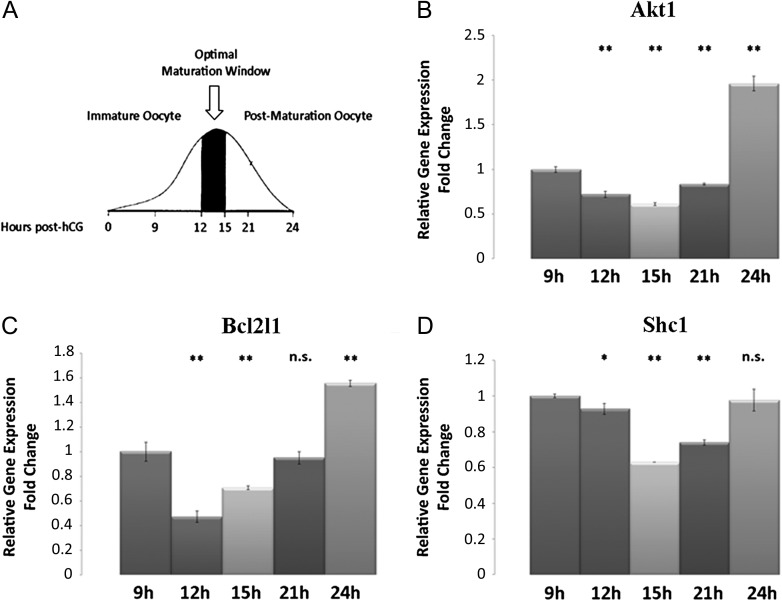
Data showed that the selective modulation of a set of transcripts involved in the PI3K/AKT signalling pathway in CCs related to the IVF outcome. In particular, the analysis highlighted 11 transcripts (namely: AKT1, ARHGEF7, BCL2L1, CCND1, E2F1, HRAS, KCNH2, PIK3C2A, SHC1, SOS1, SPP1) in this pathway significantly (P < 0.005) down-regulated in all samples of CCs from oocytes which led to clinical pregnancy (positive group) when compared to CCs associated to oocytes which gave a positive fertilization and transfer but did not result in a clinical pregnancy (negative group) (Fig. 2). This expression pattern likely reflects the oocyte maturation and quality in the positive group. To further validate this hypothesis, the expression of three genes included in the significant data set for biological relevance (AKT1) or highest fold changes (BCL2L1 and SHC1) were analysed in mouse CCs collected from oocytes with different levels of competence based on their post-ovulatory age as shown in Fig. 3A (9, 12, 15, 21 and 24 h post-hCG administration). A lower significant expression of these transcripts in CCs associated with oocytes in the correct window of maturation (12 and 15 h) was revealed, whereas higher expression levels were detected in CCs associated with not fully competent (9 h) or aged (21 and 24 h) MII oocytes (Fig. 3B, C, D). These results were in line with data from human samples.
Figure 2.
Genes down-regulated in CCs associated with positive versus negative outcome. Bar charts show the 11 transcripts down-expressed in CCs associated with positive outcome versus negative outcome (control) ± SD. Raw data were analysed by DataAssist software under global normalization using GAPDH, 18 s and GUSB as selected internal controls. A gene was considered differentially expressed in positive CCs versus negative CCs when showing a fold change >1.4 or <0.7 and a P-value < 0.05 (ANOVA). P-values were adjusted using Benjamini–Hochberg FDR correction.
Figure 3.
qRT-PCR for Akt1, Bcl2l1 and Shc in mouse CCs from oocytes at different maturation stages after hCG administration. (A) The curve draws the oocytes maturation profile after hCG administration. 12–15 h post-hCG (arrow) represents the optimal maturation window, 9 h represents the immature oocyte and 21–24 h the post maturation stage. (B) Relative Fold Change of Akt1 gene. (C) Relative Fold Change of Bcl2l1 gene. (D) Relative Fold Change of Shc1. Fold changes are calculated using ΔΔCt method to compare mRNA levels between samples (12, 15, 21, 24 h post-hCG) and control (9 h post-hCG). ANOVA test was used to assess the P-value, considering data significant when P < 0.05 (*P < 0.05, **P < 0.005).
