Abstract
Viruses have evolved various mechanisms to evade host immunity and ensure efficient viral replication and persistence. Several DNA tumor viruses modulate host DNA methyltransferases for epigenetic dysregulation of immune-related gene expression in host cells. The host immune responses suppressed by virus-induced aberrant DNA methylation are also frequently involved in antitumor immune responses. Here, we describe viral mechanisms and virus–host interactions by which DNA tumor viruses regulate host DNA methylation to evade antiviral immunity, which may contribute to the generation of an immunosuppressive microenvironment during cancer development. Recent trials of immunotherapies have shown promising results to treat multiple cancers; however, a significant number of non-responders necessitate identifying additional targets for cancer immunotherapies. Thus, understanding immune evasion mechanisms of cancer-causing viruses may provide great insights for reversing immune suppression to prevent and treat associated cancers.
Keywords: DNA methylation, DNMT, antiviral immunity, immune evasion, herpesvirus, papillomavirus, KSHV, EBV, HBV, HPV
1. Introduction
Recent studies have revealed that DNA methylation is associated with many different diseases including microbial infections and cancers (reviewed in [1,2]). DNA methylation is a potent epigenetic mechanism to regulate gene expression without altering DNA sequences. Methylation of cytosines in CpG motifs frequently occurs in promoter regions but is also found in enhancers, insulators, gene bodies, transposable elements, and repetitive DNA elements (reviewed in [3]). DNA methylation is most dynamic in CpG islands near transcription start sites. CpG islands, which are typically hypomethylated, are DNA regions with a greater abundance of CpG dinucleotides compared to the remainder of the genome. Generally, promoter methylation represses gene transcription, while gene body methylation induces gene transactivation (reviewed in [3,4]).
DNA methylation is catalyzed by six DNA methyltransferases (DNMTs) that have been characterized to date: DNMT1, DNMT2, DNMT3A, DNMT3B, DNMT3C, and DNMT3L. Among them, DNMT3A and DNMT3B produce multiple isoforms by an alternative promoter and an alternative splicing, respectively, for further regulation of their enzymatic activity (reviewed in [1]). Each DNMT has distinct functions in its role in gene expression regulation. DNMT1 is responsible for maintaining heritable DNA methylation by copying methylation patterns from a parental cell to a daughter cell shortly after mitosis (reviewed in [1]). Whereas DNMT2 is a tRNA methyltransferase [5], DNMT3A and DNMT3B are de novo methyltransferases that generate new methylation marks on unmethylated CpG DNA sites (reviewed in [1]). DNMT3C has been recently discovered in mice as a DNA methyltransferase involved in fertility [6]. DNMT3L is a catalytically inactive DNMT3 variant that interacts with and amplifies DNMT3A and DNMT3B activities [7,8,9,10]. While the mechanisms by which DNMTs methylate DNA have been well characterized, no specific DNA demethylase has been identified to date that reverses DNA methylation. Instead, it has been suggested that methylated cytosines are removed during DNA repair after the conversion of 5-methylcytosine to 5-hydroxymethylcytosine by the methylcytosine dioxygenases ten-eleven translocases (TET) (reviewed in [1,3]).
Gene expression regulation by DNA methylation is intimately linked to chromatin arrangement (reviewed in [3,11,12]). In fact, chromatin structure is altered when DNA is methylated [11,13,14], and histone deacetylases (HDAC) interact with DNMT1 [15,16,17], DNMT3A [18,19], and DNMT3B [18]. Hypermethylated DNA is often associated with hypoacetylated histones and condensed chromatin for transcriptional repression [11,12].
Previous studies have suggested that DNA methylation functions as an antiviral defense mechanism by inactivating viral gene transcription and replication. It is well known that most endogenous retroviruses and retrotransposons in the human genome are inactivated by DNA hypermethylation [20,21]. Roulois et al. and Chiappinelli et al. have concurrently reported that treating colon and ovarian cancer cells with demethylating agents activates viral RNA transcription from dormant endogenous retroviruses and stimulates antiviral interferon (IFN) signaling, which subsequently activates antitumor immune responses [22,23]. DNA demethylation also activates retrotransposons and triggers antiviral signaling in zebrafish embryos [24]. In addition to endogenous retroviruses, the genomes of DNA viruses, such as human papillomavirus (HPV), herpes simplex virus 1 (HSV-1), adenovirus, and hepatitis B virus (HBV), are also frequently methylated and silenced in infected cells [25,26,27,28,29,30,31,32,33]. Interestingly, methylation of HPV DNA is commonly detected in infected cervical lesions, and its methylation levels correlate to disease progression in high-grade premalignant cervical lesions and cancer [34,35,36,37,38]. Similarly, methylation of HBV covalently closed circular DNA (cccDNA) markedly reduces viral gene transcription and genome replication during chronic infection [33,39].
Many viruses, particularly small DNA viruses, harbor genomic signatures indicating that they have evolved for millions of years to evade the antiviral effects of DNA methylation [40,41,42,43,44]. Our study has shown that the prevalence of CpG dinucleotides, the target motif of DNA methylation, is significantly lower in the genomes of papillomaviruses compared to other dinucleotide motifs [45]. These results suggest that gene expression regulation by DNA methylation may play a critical role in arms races between viruses and their hosts.
To evade detection and restriction by the host immune response, viruses also employ various mechanisms to control gene expression related to immunity, including hijacking epigenetic machinery (reviewed in [46,47]). A recently described mechanism for viruses to epigenetically subvert host immunity is repression of immune-related gene expression by induction of DNA hypermethylation. In particular, DNA tumor viruses utilize this mechanism to manipulate host DNA methylation to alter expression of immune-related genes [48,49,50,51,52,53,54]. Indeed, several DNA tumor viruses have been found to regulate multiple DNMTs, suggesting that aberrant DNA methylation caused by viruses may be linked to virus-associated carcinogenesis [55]. Particularly in tumor virus infections, dysregulation of antiviral immune gene expression can have dual consequences. While a virus evades antiviral immune surveillance to establish a persistent infection, immune impairment induced by the virus can result in cancer cell evasion from antitumor immune responses during oncogenesis, as antiviral and antitumor immunity share similar immune mechanisms (reviewed in [56]). Since recent immunotherapies have shown promising efficacy to treat late-stage cancers [57,58,59,60,61], research regarding immune dysregulation by tumor virus-induced DNA methylation is of critical importance but largely understudied. Here, we discuss several compelling studies showing that DNA tumor virus regulation of host immune-related genes by DNA methylation contributes to cancer progression and is likely a result of virus-driven immune suppression to evade host antiviral responses.
2. DNA Tumor Viruses and DNA Methylation of Host Genes
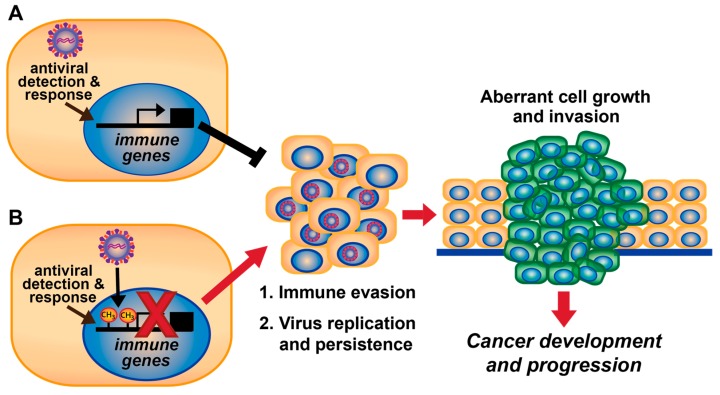
Hijacking DNA methylation machinery by DNA tumor viruses is likely a viral mechanism to promote virus replication by evading antiviral immunity. Immune suppression caused by aberrant DNA methylation over time may contribute to cancer development and progression associated with DNA tumor viruses (Figure 1). In fact, tumorigenesis is enhanced when antiviral immune responses are dampened [62,63,64,65] (reviewed in [56]). Kaposi’s sarcoma-associated herpesvirus (KSHV; also known as human herpesvirus 8), Epstein-Barr virus (EBV; also known as human herpesvirus 4), HBV, and HPV induce promoter methylation which downregulates expression of host immune-related genes, as will be discussed herein. Although here we focus on DNA tumor viruses, regulation of host immune genes by DNA methylation has also been demonstrated for the RNA virus human immunodeficiency virus (HIV) [66,67,68,69].
Figure 1.
Model for DNA tumor virus-mediated DNA methylation to evade antiviral and antitumor immunity during viral persistence and carcinogenesis. (A) Cellular detection of viruses activates immune gene expression to induce an antiviral immune response. Proliferation of infected and neighboring cells can be blocked (black T bar) by immune-mediated apoptosis and/or cell cycle inhibition, which prevent cancer development. (B) DNA tumor viruses induce hypermethylation of immune genes that inhibit expression of antiviral immune genes (denoted by red “×”), resulting in immune evasion, which promotes (long red arrow) viral replication and persistence. Over long periods of time (multiple years), immune evasion and viral persistence can promote (short red arrow) cell proliferation and carcinogenesis. In addition, downregulation of immune gene expression by viral-induced DNA methylation may also contribute to host cell evasion of antitumor immune responses.
Many genome-wide methylome and transcriptome analyses have linked DNA tumor virus infection to the dysregulation of host gene hypermethylation during viral persistence and carcinogenesis. These DNA tumor viruses include gammaherpesviruses (EBV [70,71,72,73,74,75,76,77] and KSHV [78]), a hepadnavirus (HBV) [79,80], alphapapillomaviruses (HPV) [53,54,81,82,83,84,85], and polyomaviruses (simian virus 40 (SV40) [86,87,88,89], Merkel cell carcinoma virus (MCPyV) [90,91], JC virus (JCV) [92,93]) (Table 1). Frequently, virus-associated cancers show highly increased levels of DNMT expression [75,76,77,85,94,95,96,97,98,99]. In HBV-associated hepatocellular carcinoma (HCC), DNMT expression is inversely correlated with levels of tumor suppressor microRNAs (miRNAs), including miR-152 targeting DNMT1 [97] and miR-101 targeting DNMT3A [99]. Virus-induced DNA hypermethylation is commonly found on several tumor suppressor genes including RASSF1A, p16 (also known as cyclin dependent kinase inhibitor 2A (CDKN2A)), TP73, p21 (also known as CDKN1A), and retinoblastoma-associated protein (pRb) (Table 1). These findings suggest that induction of DNA methylation is likely a viral mechanism to promote cell proliferation that supports efficient viral replication, particularly for DNA viruses. Thus, downregulation of these tumor suppressors by promoter hypermethylation during virus infection could be a determinant of virus-driven tumorigenesis. We summarize host genes with diverse functions regulated by DNA methylation in cancers associated with DNA tumor viruses in Table 1. Additionally, DNA methylation-associated pathogenesis for EBV-associated gastric carcinoma (EBVaGC) [70] and HBV-associated HCC [79,80] have been previously reviewed in detail. Although definitive identification of particular hypermethylated genes that directly promote oncogenesis remains elusive, these studies have shown that increased DNA hypermethylation strongly correlates with disease progression of various virus-induced cancers.
Table 1.
DNA methylation associated with DNA tumor virus-induced carcinomas.
| Virus | Cancer Type | Tumor Tissue (TT) or Cell Line (CL) | Methylated DNA/Gene | Reference |
|---|---|---|---|---|
| Herpesviridae | ||||
| EBV | Gastric carcinoma | Reviewed in [70] | ||
| NPC | TT, CL | miR-31 | [71] | |
| TT |
DAPK, RASSF1A, p16 (CDKN2A) |
[72] | ||
| TT | RASSF1A | [73] | ||
| CL | Retinoic acid receptor-β2 (RAR-β2) | [76] | ||
| CL | E-cadherin | [77] | ||
| Burkitt’s lymphoma (BL) | CL | 4712 differentially methylated genes | [74] | |
| Germinal center (GC) B cell malignancies, Hodgkin’s lymphoma (HL) | TT (GC) CL (HL) |
1745 DMPs | [75] | |
| KSHV | Primary effusion lymphoma | CL | p16 (CDKN2A) | [78] |
| Hepadnaviridae | ||||
| HBV | HBV-associated HCC | Reviewed in [79,80] | ||
| Papillomaviridae | ||||
| HPV | Head and neck SCC | TT | NSD1, NOTCH1 | [81] |
| Cervical squamous intraepithelial lesions | TT | SIM1, DLX4 | [82] | |
| E6/E7 immortalized keratinocytes | CL | hTERT, miR124-2, PRDM14, FAM19A4, SFRP2, PHACTR3, MAL, CYGB, ROBO3 | [83] | |
| HPV16/18 keratinocytes, cervical cancer cells | CL | hTERT | [84] | |
| Head and neck SCC, cervical carcinoma | TT | CXCL14 | [54] | |
| HPV16/18 immortalized keratinocytes | TT | HLA-E, CCNA1, TERT; 5190 DMPs | [53] | |
| SCC | CL | 75 differentially methylated genes | [85] | |
| Polyomaviridae | ||||
| SV40 | Diffuse large B cell lymphoma | TT |
DAPK, CDH1, GSTP1, p16 (CDKN2A), SHP1 |
[86] |
| Non-Hodgkin’s lymphoma/leukemia | TT |
CDH1, CDH13, CRBP, p16 (CDKN2A), DAPK, DcR1, DcR2 |
[87] | |
| Malignant mesothelioma | TT | RASSF1A | [88,89] | |
| Lung adenocarcinoma | TT | RASSF1A | [88] | |
| MCPyV | Merkel cell carcinoma, small cell lung cancer | TT | RASSF1A | [90,91] |
| JC virus | Gastric carcinoma | TT | p16 (CDKN2A), p14 | [92] |
| Colorectal cancer | TT | hMLH1, PTEN, RUNX3, p16 (CDKN2A) | [93] |
NPC: Nasopharyngeal carcinoma; SCC: Squamous cell carcinoma; DMP: Differentially methylated position.
