Abstract
Methylation of the adenosine base at the nitrogen-6 position (m6A) is the most prevalent internal posttranscriptional modification of mRNAs in many eukaryotes. Despite the rapid progress in the transcriptome-wide mapping of m6As, identification of proteins responsible for writing, reading, and erasing m6As, and elucidation of m6A functions in splicing, RNA stability, translation, and other processes, it is unknown whether most observed m6A modifications are functional. To address this question, we respectively analyze the evolutionary conservation of yeast and human m6As in protein-coding regions. Relative to comparable unmethylated As, m6As are overall no more conserved in yeasts and only slightly more conserved in mammals. Furthermore, yeast m6As and comparable unmethylated As have no significant difference in single nucleotide polymorphism (SNP) density or SNP site frequency spectrum. The same is true in human. The methylation status of a gene, not necessarily the specific sites methylated in the gene, is subject to purifying selection for no more than ∼20% of m6A-modified genes. These observations suggest that most m6A modifications in protein-coding regions are nonfunctional and nonadaptive, probably resulting from off-target activities of m6A methyltransferases. In addition, our reanalysis invalidates the recent claim of positive selection for newly acquired m6A modifications in human evolution. Regarding the small number of evolutionarily conserved m6As, evidence suggests that a large proportion of them are likely functional; they should be prioritized in future functional characterizations of m6As. Together, these findings have important implications for understanding the biological significance of m6A and other posttranscriptional modifications.
Keywords: evolution, human, posttranscriptional modification, RNA methylation, yeast
Introduction
RNAs are frequently chemically modified co and posttranscriptionally (Cantara et al. 2011; Machnicka et al. 2013; Li and Mason 2014; Sun et al. 2016). Among over 100 distinct chemical modifications characterized to date, methylation of the adenosine base at the nitrogen-6 position (m6A) is the most prevalent internal modification of mRNAs (Fu et al. 2014; Meyer and Jaffrey 2014). First reported in the 1970s (Desrosiers et al. 1974), m6A modification was recently found to be widespread in both prokaryotes and eukaryotes (Dominissini et al. 2012; Meyer et al. 2012; Schwartz et al. 2013; Luo et al. 2014; Deng et al. 2015). Disrupting m6A modification affects mRNA stability (Wang et al. 2014), translational efficiency (Wang et al. 2015; Slobodin et al. 2017), cell fate (Batista et al. 2014; Geula et al. 2015), spermatogenesis (Hsu et al. 2017; Xu et al. 2017), sex determination (Lence et al. 2016; Kan et al. 2017), and other processes (Roignant and Soller 2017; Xiang et al. 2017; Zhao et al. 2017). Although challenges remain in precisely identifying m6As (Li et al. 2016), advances in methyl-RNA immunoprecipitation coupled with next-generation sequencing have provided transcriptome-wide coarse-grained maps of m6As (Dominissini et al. 2012; Meyer et al. 2012; Schwartz et al. 2013). m6As appear to be restricted to adenosines in particular sequence contexts. For example, the consensus sequence is RGAC (R = A or G; A = methylatable A) in the yeast Saccharomyces cerevisiae and related species (Schwartz et al. 2013) and DRACH (D = A, G, or U; H = A, C, or U) in mammals (Dominissini et al. 2012; Meyer et al. 2012). Recent applications of cross-linking allowed for near-single-nucleotide-resolution determination of m6As in mRNAs (Ke et al. 2015; Linder et al. 2015).
Despite unambiguous evidence for the functional importance of some m6As aforementioned, it is unknown whether the vast majority of m6As are functional. This question is relevant, because if only a small fraction of m6As are functional, 1) a search for m6A function from randomly picked m6As would be inefficient, wasteful, and futile; 2) automatically assigning functions detected from a small number of m6As to other m6As would be error-prone; 3) the functional importance of m6A modification would need a reassessment; and 4) most importantly, the tendency of many molecular biologists to claim or assume almost any cellular and molecular process or feature as adaptive would require reflection at the minimum.
A biological process or feature is functional only if it increases organismal fitness, which can be detected from signals of positive or purifying selection (Doolittle et al. 2014; Graur et al. 2015). Here, we focus on purifying selection that maintains functional m6As during evolution, because purifying selection is much more common and hence much more readily detectable than positive selection in molecular evolution (Zhang 2010). We provide evidence that only a minority of m6As in protein-coding regions are evolutionarily conserved, suggesting that most m6As in protein-coding regions are likely nonfunctional. Furthermore, our reanalysis invalidates the recent claim of positive selection for the acquisition of new m6A modifications during human evolution (Ma et al. 2017). We discuss the implications of these findings for understanding the biological significance of m6A and other posttranscriptional modifications.
Results
Are m6As More Conserved than Unmethylated As?
If a substantial proportion of m6A modifications are functional, m6As should be evolutionarily more conserved than comparable unmethylated As due to the additional selective constraint associated with m6A-specific functions. Hence, comparing the evolutionary conservation of m6As with that of unmethylated As allows assessing the fraction of functional m6As. We focused on protein-coding regions in our analysis, because the majority of high-confidence m6As mapped to date are in mRNAs, especially in protein-coding regions (Dominissini et al. 2012; Meyer et al. 2012; Schwartz et al. 2013) and because alignment-dependent sequence analysis is more reliable for protein-coding regions than other genomic regions. We compared m6As and unmethylated As from the same genes to avoid the confounding factor of gene expression level, which is a primary determinant of protein sequence conservation (Zhang and Yang 2015).
