Abstract
Amoebozoa is the eukaryotic supergroup sister to Obazoa, the lineage that contains the animals and Fungi, as well as their protistan relatives, and the breviate and apusomonad flagellates. Amoebozoa is extraordinarily diverse, encompassing important model organisms and significant pathogens. Although amoebozoans are integral to global nutrient cycles and present in nearly all environments, they remain vastly understudied. We present a robust phylogeny of Amoebozoa based on broad representative set of taxa in a phylogenomic framework (325 genes). By sampling 61 taxa using culture-based and single-cell transcriptomics, our analyses show two major clades of Amoebozoa, Discosea, and Tevosa. This phylogeny refutes previous studies in major respects. Our results support the hypothesis that the last common ancestor of Amoebozoa was sexual and flagellated, it also may have had the ability to disperse propagules from a sporocarp-type fruiting body. Overall, the main macroevolutionary patterns in Amoebozoa appear to result from the parallel losses of homologous characters of a multiphase life cycle that included flagella, sex, and sporocarps rather than independent acquisition of convergent features.
Keywords: phylogenomics, transcriptomes, Amoebozoa, reductive evolution, phylotranscriptomics
Introduction
Amoeboid cells (amoebae) are morphologically dynamic eukaryotic cell types that move and/or feed using transient extensions of the cell called pseudopodia. Classically, all amoeboid microbes were included in the taxon Sarcodina (Schmarda 1871; Page 1976). However, molecular phylogenetic studies have shown that they are scattered among the eukaryotes, with the bulk being in the taxa Amoebozoa and Rhizaria, and a few in the taxa Heterolobosea, Stramenopila, and the lineage containing Metazoa and Fungi (Obazoa; Pawlowski 2008; Brown et al. 2013). Amoebozoa is morphologically and ecologically very diverse including important pathogens to metazoans (e.g., Entamoeba and Acanthamoeba; Visvesvara et al. 2007) and the model organisms Dictyostelium discoideum and Physarum polycephalum (Eichinger et al. 2005; Schaap et al. 2015). Amoebozoa occupies an evolutionarily important position as the sister lineage to Obazoa (Brown et al. 2013). A deeper understanding of the group will help resolve the evolutionary histories and trajectories of Amoebozoa and Obazoa at both a morphological and genome complexity level.
Nearly all species in Amoebozoa have amoeboid cells in their life cycles, including the well-known taxon, Amoeba proteus. However, most amoebozoans are not as simple as A. proteus whose only known morphlological state is a “naked” amoeba. Rather, amoebozoans are exceptionally diverse in cell form and life cycle (figs. 1,2), including naked amoebae (amoebae without a shell; fig. 1), testate amoebae (i.e., amoebae with a shell, also known as a test; fig. 1A–C, J–L), amoeboid flagellates (fig. 1Q–T,V–X,AA), and those with life cycles that include cysts (sessile, walled, dormant states not formed through the development of fruiting bodies, further defined in supplementary table S1, Supplementary Material online), sexual states (Brown et al. 2007; Lahr et al. 2011b; Spiegel 2011; Tekle et al. 2017), and/or spore-bearing structures called fruiting bodies (fig. 2; Shadwick et al. 2009, 2016; Kudryavtsev et al. 2014).
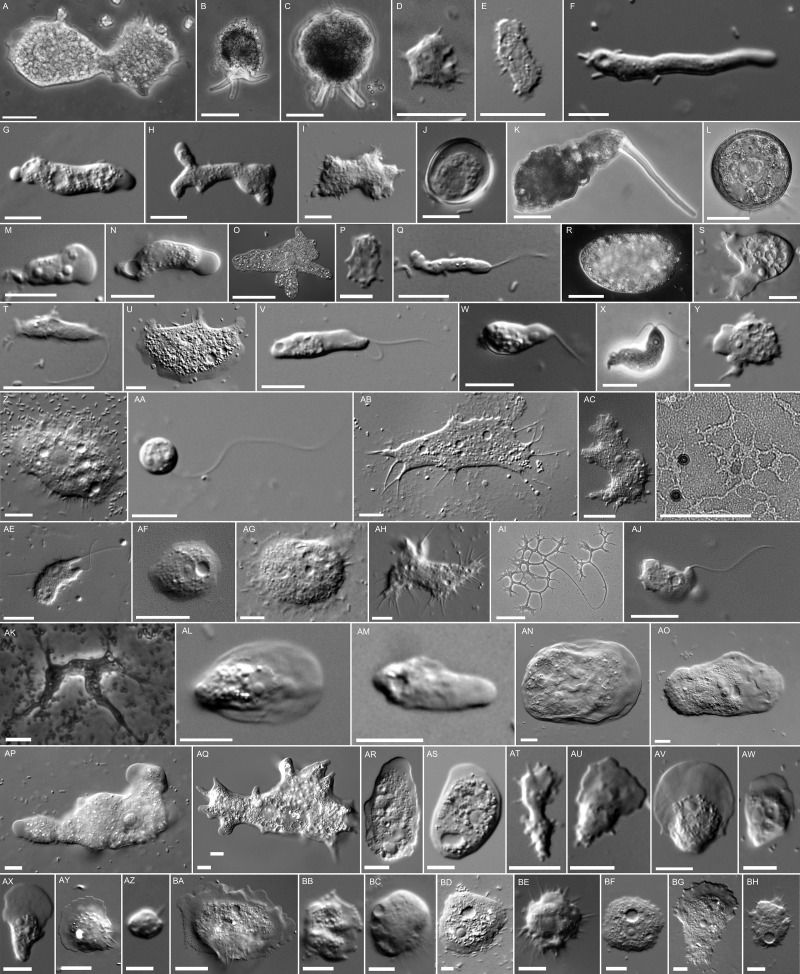
Fig. 1.