In contrast to virus-induced DNA hypermethylation, viruses can also decrease host DNA methylation to regulate host gene expression. DNA hypomethylation usually results in increased gene expression. For example, Kaposi’s sarcoma (KS) cell lines display hypomethylation of AXL receptor tyrosine kinase (AXL), which is linked to oncogenesis [100]. The KSHV viral FLICE-inhibitory protein (vFLIP) induces AXL expression potentially through AXL gene hypomethylation [100]. Alterations in DNA methylation status of particular genes by viruses may have profound effects on cancer development and progression.
3. Herpesviridae
3.1. Oncogenesis by Herpesviruses
Herpesviruses are large double-stranded DNA viruses that persistently infect their hosts, often for an entire lifetime. Herpesvirus infections generally do not cause any significant disease unless host immune responses are suppressed [101]. Herpesviruses have both lytic and latent cycles of infection. Once a lytic infection ensues, herpesviruses undergo a dormant cycle (latency) and occasionally reactivate from latency to undergo lytic replication. One genus of herpesviruses, gammaherpesviruses, such as KSHV and EBV, have oncogenic potential [101]. KSHV causes KS, primary effusion lymphomas (PEL) and multicentric Castleman disease in immunocompromised individuals (reviewed in [102]). EBV infection is associated with various lymphomas, including Burkitt’s and Hodgkin’s lymphomas, in addition to carcinomas of the nasopharynx and stomach (reviewed in [103]).
KSHV encodes multiple oncoproteins and oncogenic miRNAs that dysregulate host functions and promote cancer progression of endothelial and B cells (reviewed in [102,104]). A primary mechanism underlying KSHV-induced cancers is activation of angiogenesis by KSHV miRNAs and the oncoproteins vIRF3, K1, K8.1, K15, glycoprotein B (gB), latency-associated nuclear antigen (LANA), viral G-protein coupled receptor (vGPCR), vFLIP, and viral chemokines (reviewed in [104]). In addition, KSHV K1, vFLIP, LANA, and viral interleukin-6 (vIL-6) inhibit apoptosis in KSHV-infected cells to support cell proliferation as well as viral replication and persistence (reviewed in [102,104]). KSHV also activates the host cell cycle, cell proliferation, migration, and invasion, which contribute to viral oncogenesis (reviewed in [104]). LANA is consistently expressed in KSHV-positive cancers and known to induce angiogenesis and activate the host cell cycle by degrading p53 and stabilizing c-MYC (reviewed in [105]). Thus, KSHV has evolved mechanisms to modulate various aspects of host biology to drive oncogenesis.
EBV also encodes several proteins and miRNAs that promote transformation of B cells and epithelial cells, such as latent membrane proteins (LMP1 and LMP2), EBV nuclear antigens (EBNA1-3, leader protein (LP)), BamHI fragment H rightward open reading frame 1 (BHRF1), BamHI A reading frame 1 (BARF1), and BamHI A rightward transcript miRNAs (miR-BART) (reviewed in [103,106]). Several miR-BARTs, BHRF1, and BARF1 block pro-apoptotic proteins in host cells (reviewed in [103,106]). EBV LMP activates oncogenic signaling including mitogen-activated protein kinase, c-Jun N-terminal kinase, phosphatidylinositol 3-kinase, and NF-κB pathways (reviewed in [106]). EBNAs stimulate cell proliferation through multiple mechanisms. EBNAs inhibit p53 and pRb tumor suppressors, hijack RBP-Jκ and NF-κB transcription factors to express oncogenes, usurp HDACs (reviewed in [107]), and induce DNA damage by activating recombinase-activating genes (RAG1 and RAG2) [108].
3.2. Herpesviruses Modulate DNMTs
Both KSHV and EBV have been shown to stimulate DNA hypermethylation of host genes, which likely contributes to virus-driven tumorigenesis. KSHV LANA interacts with DNMTs and recruits DNMT3A to host chromatin [109] (Figure 2A). DNMT3A mRNA expression is also increased in KSHV-infected cells, which likely results in DNMT3A-mediated repression of host genes [110]. KSHV vIL-6 enhances DNMT1 expression levels and activity, resulting in global DNA hypermethylation in endothelial cells [111]. Treatment of vIL-6-expressing cells with the demethylating agent 5-aza-2′-deoxycytidine reduced aberrant cell proliferation and migration induced by vIL-6. These results suggest that vIL-6-induced host DNA hypermethylation increases host cell proliferation and migration [111].
Figure 2.
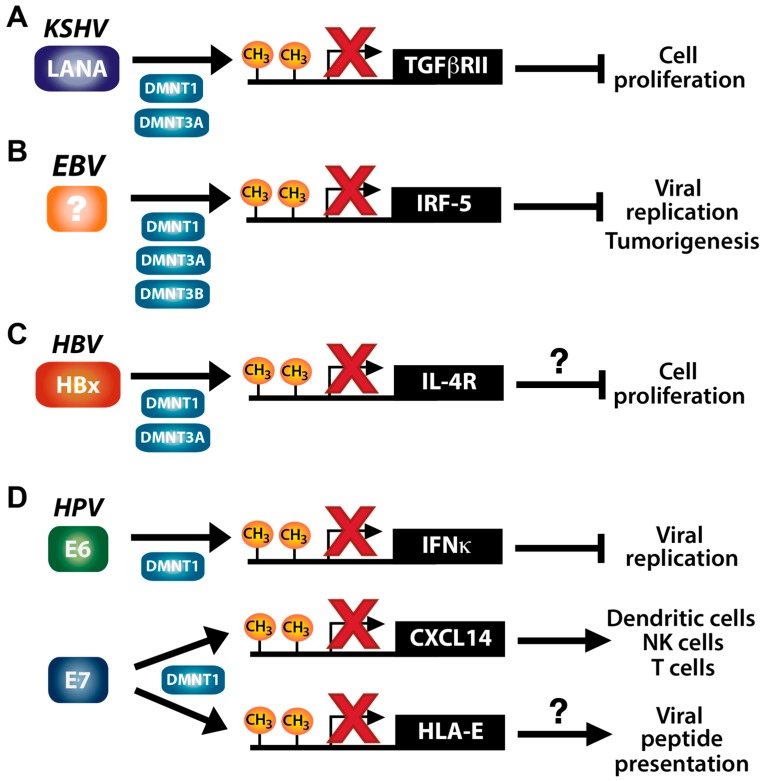
DNA tumor viruses that promote DNA hypermethylation of immune-related genes. (A) KSHV LANA; (B) an unidentified EBV protein; (C) HBV HBx; and (D) HPV E6 and E7 proteins upregulate the DNMTs shown to induce DNA methylation and transcription inhibition (indicated by red “×”) of the denoted immune-related genes. The outcome of immune gene suppression by the viruses promotes virus replication and host cell proliferation. Circled question marks indicate potential mechanisms that have not yet been fully defined.
EBV can infect and modulate DNMT levels in both epithelial and B cells. Expression of distinct DNMTs is regulated differently by EBV based on cell/tissue types and latency programs. EBV has three latency stages (I-III) determined by differential viral gene expression, which influences host gene expression, including DNMTs [112]. DNMT1 and DNMT3B are upregulated in EBVaGC (latency I) [94,95] by LMP2A [94]; DNMT1, DNMT3A, and DNMT3B are upregulated in EBV-associated nasopharyngeal carcinoma (NPC; latency II) by LMP1 [76,77,113]; lastly, DNMT3A levels are increased in Hodgkin’s lymphoma cell lines (latency II) as well as lymphoblastoid cell lines derived from EBV-positive germinal center B cells (latency III) by an unknown viral gene [75] (Figure 2B). DNMT1 and DNMT3B expression was shown to be downregulated in EBV-infected lymphoblastoid cells and Hodgkin’s lymphoma cell lines, which is distinct from that observed for EBVaGC and EBV-positive NPC [75]. Ectopic expression of EBV LMP1 reduces DNMT1 levels in lymphoblastoid cells [75]. LMP1 effects on DNMT1 levels in lymphoblastoid cells compared to EBVaGC and NPC could be due to differing EBV latency stages, which are dictated by distinct EBV gene expression patterns. Upregulation of DNMT1 expression by EBV LMP2a in EBVaGC [94] may be compensatory for the lack of LMP1 expression in latency I EBVaGC cells. Taken together, these results suggest that EBV upregulates different DNMT proteins in varying host cell/tissue types to induce hypermethylation of host genes (Figure 2B). In summary, DNA methyltransferase activity is altered in both KSHV- and EBV-infected cells, changing the landscape of promoter methylation and host gene expression.
3.3. KSHV and TGF-β Signaling
Downregulation of the transforming growth factor β (TGF-β) type II receptor (TβRII) by LANA-mediated promoter methylation contributes to development of KSHV-induced PEL [48] (Figure 2A). Upon binding of the ligand TGF-β, TGF-β type I receptor (TβRI) is recruited to and heterodimerizes with TβRII to initiate downstream signaling. TβR signaling contributes to embryo and organ development by regulating cell proliferation, differentiation, apoptosis, homeostasis, and other cellular processes (reviewed in [114,115,116]). Furthermore, the TβR signaling pathway plays important roles in cancer development and progression [117,118]. For example, TGF-β is considered a tumor suppressor as it inhibits proliferation of colon cancer cells [119], pancreatic ductal adenocarcinoma cells [120], and hepatocytes [118,121,122]. Interestingly, previous studies using cancer cell lines and patient tissue samples showed that loss of TGF-β signaling is often strongly correlated with hypermethylation of the TβRII promoter and poor prognosis for patients with different types of cancers [123,124,125,126,127,128,129]. TGF-β signaling is downregulated by KSHV, which stimulates cell proliferation and could promote cancer development [104]. KSHV-positive PEL cells were found to be unresponsive to TGF-β stimulation [48]. TβRII expression and TGF-β signaling activation was restored in KSHV-positive PEL cells by treatment with 5-aza-2′-deoxycytidine in combination with an HDAC inhibitor MS-275. Furthermore, reversing the epigenetic silencing of TGF-β signaling decreases cell proliferation and increases apoptosis [48]. These results suggest that downregulation of TβRII expression through KSHV-induced DNA methylation abrogates TGF-β signaling and drives transformation of KSHV-infected cells.
Several studies have revealed that TGF-β signaling is detrimental to KSHV infection, as KSHV employs various mechanisms to avoid host restriction mediated by TGF-β signaling. These mechanisms include: (1) vFLIP and viral cyclin (vCyclin) activation of oncogenic host miRNAs that target SMAD family member 2 (SMAD2), a downstream component of TGF-β signaling [130]; (2) virally-encoded miRNA targeting of TβRII [131], SMAD5 [132], and thrombospondin 1, a mediator of latent TGF-β activation [133]; (3) viral IFN regulatory factor 1 (vIRF1) binding to and inhibition of SMAD3-SMAD4 functions in the TGF-β signaling complex [134]; (4) viral K-bZIP disruption of the SMAD3 interaction with CREB-binding protein (CBP) [135], which is important for transcriptional activation of TGF-β [136,137,138,139,140]; and (5) cytokine receptor gp130 activation by vIL-6 that leads to downregulation of TGF-β2 expression [141,142]. These findings strongly indicate that TGF-β signaling activation potently restricts productive KSHV infection. Taken together, epigenetic downregulation of TβRII by KSHV plays an important role for virus evasion of TGF-β signaling-mediated host restriction during virus persistence and disease progression (Figure 2A).
3.4. EBV and the Antiviral Protein IRF5
EBV induces promoter hypermethylation of IFN regulatory factor 5 (IRF5) during oncogenesis of Burkitt’s lymphoma and EBVaGC [49] (Figure 2B). IRF5 functions as both an antiviral signaling factor and tumor suppressor by inducing apoptosis in response to viral infection or DNA damage [143]. Activation of toll-like receptor (TLR)-myeloid differentiation primary response 88 (MyD88) signaling induces IRF5 phosphorylation, which translocates IRF5 into the nucleus and transactivates pro-inflammatory cytokines and DNA damage response genes [143,144,145]. IRF5 is a potent tumor suppressor. H-Ras transformed mouse embryonic fibroblasts (MEF) expressing IRF5 do not develop tumors in vivo, whereas cells lacking IRF5 readily form tumors [143]. In this model, IRF5 tumor suppression is likely mediated through induction of apoptosis, as cells lacking IRF5 are resistant to apoptosis compared to IRF5-expressing cells [143]. Additionally, IRF5 expression is decreased in breast cancer tissues, and overexpression of IRF5 in breast cancer cell lines results in DNA damage-induced cell death and tumor suppression [146]. A previous study showed that EBV-induced promoter methylation and repression of IRF5 transcription are linked to gastric carcinoma development [147]. Dong et al. demonstrated that hypermethylation of the IRF5 promoter was 5-fold higher in EBVaGC cell lines compared to EBV-negative gastric carcinoma cell lines. In addition, IRF5 expression in EBVaGC cells was rescued by treatment with a demethylating agent [49]. While the EBV protein that mediates IRF5 DNA methylation remains unknown, these results imply that downregulation of IRF5 expression by EBV blocks apoptosis of infected cells and contributes to cell transformation and oncogenesis.
