We assessed and found cross-protection between the influenza pandemic and 2 seasonal viruses that circulated in Singapore. Age-related differences in disease severity between the subtypes were investigated, and we demonstrate birth cohort effects consistent with the theory of antigenic sin.
Keywords: H1N1pdm09, seroepidemiology, cross-protection, severity, birth cohort effect
Abstract
Background
After 2009, pandemic influenza A(H1N1) [A(H1N1)pdm09] cocirculated with A(H3N2) and B in Singapore.
Methods
A cohort of 760 participants contributed demographic data and up to 4 blood samples each from October 2009 to September 2010. We compared epidemiology of the 3 subtypes and investigated evidence for heterotypic immunity through multivariable logistic regression using a generalized estimating equation. To examine age-related differences in severity between subtypes, we used LOESS (locally weighted smoothing) plots of hospitalization to infection ratios and explored birth cohort effects referencing the pandemic years (1957; 1968).
Results
Having more household members aged 5–19 years and frequent public transport use increased risk of infection, while preexisting antibodies against the same subtype (odds ratio [OR], 0.61; P = .002) and previous influenza infection against heterotypic infections (OR, 0.32; P = .045) were protective. A(H1N1)pdm09 severity peaked in those born around 1957, while A(H3N2) severity was least in the youngest individuals and increased until it surpassed A(H1N1)pdm09 in those born in 1952 or earlier. Further analysis showed that severity of A(H1N1)pdm09 was less than that for A(H3N2) in those born in 1956 or earlier (P = .021) and vice versa for those born in 1968 or later (P < .001), with no difference in those born between 1957 and 1967 (P = .632).
Conclusions
Our findings suggest that childhood exposures had long-term impact on immune responses consistent with the theory of antigenic sin. This, plus observations on short-term cross-protection, have implications for vaccination and influenza epidemic and pandemic mitigation strategies.
A novel pandemic influenza A virus of swine origin emerged in the United States and Mexico in early 2009 and spread globally [1]. Singapore detected its first case in May 2009 [2]. The initial epidemic of influenza A(H1N1)pdm09 peaked in early August 2009 [3], subsided by September [4], and was followed by 2 additional epidemics that overlapped temporally with an epidemic of influenza A(H3N2) and continuous influenza B circulation [5].
The A(H1N1)pdm09 pandemic provided a rare opportunity to compare the epidemiology between emergent and endemic influenza viruses and to investigate potential interactions. The influenza pandemic viruses of 1957 and 1968 replaced their seasonal influenza A counterparts; circulating A(H1N1) was displaced by the emergence of A/Singapore/1/57(H2N2), which in turn was displaced by A/HongKong/1/68(H3N2) [6]. After 2009, A(H1N1)pdm09 pandemic virus displaced the circulating seasonal A(H1N1) strain but not the A(H3N2) subtype. These ecological observations suggest that infection may confer limited cross-protection from other subtypes. However, in addition to 2 studies during the 1957 pandemic that reported how prior infection with seasonal influenza subtypes correlated with protection against the pandemic influenza A/Singapore/1/57(H2N2) virus [7, 8], the strength of homo- or heterotypic cross-protective effects remains poorly characterized. Also, past influenza pandemics have been associated with an “age shift” in mortality patterns relative to seasonal influenza [9, 10]. During the 2009 pandemic, there were more hospitalizations in younger age groups than for seasonal influenza and vice versa for the elderly [11, 12], but objective assessments are lacking as to whether these reflect differences in age-specific incidence rates or underlying differences in age-related severity.
Using a sero-incidence cohort, we investigated risk factors for infection with the predominant circulating influenza A(H1N1)pdm09, A(H3N2), and B strains after the initial epidemic of A(H1N1)pdm09 in Singapore and sought evidence for possible cross-protection. We also combined sero-incidence with hospitalization data to explore age-related differences in the severity of infection between influenza subtypes.
MATERIALS AND METHODS
Overview of Study Design
Participants of the Multi-ethnic Cohort study hosted at the National University of Singapore (NUS) [13] aged 21–75 years were invited to participate in a study of the influenza A(H1N1)pdm09 pandemic [14]. Consenting participants were re-enrolled, and follow-up continued during the post-pandemic period in late 2009 to 2010 [15]. The NUS ethics review board approved the study.
Data and Sample Collection, and Laboratory Testing
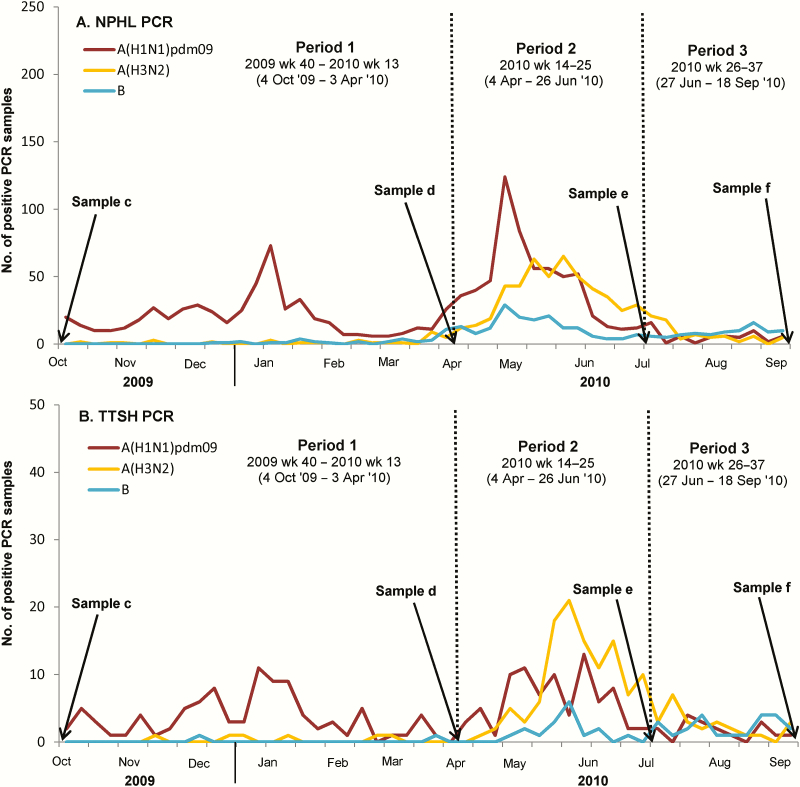
Each consenting participant contributed up to 6 blood samples, samples a to f, taken at various time points up to 27 September 2010 (Figure 1A); this analysis focuses on the latter 4 samples. Demographic, household, and lifestyle data were collected at enrollment in 2009, with additional questionnaires coinciding with later samples to detect interval influenza vaccination events.