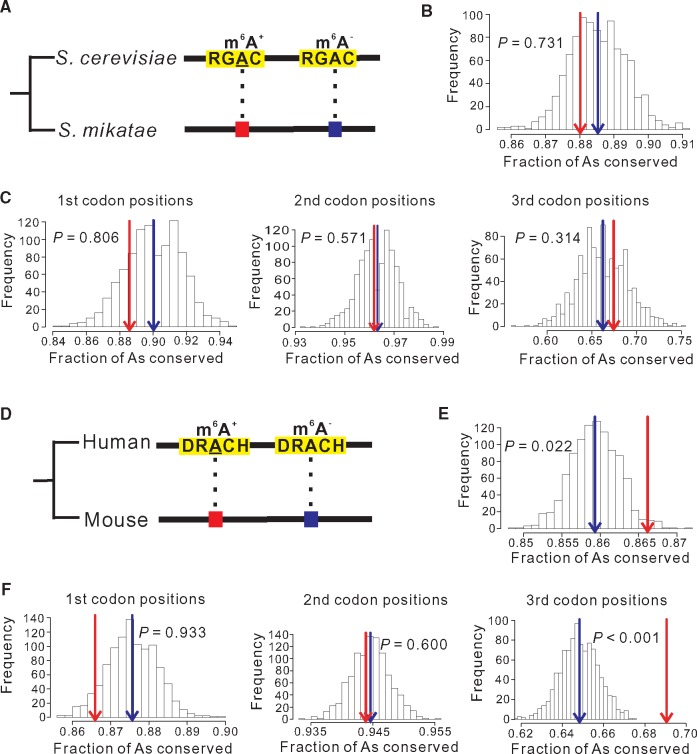
Schwartz et al. (2013) reported 1,308 m6As in 1,183 genes from S. cerevisiae during meiosis, the only life stage in yeast with extensive m6A modifications. To study sequence conservation, we concentrated on the aligned nongapped regions of one-to-one orthologous genes between S. cerevisiae and S. mikatae, a species in the Saccharomyces sensu stricto complex that is relatively closely related to S. cerevisiae. We asked whether S. cerevisiae m6As are more likely than unmethylated As to remain As in S. mikatae (i.e., conservation). Because m6As occur in the consensus motif RGAC in yeasts, to control for the potential impact of neighboring sites on the conservation of As, we considered only those unmethylated As that are also in the motif RGAC. We refer to these methylated and unmethylated As as m6A+ and m6A− sites, respectively (fig. 1A). To ensure a fair comparison, we randomly picked the same number of m6A− sites as the number of observed m6A+ sites at each of the three codon positions of each gene concerned. In total, 776 m6A+ sites from 718 genes were compared with the same number of m6A− sites. The reason that only 776 of the 1,308 S. cerevisiae m6As were compared is that the rest of m6As do not have one-to-one orthologous positions in S. mikatae. The above comparison was repeated 1,000 times by using different random sets of 776 m6A− sites chosen following the above rules. We found the mean conservation to be similar between m6A+ (0.880) and m6A− (0.885) sites (P = 0.731; fig. 1B). Relative to m6A− sites, m6A+ sites are significantly enriched at second codon positions for unknown reasons (supplementary fig. S1A, Supplementary Material online), but this bias does not affect the above comparison because of the controls applied. Regardless, none of the three codon positions shows a significant difference in conservation between m6A+ and m6A− sites in yeasts (fig. 1C).
Fig. 1.
Comparison in evolutionary conservation between m6A+ and m6A- sites of the same genes. (A) A schematic diagram illustrating the comparison between Saccharomyces cerevisiae m6A+ and m6A- sites in terms of their evolutionary conservations in Saccharomyces mikatae. The yeast consensus m6A motifs are highlighted in yellow. The underlined A indicates m6A modification. (B, C) Frequency distribution of the fraction of conserved S. cerevisiae m6A- sites in 1,000 random sets with the sample size equal to the number of m6A+ sites at all positions (B) or at three codon positions separately (C). Red and blue arrows respectively indicate the fraction of conserved m6A+ sites and the mean fraction of conserved m6A- sites in 1,000 random sets for all positions or individual codon positions concerned. P-value is the fraction of the distribution on the right side of the red arrow. (D) A schematic diagram illustrating the comparison between human m6A+ and m6A- sites in terms of their evolutionary conservations in mouse. The mammalian consensus m6A motifs are highlighted in yellow. The underlined A indicates m6A modification. (E, F) Frequency distribution of the fraction of conserved human m6A- sites in 1,000 random sets with the sample size equal to the number of m6A+ sites at all positions (E) or at three codon positions separately (F). All symbols have the same meanings as in panels B and C.
To examine the general validity of the above results, we turned to mammals. We compiled a data set of human m6As and examined their conservation in mouse (see Materials and Methods). We applied the yeast method except that the mammalian consensus motif DRACH was used (fig. 1D). In comparing 9,077 m6A+ sites and the same number of m6A− sites from 3,296 human genes, we found the conservation of the former (0.866) to be slightly, but significantly higher than that of the latter (0.859) (P = 0.022; fig. 1E). This significant difference in conservation appears to be entirely attributable to third codon positions, because no significant difference is observed at the first and second codon positions (fig. 1F). The reason for this variation among codon positions is unknown. Similar to what was observed in yeast (supplementary fig. S1A, Supplementary Material online), human m6A+ sites are enriched at second codon positions and underrepresented at first and third codon positions, relative to m6A− sites (supplementary fig. S1B, Supplementary Material online).
Thus, compared with unmethylated As, m6As are no more conserved in yeasts but are more conserved in mammals. Despite this qualitative difference between taxa, we note that the evolutionary rate at m6A sites (1 – 0.866 = 0.134) is <5% lower than that at unmethylated A sites (1 – 0.859 = 0.141) in mammals, suggesting that, even in mammals, only a minority of m6A modifications may be selectively constrained.
Are m6A+ Sites Less Polymorphic than m6A− Sites?
That most m6As are not evolutionarily conserved suggests that they either are nonfunctional or have lineage/species-specific functions. To examine the latter possibility, we respectively computed single nucleotide polymorphism (SNP) density at m6A+ and m6A− sites in m6A-modified genes. If a large fraction of m6As have lineage/species-specific functions such that they are subject to purifying selection within species albeit unconserved between species, m6A+ sites should show reduced intraspecific polymorphism when compared with m6A− sites. However, no significant difference in SNP density was observed at 776 m6A+ and 8,229 m6A− sites in the 718 m6A-modified S. cerevisiae genes (table 1). The same is true when the three codon positions are separately analyzed (table 1). Similarly, there is no significant difference in SNP density at 9,077 m6A+ and 137,872 m6A− sites in the 3,296 human genes with m6A modifications, with or without separately considering different codon positions (table 1). We also found no significant difference in minor allele frequency between m6A+ and m6A− SNPs (P = 0.62 for yeast and 0.88 for human; Mann–Whitney U test). These consistent findings in yeast and human do not support the hypothesis of lineage/species-specific function for any sizable fraction of m6As.