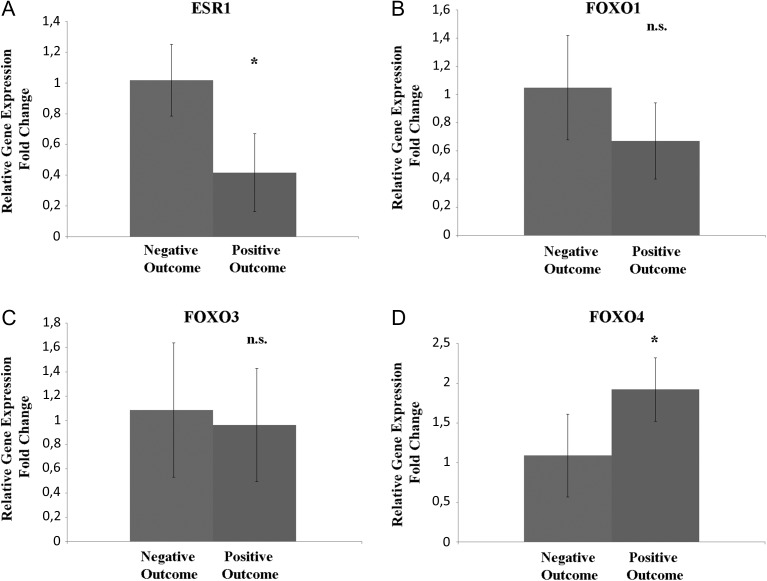
Moreover, we analysed the expression of other genes affected by PI3K/Akt but not included in the array used, namely ESR1, FOXO1, FOXO3 and FOXO4 in mouse and human CCs (Figs 4 and 5).
Figure 4.
qRT-PCR ESR1, FOXO1, FOXO3 and FOXO4 human CCs from oocytes leading to a positive outcome versus a negative outcome. (A) Relative fold change of ESR1 gene. (B) Relative fold change of FOXO1 gene. (C) Relative fold change of FOXO3 gene. (D) Relative fold change of FOXO4 gene. ANOVA test was used to assess the P-value, considering data significant when P < 0.05 (*P < 0.05).
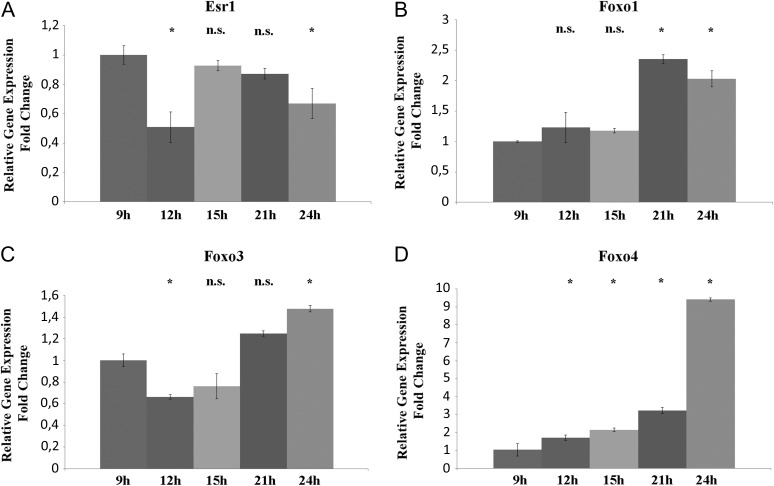
Figure 5.
qRT-PCR for Esr1, Foxo1, Foxo3 and Foxo4 in mouse CCs from oocytes at different maturation stages after hCG administration. (A) Relative Fold Change of Esr1 gene. (B) Relative Fold Change of Foxo1 gene. (C) Relative Fold Change of Foxo3 gene. (D) Relative Fold Change of Foxo4 gene. ANOVA test was used to assess the P-values, considering data significant when P < 0.05 (*P < 0.05).
In human CCs, ESR1 transcript revealed a significant down-expression in CCs from positive group when compared to negative group. FOXO1 and FOXO3 did not show any significant variation between the groupss. FOXO4 showed a higher expression in CCs from the positive group.
In mouse, CCs a significantly lower expression of Esr1 was seen in oocytes at 12 and 24 h post-hCG compared to 9 h whereas no significant difference was detected in CCs associated with oocytes at 15 and 21 h.
Foxo1, Foxo3 and Foxo4 expression modulation in mouse CCs was variable at the different maturation times as reported in Fig. 5.
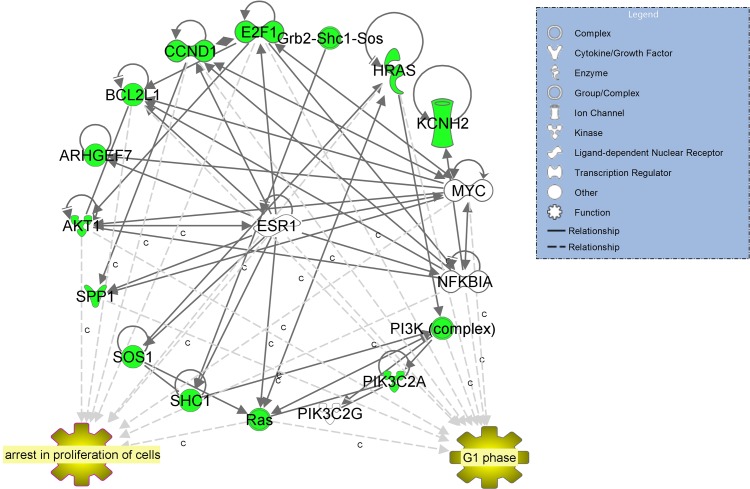
IPA analysis of the 11 genes in the data set showed that they are connected in one network with a score = 33 centred around oestrogen receptor ESR1 (Fig. 6). IPA upstream regulators analysis predicted a down-regulation of ESR1 expression (z-score −0.644, P-value < 0.000004) as the driving molecule of the reported changes.