The antiviral activity of IRF5 was first demonstrated in vesicular stomatitis virus (VSV) and HSV-1 infections [143]. IRF5 inhibits proliferation of EBV-infected cells [148] and downregulates expression of both EBV LMP1 mRNA [149] and the latency BART mRNAs [150]. In fact, LMP1 was recently shown to inhibit IRF5-mediated apoptosis during infection [151]. Interestingly, another study showed that although TLR7 signaling is activated during EBV infection, downstream IRF5 expression is repressed by induction of an IRF5 dominant-negative splice variant [152]. Overall, these results suggest that EBV has developed several mechanisms to block IRF5 induction of apoptosis in EBV infected cells. Thus, EBV-induced DNA methylation of IRF5 may play an important role in evasion of host immunity during virus persistence and oncogenesis (Figure 2B).
4. Hepadnaviridae
4.1. HBV Oncogenesis
HBV is a small, partially double-stranded DNA virus that infects hepatocytes and causes hepatitis, cirrhosis, and HCC. Nearly 4% of the worldwide population is chronically infected with HBV, which contributes to HCC being the fifth most common cancer worldwide (reviewed in [153]). HBV persistently infects hepatocytes as cccDNA, which is the genomic template for viral replication. HBV-driven HCC development is thought to occur through multiple oncogenic mechanisms including: (1) HBV DNA integration into the host genome; (2) cellular stress induced by accumulation of HBV surface antigen (HBsAg) in the endoplasmic reticulum; and (3) the multiple oncogenic functions of HBV X-protein (HBx). HBx interferes with proteasomal protein degradation, induces host miRNA expression, dysregulates host epigenetics, activates oncogenic signaling (e.g., Ras, Src and Wnt signaling), and stimulates the host cell cycle by inhibiting tumor suppressors such as p53 (reviewed in [153,154]). Altogether, persistent HBV infection presents a myriad of mechanisms that predispose cells to transformation. Here, we describe the roles of host DNA methylation in HBV infection and its impact on oncogenesis.
4.2. HBx Modulation of DNMTs
The HBV oncoprotein HBx upregulates expression of DNMT1 and DNMT3A, which leads to promoter methylation and transcriptional repression of several tumor suppressor genes [96,155,156,157,158,159,160,161] (Figure 2C). HBx activates the host cell cycle by upregulating DNMT1 through a positive feedback mechanism [158]. HBx represses expression of the cyclin dependent kinase (CDK) inhibitor p16 by DNMT1-mediated promoter methylation. Downregulation of p16 expression leads to cell cycle activation through inhibition of pRb and upregulation of E2F1. This ultimately results in increased DNMT1 levels and creates a positive feedback loop to further reduce pRb expression by p16 promoter methylation [158]. HBx also promotes cell cycle progression through hypermethylation of other CDK inhibitors, p21 and p27 [157,162]. These results suggest that manipulation of the cell cycle by HBV HBx through enhanced DNA methylation may contribute to HCC development [157,158,161,162].
Several studies have demonstrated that expression of DNMT1 and DNMT3A/B are upregulated in HBV-associated HCC tissues compared to adjacent normal liver tissues from patients [96,97,98,99]. On the other hand, studies using mice expressing HBx in hepatocytes revealed roles for HBx in DNA hypomethylation, which involve altered DNMT expression or promoter binding and may promote the development of HCC [163,164,165]. HBx repressed expression of DNMT3A and DNMT3L in HBx-expressing mice by binding their promoters in conjunction with HDAC1, which leads to global hypomethylation of CpG regions in the host genome [163]. Additionally, epithelial cell adhesion molecule (EpCAM) expression is upregulated in HBV-associated HCC by HBx-induced hypomethylation of the EpCAM gene by an uncharacterized mechanism, which involves DNMT3L [164]. Despite the well established role of DNMT3L in enhancing DNMT3A and DNMT3B activity [7,8,9,10], DNMT3L can also negatively regulate DNA methylation by competing with DNMT3A and DNMT3B binding to polycomb-repressive complex 2 (PRC2) to prevent de novo DNA methylation at histone 3 lysine 27 trimethylation (H3K27me3) sites [166]; this may explain the role of DNMT3L in HBx-mediated upregulation of EpCAM expression. In addition, the cyclooxygenase-2 (COX-2) promoter is hypomethylated in HBV-positive cells with reduced binding of DNMT3B to the COX-2 promoter. HBx transgenic mice display elevated COX-2 expression as compared to mice lacking HBx, indicating that HBx is critical for increasing COX-2 expression in hepatocytes [165]. These results suggest that HBV HBx employs multiple mechanisms to induce or inhibit DNA methylation on different genes during hepatocarcinogenesis.
4.3. HBV and IL-4R Signaling
In addition to hypermethylation and downregulation of the tumor suppressors p16, p21 and p27, HBx also induces methylation of the IL-4 receptor (IL-4R) gene, leading to downregulation of its expression [50]. The ligand of IL-4R, IL-4, is an anti-inflammatory cytokine that suppresses host cell growth and induces apoptosis [167]. IL-4R signaling primarily functions in hematopoietic cells; however, its activity has been observed in hepatocytes as well [168,169,170]. Interestingly, Zheng et al. found that expression of several genes downregulated by HBx is restored when DNA methylation is inhibited by treatment with 5-aza-2′-deoxycytidine [50]. The authors further revealed that HBx binds DNMT1 and DNMT3A. Notably, HBx binds the IL-4R promoter to facilitate its DNA methylation and silencing of IL-4R expression is DNMT3A-dependent [50]. These results indicate that IL-4R expression is repressed by HBx in HBV-infected hepatocytes through promoter methylation (Figure 2C).
IL-4R signaling limits HBV infection [171,172]. Activation of IL-4R signaling in HBV-infected hepatocytes inhibits viral replication and reduces HBsAg and HBV e antigen (HBeAg) expression. IL-4 represses expression of C/EBPα, a transactivator of the HBV genome core promoter, to inhibit HBV replication [171]. Consistently, production of both HBsAg and HBeAg is decreased by IL-4 treatment [172]. Altogether, these results suggest that HBV replication and viral gene expression is inhibited by IL-4. Thus, downregulation of IL-4R expression by HBx-induced promoter methylation is likely an immune evasion mechanism of HBV [50]. Since IL-4-mediated signaling is pro-apoptotic [173], HBV downregulation of IL-4R expression via promoter methylation may contribute to cell proliferation and HCC development.
5. Papillomaviridae
5.1. HPV Oncogenesis
HPVs are small double-stranded DNA viruses that infect cutaneous and mucosal keratinocytes. While infection with low-risk HPV genotypes (e.g., HPV6 and -11) leads to development of benign skin lesions such as warts, several high-risk genotypes (e.g., HPV16 and -18) are causally associated with cervical, anogenital, or head and neck cancers (HNC). HPV associated cancers account for over 5% of all cancers worldwide [174]. While the majority of initial HPV infections are cleared within a few years, about 10% of infected people establish persistent HPV that likely exists for their lifetime [175,176]. Persistent infection with high-risk HPV genotypes and continuous expression of the HPV oncogenes, E6 and E7, are required for HPV-associated cancer progression and maintenance [177,178]. E6 and E7 contribute to cancer progression through various oncogenic mechanisms including inactivation of the tumor suppressors p53 and pRb, respectively [179]. Recent studies have suggested that dysregulation of DNA methyltransferase activity may also affect HPV-associated carcinogenesis.
5.2. HPV Modulation of DNMTs
HPV E6 and E7 enhance promoter methylation by upregulation of DNMT1 expression through p53 degradation and a direct interaction with the DNMT1 protein, respectively [180,181] (Figure 2D). Our studies have shown that dysregulation of host DNA methylation by HPV16 E7 is associated with host immune suppression during HPV-associated cancer progression [53,54,182]. Interestingly, a recent clinical trial revealed that treatment with DNA methylation inhibitors suppressed HPV-positive HNC growth. Notably, HPV-positive HNC is more sensitive to treatment with the DNA demethylating agent 5-aza-2′-deoxycytidine compared to HPV-negative HNC [183]. These findings suggest that HPV dysregulation of DNA methylation can be reversed using demethylating agents as a targeted therapy for HPV-associated cancers. Here, we discuss several immune genes regulated by HPV through promoter hypermethylation.
5.3. High-Risk HPV and IFNκ Signaling
Dysregulation of immune-related gene expression by high-risk HPV-mediated DNA methylation was first demonstrated with IFN-kappa (IFNκ) [51,52] (Figure 2D). IFNκ is a type I IFN that is constitutively expressed in human keratinocytes, the natural host cell type for HPV infection [184]. IFNκ is an antiviral factor that restricts HPV replication in keratinocytes [185]. Previous studies showed that IFNκ expression is significantly downregulated in cells harboring high-risk HPV genomes (HPV16, -18 or -31) or expressing HPV16 E6 [51,52]. IFNκ expression in HPV-positive cells is restored by treatment with the demethylating agent 5-aza-2′-deoxycytidine, indicating that HPV induces methylation of IFNκ to reduce its expression. HPV16 E6, but not E7, is necessary and sufficient for induction of IFNκ promoter methylation [51,52]. Consistently, HPV16-positive cervical intraepithelial neoplasia and cervical cancer tissues are devoid of IFNκ expression, whereas HPV16-negative normal mucosal tissues display strong IFNκ expression [51]. Furthermore, ectopic expression of IFNκ in HPV16-positive cells restores antiviral signaling as determined by induction of IFN-stimulated gene expression and suppression of VSV replication. This indicates that the downstream signaling of IFNκ is still intact despite decreased IFNκ expression by HPV16 E6 [51]. These results suggest that high-risk HPV E6 interferes with expression of type I IFN to promote HPV persistence in host cells.
5.4. High-Risk HPV and CXCL14 Expression
In previous gene expression studies using cervical and HNC patient tissue samples, we have revealed that numerous immune-related genes are dysregulated in HPV-positive cancers compared to normal tissue and HPV-negative cancers [54,186,187]. To determine if HPV directly affects expression of these immune-related genes, we recently performed a global gene expression analysis using normal keratinocytes with and without the HPV16 genome [53,54]. The two most downregulated groups of genes were those involved in immune regulation and extracellular matrix organization. Furthermore, many of these immune-related genes were specifically downregulated by the HPV16 oncoprotein E7, which was previously suggested to suppress antitumor immune responses [53,54,188]. Interestingly, our recent study showed that HPV16 E7 significantly downregulates the chemokine (C-X-C motif) ligand 14 (CXCL14) through promoter hypermethylation [54].
CXCL14 is a relatively novel chemokine, and its native receptor is still unidentified. CXCL14 inhibits angiogenesis and directly recruits several types of immune cells such as dendritic, natural killer (NK), and T cells [54,189,190]. We and other groups have shown antitumor activity of CXCL14 in cancers of the lung, head and neck, colon, and liver [54,191,192,193,194,195]. Consistently, the levels of CXCL14 expression are reduced in these and other cancers [54,187,193,195,196,197]. Our study revealed that HPV16 E7 is responsible for CXCL14 downregulation by facilitating hypermethylation of the CXCL14 promoter, which is reversed by 5-aza-2′-deoxycytidine treatment [54] (Figure 2D). Restoration of CXCL14 expression in HPV-positive cancer cells significantly increases NK and T cell recruitment and dramatically suppresses tumor cell growth in vivo. These results suggest that CXCL14 is a tumor suppressing chemokine, which is downregulated by HPV E7-induced promoter hypermethylation [54]. Similarly, CXCL14 is also downregulated by promoter methylation in HCC cells and patient tumors [195,198]. Consistently, ectopic expression of CXCL14 in HCC cells decreases colony formation, cell viability, cell invasion, and tumor growth in vitro and in vivo [195]. As HBV infection is a major driver of HCC, this suggests that HBV might similarly downregulate CXCL14 expression during persistence and HCC development.
Although a direct antiviral role of CXCL14 has not been demonstrated, CXCL14 may play a protective role at the cutaneous and mucosal skin layers to prevent HPV infection. CXCL14 is highly expressed in normal keratinocytes and structurally similar to antimicrobial proteins such as defensins [199], which restrict HPV infection [200,201]. Taken together, these results imply that downregulation of CXCL14 by HPV E7-induced DNA methylation to evade host immunity contributes to suppression of host antitumor immune responses during HPV persistence and cancer progression.
5.5. High-Risk HPV and HLA-E Expression
We have recently discovered that high-risk HPV E7s, but not low-risk HPV E7s, downregulate HLA-E expression in keratinocytes by promoter hypermethylation, as shown by restoration of HLA-E expression using a demethylating agent (Figure 2D) [53]. HLA-E is a non-classical major histocompatibility complex I (MHC-I) protein that presents T cell epitopes on the cell surface and regulates NK and CD8+ T cell activation (reviewed in [202]). Peptide presentation, usually of self antigens, to NK cells by HLA-E inhibits NK cell-mediated cytolysis; however, pathogen antigen presentation by HLA-E typically prompts the killing activity of CD8+ T cells, including NK T cells, a subset of CD8+ T cells (reviewed in [202,203]). Therefore, downregulation of HLA-E by high-risk HPV E7 [53] implies that HLA-E may present HPV peptides to CD8+ T cells, resulting in elimination of infected cells. There is precedence for HLA-E presentation of viral peptides, but a clear antiviral mechanism has not been studied. A recent study showed that HLA-E interacts with and presents a conserved HIV-1 envelope peptide to activate NK cells to kill virus-infected T cells [204]. Thus, it is possible that downregulation of HLA-E expression by HPV-induced promoter hypermethylation is a viral mechanism of avoiding immune detection and cell-mediated cytotoxicity by decreasing viral peptide presentation to CD8+ T cells. This may lead to viral persistence and HPV-associated cancer development.