Table 1.
SNP Densities at m6A+ and m6A− Sites of the Same Genes.
| Type of Sites | # of Sites | # of SNPs | SNP Density | P-Valuea | |
|---|---|---|---|---|---|
| Yeast (S. cerevisiae) | |||||
| 1st codon positions | m6A+ | 184 | 4 | 0.0217 | |
| m6A− | 1,995 | 55 | 0.0276 | 0.65 | |
| 2nd codon positions | m6A+ | 420 | 14 | 0.0333 | |
| m6A− | 4,151 | 93 | 0.0224 | 0.17 | |
| 3rd codon positions | m6A+ | 172 | 16 | 0.0930 | |
| m6A− | 2,083 | 151 | 0.0725 | 0.36 | |
| Total | m6A+ | 776 | 34 | 0.0438 | |
| m6A− | 8,229 | 299 | 0.0363 | 0.31 | |
| Human (H. sapiens) | |||||
| 1st codon positions | m6A+ | 2,350 | 3 | 0.00128 | |
| m6A− | 40,396 | 63 | 0.00156 | 0.89 | |
| 2nd codon positions | m6A+ | 4,665 | 6 | 0.00129 | |
| m6A− | 63,935 | 93 | 0.00145 | 0.77 | |
| 3rd codon positions | m6A+ | 2,062 | 4 | 0.00194 | |
| m6A− | 33,541 | 48 | 0.00143 | 0.56 | |
| Total | m6A+ | 9,077 | 13 | 0.00143 | |
| m6A− | 137,872 | 204 | 0.00148 | 0.92 | |
Chi-squared test of the null hypothesis of equal SNP densities at m6A+ and m6A− sites.
Do Different Species Share More m6As than Expected by Chance?
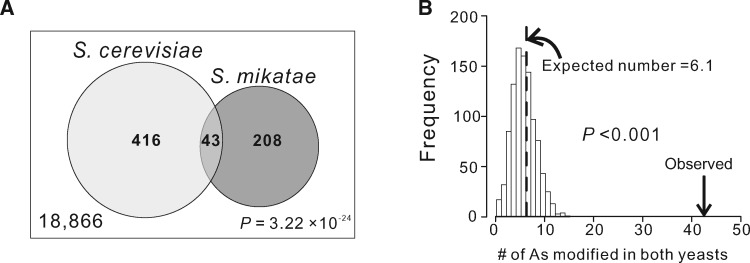
Another prediction of the functionality of m6A modification is that m6As of different species should overlap more than expected by chance. To verify this prediction, we analyzed m6As respectively identified in S. cerevisiae and S. mikatae during meiosis (Schwartz et al. 2013). Given the divergence between these two yeasts, it is reasonable to assume that an m6A modification in one of them is lost in the other unless the modification is functional and hence conserved. In the one-to-one orthologous protein-coding regions of the two species (after the removal of alignment gaps), there are 18,866 RGAC motifs that are shared by the two yeasts. Within these motifs, 459 and 251 m6As are respectively observed in S. cerevisiae and S. mikatae. The number of m6As shared by the two species is 43, which is significantly greater than the random expectation of 6.11 (P = 3.22 × 10−24, hypergeometric test; fig. 2A), suggesting purifying selection acting on some m6As.
Fig. 2.
Evolutionarily conserved m6As between two yeasts. (A) Venn diagram showing the number of shared m6A consensus motifs in one-to-one orthologous genes under comparison (large box, with the number indicated at the lower left corner), as well as the number of m6As observed in each yeast species and the number of shared m6As. P-value from a hypergeometric test indicates the probability of observation under the null hypothesis that m6A sites in the two yeasts are independent from each other. (B) Frequency distribution of the number of shared m6As expected by chance in yeasts upon the control for potential detection bias. The dotted line shows the mean of the distribution, whereas the arrow indicates the number of observed shared m6As. P-value is the fraction of the distribution on the right side of the arrow that indicates the observed number of shared m6As.
One caveat in the above hypergeometric test is the implicit assumption that m6A modifications are equally detectable in all genes, despite that in principle m6As are less detectable in lowly expressed genes than in highly expressed genes. Indeed, median expression level is significantly higher for m6A-modified genes than m6A-absent genes in yeast (P = 7.44 × 10−6, Mann–Whitney U test; supplementary fig. S2A, Supplementary Material online) and human (P < 2.2 × 10−16; supplementary fig. S2B, Supplementary Material online). To control for this potential detection bias, we devised the following test. We ranked the 18,866 RGAC motifs according to the S. cerevisiae expression levels of the genes where the motifs are found and divided them into 10 equal-sized bins. In each bin, we randomly picked k motifs, where k is the number of S. cerevisiae m6As in the bin, and counted the number of S. mikatae m6As within these k motifs; this number represents the expected number of sites in the bin that are modified in both species by chance when m6A modifications in the two species are independent. This was performed for all bins and the total number (T) of As modified in both species by chance was obtained. We repeated this process 1,000 times to acquire 1,000 T values. The actual number of orthologous m6As in the yeasts (43) is significantly greater than all 1,000 T values (fig. 2B). Hence, controlling for the potential detection bias does not alter the conclusion that some m6As are evolutionarily conserved. We were surprised that the mean T of 6.1 is not >6.11, the random expectation without correction for the potential detection bias. We subsequently discovered that the expression levels of m6A-modified genes and unmodified genes are not significantly different (P = 0.34) for one-to-one orthologous genes between the two yeasts, which are the relevant gene set in the present analysis. Because there are 928 S. cerevisiae m6As in one-to-one orthologs of the two yeasts, but only 43 - 6.1 = 36.9 of them are shared between the two yeasts beyond the chance expectation, the fraction of S. cerevisiae m6As that are selectively conserved is 36.9/926 = 4.0%. The corresponding fraction in S. mikatae is 36.9/484 = 7.6%. These results indicate that only a small fraction of m6As are selectively conserved in yeasts. This finding explains why no significant difference in overall conservation is detected between methylated and unmethylated As in yeasts. Unfortunately, a similar analysis cannot be conducted between human and mouse, because of the lack of nucleotide-resolution transcriptomic m6A data from the same tissues of the two mammals.