Figure 6.
IPA-inferred network analysis for the 11 genes down-regulated in CCs from oocytes leading to a positive outcome versus a negative outcome. IPA-inferred network analysis of the 11 genes revealed one network with a score = 33 centred around oestrogen receptor ERβ (central node). IPA upstream regulators analysis predicted a down-regulation of ERβ expression (z-score −0.644, P-value < 4.01E-07) as the driving molecule of the reported changes in our data set.
Discussion
The objective of this study was to investigate a PI3K/AKT pathway gene expression signature in human CCs as a predictive marker of successful embryo implantation. The expression of 92 genes was evaluated in the PI3K/AKT pathway in CCs associated with negative or positive reproductive outcome.
To increase the predictive power of the analysis, healthy women aged <35 years were enroled to minimize the potential age-induced bias (Ebner et al., 2014; Lourenço et al., 2014). In addition, patients receiving different stimulation preparations (see Table I and Fig. 1) were recruited to obtain indicators of oocyte quality independent from the pharmaceutical treatment used since it is well known that r-hFSH, r-LH and hCG molecules are able to modulate CCs transcriptome (Gatta et al., 2013). Data analysis demonstrates that 11 transcripts included in the PI3K/AKT pathway are significantly down-regulated in all samples from the positive group when compared to the negative group. Accumulating evidence points to the PI3K signalling pathway as a critical regulator of COC function modulating oocyte maintenance or activation, and the proliferation, differentiation and stress response in granulosa cells (GCs) (Makker et al., 2014).
The de-regulation of this pathway contributes to infertility status caused by impaired follicular development, intra-follicular oocyte development and ovulation (Zheng et al., 2012; Makker et al., 2014).
The 11 transcripts (Table II) differentially expressed between the two analysed groups are involved in the control of proliferation, with anti-apoptotic functioning. In particular, AKT1 is a member of the serine/threonine-protein kinase family (AKT1, AKT2 and AKT3) which regulates many cellular processes including metabolism, proliferation, cell survival, growth and angiogenesis. AKT1 malfunction produces growth retardation and increased apoptosis in several tissue types (Chen et al., 2002; Brown et al., 2010). AKT1 expression was found in follicles at each developmental stage, in oocytes, and in GCs (Cecconi et al., 2010). Data from mice highlighted that Akt1 disruption leads to a reduced number of mature follicles, reduced proliferation of GCs and increased number of degenerated oocytes. In the absence of Akt1 gene, a concomitant decreased expression of the anti-apoptotic factor, Bcl2l1 was reported (Brown et al., 2010). BCL2L1 is widely expressed in human ovaries with an active role in protecting cells from apoptosis, as also demonstrated in rodents (Jin et al., 2005). Moreover, oocytes and GCs express BCL2L1 during follicular development contrasting external apoptotic signals and promoting follicle growth (Rucker et al., 2000; Jääskeläinen et al., 2010). GCs and follicle proliferation and survival are also regulated by the activation of ARHGEF7 and CCND1, by respectively acting on FOXO3A action and cell cycle progression (G1/S phase transition) (Stolk et al., 2009; Shimizu et al., 2013). E2F1, HRAS and SSP1 are involved in proliferation control, and their down-regulation has been observed in bovine granulosa cells during the shift from the growing follicle to the mature follicle (Douville and Sirard, 2014).
Table II.
The 11 transcripts down-regulated in CCs from the positive outcome group compared to CCs in the negative outcome group.
| Symbol | Entrez gene name | Fold change | Location | Family |
|---|---|---|---|---|
| AKT1 | v-akt murine thymoma viral oncogene homologue 1 | −6.53 | Cytoplasm | kinase |
| ARHGEF7 | Rho guanine nucleotide exchange factor 7 | −4.24 | Cytoplasm | other |
| BCL2L1 | BCL2 like 1 | −9.72 | Cytoplasm | other |
| CCND1 | cyclin D1 | −10.69 | Nucleus | transcription regulator |
| E2F1 | E2F transcription factor 1 | −4.58 | Nucleus | transcription regulator |
| HRAS | Harvey rat sarcoma viral oncogene homologue | −5.69 | Plasma Membrane | enzyme |
| KCNH2 | potassium voltage-gated channel subfamily H member 2 | −3.53 | Plasma Membrane | ion channel |
| PIK3C2A | phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 alpha | −13.45 | Cytoplasm | kinase |
| SHC1 | SHC (Src homology two domain containing) transforming protein 1 | −9.19 | Cytoplasm | other |
| SOS1 | SOS Ras/Rac guanine nucleotide exchange factor 1 | −6.55 | Cytoplasm | other |
| SPP1 | Secreted phosphoprotein 1 | −5.67 | Extracellular Space | cytokine |
CC, cumulus cells.