6. Other DNA Tumor Viruses
In addition to the viruses described above, other DNA tumor viruses alter promoter methylation of host genes. Adenovirus-5 E1A upregulates DNMT1 expression and also directly interacts with DNMT1 protein. Interestingly, a transformation-deficient E1A mutant that cannot bind DNMT1 abrogates virus-induced DNA methylation [181]. This suggests that the interaction of E1A with DNMT1 and induction of DNA methylation is linked to cellular transformation. Additionally, infection with the polyomavirus SV40 stabilizes DNMT activity and increases host DNA methylation in immortalized fibroblasts by unknown mechanisms [205]. Moreover, SV40 antigens are associated with aberrant DNA methylation in tumor tissues (Table 1) [86,87,88,89], suggesting that its stabilization of DNMT activity might promote tumor progression. Similarly, the presence of antigens from two other polyomaviruses, Merkel cell polyomavirus and JC virus, correlated with DNA hypermethylation in tumor tissues [90,91,92,93]. Polyomavirus-induced DNA hypermethylation is often found on tumor suppressor genes (Table 1). Altogether, these results suggest that viral-induced host DNA methylation may be a common mechanism to repress host gene expression to facilitate persistent viral infection and potentiate virus-induced cancer progression (Figure 1).
7. Conclusions
Recent studies in virus-driven dysregulation of host immune-related gene expression through DNA methylation presents a novel viral mechanism to inhibit immune responses. This field is largely understudied, and several important questions remain: (1) Is alteration of host DNA methylation a major mechanism generally employed by diverse viruses, including RNA viruses, to regulate immune responses? (2) Are specific antiviral immune genes prone to virus-driven DNA methylation? (3) Are there specific hot spots in host genomes that viruses target utilizing DNA methylation to alter gene expression? Parallel analyses of global gene expression and the cellular methylome altered by virus infection may be useful to determine whether viral evasion of host immune responses is associated with aberrant DNA methylation induced by diverse viruses. These analyses would also reveal whether certain immune-related genes are commonly targeted by different viruses to evade host immunity.
One factor that increases the probability of DNA methylation in a promoter region is the presence and abundance of CpG islands. Additionally, DNA methylation site specificity can be facilitated through multiple mechanisms: (1) specific transcription factors and DNA binding proteins that recruit DNMTs to distinct genomic regions; (2) DNMT interactions with HDACs to enhance chromatin packaging and gene silencing; (3) three-dimensional DNA structural changes that alter DNA binding protein accessibility; and (4) nucleosome stability and positioning in the nucleus (reviewed in [11,12]). However, the exact signals or mechanisms that drive specificity of genes affected by DNA methylation are mostly unknown. Defining how particular genes are targeted by virus-induced DNA methylation would significantly impact our understanding of cellular gene regulation not only by viruses, but also different cellular stimuli or processes, through DNA methylation. In addition, the majority of viruses discussed above upregulate DNMT expression and/or activity; however, these viruses can also modulate other factors involved in epigenetic reprogramming linked to DNA methylation, such as histone modifications (reviewed in [206]). This may provide additional means to control host gene transcription through DNA methylation independently of DNMT upregulation. It is of interest to understand how cellular gene transcription is affected by the interplay between DNA methylation and other epigenetic factors. For instance, KSHV-induced downregulation of TβRII was not fully reversed using a demethylating agent or HDAC inhibitor alone, but a combination of the two fully restored TβRII expression [48]. These results suggest that some host genes are not silenced simply through promoter hypermethylation or histone deacetylation alone, and therefore, viruses may have evolved mechanisms to ensure host gene downregulation through multiple epigenetic modifications. Similar transcriptional regulation occurs in cells to regulate gene expression, but the mechanisms remain elusive [3]. Using viruses, or viral proteins mediating host epigenetic changes, could also be useful in dissecting how DNA methylation alters chromatin structure, or vice versa. In addition, viral studies on transcriptional regulation by DNA methylation and chromatin modifications may help reveal which epigenetic change initiates gene silencing, as this is still largely unclear.
Further mechanistic understanding is necessary to define the role and result of virus-driven aberrant DNA methylation of particular genes in oncogenesis. These studies would be of great interest and may provide useful targets for novel treatments for these virus-associated cancers. Since many DNA tumor viruses stimulate DNA methylation of host genes, including tumor suppressors and immune regulators, demethylating agents could be used to treat virus-associated cancers. In fact, 5-aza-2′-deoxycytidine treatment of HPV-positive HNC cells resulted in cell cycle arrest, p53-dependent apoptosis, activation of IFN signaling, and inhibition of metastasis [183]. 5-aza-2′-deoxycytidine treatment also decreased HPV gene expression within infected cells, which may result from the aforementioned effects of 5-aza-2′-deoxycytidine treatment on cell proliferation and IFN signaling [183]. Additionally, recent studies have shown that inhibition of DNA methylation significantly induces antitumor immune responses in colon and ovarian cancers [22,23]. Treatment with 5-aza-2′-deoxycytidine reactivates endogenous retroviruses that are recognized by cellular innate immune receptors and stimulate antiviral IFN responses [22,23]. Accordingly, demethylating drugs are generally being considered for cancer treatment in combination with other therapeutics to combat aberrant DNA methylation in oncogenesis (reviewed in [1]) and may be a highly effective strategy to treat virus-associated cancers. Therefore, a better understanding of virus-mediated dysregulation of host DNA methylation is of critical importance.
Acknowledgments
This work was supported in part by the National Institutes of Health (R01 AI091968 and R01 DE026125 to Dohun Pyeon and T32 AI052066 to Joseph A. Westrich). We thank Breck Duerkop for providing comments on the manuscript.
Author Contributions
Sharon K. Kuss-Duerkop and Dohun Pyeon reviewed the literature and wrote the manuscript. Sharon K. Kuss-Duerkop generated the figures. Joseph A. Westrich provided edits and approved the final version.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Liang G., Weisenberger D.J. DNA methylation aberrancies as a guide for surveillance and treatment of human cancers. Epigenetics. 2017;12:416–432. doi: 10.1080/15592294.2017.1311434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silmon de Monerri N.C., Kim K. Pathogens hijack the epigenome: A new twist on host-pathogen interactions. Am. J. Pathol. 2014;184:897–911. doi: 10.1016/j.ajpath.2013.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 4.Illingworth R.S., Bird A.P. CpG islands—‘A rough guide’. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 5.Goll M.G., Kirpekar F., Maggert K.A., Yoder J.A., Hsieh C.L., Zhang X., Golic K.G., Jacobsen S.E., Bestor T.H. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- 6.Barau J., Teissandier A., Zamudio N., Roy S., Nalesso V., Herault Y., Guillou F., Bourc’his D. The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science. 2016;354:909–912. doi: 10.1126/science.aah5143. [DOI] [PubMed] [Google Scholar]
- 7.Hata K., Okano M., Lei H., Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- 8.Chedin F., Lieber M.R., Hsieh C.L. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc. Natl. Acad. Sci. USA. 2002;99:16916–16921. doi: 10.1073/pnas.262443999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suetake I., Shinozaki F., Miyagawa J., Takeshima H., Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J. Biol. Chem. 2004;279:27816–27823. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- 10.Bourc’his D., Xu G.L., Lin C.S., Bollman B., Bestor T.H. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 11.Dantas Machado A.C., Zhou T., Rao S., Goel P., Rastogi C., Lazarovici A., Bussemaker H.J., Rohs R. Evolving insights on how cytosine methylation affects protein-DNA binding. Brief. Funct. Genom. 2015;14:61–73. doi: 10.1093/bfgp/elu040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robertson K.D. DNA methylation and chromatin—Unraveling the tangled web. Oncogene. 2002;21:5361–5379. doi: 10.1038/sj.onc.1205609. [DOI] [PubMed] [Google Scholar]
- 13.Perez A., Castellazzi C.L., Battistini F., Collinet K., Flores O., Deniz O., Ruiz M.L., Torrents D., Eritja R., Soler-Lopez M., et al. Impact of methylation on the physical properties of DNA. Biophys. J. 2012;102:2140–2148. doi: 10.1016/j.bpj.2012.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Severin P.M., Zou X., Gaub H.E., Schulten K. Cytosine methylation alters DNA mechanical properties. Nucleic Acids Res. 2011;39:8740–8751. doi: 10.1093/nar/gkr578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuks F., Burgers W.A., Brehm A., Hughes-Davies L., Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat. Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 16.Robertson K.D., Ait-Si-Ali S., Yokochi T., Wade P.A., Jones P.L., Wolffe A.P. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 17.Rountree M.R., Bachman K.E., Baylin S.B. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 18.Bachman K.E., Rountree M.R., Baylin S.B. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J. Biol. Chem. 2001;276:32282–32287. doi: 10.1074/jbc.M104661200. [DOI] [PubMed] [Google Scholar]
- 19.Fuks F., Burgers W.A., Godin N., Kasai M., Kouzarides T. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 2001;20:2536–2544. doi: 10.1093/emboj/20.10.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bestor T.H., Tycko B. Creation of genomic methylation patterns. Nat. Genet. 1996;12:363–367. doi: 10.1038/ng0496-363. [DOI] [PubMed] [Google Scholar]
- 21.Walsh C.P., Chaillet J.R., Bestor T.H. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- 22.Roulois D., Loo Yau H., Singhania R., Wang Y., Danesh A., Shen S.Y., Han H., Liang G., Jones P.A., Pugh T.J., et al. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell. 2015;162:961–973. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiappinelli K.B., Strissel P.L., Desrichard A., Li H., Henke C., Akman B., Hein A., Rote N.S., Cope L.M., Snyder A., et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell. 2015;162:974–986. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chernyavskaya Y., Mudbhary R., Zhang C., Tokarz D., Jacob V., Gopinath S., Sun X., Wang S., Magnani E., Madakashira B.P., et al. Loss of DNA methylation in zebrafish embryos activates retrotransposons to trigger antiviral signaling. Development. 2017;144:2925–2939. doi: 10.1242/dev.147629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sutter D., Doerfler W. Methylation of integrated adenovirus type 12 DNA sequences in transformed cells is inversely correlated with viral gene expression. Proc. Natl. Acad. Sci. USA. 1980;77:253–256. doi: 10.1073/pnas.77.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barletta J., Greer S.B. Methylation of HSV-1 DNA as a mechanism of viral inhibition: Studies of an analogue of methyldeoxycytidine: Trifluoromethyldeoxycytidine (F3mdCyd) Antiviral Res. 1992;18:1–25. doi: 10.1016/0166-3542(92)90002-M. [DOI] [PubMed] [Google Scholar]
- 27.Toth M., Muller U., Doerfler W. Establishment of de novo DNA methylation patterns. Transcription factor binding and deoxycytidine methylation at CpG and non-CpG sequences in an integrated adenovirus promoter. J. Mol. Biol. 1990;214:673–683. doi: 10.1016/0022-2836(90)90285-T. [DOI] [PubMed] [Google Scholar]