Do Different Species Share More m6A-Modified Genes than the Chance Expectation?
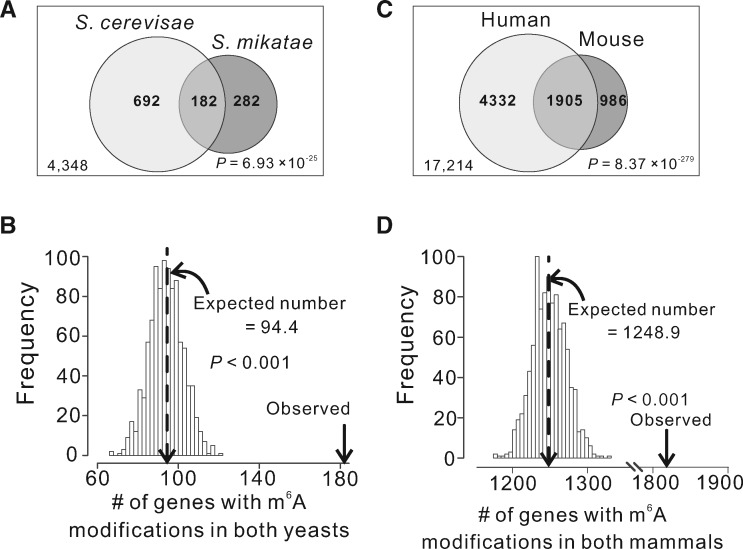
In the study of protein phosphorylation, it was discovered that the specific sites phosphorylated may not be important as long as the protein or segment of protein is phosphorylated (Moses et al. 2007; Nguyen Ba and Moses 2010; Landry et al. 2014). By analogy, one possibility why most m6As are evolutionarily unconserved is the potential functional importance of the methylation of a gene rather than particular sites in the gene. Under this scenario, a gene that is m6A-modified in one species tends to be m6A-modified in other species, but the specific methylated site(s) in the gene may vary. Among the 4,348 one-to-one orthologous genes of the two yeasts, 874 S. cerevisiae genes and 464 S. mikatae genes are m6A-modified in protein-coding regions. They overlap by 182 genes, significantly more than the chance expectation of 93.3 (P = 6.93 × 10−25, hypergeometric test; fig. 3A). The number of genes modified in both yeasts (182) remains significantly greater than the chance expectation (94.4) even after the control of the potential detection bias (P < 0.001; fig. 3B). Notwithstanding, only 182 - 94.4 = 87.6 gene-level m6A-modifications may be considered functional, which constitute 87.6/874 = 10.0% of m6A-modified genes in S. cerevisiae and 87.6/464 = 18.9% in S. mikatae.
Fig. 3.
Sharing of m6A-modified genes between species. (A) Venn diagram showing the number of one-to-one orthologous genes under comparison (large box, with the number indicated at the lower left corner), as well as the number of m6A-modified genes observed in each yeast species and the number of shared m6A-modified genes. P-value from a hypergeometric test indicates the probability of observation under the null hypothesis of independent m6A modifications of genes in the two yeasts. (B) Frequency distribution of the number of shared m6A-modified genes expected by chance in yeasts upon the control for potential detection bias. The dotted line shows the mean of the distribution, whereas the arrow indicates the number of observed shared m6A-modified genes. P-value is the fraction of the distribution on the right side of the arrow that indicates the observed number. (C) Venn diagram showing the number of one-to-one orthologous genes under comparison (large box, with the number indicated at the lower left corner), as well as the number of m6A-modified genes observed in each mammalian species and the number of shared m6A-modified genes. P-value is from a hypergeometric test. (D) Frequency distribution of the number of shared m6A-modified genes expected by chance in mammals upon the control for potential detection bias. All symbols have the same meanings as in panel B.
We similarly analyzed the sharing of m6A-modified genes between human and mouse, using coarse-grained m6A data from a human hepatocellular carcinoma cell line and mouse liver (Dominissini et al. 2012). These data do not identify nucleotide-resolution m6As but provide information on the genes that are m6A-modified. The data include 6,237 m6A-modified human genes and 2,891 m6A-modified mouse genes among 17,214 one-to-one orthologs between the two species (Dominissini et al. 2012). The number of genes modified in both species is 1,905, significantly more than the chance expectation of 1,047.5 (P = 8.37 × 10−279, hypergeometric test; fig. 3C). After controlling for the potential detection bias, we found that 1,248.9 genes are expected to be modified in both species under the hypothesis of independent modifications in the two mammals, which is still significantly fewer than the observed number of 1,905 (P < 0.001, randomization test; fig. 3D). This result suggests that (1905 - 1248.9)/6237 = 10.5% of m6A-modified genes in human and (1905 - 1248.9)/2891 = 22.7% in mouse are subject to purifying selection for maintaining gene-level m6A modification.
Positive Selection for New m6A Modifications in Human Evolution?