Although, the remaining genes in our significant data set (KCNH2, PIK3C2A, SOS1 and SHC1) have not been specifically described in the ovary, they are known to have anti-apoptotic activity in other cellular types (Afrasiabi et al., 2010; Zhang et al., 2015). Intriguingly, SOS1 is an upstream regulator of the PI3K/AKT pathway, inducing HRAS cascade and resulting in the nuclear translocation of AKT and BCL2 (Liu et al., 2015) whereas SHC1 alterations cause embryo developmental arrest (Favetta et al., 2004).
Based on the above observations, the identified down-expression pattern suggests that CCs from oocytes with high developmental competence undergo proliferation arrest and a consequent increase in apoptotic activity. These results are in line with a recent study by Borup (Borup et al., 2016) who defined a signature of 30 genes in CCs predictive of live birth. The authors found that the majority of these genes were down-regulated (Borup et al., 2016). The main downstream effect of these genes is the increase of the apoptosis processes in the CCs surrounding competent oocytes capable to develop, implant and lead to live births (Borup et al., 2016). This hypothesis is supported by another study which shows an increase of the apoptotic activity in CCs from MII oocytes when compared with GV oocytes (Ouandaogo et al., 2012). Noteworthy, the apoptotic activity in follicular cells has been also investigated revealing that several apoptotic follicular cells could be found in healthy follicles (Yuan et al., 2005). Raman et al. demonstrated a positive association between apoptotic CCs having increased DNA fragmentation and a higher fertilization rate during ICSI procedures (Raman et al., 2001). In contrast, many studies suggested inconsistent correlations between apoptosis in CCs and oocyte maturity (Ruvolo et al., 2013; Lourenço et al., 2014) or evidenced a correlation between oocyte competence and the presence of CC apoptotic processes (Janowski et al., 2012).
Results from the animal model fit with the hypothesis that the down-expression signature of genes included in the PI3K/AKT pathway (Akt1, Bcl2l1 and Shc1) is correlated with the maturation and competence of oocytes. The lower expression of these transcripts in CCs associated with oocytes having correct timing of maturation (12–15 h) was evidenced, whereas higher levels in CCs associated with non-mature or aged oocytes were detected. In addition, results from the mouse model represent evidence that the genes are differentially expressed in CCs during the periovulatory and post-ovulatory periods. The expression changes in the analyzed transcripts are thereby associated with the optimal fertilization window and could represent potential markers of oocyte competence to be further explored in humans.
Finally, it can be speculated that the reported differential CC gene expression in the PI3K/AKT signalling is driven by hormonal changes. It has been demonstrated that the FSH-driven PI3K pathway activation in GCs, results in phosphorylation/activation of the downstream branch-point kinase AKT and its targets. This leads to the modulation of FSH target genes, namely the forkhead box-containing proteins in the O subfamily (FOXO1, FOXO3a and FOXO4) and Tuberin gene (Douville and Sirard, 2014). IPA upstream analysis supports this idea highlighting that the oestrogen receptor ESR1 is the driver of the expression modulation of the 11 evidenced genes (Fig. 4). ESR1 is predicted to be down-regulated (z-score −0644) likely due to the decreasing FSH support at the end of the follicle maturation phase. This could lead to PI3K/AKT signalling switch off and the determination of the CCs fate including long-term survival in a quiescent state or programmed cell death (Burgering and Medema, 2003). To better investigate our hypothesis, the expression of ESR1, FOXO1, FOXO3 and FOXO4 was analysed in human and mouse CCs. Consistent with the IPA upstream analysis prediction, ESR1 was found to be down-regulated in both human and mouse CCs from competent oocytes. Thus, this drop seems to be associated with oocyte competence and is likely to result from follicle luteinisation. Nevertheless, in mouse CCs, Esr1 expression levels are partially recovered as early as 15 h post hCG and maintained during post-ovulatory ageing. This behaviour could be explained by considering that Esr1 can also act as an inhibitor of steroidogenesis throughout its ability to interact with multiple regulatory elements of oestrogen-dependent genes (Hosseini et al., 2016). Moreover, high ESR1 levels may be ascribed to ROS generation during post-ovulatory aging (Tamir et al. 2002; Tatone et al., 2011).