- 28.Lieberman P.M. Epigenetics and Genetics of Viral Latency. Cell Host Microbe. 2016;19:619–628. doi: 10.1016/j.chom.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim K., Garner-Hamrick P.A., Fisher C., Lee D., Lambert P.F. Methylation patterns of papillomavirus DNA, its influence on E2 function, and implications in viral infection. J. Virol. 2003;77:12450–12459. doi: 10.1128/JVI.77.23.12450-12459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Badal S., Badal V., Calleja-Macias I.E., Kalantari M., Chuang L.S., Li B.F., Bernard H.U. The human papillomavirus-18 genome is efficiently targeted by cellular DNA methylation. Virology. 2004;324:483–492. doi: 10.1016/j.virol.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 31.Ding D.C., Chiang M.H., Lai H.C., Hsiung C.A., Hsieh C.Y., Chu T.Y. Methylation of the long control region of HPV16 is related to the severity of cervical neoplasia. Eur. J. Obstet. Gynecol. Reprod. Biol. 2009;147:215–220. doi: 10.1016/j.ejogrb.2009.08.023. [DOI] [PubMed] [Google Scholar]
- 32.Vinokurova S., von Knebel Doeberitz M. Differential methylation of the HPV 16 upstream regulatory region during epithelial differentiation and neoplastic transformation. PLoS ONE. 2011;6:e24451. doi: 10.1371/journal.pone.0024451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y., Mao R., Yan R., Cai D., Zhang Y., Zhu H., Kang Y., Liu H., Wang J., Qin Y., et al. Transcription of hepatitis B virus covalently closed circular DNA is regulated by CpG methylation during chronic infection. PLoS ONE. 2014;9:e110442. doi: 10.1371/journal.pone.0110442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bryant D., Tristram A., Liloglou T., Hibbitts S., Fiander A., Powell N. Quantitative measurement of Human Papillomavirus type 16 L1/L2 DNA methylation correlates with cervical disease grade. J. Clin. Virol. 2014;59:24–29. doi: 10.1016/j.jcv.2013.10.029. [DOI] [PubMed] [Google Scholar]
- 35.Cornall A.M., Roberts J.M., Molano M., Machalek D.A., Phillips S., Hillman R.J., Grulich A.E., Jin F., Poynten I.M., Templeton D.J., et al. Laser capture microdissection as a tool to evaluate human papillomavirus genotyping and methylation as biomarkers of persistence and progression of anal lesions. BMJ Open. 2015;5:e008439. doi: 10.1136/bmjopen-2015-008439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turan T., Kalantari M., Cuschieri K., Cubie H.A., Skomedal H., Bernard H.U. High-throughput detection of human papillomavirus-18 L1 gene methylation, a candidate biomarker for the progression of cervical neoplasia. Virology. 2007;361:185–193. doi: 10.1016/j.virol.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu P., Iden M., Fye S., Huang Y.W., Hopp E., Chu C., Lu Y., Rader J.S. Targeted, Deep Sequencing Reveals Full Methylation Profiles of Multiple HPV Types and Potential Biomarkers for Cervical Cancer Progression. Cancer Epidemiol. Biomark. Prev. 2017;26:642–650. doi: 10.1158/1055-9965.EPI-16-0368. [DOI] [PubMed] [Google Scholar]
- 38.Niyazi M., Sui S., Zhu K., Wang L., Jiao Z., Lu P. Correlation between Methylation of Human Papillomavirus-16 L1 Gene and Cervical Carcinoma in Uyghur Women. Gynecol. Obstet. Investig. 2017;82:22–29. doi: 10.1159/000444585. [DOI] [PubMed] [Google Scholar]
- 39.Hong X., Kim E.S., Guo H. Epigenetic Regulation of Hepatitis B Virus Covalently Closed Circular DNA: Implications for Epigenetic Therapy against Chronic Hepatitis B. Hepatology. 2017;66:2066–2077. doi: 10.1002/hep.29479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karlin S., Doerfler W., Cardon L.R. Why is CpG suppressed in the genomes of virtually all small eukaryotic viruses but not in those of large eukaryotic viruses? J. Virol. 1994;68:2889–2897. doi: 10.1128/jvi.68.5.2889-2897.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shackelton L.A., Parrish C.R., Holmes E.C. Evolutionary basis of codon usage and nucleotide composition bias in vertebrate DNA viruses. J. Mol. Evol. 2006;62:551–563. doi: 10.1007/s00239-005-0221-1. [DOI] [PubMed] [Google Scholar]
- 42.Hoelzer K., Shackelton L.A., Parrish C.R. Presence and role of cytosine methylation in DNA viruses of animals. Nucleic Acids Res. 2008;36:2825–2837. doi: 10.1093/nar/gkn121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Upadhyay M., Samal J., Kandpal M., Vasaikar S., Biswas B., Gomes J., Vivekanandan P. CpG dinucleotide frequencies reveal the role of host methylation capabilities in parvovirus evolution. J. Virol. 2013;87:13816–13824. doi: 10.1128/JVI.02515-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Upadhyay M., Vivekanandan P. Depletion of CpG Dinucleotides in Papillomaviruses and Polyomaviruses: A Role for Divergent Evolutionary Pressures. PLoS ONE. 2015;10:e0142368. doi: 10.1371/journal.pone.0142368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warren C.J., Van Doorslaer K., Pandey A., Espinosa J.M., Pyeon D. Role of the host restriction factor APOBEC3 on papillomavirus evolution. Virus Evol. 2015;1:vev015. doi: 10.1093/ve/vev015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adhya D., Basu A. Epigenetic modulation of host: New insights into immune evasion by viruses. J. Biosci. 2010;35:647–663. doi: 10.1007/s12038-010-0072-9. [DOI] [PubMed] [Google Scholar]
- 47.Paschos K., Allday M.J. Epigenetic reprogramming of host genes in viral and microbial pathogenesis. Trends Microbiol. 2010;18:439–447. doi: 10.1016/j.tim.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Di Bartolo D.L., Cannon M., Liu Y.F., Renne R., Chadburn A., Boshoff C., Cesarman E. KSHV LANA inhibits TGF-beta signaling through epigenetic silencing of the TGF-beta type II receptor. Blood. 2008;111:4731–4740. doi: 10.1182/blood-2007-09-110544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dong S.M., Lee H.G., Cho S.G., Kwon S.H., Yoon H., Kwon H.J., Lee J.H., Kim H., Park P.G., Kim H., et al. Hypermethylation of the interferon regulatory factor 5 promoter in Epstein-Barr virus-associated gastric carcinoma. J. Microbiol. 2015;53:70–76. doi: 10.1007/s12275-014-4654-3. [DOI] [PubMed] [Google Scholar]
- 50.Zheng D.L., Zhang L., Cheng N., Xu X., Deng Q., Teng X.M., Wang K.S., Zhang X., Huang J., Han Z.G. Epigenetic modification induced by hepatitis B virus X protein via interaction with de novo DNA methyltransferase DNMT3A. J. Hepatol. 2009;50:377–387. doi: 10.1016/j.jhep.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 51.Rincon-Orozco B., Halec G., Rosenberger S., Muschik D., Nindl I., Bachmann A., Ritter T.M., Dondog B., Ly R., Bosch F.X., et al. Epigenetic silencing of interferon-kappa in human papillomavirus type 16-positive cells. Cancer Res. 2009;69:8718–8725. doi: 10.1158/0008-5472.CAN-09-0550. [DOI] [PubMed] [Google Scholar]
- 52.Reiser J., Hurst J., Voges M., Krauss P., Munch P., Iftner T., Stubenrauch F. High-risk human papillomaviruses repress constitutive kappa interferon transcription via E6 to prevent pathogen recognition receptor and antiviral-gene expression. J. Virol. 2011;85:11372–11380. doi: 10.1128/JVI.05279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cicchini L., Blumhagen R.Z., Westrich J.A., Myers M.E., Warren C.J., Siska C., Raben D., Kechris K.J., Pyeon D. High-Risk Human Papillomavirus E7 Alters Host DNA Methylome and Represses HLA-E Expression in Human Keratinocytes. Sci. Rep. 2017;7:3633. doi: 10.1038/s41598-017-03295-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cicchini L., Westrich J.A., Xu T., Vermeer D.W., Berger J.N., Clambey E.T., Lee D., Song J.I., Lambert P.F., Greer R.O., et al. Suppression of Antitumor Immune Responses by Human Papillomavirus through Epigenetic Downregulation of CXCL14. MBio. 2016;7 doi: 10.1128/mBio.00270-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hattori N., Ushijima T. Epigenetic impact of infection on carcinogenesis: Mechanisms and applications. Genome Med. 2016;8:10. doi: 10.1186/s13073-016-0267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zitvogel L., Galluzzi L., Kepp O., Smyth M.J., Kroemer G. Type I interferons in anticancer immunity. Nat. Rev. Immunol. 2015;15:405–414. doi: 10.1038/nri3845. [DOI] [PubMed] [Google Scholar]
- 57.Gildener-Leapman N., Ferris R.L., Bauman J.E. Promising systemic immunotherapies in head and neck squamous cell carcinoma. Oral Oncol. 2013;49:1089–1096. doi: 10.1016/j.oraloncology.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ribas A., Tumeh P.C. The future of cancer therapy: Selecting patients likely to respond to PD1/L1 blockade. Clin. Cancer Res. 2014;20:4982–4984. doi: 10.1158/1078-0432.CCR-14-0933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Powles T., Eder J.P., Fine G.D., Braiteh F.S., Loriot Y., Cruz C., Bellmunt J., Burris H.A., Petrylak D.P., Teng S.L., et al. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature. 2014;515:558–562. doi: 10.1038/nature13904. [DOI] [PubMed] [Google Scholar]
- 60.Herbst R.S., Soria J.C., Kowanetz M., Fine G.D., Hamid O., Gordon M.S., Sosman J.A., McDermott D.F., Powderly J.D., Gettinger S.N., et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tumeh P.C., Harview C.L., Yearley J.H., Shintaku I.P., Taylor E.J., Robert L., Chmielowski B., Spasic M., Henry G., Ciobanu V., et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Katlinski K.V., Gui J., Katlinskaya Y.V., Ortiz A., Chakraborty R., Bhattacharya S., Carbone C.J., Beiting D.P., Girondo M.A., Peck A.R., et al. Inactivation of Interferon Receptor Promotes the Establishment of Immune Privileged Tumor Microenvironment. Cancer Cell. 2017;31:194–207. doi: 10.1016/j.ccell.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Diamond M.S., Kinder M., Matsushita H., Mashayekhi M., Dunn G.P., Archambault J.M., Lee H., Arthur C.D., White J.M., Kalinke U., et al. Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J. Exp. Med. 2011;208:1989–2003. doi: 10.1084/jem.20101158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fuertes M.B., Kacha A.K., Kline J., Woo S.R., Kranz D.M., Murphy K.M., Gajewski T.F. Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8{alpha}+ dendritic cells. J. Exp. Med. 2011;208:2005–2016. doi: 10.1084/jem.20101159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen H.M., Tanaka N., Mitani Y., Oda E., Nozawa H., Chen J.Z., Yanai H., Negishi H., Choi M.K., Iwasaki T., et al. Critical role for constitutive type I interferon signaling in the prevention of cellular transformation. Cancer Sci. 2009;100:449–456. doi: 10.1111/j.1349-7006.2008.01051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang X., Justice A.C., Hu Y., Wang Z., Zhao H., Wang G., Johnson E.O., Emu B., Sutton R.E., Krystal J.H., et al. Epigenome-wide differential DNA methylation between HIV-infected and uninfected individuals. Epigenetics. 2016:1–11. doi: 10.1080/15592294.2016.1221569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abdel-Hameed E.A., Ji H., Shata M.T. HIV-Induced Epigenetic Alterations in Host Cells. Adv. Exp. Med. Biol. 2016;879:27–38. doi: 10.1007/978-3-319-24738-0_2. [DOI] [PubMed] [Google Scholar]
- 68.Nakayama-Hosoya K., Ishida T., Youngblood B., Nakamura H., Hosoya N., Koga M., Koibuchi T., Iwamoto A., Kawana-Tachikawa A. Epigenetic repression of interleukin 2 expression in senescent CD4+ T cells during chronic HIV type 1 infection. J. Infect. Dis. 2015;211:28–39. doi: 10.1093/infdis/jiu376. [DOI] [PubMed] [Google Scholar]
- 69.Mikovits J.A., Young H.A., Vertino P., Issa J.P., Pitha P.M., Turcoski-Corrales S., Taub D.D., Petrow C.L., Baylin S.B., Ruscetti F.W. Infection with human immunodeficiency virus type 1 upregulates DNA methyltransferase, resulting in de novo methylation of the gamma interferon (IFN-gamma) promoter and subsequent downregulation of IFN-gamma production. Mol. Cell. Biol. 1998;18:5166–5177. doi: 10.1128/MCB.18.9.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Choi S.J., Shin Y.S., Kang B.W., Kim J.G., Won K.J., Lieberman P.M., Cho H., Kang H. DNA hypermethylation induced by Epstein-Barr virus in the development of Epstein-Barr virus-associated gastric carcinoma. Arch. Pharm. Res. 2017;40:894–905. doi: 10.1007/s12272-017-0939-5. [DOI] [PubMed] [Google Scholar]
- 71.Cheung C.C., Chung G.T., Lun S.W., To K.F., Choy K.W., Lau K.M., Siu S.P., Guan X.Y., Ngan R.K., Yip T.T., et al. miR-31 is consistently inactivated in EBV-associated nasopharyngeal carcinoma and contributes to its tumorigenesis. Mol. Cancer. 2014;13:184. doi: 10.1186/1476-4598-13-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tong J.H., Tsang R.K., Lo K.W., Woo J.K., Kwong J., Chan M.W., Chang A.R., van Hasselt C.A., Huang D.P., To K.F. Quantitative Epstein-Barr virus DNA analysis and detection of gene promoter hypermethylation in nasopharyngeal (NP) brushing samples from patients with NP carcinoma. Clin. Cancer Res. 2002;8:2612–2619. [PubMed] [Google Scholar]