Figure 1.
A, Time course of serological analyses and polymerase chain reaction (PCR)–positive influenza cases detected by the National Public Health Laboratory (NPHL) surveillance system. B, Admissions to Tan Tock Seng Hospital (TTSH). Line graph denotes the weekly number of A(H1N1)pdm09 (brown–red), A(H3N2) (yellow), and influenza B (blue) PCR-positive cases among influenza-like illness samples submitted by general practitioners and polyclinics to the NPHL or from TTSH hospital. Sample a: 29 June 2005–27 June 2009; mostly banked samples from prior participation in the multi-ethnic cohort; not shown in figure. Sample b: 20 August 2009–29 August 2009; 3–4 weeks after the first peak of the pandemic; not shown in figure. Sample c: 6 October 2009–11 October 2009; 3–4 weeks after the first period of H1N1pdm09 epidemic activity had subsided. Sample d: 8 April 2010–22 April 2010; before the month of May, the most common influenza epidemic period in Singapore, and after the second most common epidemic period (typically between December and February [26]). Sample e: 2 July 2010–8 July 2010; 10–12 weeks after sample d. Sample f: 19 September 2010–27 September 2010; 10–12 weeks after sample e. Abbreviations: PHL, National Public Health Laboratory; PCR, polymerase chain reaction; TTSH, Tan Tock Seng Hospital.
Participants contributed up to 10 mL of venous blood at each time point. Hemagglutination inhibition (HI) assays were performed following standard protocols at the World Health Organization (WHO) Collaborating Centre for Reference and Research on Influenza in Melbourne, Australia, as previously described [16, 17]. HI titers were expressed as the reciprocal of the highest dilution of serum where hemagglutination was prevented (from 1:10 to a maximum of 1:1280) and analyzed on a log scale (with titers <10 and ≥1280 assigned a value of 5 and 1280, respectively).
To detect infection, we used the following strains, which corresponded to those in the Southern Hemisphere 2010 vaccine [18]: A/California/7/2009(H1N1), A/Wisconsin/15/2009(H3N2), an A/Perth/16/2009(H3N2)-like virus, and B/Brisbane/60/2008 (B/Victoria/2/87-lineage) [19].
Data Analyses
Though influenza A and B are technically different influenza types, for convenience we subsequently reference A(H1N1)pdm09 and A(H3N2) and B as different influenza subtypes, with cross-protection between different subtypes and protection against the same subtype as heterotypic and homotypic protection, respectively. We defined a 4-fold or greater increase in HI antibody titers as seroconversion to the corresponding influenza subtype between any successive pair of available samples. When determining infection, observations from specific intervals with self-reported influenza vaccination were excluded, since vaccination (which in Singapore included the A/California/7/2009(H1N1pdm09) strain after October 2009) would potentially induce seroconversion indistinguishable from infection.
Analysis focused on seroconversion events between samples collected in 2010 (d to f) and the last available sample of 2009 (either b or c). Seroconversions between samples a to c (which overlaps with the initial epidemic of A(H1N1)pdm09 in Singapore) were used mainly to assess prior infection with A(H1N1)pdm09. Since some factors of interest (eg, prior infection with the same and different subtypes, antibody titers) could change over the course of the study, we defined 3 time periods during which each participant could be observed for seroconversion events: period 1 between samples c to d, period 2 from d to e, and period 3 from e to f (Figure 1); the earlier of each pair demarcating a period was defined as the antecedent sample. This was typically sample c for period 1 (except in 28 participants who were missing sample c where sample b was used) and samples d and e for periods 2 and 3, respectively. A participant could therefore be observed for the binary outcome of serologically detected infection to each of the 3 subtypes for each period.
In the stratified analysis by subtype, the unit of analysis was the participant period, with each participant contributing up to 3 observations. We also performed an analysis that combined observations for all 3 subtypes, where each participant contributed up to 9 participant-period-subtype observations. To account for potential clustering of observations from the same participant, we used a logistic model with generalized estimating equation (GEE). Using univariable and multivariable analyses, we assessed associations between serologically detected infection and various factors, including demographic, lifestyle, and household characteristics, and for each period of assessment, self-reported vaccination, antecedent antibody titer to the corresponding subtype, and history of infection (in the period immediately preceding the one assessed, or 2 or more periods ago). In some instances, infection history could not be assessed because vaccination occurred in a preceding period. Also, because of the use of banked specimens for sample a (mostly from before June 2009), HI assays between samples a and c could not be used to accurately assess A(H3N2) and influenza B infections immediately preceding period 1 observations. These observations were, for convenience, assumed to have no infections in the preceding period in the multivariable analysis; additional sensitivity analysis with these coded as a category denoting missing observations gave similar results (see Supplementary Table 2).