Detection of positive selection acting on m6A modification would be a strong indication of its functionality. A recent study of primate m6As claimed that positive selection drove the emergences of new m6A motifs and modifications during human evolution (Ma et al. 2017). This conclusion was based on the finding that, for m6A modifications gained in human evolution, the number of substitutions per site within the m6A motif GGACT (Kb) in the human lineage since the human–chimpanzee split is significantly greater than the corresponding number of substitutions per site at the rest of the sites covered by m6A peaks (Ki). Here, m6A peaks refer to the regions where modified sites reside; the exact sites modified could not be experimentally determined due to insufficient resolutions. But, this comparison is inappropriate because the gain of an m6A peak may imply at least one nucleotide substitution in the motif whereas no substitution is required in nonmotif regions. Consequently, Kb/Ki may exceed 1 because of this ascertainment bias rather than positive selection for m6A modification. To be fair, we shall mention that Ma and colleagues included a negative control in their analysis, which is the Kb/Ki ratio computed from m6A motifs gained in human evolution and their flanking regions (50 nucleotides on each side of a motif) found in untranscribed regions. They reported that Kb/Ki = 1.02 for the negative control, which appears to validate their approach. However, this finding from the negative control is highly unexpected given the ascertainment bias mentioned. We thus conducted an independent analysis following their method. From untranscribed regions, we picked 201 m6A motifs that have been gained in human evolution. In these motifs and their flanking regions, we found Kb = 0.2, Ki = 0.0066, and Kb/Ki = 30.45 (table 2). These values conform to our expectation, because 1) m6A motifs gained in human evolution should have at least one substitution in the five-nucleotide motif, resulting in a minimum Kb value of 0.2, and 2) neutral divergence between human and chimpanzee is ∼1.24% (Chen and Li 2001), resulting in a Ki of ∼0.00124/2 = 0.0062. Positive selection for m6A modification would be supported if Kb/Ki is significantly higher for the experimental group (i.e., m6A peaks gained in human) than the negative control. Our reanalysis of their experimental group found Kb/Ki = 4.84 (table 2), virtually identical to their reported value of 4.82 (Ma et al. 2017). Hence, Kb/Ki is in fact significantly lower for the experimental group than the negative control (P < 0.001, bootstrap test). To understand the cause of this result, we examined the 91 human-gained m6A peaks reported (Ma et al. 2017). Somewhat unexpectedly, only six of them have human-gained motifs, whereas the rest all have motifs shared at least between human and chimpanzee. For the six human-gained m6A peaks with human-gained motifs, Kb/Ki = 33.75, not significantly greater than that (30.45) for the negative control (P = 0.15, bootstrap test). Taken together, our analysis found no evidence supporting the claim of positive selection for newly gained m6A modifications in human evolution.
Table 2.
Number of Nucleotide Substitutions Per Site in the m6A Motif GGACT (Kb) and That in its Flanking Regions (Ki) During Human Evolution.
| Types of m6A Motifs | # of Motifs | Kb | Ki | Kb/Ki | Comparison | P-Valuea |
|---|---|---|---|---|---|---|
| A. Motifs in human-gained m6A peaks | 91 | 0.0135 | 0.0028 | 4.84 | A vs. C | <0.001 |
| B. Human-gained motifs in human-gained m6A peaks | 6 | 0.2000 | 0.0059 | 33.75 | B vs. C | 0.15 |
| C. Human-gained motifs in untranscribed regions | 201 | 0.2000 | 0.0066 | 30.45 |
One-tailed bootstrap test from 1,000 bootstrap samples.
Which m6As Are Potentially Functional?
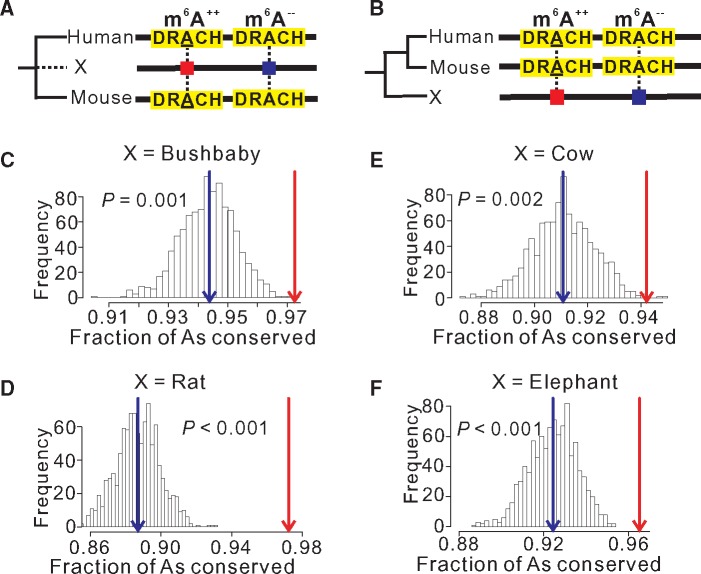
Because our results suggest that only a small fraction of m6As in protein-coding regions are functional, identifying these functional m6As and their functions becomes a high priority in m6A research. Evolutionary principles indicate that evolutionarily conserved m6As are much more likely than other m6As to be functional. However, this prediction is not easy to confirm directly due to the lack of a substantial group of m6As with known functions. As an indirect test, we examined whether sites that are m6A-modified in both human and mouse (referred to as m6A++ sites) are more conserved in a third species (as As) than sites that remain unmethylated As in both human and mouse (referred to as m6A−− sites), where the third species may be an ingroup (fig. 4A) or outgroup (fig. 4B) of the human–mouse clade. As before, we considered only m6As and the same number of unmethylated As that are in DRACH motifs of the same genes. Two ingroup (bushbaby and rat) and two outgroup (cow and elephant) mammalian species were considered individually as the third species in the test, and our prediction is significantly supported in each case (fig. 4C–F). For instance, 97.2% of m6A++ sites exhibit As in bushbaby, significantly greater than the corresponding value of 94.4% for m6A−− sites (P = 0.001, randomization test; fig. 4C). The evolutionary rate at m6A++ sites (1 − 0.972 = 0.028) is 50% lower than that at m6A−− sites (1 − 0.944 = 0.056), suggesting that at least one half of m6A++ sites are likely functional. The corresponding percentage reduction in evolutionary rate is 68, 67, and 46 when rat, cow, and elephant were used as the third species in the test, respectively (fig. 4D–F). Furthermore, m6A++ sites are generally more conserved than m6A−− sites in these species at all three codon positions (supplementary fig. S3, Supplementary Material online).
Fig. 4.
Sites subject to m6A modification in both human and mouse (denoted as m6A++) are more likely to be conserved as As in a third species than As modified in neither human nor mouse (denoted as m6A−−). (A, B) A schematic diagram illustrating the comparison when the third species X is an ingroup (A) or outgroup (B) of the human–mouse clade. Mammalian m6A consensus motifs are highlighted in yellow. The underlined A indicates m6A modification. (C–F) Frequency distribution of the fraction of m6A−− sites conserved in the bushbaby (C), rat (D), cow (E), or elephant (F) in 1,000 random sets with the sample size equal to the number of m6A++ sites. Red and blue arrows respectively indicate the fraction of conserved m6A++ sites and the mean fraction of conserved m6A−− sites in 1,000 random sets. P-value is the fraction of the distribution on the right side of the red arrow.