- 73.Zhou L., Jiang W., Ren C., Yin Z., Feng X., Liu W., Tao Q., Yao K. Frequent hypermethylation of RASSF1A and TSLC1, and high viral load of Epstein-Barr Virus DNA in nasopharyngeal carcinoma and matched tumor-adjacent tissues. Neoplasia. 2005;7:809–815. doi: 10.1593/neo.05217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hernandez-Vargas H., Gruffat H., Cros M.P., Diederichs A., Sirand C., Vargas-Ayala R.C., Jay A., Durand G., Le Calvez-Kelm F., Herceg Z., et al. Viral driven epigenetic events alter the expression of cancer-related genes in Epstein-Barr-virus naturally infected Burkitt lymphoma cell lines. Sci. Rep. 2017;7:5852. doi: 10.1038/s41598-017-05713-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leonard S., Wei W., Anderton J., Vockerodt M., Rowe M., Murray P.G., Woodman C.B. Epigenetic and transcriptional changes which follow Epstein-Barr virus infection of germinal center B cells and their relevance to the pathogenesis of Hodgkin’s lymphoma. J. Virol. 2011;85:9568–9577. doi: 10.1128/JVI.00468-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Seo S.Y., Kim E.O., Jang K.L. Epstein-Barr virus latent membrane protein 1 suppresses the growth-inhibitory effect of retinoic acid by inhibiting retinoic acid receptor-beta2 expression via DNA methylation. Cancer Lett. 2008;270:66–76. doi: 10.1016/j.canlet.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 77.Tsai C.N., Tsai C.L., Tse K.P., Chang H.Y., Chang Y.S. The Epstein-Barr virus oncogene product, latent membrane protein 1, induces the downregulation of E-cadherin gene expression via activation of DNA methyltransferases. Proc. Natl. Acad. Sci. USA. 2002;99:10084–10089. doi: 10.1073/pnas.152059399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Platt G., Carbone A., Mittnacht S. p16INK4a loss and sensitivity in KSHV associated primary effusion lymphoma. Oncogene. 2002;21:1823–1831. doi: 10.1038/sj.onc.1205360. [DOI] [PubMed] [Google Scholar]
- 79.Tian Y., Yang W., Song J., Wu Y., Ni B. Hepatitis B virus X protein-induced aberrant epigenetic modifications contributing to human hepatocellular carcinoma pathogenesis. Mol. Cell. Biol. 2013;33:2810–2816. doi: 10.1128/MCB.00205-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hensel K.O., Rendon J.C., Navas M.C., Rots M.G., Postberg J. Virus-host interplay in hepatitis B virus infection and epigenetic treatment strategies. FEBS J. 2017;284:3550–3572. doi: 10.1111/febs.14094. [DOI] [PubMed] [Google Scholar]
- 81.Cancer Genome Atlas Network Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sakane J., Taniyama K., Miyamoto K., Saito A., Kuraoka K., Nishimura T., Sentani K., Oue N., Yasui W. Aberrant DNA methylation of DLX4 and SIM1 is a predictive marker for disease progression of uterine cervical low-grade squamous intraepithelial lesion. Diagn. Cytopathol. 2015;43:462–470. doi: 10.1002/dc.23256. [DOI] [PubMed] [Google Scholar]
- 83.Schutze D.M., Kooter J.M., Wilting S.M., Meijer C.J., Quint W., Snijders P.J., Steenbergen R.D. Longitudinal assessment of DNA methylation changes during HPVE6E7-induced immortalization of primary keratinocytes. Epigenetics. 2015;10:73–81. doi: 10.4161/15592294.2014.990787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.De Wilde J., Kooter J.M., Overmeer R.M., Claassen-Kramer D., Meijer C.J., Snijders P.J., Steenbergen R.D. hTERT promoter activity and CpG methylation in HPV-induced carcinogenesis. BMC Cancer. 2010;10:271. doi: 10.1186/1471-2407-10-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sartor M.A., Dolinoy D.C., Jones T.R., Colacino J.A., Prince M.E., Carey T.E., Rozek L.S. Genome-wide methylation and expression differences in HPV(+) and HPV(−) squamous cell carcinoma cell lines are consistent with divergent mechanisms of carcinogenesis. Epigenetics. 2011;6:777–787. doi: 10.4161/epi.6.6.16216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Amara K., Trimeche M., Ziadi S., Laatiri A., Hachana M., Sriha B., Mokni M., Korbi S. Presence of simian virus 40 DNA sequences in diffuse large B-cell lymphomas in Tunisia correlates with aberrant promoter hypermethylation of multiple tumor suppressor genes. Int. J. Cancer. 2007;121:2693–2702. doi: 10.1002/ijc.23038. [DOI] [PubMed] [Google Scholar]
- 87.Shivapurkar N., Takahashi T., Reddy J., Zheng Y., Stastny V., Collins R., Toyooka S., Suzuki M., Parikh G., Asplund S., et al. Presence of simian virus 40 DNA sequences in human lymphoid and hematopoietic malignancies and their relationship to aberrant promoter methylation of multiple genes. Cancer Res. 2004;64:3757–3760. doi: 10.1158/0008-5472.CAN-03-3307. [DOI] [PubMed] [Google Scholar]
- 88.Toyooka S., Pass H.I., Shivapurkar N., Fukuyama Y., Maruyama R., Toyooka K.O., Gilcrease M., Farinas A., Minna J.D., Gazdar A.F. Aberrant methylation and simian virus 40 tag sequences in malignant mesothelioma. Cancer Res. 2001;61:5727–5730. [PubMed] [Google Scholar]
- 89.Toyooka S., Carbone M., Toyooka K.O., Bocchetta M., Shivapurkar N., Minna J.D., Gazdar A.F. Progressive aberrant methylation of the RASSF1A gene in simian virus 40 infected human mesothelial cells. Oncogene. 2002;21:4340–4344. doi: 10.1038/sj.onc.1205381. [DOI] [PubMed] [Google Scholar]
- 90.Helmbold P., Lahtz C., Enk A., Herrmann-Trost P., Marsch W., Kutzner H., Dammann R.H. Frequent occurrence of RASSF1A promoter hypermethylation and Merkel cell polyomavirus in Merkel cell carcinoma. Mol. Carcinog. 2009;48:903–909. doi: 10.1002/mc.20540. [DOI] [PubMed] [Google Scholar]
- 91.Helmbold P., Lahtz C., Herpel E., Schnabel P.A., Dammann R.H. Frequent hypermethylation of RASSF1A tumour suppressor gene promoter and presence of Merkel cell polyomavirus in small cell lung cancer. Eur. J. Cancer. 2009;45:2207–2211. doi: 10.1016/j.ejca.2009.04.038. [DOI] [PubMed] [Google Scholar]
- 92.Ksiaa F., Ziadi S., Mokni M., Korbi S., Trimeche M. The presence of JC virus in gastric carcinomas correlates with patient’s age, intestinal histological type and aberrant methylation of tumor suppressor genes. Mod. Pathol. 2010;23:522–530. doi: 10.1038/modpathol.2009.184. [DOI] [PubMed] [Google Scholar]
- 93.Goel A., Li M.S., Nagasaka T., Shin S.K., Fuerst F., Ricciardiello L., Wasserman L., Boland C.R. Association of JC virus T-antigen expression with the methylator phenotype in sporadic colorectal cancers. Gastroenterology. 2006;130:1950–1961. doi: 10.1053/j.gastro.2006.02.061. [DOI] [PubMed] [Google Scholar]
- 94.Hino R., Uozaki H., Murakami N., Ushiku T., Shinozaki A., Ishikawa S., Morikawa T., Nakaya T., Sakatani T., Takada K., et al. Activation of DNA methyltransferase 1 by EBV latent membrane protein 2A leads to promoter hypermethylation of PTEN gene in gastric carcinoma. Cancer Res. 2009;69:2766–2774. doi: 10.1158/0008-5472.CAN-08-3070. [DOI] [PubMed] [Google Scholar]
- 95.Ksiaa F., Ziadi S., Gacem R.B., Dhiab M.B., Trimeche M. Correlation between DNA methyltransferases expression and Epstein-Barr virus, JC polyomavirus and Helicobacter pylori infections in gastric carcinomas. Neoplasma. 2014;61:710–717. doi: 10.4149/neo_2014_086. [DOI] [PubMed] [Google Scholar]
- 96.Xie Q., Chen L., Shan X., Shan X., Tang J., Zhou F., Chen Q., Quan H., Nie D., Zhang W., et al. Epigenetic silencing of SFRP1 and SFRP5 by hepatitis B virus X protein enhances hepatoma cell tumorigenicity through Wnt signaling pathway. Int. J. Cancer. 2014;135:635–646. doi: 10.1002/ijc.28697. [DOI] [PubMed] [Google Scholar]
- 97.Huang J., Wang Y., Guo Y., Sun S. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology. 2010;52:60–70. doi: 10.1002/hep.23660. [DOI] [PubMed] [Google Scholar]
- 98.Zhu Y.Z., Zhu R., Fan J., Pan Q., Li H., Chen Q., Zhu H.G. Hepatitis B virus X protein induces hypermethylation of p16(INK4A) promoter via DNA methyltransferases in the early stage of HBV-associated hepatocarcinogenesis. J. Viral Hepat. 2010;17:98–107. doi: 10.1111/j.1365-2893.2009.01156.x. [DOI] [PubMed] [Google Scholar]
- 99.Wei X., Xiang T., Ren G., Tan C., Liu R., Xu X., Wu Z. miR-101 is down-regulated by the hepatitis B virus x protein and induces aberrant DNA methylation by targeting DNA methyltransferase 3A. Cell. Signal. 2013;25:439–446. doi: 10.1016/j.cellsig.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 100.Liu R., Gong M., Li X., Zhou Y., Gao W., Tulpule A., Chaudhary P.M., Jung J., Gill P.S. Induction, regulation, and biologic function of Axl receptor tyrosine kinase in Kaposi sarcoma. Blood. 2010;116:297–305. doi: 10.1182/blood-2009-12-257154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Damania B. Oncogenic gamma-herpesviruses: Comparison of viral proteins involved in tumorigenesis. Nat. Rev. Microbiol. 2004;2:656–668. doi: 10.1038/nrmicro958. [DOI] [PubMed] [Google Scholar]
- 102.Wen K.W., Damania B. Kaposi sarcoma-associated herpesvirus (KSHV): Molecular biology and oncogenesis. Cancer Lett. 2010;289:140–150. doi: 10.1016/j.canlet.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li H., Liu S., Hu J., Luo X., Li N., A M.B., Cao Y. Epstein-Barr virus lytic reactivation regulation and its pathogenic role in carcinogenesis. Int. J. Biol. Sci. 2016;12:1309–1318. doi: 10.7150/ijbs.16564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Purushothaman P., Uppal T., Sarkar R., Verma S.C. KSHV-Mediated Angiogenesis in Tumor Progression. Viruses. 2016;8:198. doi: 10.3390/v8070198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wei F., Gan J., Wang C., Zhu C., Cai Q. Cell Cycle Regulatory Functions of the KSHV Oncoprotein LANA. Front. Microbiol. 2016;7:334. doi: 10.3389/fmicb.2016.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Raab-Traub N. Novel mechanisms of EBV-induced oncogenesis. Curr. Opin. Virol. 2012;2:453–458. doi: 10.1016/j.coviro.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Young L.S., Murray P.G. Epstein-Barr virus and oncogenesis: From latent genes to tumours. Oncogene. 2003;22:5108–5121. doi: 10.1038/sj.onc.1206556. [DOI] [PubMed] [Google Scholar]
- 108.Kuhn-Hallek I., Sage D.R., Stein L., Groelle H., Fingeroth J.D. Expression of recombination activating genes (RAG-1 and RAG-2) in Epstein-Barr virus-bearing B cells. Blood. 1995;85:1289–1299. [PubMed] [Google Scholar]
- 109.Shamay M., Krithivas A., Zhang J., Hayward S.D. Recruitment of the de novo DNA methyltransferase Dnmt3a by Kaposi’s sarcoma-associated herpesvirus LANA. Proc. Natl. Acad. Sci. USA. 2006;103:14554–14559. doi: 10.1073/pnas.0604469103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sun F., Xiao Y., Qu Z. Oncovirus Kaposi sarcoma herpesvirus (KSHV) represses tumor suppressor PDLIM2 to persistently activate nuclear factor kappaB (NF-kappaB) and STAT3 transcription factors for tumorigenesis and tumor maintenance. J. Biol. Chem. 2015;290:7362–7368. doi: 10.1074/jbc.C115.637918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wu J., Xu Y., Mo D., Huang P., Sun R., Huang L., Pan S., Xu J. Kaposi’s sarcoma-associated herpesvirus (KSHV) vIL-6 promotes cell proliferation and migration by upregulating DNMT1 via STAT3 activation. PLoS ONE. 2014;9:e93478. doi: 10.1371/journal.pone.0093478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kang M.S., Kieff E. Epstein-Barr virus latent genes. Exp. Mol. Med. 2015;47:e131. doi: 10.1038/emm.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tsai C.L., Li H.P., Lu Y.J., Hsueh C., Liang Y., Chen C.L., Tsao S.W., Tse K.P., Yu J.S., Chang Y.S. Activation of DNA methyltransferase 1 by EBV LMP1 Involves c-Jun NH(2)-terminal kinase signaling. Cancer Res. 2006;66:11668–11676. doi: 10.1158/0008-5472.CAN-06-2194. [DOI] [PubMed] [Google Scholar]
- 114.Kitisin K., Saha T., Blake T., Golestaneh N., Deng M., Kim C., Tang Y., Shetty K., Mishra B., Mishra L. Tgf-Beta signaling in development. Sci. STKE. 2007;2007:cm1. doi: 10.1126/stke.3992007cm1. [DOI] [PubMed] [Google Scholar]