Potential interactions between time period and other variables were considered in subtype-specific analyses and also interactions that indicated differences in risk factors between subtypes significant at P < .10 (using the Wald test). In all other instances, statistical significance was defined as P < .05.
Hospitalization-infection ratios (ie, number of hospitalizations per 1000 infections) were used as a proxy of disease severity, with infections in the community estimated from multiplying cohort sero-incidence with the 2010 Singapore census population, and hospitalization burden extrapolated from all polymerase chain reaction (PCR)-positive influenza admissions during the study period to Tan Tock Seng Hospital (TTSH), a large acute tertiary care general hospital in central Singapore (covering 16% of adult inpatients; Figure 1B). We plotted incidence, hospitalization burden, and hospitalization-infection ratios for different influenza subtypes using smoothed LOESS (locally weighted smoothing) curves (with a span of 0.29) to investigate age-dependent variations. For selected birth cohorts, subtype-specific hospitalization-infection ratios were also calculated using both crude sero-incidence and adjusted sero-incidence (from the logistic GEE model, see Supplementary Material).
We used R Statistical Software v3.2.3 (R Foundation for Statistical Computing, Vienna, Austria) for all analyses [20].
RESULTS
We enrolled 894 individuals aged 21 to 75 years from 1296 randomly selected participants (69%) of the Multi-Ethnic Cohort study. From this initial study, 760 were re-enrolled, and 675 contributed 1 or more paired serum samples for assessment of seroconversion events. We assigned 652 paired samples to period 1, 623 to period 2, and 556 to period 3. Period 1 included 1 observation that was missing sample d and another that was missing both d and e, where seroconversion was assessed between c to e and c to f, respectively. Period 3 included 26 observations missing sample e, with seroconversion assessed between d to f. Period assignments for these observations with skipped samples were based on the midpoint between the earlier and latter follow-up dates for the sample pairs involved.
There were no significant changes in demographic and household factors across the periods (Table 1). After excluding self-reported influenza vaccination, we identified 82, 44, and 33 serologically detected infections with influenza A(H1N1)pdm09, A(H3N2), and A(H3N2)B, respectively, in 149 individuals, 9 of whom seroconverted to 2 or more subtypes across the study period. Of these, 5 individuals seroconverted to 2 or more subtypes in the same period and were excluded from all subsequent analyses (see Discussion).
Table 1.
Comparison of Study Variables in Study Population at Enrollment and Over Successive Time Periods
| Study Variable | No. of Participants | |||
|---|---|---|---|---|
| Enrollment, n = 760 | Period 1, n = 652 | Period 2, n = 623 | Period 3, n = 556 | |
| Age as of 1 Jan 2010, y | ||||
| 20–24 | 65 (8.6) | 48 (7.4) | 48 (7.7) | 33 (5.9) |
| 25–34 | 117 (15.4) | 100 (15.3) | 92 (14.8) | 83 (14.9) |
| 35–44 | 198 (26.1) | 172 (26.4) | 164 (26.3) | 154 (27.7) |
| 45–54 | 248 (32.6) | 212 (32.5) | 206 (33.1) | 190 (34.2) |
| 55–64 | 109 (14.3) | 99 (15.2) | 92 (14.8) | 80 (14.4) |
| ≥65 | 23 (3.0) | 21 (3.2) | 21 (3.4) | 16 (2.9) |
| Gender | ||||
| Male | 311 (40.9) | 259 (39.7) | 249 (40.0) | 216 (38.9) |
| Female | 449 (59.1) | 393 (60.3) | 374 (60.0) | 340 (61.2) |
| Race | ||||
| Chinese | 89 (11.7) | 78 (12.0) | 74 (11.9) | 65 (11.7) |
| Malay | 346 (45.5) | 299 (45.9) | 294 (47.2) | 265 (47.7) |
| Indian and others | 325 (42.8) | 275 (42.2) | 255 (40.9) | 226 (40.7) |
| Housing type | ||||
| 3 rooms or smaller public housing | 187 (24.6) | 158 (24.2) | 146 (23.4) | 133 (23.9) |
| 4 rooms public housing | 325 (42.8) | 285 (43.7) | 273 (43.8) | 245 (44.1) |
| 5 rooms public housing or private housing | 248 (32.6) | 209 (32.1) | 204 (32.7) | 178 (32.0) |
| Working | ||||
| No | 287 (37.8) | 250 (38.3) | 241 (38.7) | 214 (38.5) |
| Yes | 473 (62.2) | 402 (61.7) | 382 (61.3) | 342 (61.5) |
| Frequency of public transport use | ||||
| Low | 260 (34.2) | 223 (34.2) | 217 (34.8) | 190 (34.2) |
| High | 500 (65.8) | 429 (65.8) | 406 (65.2) | 366 (65.8) |
| No. of household members, mean (range) | ||||
| <5 years old | 0.28 (0–3) | 0.28 (0–3) | 0.28 (0–3) | 0.27 (0–3) |
| 5–19 years old | 1.26 (0–6) | 1.25 (0–5) | 1.25 (0–5) | 1.28 (0–5) |
| >19 years old | 3.14 (1–9) | 3.11 (1–9) | 3.12 (1–9) | 3.10 (1–9) |
| Self-reported vaccination | ||||
| No | N.A. | 636 (97.6) | 577 (92.6) | 513 (92.3) |
| Vaccinated ≥2 periods ago | N.A. | 0 (0.0) | 10 (1.6) | 36 (6.5) |
| Vaccinated in previous period | N.A. | 16 (2.5) | 36 (5.8) | 7 (1.3) |
| Seroconversion eventsa | ||||
| Influenza A(H1N1)pdm09 | N.A. | 56 (8.6) | 34 (5.5) | 14 (2.5) |
| Influenza A(H3N2) | N.A. | 10 (1.5) | 31 (5.0) | 12 (2.2) |
| Influenza B | N.A. | 19 (2.9) | 14 (2.3) | 12 (2.2) |
| Serologic evidence of infectionb | ||||
| Influenza A(H1N1)pdm09 | N.A. | 40 (6.5) | 30 (4.9) | 12 (2.2) |
| Influenza A(H3N2) | N.A. | 7 (1.1) | 27 (4.4) | 10 (1.8) |
| Influenza B | N.A. | 12 (2.0) | 10 (1.6) | 11 (2.0) |
Unless otherwise stated, cells give number of observations with column percentages in brackets.