FOXOs participate in various cellular processes, including cell proliferation, cycle and apoptosis, whose functions are regulated by Akt (Salih and Brunet, 2008; Pisarska et al., 2009). FOXO1 is known to induce cell apoptosis throughout oxidative stress (Weng et al., 2016). Thus, it is not surprising to find an increase in its expression in mouse CCs at 21 and 24 h post hCG, whereas its expression does not change in human CCs. Among FOXOs, FOXO3 is another marker of oxidative stress in CCs probably related to apoptosis. In fact, its expression profile in mouse CCs shows a minimum expression level at 12 and 15 h post hCG and a significant increase at 21 and 24 h post hCG. Similarly to FOXO1, FOXO3 is not differentially expressed in human CCs from the two analyzed groups.
In addition to its role in the process of follicle activation, Foxo3 is known to induce apoptosis in several systems including granulosa cells of mice (Li et al., 2014).
FOXO4 role in ovaries is unclear and rarely studied. Richards et al. (2002) showed that FOXO4 mRNA was elevated in ovaries after luteinisation and, consistently we found it significantly higher in the positive CCs group highlighting that a condition of oocyte competence is related to correct follicle maturation. On the other hand, here we demonstrate in the mouse model that this gene reaches ~10-fold increase at 24 h post hCG when compared with CCs from competent COCs suggesting that it is also involved in the regulation of oocyte aging.
Taken together, these data demonstrate that decreased expression of ESR1 and increased expression of FOXO4 could be considered markers of oocyte competence likely correlated with a correct luteinisation, whereas FOXO1 and FOXO3 seems to be markers of oocyte post-ovulatory aging showing expression changes only on post-ovulatory aged oocytes.
Prospective studies on a larger population are needed, correlating also gene expression to morpho-kinetics evaluations.
Conclusions
The study reveal that specific CC genes in the PI3K/AKT pathway are strictly related to oocyte competence. Data show similar results in both humans and animal indicating that CCs from oocytes with a positive outcome have reduced expression of specific genes involved in cell proliferation, whereas higher expression of these transcripts could reflect oocytes with low rate of maturation and quality.
It can be speculated that CCs associated with competent oocytes have completed their role in sustaining oocyte development and are deciding on their fate in response to metabolic and hormonal changes.
Taken together, these data might be helpful to develop new useful tools to optimize oocyte selection strategies in ART protocols and to shed light on the molecular mechanisms regulating the cross-talk between the oocyte and the CCs.
Supplementary data
Supplementary data are available at Human Reproduction online.
Supplementary Material
Authors’ roles
P.G.A. participated in study design, samples collection and wrote the manuscript; C.T. participated in study design and data interpretation; S.S., M.D.A. and S.F. conducted the experiments and data analysis; G.D.E. collected mouse samples; R.C. and M.V. participated in samples collection; C.D.P. participated in data analysis; L.S. revised the manuscript critically for important intellectual content; V.G. participated in study design and coordination, data analysis and interpretation and wrote the manuscript.
Funding
Supported by Merck Serono an affiliate of Merck KGaA, Darmstadt, Germany (research grant for the laboratory session; Merck KGaA reviewed the manuscript for medical accuracy only before journal submission. The authors are fully responsible for the content of this manuscript, and the views and opinions described in the publication reflect solely those of the authors).
Conflict of interest
The authors declare no conflict of interest.