- 115.Derynck R., Zhang Y.E. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 116.Massague J., Blain S.W., Lo R.S. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/S0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 117.Zhang Y.E. Mechanistic insight into contextual TGF-beta signaling. Curr. Opin. Cell Biol. 2017;51:1–7. doi: 10.1016/j.ceb.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Colak S., Ten Dijke P. Targeting TGF-beta Signaling in Cancer. Trends Cancer. 2017;3:56–71. doi: 10.1016/j.trecan.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 119.Tauriello D.V.F., Batlle E. Targeting the Microenvironment in Advanced Colorectal Cancer. Trends Cancer. 2016;2:495–504. doi: 10.1016/j.trecan.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 120.David C.J., Huang Y.H., Chen M., Su J., Zou Y., Bardeesy N., Iacobuzio-Donahue C.A., Massague J. TGF-beta Tumor Suppression through a Lethal EMT. Cell. 2016;164:1015–1030. doi: 10.1016/j.cell.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Oberhammer F.A., Pavelka M., Sharma S., Tiefenbacher R., Purchio A.F., Bursch W., Schulte-Hermann R. Induction of apoptosis in cultured hepatocytes and in regressing liver by transforming growth factor beta 1. Proc. Natl. Acad. Sci. USA. 1992;89:5408–5412. doi: 10.1073/pnas.89.12.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gressner A.M., Lahme B., Mannherz H.G., Polzar B. TGF-beta-mediated hepatocellular apoptosis by rat and human hepatoma cells and primary rat hepatocytes. J. Hepatol. 1997;26:1079–1092. doi: 10.1016/S0168-8278(97)80117-1. [DOI] [PubMed] [Google Scholar]
- 123.Matsumura N., Huang Z., Mori S., Baba T., Fujii S., Konishi I., Iversen E.S., Berchuck A., Murphy S.K. Epigenetic suppression of the TGF-beta pathway revealed by transcriptome profiling in ovarian cancer. Genome Res. 2011;21:74–82. doi: 10.1101/gr.108803.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dong Z., Guo W., Guo Y., Kuang G., Yang Z. Concordant promoter methylation of transforming growth factor-beta receptor types I and II occurs early in esophageal squamous cell carcinoma. Am. J. Med. Sci. 2012;343:375–381. doi: 10.1097/MAJ.0b013e3182253430. [DOI] [PubMed] [Google Scholar]
- 125.De Carvalho F., Colleoni G.W., Almeida M.S., Carvalho A.L., Vettore A.L. TGFbetaR2 aberrant methylation is a potential prognostic marker and therapeutic target in multiple myeloma. Int. J. Cancer. 2009;125:1985–1991. doi: 10.1002/ijc.24431. [DOI] [PubMed] [Google Scholar]
- 126.Yamashita S., Takahashi S., McDonell N., Watanabe N., Niwa T., Hosoya K., Tsujino Y., Shirai T., Ushijima T. Methylation silencing of transforming growth factor-beta receptor type II in rat prostate cancers. Cancer Res. 2008;68:2112–2121. doi: 10.1158/0008-5472.CAN-07-5282. [DOI] [PubMed] [Google Scholar]
- 127.Zhang Q., Rubenstein J.N., Jang T.L., Pins M., Javonovic B., Yang X., Kim S.J., Park I., Lee C. Insensitivity to transforming growth factor-beta results from promoter methylation of cognate receptors in human prostate cancer cells (LNCaP) Mol. Endocrinol. 2005;19:2390–2399. doi: 10.1210/me.2005-0096. [DOI] [PubMed] [Google Scholar]
- 128.Zhang Q., Rubenstein J.N., Liu V.C., Park I., Jang T., Lee C. Restoration of expression of transforming growth factor-beta type II receptor in murine renal cell carcinoma (renca) cells by 5-Aza-2′-deoxycytidine. Life Sci. 2005;76:1159–1166. doi: 10.1016/j.lfs.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 129.Zhao H., Shiina H., Greene K.L., Li L.C., Tanaka Y., Kishi H., Igawa M., Kane C.J., Carroll P., Dahiya R. CpG methylation at promoter site -140 inactivates TGFbeta2 receptor gene in prostate cancer. Cancer. 2005;104:44–52. doi: 10.1002/cncr.21135. [DOI] [PubMed] [Google Scholar]
- 130.Choi H.S., Jain V., Krueger B., Marshall V., Kim C.H., Shisler J.L., Whitby D., Renne R. Kaposi’s Sarcoma-Associated Herpesvirus (KSHV) Induces the Oncogenic miR-17-92 Cluster and Down-Regulates TGF-beta Signaling. PLoS Pathog. 2015;11:e1005255. doi: 10.1371/journal.ppat.1005255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lei X., Zhu Y., Jones T., Bai Z., Huang Y., Gao S.J. A Kaposi’s sarcoma-associated herpesvirus microRNA and its variants target the transforming growth factor beta pathway to promote cell survival. J. Virol. 2012;86:11698–11711. doi: 10.1128/JVI.06855-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liu Y., Sun R., Lin X., Liang D., Deng Q., Lan K. Kaposi’s sarcoma-associated herpesvirus-encoded microRNA miR-K12-11 attenuates transforming growth factor beta signaling through suppression of SMAD5. J. Virol. 2012;86:1372–1381. doi: 10.1128/JVI.06245-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Samols M.A., Skalsky R.L., Maldonado A.M., Riva A., Lopez M.C., Baker H.V., Renne R. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 2007;3:e65. doi: 10.1371/journal.ppat.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Seo T., Park J., Choe J. Kaposi’s sarcoma-associated herpesvirus viral IFN regulatory factor 1 inhibits transforming growth factor-beta signaling. Cancer Res. 2005;65:1738–1747. doi: 10.1158/0008-5472.CAN-04-2374. [DOI] [PubMed] [Google Scholar]
- 135.Tomita M., Choe J., Tsukazaki T., Mori N. The Kaposi’s sarcoma-associated herpesvirus K-bZIP protein represses transforming growth factor beta signaling through interaction with CREB-binding protein. Oncogene. 2004;23:8272–8281. doi: 10.1038/sj.onc.1208059. [DOI] [PubMed] [Google Scholar]
- 136.Feng X.H., Zhang Y., Wu R.Y., Derynck R. The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for smad3 in TGF-beta-induced transcriptional activation. Genes Dev. 1998;12:2153–2163. doi: 10.1101/gad.12.14.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Janknecht R., Wells N.J., Hunter T. TGF-beta-stimulated cooperation of smad proteins with the coactivators CBP/p300. Genes Dev. 1998;12:2114–2119. doi: 10.1101/gad.12.14.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pouponnot C., Jayaraman L., Massague J. Physical and functional interaction of SMADs and p300/CBP. J. Biol. Chem. 1998;273:22865–22868. doi: 10.1074/jbc.273.36.22865. [DOI] [PubMed] [Google Scholar]
- 139.Shen X., Hu P.P., Liberati N.T., Datto M.B., Frederick J.P., Wang X.F. TGF-beta-induced phosphorylation of Smad3 regulates its interaction with coactivator p300/CREB-binding protein. Mol. Biol. Cell. 1998;9:3309–3319. doi: 10.1091/mbc.9.12.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Topper J.N., DiChiara M.R., Brown J.D., Williams A.J., Falb D., Collins T., Gimbrone M.A., Jr. CREB binding protein is a required coactivator for Smad-dependent, transforming growth factor beta transcriptional responses in endothelial cells. Proc. Natl. Acad. Sci. USA. 1998;95:9506–9511. doi: 10.1073/pnas.95.16.9506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.DiMaio T.A., Gutierrez K.D., Lagunoff M. Kaposi’s sarcoma-associated herpesvirus downregulates transforming growth factor beta2 to promote enhanced stability of capillary-like tube formation. J. Virol. 2014;88:14301–14309. doi: 10.1128/JVI.01696-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Morris V.A., Punjabi A.S., Wells R.C., Wittkopp C.J., Vart R., Lagunoff M. The KSHV viral IL-6 homolog is sufficient to induce blood to lymphatic endothelial cell differentiation. Virology. 2012;428:112–120. doi: 10.1016/j.virol.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yanai H., Chen H.M., Inuzuka T., Kondo S., Mak T.W., Takaoka A., Honda K., Taniguchi T. Role of IFN regulatory factor 5 transcription factor in antiviral immunity and tumor suppression. Proc. Natl. Acad. Sci. USA. 2007;104:3402–3407. doi: 10.1073/pnas.0611559104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hu G., Barnes B.J. IRF-5 is a mediator of the death receptor-induced apoptotic signaling pathway. J. Biol. Chem. 2009;284:2767–2777. doi: 10.1074/jbc.M804744200. [DOI] [PubMed] [Google Scholar]
- 145.Takaoka A., Yanai H., Kondo S., Duncan G., Negishi H., Mizutani T., Kano S., Honda K., Ohba Y., Mak T.W., et al. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature. 2005;434:243–249. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- 146.Bi X., Hameed M., Mirani N., Pimenta E.M., Anari J., Barnes B.J. Loss of interferon regulatory factor 5 (IRF5) expression in human ductal carcinoma correlates with disease stage and contributes to metastasis. Breast Cancer Res. 2011;13:R111. doi: 10.1186/bcr3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yamashita M., Toyota M., Suzuki H., Nojima M., Yamamoto E., Kamimae S., Watanabe Y., Kai M., Akashi H., Maruyama R., et al. DNA methylation of interferon regulatory factors in gastric cancer and noncancerous gastric mucosae. Cancer Sci. 2010;101:1708–1716. doi: 10.1111/j.1349-7006.2010.01581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Xu D., Meyer F., Ehlers E., Blasnitz L., Zhang L. Interferon regulatory factor 4 (IRF-4) targets IRF-5 to regulate Epstein-Barr virus transformation. J. Biol. Chem. 2011;286:18261–18267. doi: 10.1074/jbc.M110.210542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ning S., Huye L.E., Pagano J.S. Interferon regulatory factor 5 represses expression of the Epstein-Barr virus oncoprotein LMP1: Braking of the IRF7/LMP1 regulatory circuit. J. Virol. 2005;79:11671–11676. doi: 10.1128/JVI.79.18.11671-11676.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chen H., Huang J., Wu F.Y., Liao G., Hutt-Fletcher L., Hayward S.D. Regulation of expression of the Epstein-Barr virus BamHI-A rightward transcripts. J. Virol. 2005;79:1724–1733. doi: 10.1128/JVI.79.3.1724-1733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang Q., Lingel A., Geiser V., Kwapnoski Z., Zhang L. Tumor Suppressor p53 Stimulates the Expression of Epstein-Barr Virus Latent Membrane Protein 1. J. Virol. 2017;91 doi: 10.1128/JVI.00312-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Martin H.J., Lee J.M., Walls D., Hayward S.D. Manipulation of the toll-like receptor 7 signaling pathway by Epstein-Barr virus. J. Virol. 2007;81:9748–9758. doi: 10.1128/JVI.01122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ringelhan M., Protzer U. Oncogenic potential of hepatitis B virus encoded proteins. Curr. Opin. Virol. 2015;14:109–115. doi: 10.1016/j.coviro.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 154.Tarocchi M., Polvani S., Marroncini G., Galli A. Molecular mechanism of hepatitis B virus-induced hepatocarcinogenesis. World J. Gastroenterol. 2014;20:11630–11640. doi: 10.3748/wjg.v20.i33.11630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhao Z., Hu Y., Shen X., Lao Y., Zhang L., Qiu X., Hu J., Gong P., Cui H., Lu S., et al. HBx represses RIZ1 expression by DNA methyltransferase 1 involvement in decreased miR-152 in hepatocellular carcinoma. Oncol. Rep. 2017;37:2811–2818. doi: 10.3892/or.2017.5518. [DOI] [PubMed] [Google Scholar]
- 156.Fu X., Song X., Li Y., Tan D., Liu G. Hepatitis B virus X protein upregulates DNA methyltransferase 3A/3B and enhances SOCS-1CpG island methylation. Mol. Med. Rep. 2016;13:301–308. doi: 10.3892/mmr.2015.4545. [DOI] [PubMed] [Google Scholar]
- 157.Jung J.K., Park S.H., Jang K.L. Hepatitis B virus X protein overcomes the growth-inhibitory potential of retinoic acid by downregulating retinoic acid receptor-beta2 expression via DNA methylation. J. Gen. Virol. 2010;91:493–500. doi: 10.1099/vir.0.015149-0. [DOI] [PubMed] [Google Scholar]
- 158.Jung J.K., Arora P., Pagano J.S., Jang K.L. Expression of DNA methyltransferase 1 is activated by hepatitis B virus X protein via a regulatory circuit involving the p16INK4a-cyclin D1-CDK 4/6-pRb-E2F1 pathway. Cancer Res. 2007;67:5771–5778. doi: 10.1158/0008-5472.CAN-07-0529. [DOI] [PubMed] [Google Scholar]
- 159.Vivekanandan P., Daniel H.D., Kannangai R., Martinez-Murillo F., Torbenson M. Hepatitis B virus replication induces methylation of both host and viral DNA. J. Virol. 2010;84:4321–4329. doi: 10.1128/JVI.02280-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lee J.O., Kwun H.J., Jung J.K., Choi K.H., Min D.S., Jang K.L. Hepatitis B virus X protein represses E-cadherin expression via activation of DNA methyltransferase 1. Oncogene. 2005;24:6617–6625. doi: 10.1038/sj.onc.1208827. [DOI] [PubMed] [Google Scholar]
- 161.Park I.Y., Sohn B.H., Yu E., Suh D.J., Chung Y.H., Lee J.H., Surzycki S.J., Lee Y.I. Aberrant epigenetic modifications in hepatocarcinogenesis induced by hepatitis B virus X protein. Gastroenterology. 2007;132:1476–1494. doi: 10.1053/j.gastro.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 162.Park S.H., Jung J.K., Lim J.S., Tiwari I., Jang K.L. Hepatitis B virus X protein overcomes all-trans retinoic acid-induced cellular senescence by downregulating levels of p16 and p21 via DNA methylation. J. Gen. Virol. 2011;92:1309–1317. doi: 10.1099/vir.0.029512-0. [DOI] [PubMed] [Google Scholar]