aIncludes seroconversions that may be due to influenza vaccination.
bExcludes observations with self-reported history of vaccination in that period, with the number of observations for periods 1, 2, and 3 being 615, 616, and 553, respectively.
Compared with other indicators of influenza activity, sero-incidence correlated well with the number of PCR-confirmed influenza infections in individuals aged 20–74 years from primary care in surveillance data collected by the National Public Health Laboratory (Pearson coefficient correlation, R = 0.920; P < .001; see Supplementary Figure 1), as well as hospital admissions (R = 0.900; P = .001).
Decreased Risk of Reinfection With an Alternative Subtype Among Those Previously Infected
Among participants with an influenza infection with the same or an alternate subtype in the immediately preceding period, the risks of infection with A(H1N1)pdm09, A(H3N2), and influenza B (Supplementary Figure 2A–2C, respectively) were generally lower than the cohort average in each period as well as when averaged across the entire study period (see Supplementary Table 1 for calculation of infection risks). There were only 3 exceptions: 1 of 83 participants with prior A(H1N1)pdm09 reinfected with A(H3N2) in period 1; 1 of 10 participants with prior influenza B reinfected with A(H3N2) in period 2; and 1 of 21 participants with prior A(H3N2) reinfected with influenza B in period 3 (Supplementary Figure 1A–1C, respectively); the denominators were relatively small in the latter 2 instances. After pooling observations for all subtypes across all 3 periods (Figure 2), the average risk of infection per time period by a single subtype was 2.8%. However, there was a significantly lower risk of reinfection in those with previous A(H1N1)pdm09 infection (0.7%, P = .012), previous infection with an alternative subtype (0.8%, P = .024), and previous infection with any subtype (0.9%, P = .009). The risk of infection among those previously infected with the same subtype was also lower at 1.1%, although not significantly so (P = .167; Figure 2).
Figure 2.
Risk of infection with influenza A(H1N1)pdm09 (A), A(H3N2) (B), influenza B (C), and all subtypes (D) in each assessed period among participants who were infected in the previous period. Red, yellow, and blue triangles denote percentage of serologically infected participants in each assessed period who were serologically infected in the previous period with A(H1N1)pdm09, A(H3N2), influenza B, respectively. Black diamonds, orange squares, and purple circles give averaged percentages for any subtype, an alternative subtype (heterotypic), and the same subtype (homotypic), respectively. Error bars (vertical lines) are 95% confidence intervals (CIs). Green bars with a horizontal mid-bar indicate average risk of a serologically detected infection by 1 subtype, with 95% CIs for all observations from participants with or without previous infection.
Risk Factors for Infection
In multivariable analyses, antibody titers to the same subtype in the antecedent sample were inversely associated with seroconversion to all subtypes (OR, 0.61; 95% confidence interval [CI], 0.44 to 0.83; Figure 3A), A(H1N1)pdm09 (OR, 0.53; 95% CI, 0.29 to 0.97; Figure 3B), and A(H3N2) (OR, 0.53; 95% CI, 0.32 to 0.89; Figure 3C). Figure 3A also shows that infection with an alternative subtype in the preceding period reduced the odds of infection (OR, 0.32; 95% CI, 0.11 to 0.98); the protective effect was weaker (and nonsignificant) if prior infection occurred ≥2 periods ago (OR, 0.77; 95% CI, 0.34 to 1.76). Having more household members aged 5–19 years significantly increased risk of infection to all subtypes (OR, 1.24; 95% CI, 1.04 to 1.47), A(H3N2) (OR, 1.42; 95% CI, 1.08 to 1.86), and influenza B (OR, 1.39; 95% CI, 1.01 to 1.91; Figure 3D), and odds of infection increased with more household members aged <5 years for A(H3N2) (OR, 1.90; 95% CI, 1.25 to 2.88). Figure 3A also shows significant associations with age (45–54 years: OR, 0.42; 95% CI, 0.22 to 0.81) and ethnicity (Malay: OR, 2.67; 95% CI, 1.28 to 5.57; Indian and others: OR, 2.88; 95% CI, 1.38 to 5.99), and significantly increased risk of infection with more frequent public transport use (OR, 1.46; 95% CI, 1.02 to 2.10).
Figure 3.