Discussion
In this work, we studied the evolutionary conservation of posttranscriptional m6A modification of protein-coding regions in yeasts and mammals. In yeasts, we found that only 5–8% of m6A modifications are subject to purifying selection at the individual site level, while the corresponding number at the gene level is 10–19%. These findings suggest that, in yeasts, 1) most m6A modifications are nonfunctional, and 2) even when a modification is functional, it is more often the modification of a gene than the modification of specific sites in the gene that is important. In mammals, the absence of proper data prohibits us from estimating the proportion of m6A modifications subject to purifying selection at the individual site level, but we estimated that ∼10–20% of m6A modifications are subject to purifying selection at the gene level. It is notable that, at least at the gene level, the percentage of m6A modifications subject to purifying selection is similar between yeasts and mammals. We further showed in human and yeast that m6A+ sites and comparable m6A− sites are not significantly different in SNP density or minor allele frequencies at SNPs, suggesting that the lack of evolutionary conservation for most m6As cannot be explained by the presence of species-specific functions in any sizable proportion of m6As. We also showed that a previous claim of positive selection for newly gained m6A modifications in human evolution is untenable.
A trivial explanation of our results is that the m6A data were too noisy to be useful for the evolutionary analyses performed, but this scenario appears improbable to us. The yeast m6As were mapped by comparing putative m6A signals in wild-type cells with those in mutants deficient of functional RNA methyltransferase and by requiring observing the signals in at least two of three biological replicates, resulting in minimal false positive errors (Schwartz et al. 2013). Regarding the mammalian data, with the exception of the analysis of gene-level m6A modification that needs only coarse-grained modification information, we required each m6A to be reported in at least two data sets, hence ensuring a low false positive rate. Furthermore, 98.4% of the human m6As analyzed appear in at least one of the near-single-nucleotide-resolution m6A data sets generated using cross-linking methods (Chen et al. 2015; Linder et al. 2015; Ke et al. 2015). The corresponding value is 76.8% for the mouse m6As analyzed (Linder et al. 2015; Ke et al. 2015). False negative rates in yeast and human m6A data, however, are more difficult to gauge, but they are not expected to affect the analysis presented in figure 1, tables 1 and 2. Presumably, false negative errors tend to occur at sites with low probabilities of m6A modification and/or in lowly expressed genes. Hence, our results in figures 2 and 3 suggest that even highly modified sites and/or modifications of highly expressed genes are not well conserved. These considerations together suggest that our findings are most likely genuine rather than artifacts.
Taken together, our evidence suggests that the most likely explanation for the lack of evolutionary conservation of most m6As is that they are nonfunctional. It is probable that these nonfunctional m6A modifications occur by error due to the limited specificity of RNA methyltransferases. It is reasonable to assume that some erroneous m6A modifications may even be deleterious, but the moderately and strongly harmful modifications presumably have been purged by natural selection such that only slightly deleterious or neutral ones remain.
Given that most m6As are nonfunctional, the next important task is to identify those that are functional and to uncover their specific functions. Our analyses suggest that at least one half of m6As that are conserved between human and mouse are subject to purifying selection and therefore are likely to be functional. Future functional characterizations of m6As should be prioritized to this small set of candidates.
It should be emphasized that m6A modification occurs not just in protein-coding regions of mRNAs. Because our analysis focused exclusively on protein-coding regions, it remains possible that m6A modifications outside protein-coding regions are mostly functional. In yeasts, ∼95% of m6As identified from mRNAs are located in coding regions (Schwartz et al. 2013), while the corresponding number reduces to ∼50% in mammals (Dominissini et al. 2012; Meyer et al. 2012; Sun et al. 2016). In the future, it will be interesting to investigate the evolutionary conservation of m6As outside protein-coding regions.
As mentioned, m6A is but one of over 100 posttranscriptional modifications. Nevertheless, our findings about m6A evolution resemble those on another common posttranscriptional modification, A-to-I RNA editing, in mammalian coding sequences. Specifically, it was found that most A-to-I RNA editing is nonadaptive (Xu and Zhang 2014) and unconserved (Pinto et al. 2014), but the very small number of conserved editing sites between human and mouse are likely functional (Xu and Zhang 2015). Interestingly, similar patterns also exist in the few types of posttranslational modifications that have been subject to evolutionary analysis. For instance, as much as 65% of protein phosphorylations were estimated to be nonfunctional (Landry et al. 2009). A recent evolutionary study of protein phosphorylation across 18 fungal species further supports this result, showing that only a small fraction of phosphorylation sites are conserved (Studer et al. 2016). In another example, analysis of protein glycosylation showed that whereas glycosylation at solvent accessible sites are generally conserved, those at solvent inaccessible sites are not (Park and Zhang 2011), suggesting that they are probably biochemical errors and nonfunctional. These similarities suggest the possibility that many other posttranscriptional and posttranslational modifications also exhibit these properties, which awaits testing when relevant large modification data become available. At the minimum, the present findings, coupled with the existing evidence from other posttranscriptional/posttranslational modifications, argue against the implicit pan-adaptationist assumption that all biological processes or features are adaptive. While this assumption has long been abandoned by most evolutionary biologists (Gould and Lewontin 1979), it remains popular among molecular biologists, especially when they attempt to explain the very existence of a newly discovered cellular/molecular phenomenon or its high prevalence. It is ironic that it was the field of molecular biology that supplied the first examples of nonadaptive features of the living world that inspired the development of the neutral theory of molecular evolution (Kimura 1968, 1983; King and Jukes 1969). This history is to be reminded when biologists ponder upon new molecular details of life unraveled by powerful genomic tools.
Materials and Methods
We downloaded the Supplementary Material in Schwartz et al. (2013) that included the information of 1,308 m6As in 1,183 genes from the SK1 strain of S. cerevisiae and 635 m6As in 610 genes from S. mikatae detected during meiosis. These m6As are precisely mapped thanks to the contrast between wild-type and mutant strains defective in the core RNA methyltransferase complex. Unless mentioned, the m6A information from human and mouse was retrieved from RMBase, which covers nearly all high-throughput m6A data and records m6A-modified sites (Sun et al. 2016). To ensure the quality of the mammalian data used, we regarded a site from RMBase as m6A only when it has support from at least two independent data sets. In addition, we used coarse-grained data on m6A-modified genes from a human hepatocellular carcinoma cell line and mouse liver (Dominissini et al. 2012) to study shared m6A-modified genes between the two mammals.