References
- Afrasiabi E, Hietamäki M, Viitanen T, Sukumaran P, Bergelin N, Törnquist K. Expression and significance of HERG (KCNH2) potassium channels in the regulation of MDA-MB-435S melanoma cell proliferation and migration. Cell Signal 2010;22:57–64. [DOI] [PubMed] [Google Scholar]
- Assidi M, Montag M, Sirard MA. Use of both cumulus cells’ transcriptomic markers and zona pellucida birefringence to select developmentally competent oocytes in human assisted reproductive technologies. BMC Genomics 2015;16:S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assidi M, Montag M, Van der Ven K, Sirard MA. Biomarkers of human oocyte developmental competence expressed in cumulus cells before ICSI: a preliminary study. J Assist Reprod Genet 2011;28:173–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assou S, Anahory T, Pantesco V, Le Carrour T, Pellestor F, Klein B, Reyftmann L, Dechaud H, De Vos J, Hamamah S. The human cumulus–oocyte complex gene-expression profile. Hum Reprod 2006;21:1705–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assou S, Haouzi D, De Vos J, Hamamah S. Human cumulus cells as biomarkers for embryo and pregnancy outcomes. Mol Hum Reprod 2010;16:531–538. [DOI] [PubMed] [Google Scholar]
- Borgbo T, Povlsen BB, Andersen CY, Borup R, Humaidan P, Grøndahl ML. Comparison of gene expression profiles in granulosa and cumulus cells after ovulation induction with either human chorionic gonadotropin or a gonadotropin-releasing hormone agonist trigger. Fertil Steril 2013;100:994–1001. [DOI] [PubMed] [Google Scholar]
- Borup R, Thuesen LL, Andersen CY, Nyboe-Andersen A, Ziebe S, Winther O, Grøndahl ML. Competence Classification of Cumulus and Granulosa Cell Transcriptome in Embryos Matched by Morphology and Female Age. PloS One 2016;11:e0153562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C, LaRocca J, Pietruska J, Ota M, Anderson L, Smith SD, Weston P, Rasoulpour T, Hixon ML. Subfertility caused by altered follicular development and oocyte growth in female mice lacking PKB alpha/Akt1. Biol Reprod 2010;82:246–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgering BMT, Medema RH. Decisions on life and death: FOXO Forkhead transcription factors are in command when PKB/Akt is off duty. J Leukoc Biol 2003;73:689–701. [DOI] [PubMed] [Google Scholar]
- Cecconi S, Rossi G, Santilli A, Stefano LD, Hoshino Y, Sato E, Palmerini MG, Macchiarelli G. Akt expression in mouse oocytes matured in vivo and in vitro. Reprod Biomed Online 2010;20:35–41. [DOI] [PubMed] [Google Scholar]
- Chen WS, Xu PZ, Gottlob K, Chen ML, Sokol K, Shiyanova T, Roninson I, Weng W, Suzuki R, Tobe K et al. . Growth retardation and increased apoptosis in mice with homozygous disruption of the Akt 1 gene. Genes Dev 2002;15:2203–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coticchio G, Dal Canto M, Mignini Renzini M, Guglielmo MC, Brambillasca F, Turchi D, Novara PV, Fadini R. Oocyte maturation: gamete-somatic cells interactions, meiotic resumption, cytoskeletal dynamics and cytoplasmic reorganization. Hum Reprod Update 2015;21:427–454. [DOI] [PubMed] [Google Scholar]
- Coticchio G, Sereni E, Serrao L, Mazzone S, Iadarola I, Borini A. What Criteria for the Definition of Oocyte Quality? Ann N Y Acad Sci 2004;1034:132–144. [DOI] [PubMed] [Google Scholar]
- Douville G, Sirard MA. Changes in granulosa cells gene expression associated with growth, plateau and atretic phases in medium bovine follicles. J Ovarian Res 2014;7:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducibella T. Biochemical and cellular insights into the temporal window of normal fertilization. Theriogenology 1998;49:53–65. [DOI] [PubMed] [Google Scholar]
- Ebner T, Shebl O, Holzer S, Oppelt P, Petek E, Schappacher-Tilp G, Mayer RG. Viability of cumulus cells is associated with basal AMH levels in assisted reproduction. Eur J Obstet Gynecol Reprod Biol 2014;183:59–63. [DOI] [PubMed] [Google Scholar]
- Fang Y, Shang W, Wei DL, Zeng SM. Cited2 protein level in cumulus cells is a biomarker for human embryo quality and pregnancy outcome in one in vitro fertilization cycle. Fertil Steril 2016;105:1351–1359. [DOI] [PubMed] [Google Scholar]
- Favetta LA, Robert C St, John EJ, Betts DH, King WA. p66shc, but not p53, is involved in early arrest of in vitro-produced bovine embryos. Mol Hum Reprod 2004;10:383–392. [DOI] [PubMed] [Google Scholar]
- Feuerstein P, Cadoret V, Dalbies-Tran R, Guerif F, Bidault R, Royere D. Gene expression in human cumulus cells: one approach to oocyte competence. Hum Reprod 2007;22:3069–3077. [DOI] [PubMed] [Google Scholar]