Univariable and multivariable analyses of possible risk factors for serologically detected infection to all 3 subtypes combined, A(H1N1)pdm09, A(H3N2), and influenza B (panels A to D, respectively). The odds ratio for serologically detected infection on univariable analyses is represented by blue diamonds, and the odds ratio for serologically detected infection on multivariable analyses is denoted by green circles, with error bars for 95% confidence intervals. Closed symbols and open symbols denote results significant and nonsignificant at P < .05, respectively. Multivariable models include all variables shown in the figure, except for a few variables including prior homotypic infection and prior heterotypic infection across graphs B to D, where it is noncalculable. For panel A, the interaction terms included in the model are (1) subtype × working (2), period × number of household members aged 5 to 19 years (3), subtype × number of household members aged 5 to 19 years, and (4) period × subtype. For panel B, the interaction terms include (1) period × working and (2) period × number of household members aged 5 to 19 years. For panel D, the interaction terms include (1) period × number of household members aged 5 to 19 years. No interaction terms were included for the model in panel C. Abbreviation: HHM, household member.
Significant Birth Cohort Effects Suggesting Long-term Implications of Early Life Exposure
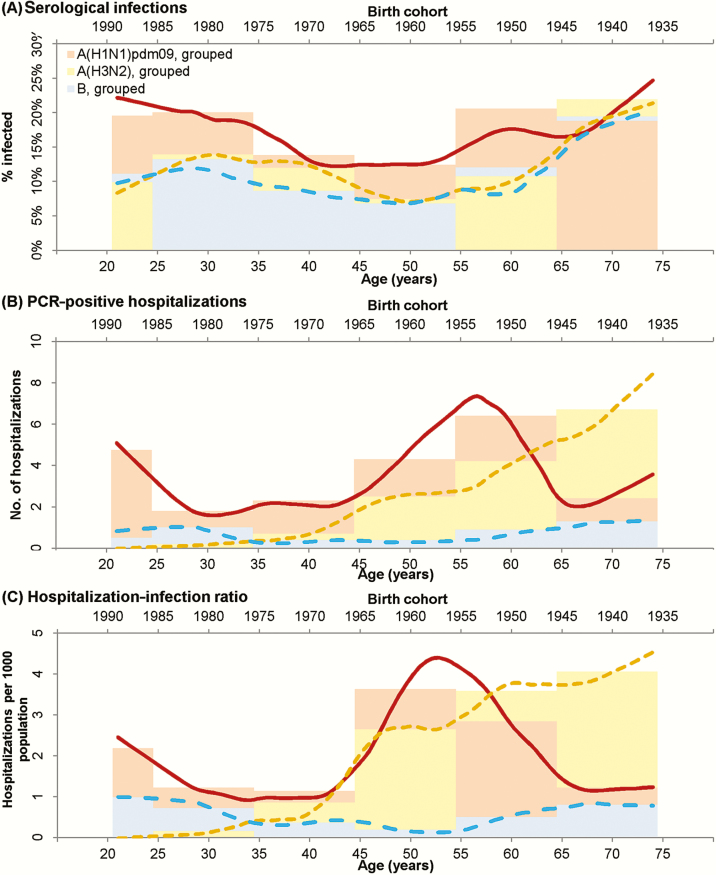
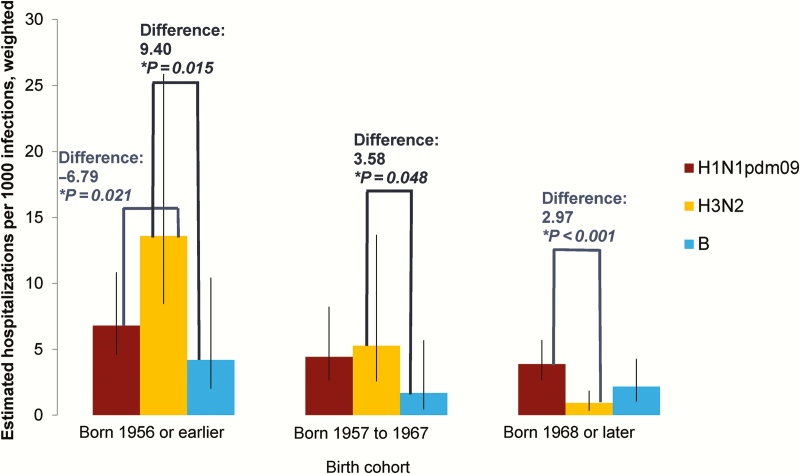
Cumulative infection rates over the study period were not markedly different by age (Figure 4A). In contrast, hospitalization with PCR-confirmed infection was more common for A(H1N1)pdm09 than A(H3N2) in younger age groups and vice versa for adults aged approximately 60 and older (Figure 4B). However, the hospitalization-infection ratios (Figure 4C) cross at various points for A(H1N1)pdm09 and A(H3N2), suggesting complex birth cohort effects. A(H3N2) was more severe than A(H1N1)pdm09 in those born before the mid-1950s, while A(H1N1)pdm09 was more severe in those born after the late 1960s. A(H1N1)pdm09 was also more severe in those born between 1952 and 1964. Since the crossing of the lines for hospitalization-infection ratios coincided approximately with the periods when the A(H2N2) and A(H3N2) pandemic viruses started circulating in Singapore (1957 and 1968, respectively) [21, 22], we referenced these calendar years in exploring birth cohort effects: born in 1956 or earlier, born between 1957 and 1967, and born in 1968 or later (Figure 5). For A(H1N1)pdm09, hospitalization-infection ratios were 6.81 (95% CI, 4.58 to 10.85), 4.45 (95% CI, 2.64 to 8.24), and 3.89 (95% CI, 2.65 to 5.72), respectively, in the 3 birth cohorts; for A(H3N2), the severity ratios were 13.60 (95% CI, 8.45 to 25.88), 5.29 (95% CI, 2.57 to 13.68), and 0.92 (95% CI, 0.34 to 1.87), respectively. Compared to A(H3N2), A(H1N1)pdm09 was less severe in those born in 1956 or earlier (difference = –6.79; 95% CI, –201.3 to 1.24) and vice versa for those born in 1968 or later (difference = 2.97; 95% CI, 1.64 to 8.35), with no difference in those born between 1957 and 1967 (difference = –0.84; 95% CI: –9.22 to 3.80). Repeating the analysis using coefficients generated from the logistic GEE model to estimate sero-incidence produced higher severity ratios (Supplementary Figure 3) and increased the birth cohort related differences in severity between A(H1N1)pdm09 and A(H3N2).