In total, 4,348 one-to-one orthologous protein-coding genes of the SK1 stain of S. cerevisiae and S. mikatae were retrieved from the Saccharomyces Genome Database (SGD) (No. JRIH00000000), whereas 17,214 one-to-one orthologous protein-coding genes of human (version GRCh38.p10) and mouse (version GRCm38.p5) were obtained from Ensembl (Ensembl genes 88). ClustalW2 (Sievers et al. 2011) with the default setting was used to align orthologous sequences upon translation to protein sequences, and the corresponding coding sequence alignments were then created accordingly using PAL2NAL (Suyama et al. 2006). In the alignment of each gene, we isolated consensus motifs (RGAC in yeasts and DRACH in mammals) using in-house Perl scripts.
To examine any potential bias in m6A detection, we downloaded from the Gene Expression Omnibus (accession numbers of GSE51583 for S. cerevisiae and GSE37005 for human) the transcriptomic raw sequencing reads of the input samples that were used as controls for calling m6A peaks in immunoprecipitation-based experiments. After using Trimmomatic (Bolger et al. 2014) to ensure the quality of the raw reads, we applied kallisto (Bray et al. 2016) with default parameters to align these reads with the relevant references of S. cerevisiae coding sequences retrieved from SGD and human coding sequences retrieved from Ensembl, respectively. We then calculated the Transcripts Per Million (TPM) value of each gene to represent its relative expression level (Li et al. 2010). Briefly, TPM of a gene is 106 times the number of reads per nucleotide for the gene, relative to the sum of the number of reads per nucleotide for all genes.
We retrieved the SNP data of S. cerevisiae from our recent population genomic analysis of the species (Maclean et al. 2017). Because the genomic sequences of SK1 (JRIH00000000) and S288c (R64-1-1) strains were respectively used as the reference to localize m6As and SNPs, we identified orthologous sites between the two genomes using in-house programs. We similarly analyzed human SNPs downloaded from the Ensembl Variation database (version 90). We recorded minor allele frequencies of SNPs when the information is available.
We reanalyzed human-gained m6A modifications following Ma et al. (2017). The longest isoforms of genes containing human-gained m6A peaks were downloaded from UCSC Genome Browser (GRCh38/hg38) according to the gene names provided in supplementary table S10, Supplementary Material online of Ma et al. (2017). We then identified 91 human-gained m6A peaks with the motif GGACT. We estimated the number of nucleotide substitutions per site within motifs (Kb) by the total number of substitutions in the 91 motifs divided by the total number of sites in these motifs. We estimated the number of nucleotide substitutions per site in flanking regions (Ki) by the total number of substitutions within the 91 m6A peaks but outside the motifs divided by the total number of sites in nonmotif regions of the m6A peaks. Following Ma et al. (2017), we chose untranscribed intergenic regions with human-gained GGACT motifs as the negative control. Briefly, we downloaded human genome annotations from UCSC (GRCh38/hg38) and identified untranscribed intergenic regions using bedtools (version 2.26.0; Quinlan and Hall 2010). After obtaining the sequences of intergenic regions from the two largest chromosomes, we picked 105-nucleotide fragments centering at the motif GGACT as queries to BLAST chimpanzee (panTro5) and macaque (rheMac8) genome sequences, respectively, and obtained orthologous fragments from these species. Using macaque as an outgroup, we identified 201 fragments with human-gained motifs in the untranscribed regions and estimated Kb and Ki from these regions. Bootstrapping the experimental and control fragments 1,000 times provided information on the variation of the Kb/Ki estimates, which allowed statistical comparison in Kb/Ki between groups.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Lijia Ma for supplying the sequences of human-gained m6A peaks in Ma et al. (2017) and Wei-Chin Ho, Wenfeng Qian, Chuan Xu, and Zhengting Zou for assistance and/or comments. This work was supported in part by a research grant from the U.S. National Institutes of Health (GM120093) to J.Z. Z.L. was supported by China Scholarship Council (201604910442).
References
- Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, et al. 2014. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 15(6):707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L.. 2016. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 34(5):525–527. [DOI] [PubMed] [Google Scholar]
- Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D, Agris PF.. 2011. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 39(Database issue):D195–D201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FC, Li WH.. 2001. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am J Hum Genet. 68(2):444–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Lu ZK, Wang X, Fu Y, Luo GZ, Liu N, Han DL, Dominissini D, Dai Q, Pan T, et al. 2015. High-resolution N-6-methyladenosine (m(6)A) map using photo-crosslinking-assisted m(6)A sequencing. Angew Chem Int Ed Engl. 54(5):1587–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Chen K, Luo GZ, Weng X, Ji Q, Zhou T, He C.. 2015. Widespread occurrence of N6-methyladenosine in bacterial mRNA. Nucleic Acids Res. 43(13):6557–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers R, Friderici K, Rottman F.. 1974. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA. 71(10):3971–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. 2012. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485(7397):201–206. [DOI] [PubMed] [Google Scholar]
- Doolittle WF, Brunet TDP, Linquist S, Gregory TR.. 2014. Distinguishing between “function” and “effect” in genome biology. Genome Biol Evol. 6(5):1234–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Dominissini D, Rechavi G, He C.. 2014. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genet. 15(5):293–306. [DOI] [PubMed] [Google Scholar]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al. 2015. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 347(6225):1002–1006. [DOI] [PubMed] [Google Scholar]
- Gould SJ, Lewontin RC.. 1979. Spandrels of San-Marco and the Panglossian Paradigm – a critique of the adaptationist program. Proc R Soc Lond B. 205(1161):581–598. [DOI] [PubMed] [Google Scholar]