- Fragouli E, Lalioti MD, Wells D. The transcriptome of follicular cells: biological insights and clinical implications for the treatment of infertility. Hum Reprod Update 2014;20:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatta V, Tatone C, Ciriminna R, Vento M, Franchi S, D’Aurora M, Sperduti S, Cela V, Borzì P, Palermo R et al. . Gene expression profiles of cumulus cells obtained from women treated with recombinant human luteinizing hormone + recombinant human follicle-stimulating hormone or highly purified human menopausal gonadotropin versus recombinant human follicle-stimulating hormone alone. Fertil Steril 2013;99:2000–8.e1. [DOI] [PubMed] [Google Scholar]
- Gunby J, Bissonnette F, Librach C, Cowan L. Assisted reproductive technologies in Canada: 2005 results from the Canadian Assisted Reproductive Technologies Register. Fertil Steril 2009;91:1721–1730. [DOI] [PubMed] [Google Scholar]
- Hamel M, Dufort I, Robert C, Gravel C, Leveille MC, Leader A, Sirard MA. Identification of differentially expressed markers in human follicular cells associated with competent oocytes. Hum Reprod 2008;23:1118–1127. [DOI] [PubMed] [Google Scholar]
- Hammond ER, Stewart B, Peek JC, Shelling AN, Cree LM. Assessing embryo quality by combining non-invasive markers: early time-lapse parameters reflect gene expression in associated cumulus cells. Hum Reprod 2015;30:1850–1860. [DOI] [PubMed] [Google Scholar]
- Hosseini E, Mehraein F, Shahhoseini M, Karimian L, Nikmard F, Ashrafi M, Afsharian P, Aflatoonian R. Epigenetic alterations of CYP19A1 gene in Cumulus cells and its relevance to infertility in endometriosis. J Assist Reprod Genet 2016;33:1105–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Wells D. The human oocyte and cumulus cells relationship: new insights from the cumulus cell transcriptome. Mol Hum Reprod 2010;16:715–725. [DOI] [PubMed] [Google Scholar]
- Jääskeläinen M, Nieminen A, Pökkylä RM, Kauppinen M, Liakka A, Heikinheimo M, Vaskivuo TE, Klefström J, Tapanainen JS. Regulation of cell death in human fetal and adult ovaries—Role of Bok and Bcl-XL. Mol Cell Endocrinol 2010;330:17–24. [DOI] [PubMed] [Google Scholar]
- Janowski D, Salilew-Wondim D, Torner H, Tesfaye D, Ghanem N, Tomek W, El-Sayed A, Schellander K, Hölker M. Incidence of apoptosis and transcript abundance in bovine follicular cells is associated with the quality of the enclosed oocyte. Theriogenology 2012;78:656–669. [DOI] [PubMed] [Google Scholar]
- Jin X, Han CS, Yu FQ, Wei P, Hu ZY, Liu YX. Anti-apoptotic action of stem cell factor on oocytes in primordial follicles and its signal transduction. Mol Reprod Dev 2005;70:82–90. [DOI] [PubMed] [Google Scholar]
- Labrecque R, Sirard MA. The study of mammalian oocyte competence by transcriptome analysis: progress and challenges. Mol Hum Reprod 2014;20:103–116. [DOI] [PubMed] [Google Scholar]
- Li L, Ji SY, Yang JL, Li XX, Zhang J, Zhang Y, Hu ZY, Liu YX. Wnt/β-catenin signaling regulates follicular development by modulating the expression of Foxo3a signaling components. Mol Cell Endocrinol 2014;382:915–925. [DOI] [PubMed] [Google Scholar]
- Liu K, Jiang T, Ouyang Y, Shi Y, Zang Y, Li N, Lu S, Chen D. Nuclear EGFR impairs ASPP2-p53 complex-induced apoptosis by inducing SOS1 expression in hepatocellular carcinoma. Oncotarget 2015;6:16507–16516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourenço B, Sousa AP, Almeida-Santos T, Ramalho-Santos J. Relation of cumulus cell status with single oocyte maturity, fertilization capability and patient age. J Reprod Infertil 2014;15:15–21. [PMC free article] [PubMed] [Google Scholar]
- Makker A, Goel MM, Mahdi AA. PI3K/PTEN/Akt and TSC/mTOR signaling pathways, ovarian dysfunction, and infertility: an update. J Mol Endocrinol 2014;53:R103–R118. [DOI] [PubMed] [Google Scholar]
- McKenzie LJ, Pangas SA, Carson SA, Kovanci E, Cisneros P, Buster JE, Amato P, Matzuk MM. Human cumulus granulosa cell gene expression: a predictor of fertilization and embryo selection in women undergoing IVF. Hum Reprod 2004;19:2869–2874. [DOI] [PubMed] [Google Scholar]
- Miao YL, Kikuchi K, Sun QY, Schatten H. Oocyte aging: cellular and molecular changes, developmental potential and reversal possibility. Hum Reprod Update 2009;15:573–585. [DOI] [PubMed] [Google Scholar]
- Ouandaogo ZG, Frydman N, Hesters L, Assou S, Haouzi D, Dechaud H, Frydman R, Hamamah S. Differences in transcriptomic profiles of human cumulus cells isolated from oocytes at GV, MI and MII stages after in vivo and in vitro oocyte maturation. Hum Reprod 2012;27:2438–2447. [DOI] [PubMed] [Google Scholar]