Figure 4.
Rates of influenza infection by age in the serological cohort (A), number of polymerase chain reaction–positive influenza admissions to Tan Tock Seng Hospital by age (B), and severity of influenza infection (ie, hospitalization-infection ratio) by age (C). Brown–red, yellow, and blue lines show smoothed results (LOESS [locally weighted smoothing] curves, span = 0.25) for A(H1N1)pdm09, A(H3N2), and influenza B, respectively. Light pink, light yellow, and light blue bars illustrate the binned results corresponding to the 6 age groups used in the generalized estimating equation model for A(H1N1)pdm09, A(H3N2), and influenza B, respectively. The infection rates and hospitalization numbers were adjusted to estimate incidence and all hospitalizations for influenza in the community. Abbreviation: PCR, polymerase chain reaction.
Figure 5.
Relative severities of influenza subtypes by selected birth cohorts using crude serological data (ie, seroconversion rates in the cohort, adjusted to reflect incidence in community). Version using adjusted serological data (ie, estimated infection rates in the community using weighted coefficients generated from the logistic generalized estimating equation model) is given in Supplementary Figure 3.
DISCUSSION
Our serological cohort study from late 2009 to 2010, when the pandemic and 2 seasonal influenza strains cocirculated in Singapore, provided a unique opportunity to assess risk factors for infection and epidemiological evidence for heterotypic immunity and to investigate birth cohort effects on their relative severity.
Having household members in pediatric age groups increased the risk of infection in adults. This influence of children on influenza transmission has been previously demonstrated in Singapore and elsewhere [23–25]. We also replicated a previous finding [26] of an increased risk of infection with more frequent public transport use, when pooling data for all 3 subtypes. Most interestingly, we documented individual-level protection from prior influenza infection in the immediate preceding period. The effect of prior infection on odds of homotypic reinfection was statistically not significant, possibly due to the small number of observed reinfection events, and not unexpectedly weakened after we adjusted for antecedent titers. However, there were significantly decreased odds of infection with an alternative subtype, with protection weakening where infection occurred 2 or more periods ago. This duration of heterotypic protection is consistent with that inferred from a study modeling influenza seasonality [27]. Mechanisms of heterotypic protection may include transient behavioral changes after symptomatic influenza infection (eg, more frequent hand washing), immunological protection through cross-reactive cytotoxic T lymphocytes [28–30], cross-neutralizing antibodies against conserved epitopes [31, 32], and innate immune pathways [33, 34]. Findings from this study support the need for detailed investigation of these mechanisms.
Our other key finding pertains to the complex effects of birth cohort on the relative severity of influenza subtypes. While age-related differences in incidence were unremarkable, those born in 1956 or earlier were protected against severe infection with A(H1N1)pdm09, those born in 1968 or later were protected against A(H3N2), with little difference in severity between the 2 influenza A subtypes for individuals born between 1957 and 1967. Early life exposure to an antigenically related virus (ie, the A(H1N1) strain that circulated after the 1918 pandemic and prior to the 1957 A(H2N2) pandemic) may mitigate against severity for the A(H1N1)pdm09 strain in older individuals. Similarly, exposure to the circulating A(H3N2) strain after the 1968 pandemic may protect those born in 1968 and later against severe infection by the contemporary A(H3N2) virus. One study reported how antibody titers in an individual are often highest for influenza viruses that are circulating when individuals were approximately aged 6 years, the time frame of first infection [35]. Our findings on relative protection against severe infection for A(H1N1)pdm09 in the elderly and A(H3N2) in the young thus supports the theory of “original antigenic sin.” This postulates that immunologic memory from the first exposure to an influenza A virus in childhood influences immune responses to subsequent infection and provides better protection against infection with the same subtype, even in older age [36]. Such birth cohort effects have implications for vaccination and surveillance strategies for future influenza pandemics. Prioritizing groups to vaccinate during a pandemic may need to take into account the likely influenza strains and subtypes that different birth cohorts were exposed to in the event when vaccine stocks are limited. Birth cohort effects may also help to explain some of the variability in influenza vaccine effectiveness, including the decline in subtype-specific vaccine effectiveness over the course of a season [37]. They also suggest that older adults may benefit from vaccines that boost influenza A(H3N2) immunity specifically, rather than the current strategy of multivalent vaccination.