- Graur D, Zheng YC, Azevedo RBR.. 2015. An evolutionary classification of genomic function. Genome Biol Evol. 7(3):642–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, Qi M, Lu Z, Shi H, Wang J, et al. 2017. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 27(9):1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan L, Grozhik AV, Vedanayagam J, Patil DP, Pang N, Lim KS, Huang YC, Joseph B, Lin CJ, Despic V, et al. 2017. The m6A pathway facilitates sex determination in Drosophila. Nat Commun. 8:15737.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, Haripal B, Zucker-Scharff I, Moore MJ, Park CY, et al. 2015. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 29(19):2037–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. 1968. Evolutionary rate at the molecular level. Nature 217(5129):624–626. [DOI] [PubMed] [Google Scholar]
- Kimura M. 1983. The neutral theory of molecular evolution. New York: Cambridge University Press. [Google Scholar]
- King J, Jukes T.. 1969. Non-Darwinian evolution. Science 164(3881):788–798. [DOI] [PubMed] [Google Scholar]
- Landry CR, Freschi L, Zarin T, Moses AM.. 2014. Turnover of protein phosphorylation evolving under stabilizing selection. Front Genet. 5:245.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CR, Levy ED, Michnick SW.. 2009. Weak functional constraints on phosphoproteomes. Trends Genet. 25(5):193–197. [DOI] [PubMed] [Google Scholar]
- Lence T, Akhtar J, Bayer M, Schmid K, Spindler L, Ho C, Kreim N, Andrade-Navarro M, Poeck B, Helm M, et al. 2016. m6A modulates neuronal functions and sex determination in Drosophila. Nature 540(7632):242–247. [DOI] [PubMed] [Google Scholar]
- Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN.. 2010. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics 26(4):493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Mason CE.. 2014. The pivotal regulatory landscape of RNA modifications. Annu Rev Genomics Hum Genet. 15:127–150. [DOI] [PubMed] [Google Scholar]
- Li X, Xiong X, Yi C.. 2016. Epitranscriptome sequencing technologies: decoding RNA modifications. Nat Methods. 14(1):23–31. [DOI] [PubMed] [Google Scholar]
- Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR.. 2015. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 12(8):767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo GZ, MacQueen A, Zheng G, Duan H, Dore LC, Lu Z, Liu J, Chen K, Jia G, Bergelson J, et al. 2014. Unique features of the m6A methylome in Arabidopsis thaliana. Nat Commun. 5:5630.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Zhao B, Chen K, Thomas A, Tuteja JH, He X, He C, White KP.. 2017. Evolution of transcript modification by N6-methyladenosine in primates. Genome Res. 27(3):385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al. 2013. 2013. MODOMICS: a database of RNA modification pathways – 2013 update. Nucleic Acids Res. 41(Database issue):D262–D267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean CJ, Metzger BPH, Yang JR, Ho WC, Moyers B, Zhang J.. 2017. Deciphering the genic basis of yeast fitness variation by simultaneous forward and reverse genetics. Mol Biol Evol. 34(10):2486–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Jaffrey SR.. 2014. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol. 15(5):313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR.. 2012. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149(7):1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses AM, Liku ME, Li JJ, Durbin R.. 2007. Regulatory evolution in proteins by turnover and lineage-specific changes of cyclin-dependent kinase consensus sites. Proc Natl Acad Sci USA. 104(45):17713–17718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Ba AN, Moses AM.. 2010. Evolution of characterized phosphorylation sites in budding yeast. Mol Biol Evol. 27(9):2027–2037. [DOI] [PubMed] [Google Scholar]
- Park C, Zhang J.. 2011. Genome-wide evolutionary conservation of N-glycosylation sites. Mol Biol Evol. 28(8):2351–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto Y, Cohen HY, Levanon EY.. 2014. Mammalian conserved ADAR targets comprise only a small fragment of the human editosome. Genome Biol. 15(1):R5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignant JY, Soller M.. 2017. m6A in mRNA: an ancient mechanism for fine-tuning gene expression. Trends Genet. 33(6):380–390. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Agarwala SD, Mumbach MR, Jovanovic M, Mertins P, Shishkin A, Tabach Y, Mikkelsen TS, Satija R, Ruvkun G, et al. 2013. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 155(6):1409–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slobodin B, Han RQ, Calderone V, Vrielink JAFO, Loayza-Puch F, Elkon R, Agami R.. 2017. Transcription impacts the efficiency of mRNA translation via co-transcriptional N6-adenosine methylation. Cell 169(2):326–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer RA, Rodriguez-Mias RA, Haas KM, Hsu JI, Vieitez C, Sole C, Swaney DL, Stanford LB, Liachko I, Bottcher R, et al. 2016. Evolution of protein phosphorylation across 18 fungal species. Science 354(6309):229–232. [DOI] [PubMed] [Google Scholar]
- Sun WJ, Li JH, Liu S, Wu J, Zhou H, Qu LH, Yang JH.. 2016. RMBase: a resource for decoding the landscape of RNA modifications from high-throughput sequencing data. Nucleic Acids Res. 44(D1):D259–D265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P.. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34(Web Server issue):W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. 2014. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505(7481):117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu ZK, Han DL, Ma HH, Weng XC, Chen K, Shi HL, He C.. 2015. N-6-methyladenosine modulates messenger RNA translation efficiency. Cell 161(6):1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Laurent B, Hsu CH, Nachtergaele S, Lu ZK, Sheng WQ, Xu CY, Hen HC, Jian OY, Wang SQ, et al. 2017. RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature 543(7646):573–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Yang Y, Feng GH, Sun BF, Chen JQ, Li YF, Chen YS, Zhang XX, Wang CX, Jiang LY, et al. 2017. Mettl3-mediated m6A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 27(9):1100–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Zhang J.. 2014. Human coding RNA editing is generally nonadaptive. Proc Natl Acad Sci USA. 111(10):3769–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Zhang J.. 2015. In search of beneficial coding RNA editing. Mol Biol Evol. 32(2):536–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. 2010. Evolutionary genetics: progress and challenges In: Bell MA, Futuyma DJ, Eanes WF, Levinton JS, editors. Evolution since Darwin: the first 150 years, Sunderland (MA: ): Sinauer; p. 87–118. [Google Scholar]
- Zhang J, Yang JR.. 2015. Determinants of the rate of protein sequence evolution. Nat Rev Genet. 16(7):409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao BS, Wang X, Beadell AV, Lu Z, Shi H, Kuuspalu A, Ho RK, He C.. 2017. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 542(7642):475–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.