- 163.Lee S.M., Lee Y.G., Bae J.B., Choi J.K., Tayama C., Hata K., Yun Y., Seong J.K., Kim Y.J. HBx induces hypomethylation of distal intragenic CpG islands required for active expression of developmental regulators. Proc. Natl. Acad. Sci. USA. 2014;111:9555–9560. doi: 10.1073/pnas.1400604111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Fan H., Zhang H., Pascuzzi P.E., Andrisani O. Hepatitis B virus X protein induces EpCAM expression via active DNA demethylation directed by RelA in complex with EZH2 and TET2. Oncogene. 2016;35:715–726. doi: 10.1038/onc.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yue X., Yang F., Yang Y., Mu Y., Sun W., Li W., Xu D., Wu J., Zhu Y. Induction of cyclooxygenase-2 expression by hepatitis B virus depends on demethylation-associated recruitment of transcription factors to the promoter. Virol. J. 2011;8:118. doi: 10.1186/1743-422X-8-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Neri F., Krepelova A., Incarnato D., Maldotti M., Parlato C., Galvagni F., Matarese F., Stunnenberg H.G., Oliviero S. Dnmt3L antagonizes DNA methylation at bivalent promoters and favors DNA methylation at gene bodies in ESCs. Cell. 2013;155:121–134. doi: 10.1016/j.cell.2013.08.056. [DOI] [PubMed] [Google Scholar]
- 167.Ul-Haq Z., Naz S., Mesaik M.A. Interleukin-4 receptor signaling and its binding mechanism: A therapeutic insight from inhibitors tool box. Cytokine Growth Factor Rev. 2016;32:3–15. doi: 10.1016/j.cytogfr.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 168.Lowenthal J.W., Castle B.E., Christiansen J., Schreurs J., Rennick D., Arai N., Hoy P., Takebe Y., Howard M. Expression of high affinity receptors for murine interleukin 4 (BSF-1) on hemopoietic and nonhemopoietic cells. J. Immunol. 1988;140:456–464. [PubMed] [Google Scholar]
- 169.Grunfeld C., Soued M., Adi S., Moser A.H., Fiers W., Dinarello C.A., Feingold K.R. Interleukin 4 inhibits stimulation of hepatic lipogenesis by tumor necrosis factor, interleukin 1, and interleukin 6 but not by interferon-alpha. Cancer Res. 1991;51:2803–2807. [PubMed] [Google Scholar]
- 170.Loyer P., Ilyin G., Abdel Razzak Z., Banchereau J., Dezier J.F., Campion J.P., Guguen-Guillouzo C., Guillouzo A. Interleukin 4 inhibits the production of some acute-phase proteins by human hepatocytes in primary culture. FEBS Lett. 1993;336:215–220. doi: 10.1016/0014-5793(93)80806-6. [DOI] [PubMed] [Google Scholar]
- 171.Lin S.J., Shu P.Y., Chang C., Ng A.K., Hu C.P. IL-4 suppresses the expression and the replication of hepatitis B virus in the hepatocellular carcinoma cell line Hep3B. J. Immunol. 2003;171:4708–4716. doi: 10.4049/jimmunol.171.9.4708. [DOI] [PubMed] [Google Scholar]
- 172.Yao Y., Li J., Lu Z., Tong A., Wang W., Su X., Zhou Y., Mu B., Zhou S., Li X., et al. Proteomic analysis of the interleukin-4 (IL-4) response in hepatitis B virus-positive human hepatocelluar carcinoma cell line HepG2.2.15. Electrophoresis. 2011;32:2004–2012. doi: 10.1002/elps.201100147. [DOI] [PubMed] [Google Scholar]
- 173.Aoudjehane L., Podevin P., Scatton O., Jaffray P., Dusanter-Fourt I., Feldmann G., Massault P.P., Grira L., Bringuier A., Dousset B., et al. Interleukin-4 induces human hepatocyte apoptosis through a Fas-independent pathway. FASEB J. 2007;21:1433–1444. doi: 10.1096/fj.06-6319com. [DOI] [PubMed] [Google Scholar]
- 174.Forman D., de Martel C., Lacey C.J., Soerjomataram I., Lortet-Tieulent J., Bruni L., Vignat J., Ferlay J., Bray F., Plummer M., et al. Global burden of human papillomavirus and related diseases. Vaccine. 2012;30(Suppl. 5):F12–F23. doi: 10.1016/j.vaccine.2012.07.055. [DOI] [PubMed] [Google Scholar]
- 175.Evander M., Edlund K., Gustafsson A., Jonsson M., Karlsson R., Rylander E., Wadell G. Human papillomavirus infection is transient in young women: A population-based cohort study. J. Infect. Dis. 1995;171:1026–1030. doi: 10.1093/infdis/171.4.1026. [DOI] [PubMed] [Google Scholar]
- 176.Winer R.L., Hughes J.P., Feng Q., Xi L.F., Cherne S., O’Reilly S., Kiviat N.B., Koutsky L.A. Early natural history of incident, type-specific human papillomavirus infections in newly sexually active young women. Cancer Epidemiol. Biomark. Prev. 2011;20:699–707. doi: 10.1158/1055-9965.EPI-10-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Jabbar S.F., Abrams L., Glick A., Lambert P.F. Persistence of high-grade cervical dysplasia and cervical cancer requires the continuous expression of the human papillomavirus type 16 E7 oncogene. Cancer Res. 2009;69:4407–4414. doi: 10.1158/0008-5472.CAN-09-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Jabbar S.F., Park S., Schweizer J., Berard-Bergery M., Pitot H.C., Lee D., Lambert P.F. Cervical cancers require the continuous expression of the human papillomavirus type 16 E7 oncoprotein even in the presence of the viral E6 oncoprotein. Cancer Res. 2012;72:4008–4016. doi: 10.1158/0008-5472.CAN-11-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Stanley M.A. Epithelial cell responses to infection with human papillomavirus. Clin. Microbiol. Rev. 2012;25:215–222. doi: 10.1128/CMR.05028-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Au Yeung C.L., Tsang W.P., Tsang T.Y., Co N.N., Yau P.L., Kwok T.T. HPV-16 E6 upregulation of DNMT1 through repression of tumor suppressor p53. Oncol. Rep. 2010;24:1599–1604. doi: 10.3892/or_00001023. [DOI] [PubMed] [Google Scholar]
- 181.Burgers W.A., Blanchon L., Pradhan S., de Launoit Y., Kouzarides T., Fuks F. Viral oncoproteins target the DNA methyltransferases. Oncogene. 2007;26:1650–1655. doi: 10.1038/sj.onc.1209950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Westrich J.A., Warren C.J., Pyeon D. Evasion of host immune defenses by human papillomavirus. Virus Res. 2017;231:21–33. doi: 10.1016/j.virusres.2016.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Biktasova A.K., Hajek M., Sewell A.B., Gary C.S., Bellinger G., Deshpande H., Bhatia A.K., Burtness B.A., Judson B.L., Mehra S., et al. Demethylation therapy as a targeted treatment for human papilloma virus-associated head and neck cancer. Clin. Cancer Res. 2017;23:7276–7287. doi: 10.1158/1078-0432.CCR-17-1438. [DOI] [PubMed] [Google Scholar]
- 184.LaFleur D.W., Nardelli B., Tsareva T., Mather D., Feng P., Semenuk M., Taylor K., Buergin M., Chinchilla D., Roshke V., et al. Interferon-kappa, a novel type I interferon expressed in human keratinocytes. J. Biol. Chem. 2001;276:39765–39771. doi: 10.1074/jbc.M102502200. [DOI] [PubMed] [Google Scholar]
- 185.Habiger C., Jager G., Walter M., Iftner T., Stubenrauch F. Interferon Kappa Inhibits Human Papillomavirus 31 Transcription by Inducing Sp100 Proteins. J. Virol. 2015;90:694–704. doi: 10.1128/JVI.02137-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Pyeon D., Newton M.A., Lambert P.F., den Boon J.A., Sengupta S., Marsit C.J., Woodworth C.D., Connor J.P., Haugen T.H., Smith E.M., et al. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res. 2007;67:4605–4619. doi: 10.1158/0008-5472.CAN-06-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.den Boon J.A., Pyeon D., Wang S.S., Horswill M., Schiffman M., Sherman M., Zuna R.E., Wang Z., Hewitt S.M., Pearson R., et al. Molecular transitions from papillomavirus infection to cervical precancer and cancer: Role of stromal estrogen receptor signaling. Proc. Natl. Acad. Sci. USA. 2015;112:E3255–E3264. doi: 10.1073/pnas.1509322112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Tindle R.W. Immune evasion in human papillomavirus-associated cervical cancer. Nat. Rev. Cancer. 2002;2:59–65. doi: 10.1038/nrc700. [DOI] [PubMed] [Google Scholar]
- 189.Shellenberger T.D., Wang M., Gujrati M., Jayakumar A., Strieter R.M., Burdick M.D., Ioannides C.G., Efferson C.L., El-Naggar A.K., Roberts D., et al. BRAK/CXCL14 is a potent inhibitor of angiogenesis and a chemotactic factor for immature dendritic cells. Cancer Res. 2004;64:8262–8270. doi: 10.1158/0008-5472.CAN-04-2056. [DOI] [PubMed] [Google Scholar]
- 190.Starnes T., Rasila K.K., Robertson M.J., Brahmi Z., Dahl R., Christopherson K., Hromas R. The chemokine CXCL14 (BRAK) stimulates activated NK cell migration: Implications for the downregulation of CXCL14 in malignancy. Exp. Hematol. 2006;34:1101–1105. doi: 10.1016/j.exphem.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 191.Ozawa S., Kato Y., Komori R., Maehata Y., Kubota E., Hata R. BRAK/CXCL14 expression suppresses tumor growth in vivo in human oral carcinoma cells. Biochem. Biophys. Res. Commun. 2006;348:406–412. doi: 10.1016/j.bbrc.2006.07.070. [DOI] [PubMed] [Google Scholar]
- 192.Tessema M., Klinge D.M., Yingling C.M., Do K., Van Neste L., Belinsky S.A. Re-expression of CXCL14, a common target for epigenetic silencing in lung cancer, induces tumor necrosis. Oncogene. 2010;29:5159–5170. doi: 10.1038/onc.2010.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ito S., Ozawa S., Ikoma T., Yajima N., Kiyono T., Hata R. Expression of a chemokine BRAK/CXCL14 in oral floor carcinoma cells reduces the settlement rate of the cells and suppresses their proliferation in vivo. Biomed. Res. 2010;31:199–206. doi: 10.2220/biomedres.31.199. [DOI] [PubMed] [Google Scholar]
- 194.Hata R., Izukuri K., Kato Y., Sasaki S., Mukaida N., Maehata Y., Miyamoto C., Akasaka T., Yang X., Nagashima Y., et al. Suppressed rate of carcinogenesis and decreases in tumour volume and lung metastasis in CXCL14/BRAK transgenic mice. Sci. Rep. 2015;5:9083. doi: 10.1038/srep09083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wang W., Huang P., Zhang L., Wei J., Xie Q., Sun Q., Zhou X., Xie H., Zhou L., Zheng S. Antitumor efficacy of C-X-C motif chemokine ligand 14 in hepatocellular carcinoma in vitro and in vivo. Cancer Sci. 2013;104:1523–1531. doi: 10.1111/cas.12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Frederick M.J., Henderson Y., Xu X., Deavers M.T., Sahin A.A., Wu H., Lewis D.E., El-Naggar A.K., Clayman G.L. In vivo expression of the novel CXC chemokine BRAK in normal and cancerous human tissue. Am. J. Pathol. 2000;156:1937–1950. doi: 10.1016/S0002-9440(10)65067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Shurin G.V., Ferris R.L., Tourkova I.L., Perez L., Lokshin A., Balkir L., Collins B., Chatta G.S., Shurin M.R. Loss of new chemokine CXCL14 in tumor tissue is associated with low infiltration by dendritic cells (DC), while restoration of human CXCL14 expression in tumor cells causes attraction of DC both in vitro and in vivo. J. Immunol. 2005;174:5490–5498. doi: 10.4049/jimmunol.174.9.5490. [DOI] [PubMed] [Google Scholar]
- 198.Niu J., Lin Y., Liu P., Yu Y., Su C., Wang X. Microarray analysis on the lncRNA expression profile in male hepatocelluar carcinoma patients with chronic hepatitis B virus infection. Oncotarget. 2016;7:76169–76180. doi: 10.18632/oncotarget.12732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lu J., Chatterjee M., Schmid H., Beck S., Gawaz M. CXCL14 as an emerging immune and inflammatory modulator. J. Inflamm. 2016;13:1. doi: 10.1186/s12950-015-0109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Buck C.B., Day P.M., Thompson C.D., Lubkowski J., Lu W., Lowy D.R., Schiller J.T. Human alpha-defensins block papillomavirus infection. Proc. Natl. Acad. Sci. USA. 2006;103:1516–1521. doi: 10.1073/pnas.0508033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wiens M.E., Smith J.G. Alpha-defensin HD5 inhibits furin cleavage of human papillomavirus 16 L2 to block infection. J. Virol. 2015;89:2866–2874. doi: 10.1128/JVI.02901-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Joosten S.A., Sullivan L.C., Ottenhoff T.H. Characteristics of HLA-E Restricted T-Cell Responses and Their Role in Infectious Diseases. J. Immunol. Res. 2016;2016:2695396. doi: 10.1155/2016/2695396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Moretta L., Romagnani C., Pietra G., Moretta A., Mingari M.C. NK-CTLs, a novel HLA-E-restricted T-cell subset. Trends Immunol. 2003;24:136–143. doi: 10.1016/S1471-4906(03)00031-0. [DOI] [PubMed] [Google Scholar]
- 204.Davis Z.B., Cogswell A., Scott H., Mertsching A., Boucau J., Wambua D., Le Gall S., Planelles V., Campbell K.S., Barker E. A Conserved HIV-1-Derived Peptide Presented by HLA-E Renders Infected T-cells Highly Susceptible to Attack by NKG2A/CD94-Bearing Natural Killer Cells. PLoS Pathog. 2016;12:e1005421. doi: 10.1371/journal.ppat.1005421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Vertino P.M., Issa J.P., Pereira-Smith O.M., Baylin S.B. Stabilization of DNA methyltransferase levels and CpG island hypermethylation precede SV40-induced immortalization of human fibroblasts. Cell Growth Differ. 1994;5:1395–1402. [PubMed] [Google Scholar]
- 206.El-Araby A.M., Fouad A.A., Hanbal A.M., Abdelwahab S.M., Qassem O.M., El-Araby M.E. Epigenetic Pathways of Oncogenic Viruses: Therapeutic Promises. Arch. Pharm. 2016;349:73–90. doi: 10.1002/ardp.201500375. [DOI] [PubMed] [Google Scholar]