Our study has several limitations. Defining seroconversion as a 4-fold or greater increase in titers is widely accepted but has imperfect sensitivity [38]. With a longer sampling interval in period 1, infections may also have been missed, as HI titers can wane rapidly [39]. Furthermore, we acknowledge that only 9 of 760 participants seroconverted to 2 or more subtypes across all 3 periods. We also discarded observations from 5 participants where seroconversions to more than 1 subtype occurred within the same period, as serological investigations are unable to determine the order of subtype infection, or if these could have reflected a cross-reactive immune response. They could have also been due to undisclosed influenza vaccination. Notably, even if all 5 instances were due to a cross-reactive immune response, the infrequency of such events relative to all seroconversions detected suggests the HI assay was reasonably specific. Since these observations were excluded, cross-reactive antibodies are not a likely explanation for the heterotypic protection we observed. However, we acknowledge that our findings should ideally be replicated in larger prospective cohorts with virologic sampling. Influenza B infections may have been slightly underestimated as we did not assess seroconversion using both lineages but only a strain from the Victoria lineage, which accounted for 90% of circulating influenza B isolates in Singapore in 2010 [19]. Finally, additional studies are needed to explore T-cell–mediated cross-protection.
In conclusion, we demonstrated a time-limited heterotypic protection across influenza strains. The observed birth cohort effects also support an “antigenic sin” phenomenon with the dominant circulating influenza subtype soon after birth, conferring long-term protection against severe disease, with implications for risk assessment in future influenza pandemics.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Acknowledgments. The World Health Organization Collaborating Centre for Reference and Research on Influenza is supported by the Australian Government Department of Health and Ageing. We acknowledge Nataline Tang Yan Ling, Dollyn Quek Liying, and Loh Pei Ling from the National Public Health Laboratory for their contribution in processing and testing laboratory samples.
Financial Support. This work was funded by the National Medical Research Council of Singapore (H1N1-005 and PPG10-09).
Potential conflicts of interest. V. J. L. has received unrelated research funding from GSK. B. Y. has received unrelated research funding from Sanofi-Pasteur. Y.-S. L. is a consultant to Sanofi-Pasteur. All other authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: 1st International Meeting on Respiratory Pathogens. Singapore, 2–4 September 2015. Abstract ASN0030; and Options IX for the Control of Influenza. Chicago, Illinois, 24–28 August 2016. Abstracts AOIX00227, AOIX00226.
References
- 1. World Health Organization. Swine Influenza 2009 (31 March 2015). Available from: http://www.who.int/mediacentre/news/statements/2009/h1n1_20090425/en/. Accessed 31 March 2015.
- 2. Mukherjee P, Lim P, Chow A et al. Epidemiology of travel-associated pandemic (H1N1) 2009 infection in 116 patients, Singapore. Electr J Surg 2010; 16:21–6.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ong JBS, Chen MIC, Cook AR et al. Real-time epidemic monitoring and forecasting of H1N1-2009 using influenza-like illness from general practice and family doctor clinics in Singapore. PLoS ONE 2010; 5:e10036–e.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tay J, Ng YF, Cutter J, James L. Influenza A (H1n1-2009) pandemic in Singapore—public health control measures implemented and lessons learnt. Ann Acad Med Singapore 2010; 39:313–12.394. [PubMed] [Google Scholar]
- 5. Win MK, Chen MI, Barkham T et al. Influenza disease burden in adults by subtypes following the initial epidemic of pandemic H1N1 in Singapore. Influenza Other Respir Viruses 2011; 5:e563–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Palese P, Wang TT. Why do influenza virus subtypes die out? A hypothesis. MBio 2011; 2:e00150–11.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Epstein SL. Prior H1N1 influenza infection and susceptibility of Cleveland Family Study participants during the H2N2 pandemic of 1957: an experiment of nature. J Infect Dis 2006; 193:49–53.1931. [DOI] [PubMed] [Google Scholar]
- 8. Slepushkin A. The effect of a previous attack of A1 influenza on susceptibility to A2 virus during the 1957 outbreak. Bull World Health Organ 1959; 20:297.202–3. [PMC free article] [PubMed] [Google Scholar]
- 9. Simonsen L, Clarke MJ, Schonberger LB, Arden NH, Cox NJ, Fukuda K. Pandemic versus epidemic influenza mortality: a pattern of changing age distribution. J Infect Dis 1998; 178:53–60. [DOI] [PubMed] [Google Scholar]
- 10. Saglanmak N, Andreasen V, Simonsen L, Mølbak K, Miller MA, Viboud C. Gradual changes in the age distribution of excess deaths in the years following the 1918 influenza pandemic in Copenhagen: using epidemiological evidence to detect antigenic drift. Vaccine 2011; 29:B42–B8.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hernandez JE, Grainger J, Simonsen L, Collis P, Edelman L, Sheridan WP. Impact of the 2009/2010 influenza A (H1N1) pandemic on trends in influenza hospitalization, diagnostic testing, and treatment. Influenza and Other Respir Viruses 2012; 6:305–8.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shrestha SS, Swerdlow DL, Borse RH et al. Estimating the burden of 2009 pandemic influenza A (H1N1) in the United States (April 2009–April 2010). Clin Infect Dis 2011; 52:S75–S82.52. [DOI] [PubMed] [Google Scholar]
- 13. National University of Singapore. Singapore Population Health Study (10 August 2016). Available at: https://www.sph.nus.edu.sg/research/sphs.
- 14. Lim W-Y, Chen C, Ma Y et al. Risk factors for pandemic (H1N1) 2009 seroconversion among adults, Singapore, 2009. Emerg Infect Dis 2011; 17:1455–62.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen MI, Cook AR, Lim WY et al. Factors influencing infection by pandemic influenza A(H1N1)pdm09 over three epidemic waves in Singapore. Influenza Other Respir Viruses 2013; 7:1380–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen MI, Barr IG, Koh GC et al. Serological response in RT-PCR confirmed H1N1-2009 influenza a by hemagglutination inhibition and virus neutralization assays: an observational study. PLoS One 2010; 5:e12474.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kendal A, Skehel J, Pereira M.. World Health Organization Collaborating Centers for Reference and Research on Influenza: concepts and procedures for laboratory-based influenza surveillance. Atlanta, GA: World Health Organization Collaborating Centers for Reference and Research in Influenza, Centers for Disease Control; B17–B35 p. 1982. [Google Scholar]
- 18. World Health Organization. Recommendations for Influenza Vaccine Composition (6 October 2015). Available at: http://www.who.int/influenza/vaccines/vaccinerecommendations1/en/.