- Ozgur K, Humaidan P, Coetzee K. Segmented ART - The new era in ART? Reprod Biol 2016;16:91–103. [DOI] [PubMed] [Google Scholar]
- Pisarska MD, Kuo FT, Tang D, Zarrini P, Khan S, Ketefian A. Expression of forkhead transcription factors in human granulosa cells. Fertil Steril 2009;91:1392–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman RS, Chan PJ, Corselli JU, Patton WC, Jacobson JD, Chan SR, King A. Comet assay of cumulus cell DNA status and the relationship to oocyte fertilization via intracytoplasmic sperm injection. Hum Reprod 2001;16:831–835. [DOI] [PubMed] [Google Scholar]
- Regassa A, Rings F, Hoelker M, Cinar U, Tholen E, Looft C, Schellander K, Tesfaye D. Transcriptome dynamics and molecular cross-talk between bovine oocyte and its companion cumulus cells. BMC Genomics 2011;12:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards JS, Sharma SC, Falender AE, Lo YH. Expression of FKHR, FKHRL1, and AFX genes in the rodent ovary: evidence for regulation by IGF-I, estrogen, and the gonadotropins. Mol Endocrinol 2002;16:580–599. [DOI] [PubMed] [Google Scholar]
- Rucker EB, Dierisseau P, Wagner KU, Garrett L, Wynshaw-Boris A, Flaws JA, Hennighausen L. Bcl-x and Bax regulate mouse primordial gem cell survival and apoptosis during embryogenesis. Mol Endocrinol 2000;14:1038–1052. [DOI] [PubMed] [Google Scholar]
- Ruvolo G, Fattouh RR, Bosco L, Brucculeri AM, Cittadini E. New molecular markers for the evaluation of gamete quality. J Assist Reprod Genet 2013;30:207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salih DA, Brunet A. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr Opin Cell Biol 2008;20:126–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Hirai Y, Miyamoto A. Expression of cyclins and cyclin-dependent kinase inhibitors in granulosa cells from bovine ovary. Reprod Domest Anim 2013;48:e65–e69. [DOI] [PubMed] [Google Scholar]
- Sirard MA. Follicle environment and quality of in vitro matured oocytes. J Assist Reprod Genet 2011;28:483–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolk L, Zhai G, van Meurs JB, Verbiest MM, Visser JA, Estrada K, Rivadeneira F, Williams FM, Cherkas L, Deloukas P et al. . Loci at chromosomes 13, 19 and 20 influence age at natural menopause. Nat Genet 2009;41:645–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamir S, Izrael S, Vaya J. The effect of oxidative stress on ERalpha and ERbeta expression. J Steroid Biochem Mol Biol 2002;81:327–332. [DOI] [PubMed] [Google Scholar]
- Tatone C, Di Emidio G, Barbaro R, Vento M, Ciriminna R, Artini PG. Effects of reproductive aging and postovulatory aging on the maintenance of biological competence after oocyte vitrification: insights from the mouse model. Theriogenology 2011;76:864–873. [DOI] [PubMed] [Google Scholar]
- Uyar A, Torrealday S, Seli E. Cumulus and granulosa cell markers of oocyte and embryo quality. Fertil Steril 2013;99:979–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Q, Liu Z, Li B, Liu K, Wu W, Liu H. Oxidative Stress Induces Mouse Follicular Granulosa Cells Apoptosis via JNK/FoxO1 Pathway. PLoS One 2016;11:e0167869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Fazleabas AT, Shikanov A, Jackson E, Barrett SL, Hirshfeld-Cytron J, Kiesewetter SE, Shea LD, Woodruff TK. In vitro oocyte maturation and preantral follicle culture from the luteal-phase baboon ovary produce mature oocytes. Biol Reprod 2011;84:689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerushalmi GM, Salmon-Divon M, Yung Y, Maman E, Kedem A, Ophir L, Elemento O, Coticchio G, Dal Canto M, Mignini Renzinu M et al. . Characterization of the human cumulus cell transcriptome during final follicular maturation and ovulation. Mol Hum Reprod 2014;20:719–735. [DOI] [PubMed] [Google Scholar]
- Yuan YQ, Van Soom A, Leroy JL, Dewulf J, Van Zeveren A, de Kruif A, Peelman LJ. Apoptosis in cumulus cells, but not in oocytes, may influence bovine embryonic developmental competence. Theriogenology 2005;63:2147–2163. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhu S, Shi X, Sha W. The silence of p66(Shc) in HCT8 cells inhibits the viability via PI3K/AKT/Mdm-2/p53 signaling pathway. Int J Clin Exp Pathol 2015;8:9097–9104. [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Nagaraju G, Liu Z, Liu K. Functional roles of the phosphatidylinositol 3-kinases (PI3Ks) signaling in the mammalian ovary. Mol Cell Endocrinol 2012;356:24–30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.