- 19. Singapore Ministry of Health. Communicable Diseases Surveillance in Singapore 2010 2010:10 (12 January 2016). Available from: https://www.moh.gov.sg/content/dam/moh_web/Publications/Reports/2011/Communicable%20Diseases%20Surveillance%20in%20Singapore%202010/Air%20droplet-born%202010.pdf.
- 20. R Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2015. [Google Scholar]
- 21. Lee VJ, Chen MI, Chan SP et al. Influenza pandemics in Singapore, a tropical, globally connected city. Emerg Infect Dis 2007; 13:1052–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee VJ, Wong CS, Tambyah PA, Cutter J, Chen MI, Goh KT. Twentieth century influenza pandemics in Singapore. Ann Acad Med Singapore 2008; 37:470–6. [PubMed] [Google Scholar]
- 23. Soh SE, Cook AR, Chen MI et al. Teacher led school-based surveillance can allow accurate tracking of emerging infectious diseases—evidence from serial cross-sectional surveys of febrile respiratory illness during the H1N1 2009 influenza pandemic in Singapore. BMC Infectious Diseases 2012; 12:336.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen MI, Lee VJ, Barr I et al. Risk factors for pandemic (H1N1) 2009 virus seroconversion among hospital staff, Singapore. Emerg Infect Dis 2010; 16:1554.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Viboud C, Boëlle PY, Cauchemez S et al. Risk factors of influenza transmission in households. Br J Gen Pract 2004; 54:684–9. [PMC free article] [PubMed] [Google Scholar]
- 26. Lee VJ, Yap J, Ong JB et al. Influenza excess mortality from 1950–2000 in tropical Singapore. PLoS One 2009; 4:e8096.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ferguson NM, Galvani AP, Bush RM. Ecological and immunological determinants of influenza evolution. Nature 2003; 422:428–33.4226930. [DOI] [PubMed] [Google Scholar]
- 28. Hayward AC, Wang L, Goonetilleke N et al. ; Flu Watch Group Natural T cell-mediated protection against seasonal and pandemic influenza. Results of the Flu Watch Cohort study. Am J Respir Crit Care Med 2015; 191:1422–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hsu J, Phoon M, Koh GC et al. Comparison of neutralizing antibody and cell-mediated immune responses to pandemic H1N1 2009 influenza virus before and after H1N1 2009 influenza vaccination of elderly subjects and healthcare workers. Int J Infect Dis 2012; 16:e621–e7.168. [DOI] [PubMed] [Google Scholar]
- 30. Lee CX, Poh WP, Sakharkar KR, Sakharkar MKI, Chow VTK. Differences between predicted cytotoxic T-lymphocyte epitopes of 2009 H1N1 swine-origin influenza virus compared with other H1N1 strains of 1918, 1934 and 2007 for HLA-A*0201, HLA* 1101, n HLA-A*2402 alleles: clinical implications. Int J Integr Biol 2010; 10: 124–31.103. [Google Scholar]
- 31. Pica N, Hai R, Krammer F et al. Hemagglutinin stalk antibodies elicited by the 2009 pandemic influenza virus as a mechanism for the extinction of seasonal H1N1 viruses. Proc Natl Acad Sci U S A 2012; 109:2573–8.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee VJ, Tay JK, Chen MI et al. Inactivated trivalent seasonal influenza vaccine induces limited cross-reactive neutralizing antibody responses against 2009 pandemic and 1934 PR8 H1N1 strains. Vaccine 2010; 28:6852–7.2842. [DOI] [PubMed] [Google Scholar]
- 33. Kelly H, Barry S, Laurie K, Mercer G. Seasonal influenza vaccination and the risk of infection with pandemic influenza: a possible illustration of non-specific temporary immunity following infection. Challenge 2010; 16:17.16. [DOI] [PubMed] [Google Scholar]
- 34. Linde A, Rotzen-Ostlund M, Zweygberg-Wirgart B, Rubinova S, Brytting M. Does viral interference affect spread of influenza? Euro Surveill 2009;14:pii: 19354.1440. [PubMed] [Google Scholar]
- 35. Fonville JM, Wilks SH, James SL et al. Antibody landscapes after influenza virus infection or vaccination. Science 2014; 346:996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Francis T. On the doctrine of original antigenic sin. Proc Am Philos Soc 1960; 104:572–8.1046. [Google Scholar]
- 37. Ferdinands JM, Fry AM, Reynolds S et al. Intraseason waning of influenza vaccine protection: evidence from the US Influenza Vaccine Effectiveness Network, 2011–12 through 2014–15. Clin Infect Dis 2017; 64:544–50. [DOI] [PubMed] [Google Scholar]
- 38. Cauchemez S, Horby P, Fox A et al. Influenza infection rates, measurement errors and the interpretation of paired serology. PLoS Pathog 2012; 8:e1003061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hsu JP, Zhao X, Mark I et al. Rate of decline of antibody titers to pandemic influenza A (H1N1-2009) by hemagglutination inhibition and virus microneutralization assays in a cohort of seroconverting adults in Singapore. BMC Infect Dis 2014; 14:414.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.