Representative trophic cells of amoebozoans examined in this study. (A) Trichosphaerium sp. leathery shell corycid amoeba. (B) Diplochlamys sp. leathery shell corycid amoeba. Scale bar = 50 µm. (C) Amphizonella sp. leathery shell corycid amoeba. Scale bar = 50 µm. (D) Micriamoeba sp. amoeba. (E) Echinamoeba exudans amoeba. (F) Vermamoeba vermiformis amoeba. (G) Vermamoeba sp. (CCAP1503-5) amoeba. (H) Rhizamoeba saxonica amoeba. (I) Flabellula citata amoeba. (J) Cryptodifflugia operculata testate amoeba. (K) Difflugia bryophila testate amoeba. Scale bar = 50 µm. (L) Arcella intermedia testate amoeba. (M) Nolandella sp. amoeba. (N) Copromyxa protea amoeba. (O) Amoeba proteus amoeba. Scale bar = 100 µm. (P) Squamamoeba japonica amoeba. Scale bar = 5 µm. (Q) Rhizomastix elongata amoeboflagellate. (R) Pelomyxa sp. amoeboflagellate. Scale bar = 50 µm. (S) Mastigella eilhardii amoeboflagellate. (T) Mastigamoeba abducta amoeboflagellate. (U) Echinosteliopsis oligospora amoeba. (V) Echinostelium minutum amoeboflagellate. (W) Echinostelium bisporum amoeboflagellate. (X) Protosporangium articulatum amoeboflagellate. (Y) Clastostelium recurvatum amoeba. (Z) Flamella aegyptia amoeba. (AA) Phalansterium solitarium flagellate. (AB) Schizoplasmodiopsis vulgaris amoeba. (AC) Tychosporium acutostipes amoeba. (AD) Schizoplasmodiopsis pseudoendospora plasmodium. Scale bar = 100 µm. (AE) Cavostelium apophysatum amoeboflagellate. (AF) Protostelium nocturnum amoeba. (AG) Soliformovum irregularis amoeba. (AH) Grellamoeba robusta amoeba. (AI) Nematostelium gracile plasmodium. Scale bar = 100 µm. (AJ) Ceratiomyxella tahitiensis amoeba. (AK) Acramoeba dendroida amoeba. (AL) Stenamoeba stenopodia amoeba. (AM) Stenamoeba limacina amoeba. (AN) Thecamoeba sp. amoeba. (AO) Sappinia pedata amoeba. (AP) Thecamoebidae isolate (RHP1-1) amoeba. (AQ) Mayorella cantabrigiensis amoeba. (AR) Paradermamoeba levis amoeba. (AS) Dermamoeba algensis amoeba. (AT) Vexillifera minutissima amoeba. (AU) Cunea sp. (JDS-Ruffled) amoeba. Scale bar = 5 µm. (AV) Vannella fimicola amoeba. (AW) Ripella sp. amoeba. (AX) Lingulamoeba sp. amoeba. (AY) Ovalopodium desertum amoeba. (AZ) Parvamoeba rugata amoeba Scale bar = 5 µm. (BA) Cochliopodium minus amoeba. (BB) Pellita catalonica amoeba. (BC) Gocevia fonbrunei amoeba. (BD) Endostelium zonatum (PRA-191) amoeba. (BE) Protacanthamoeba bohemica amoeba. (BF) Luapeleamoeba arachisporum amoeba. (BG) Luapeleamoeba hula amoeba. (BH) Acanthamoeba pyriformis amoeba. All scale bars = 10 µm unless otherwise noted.
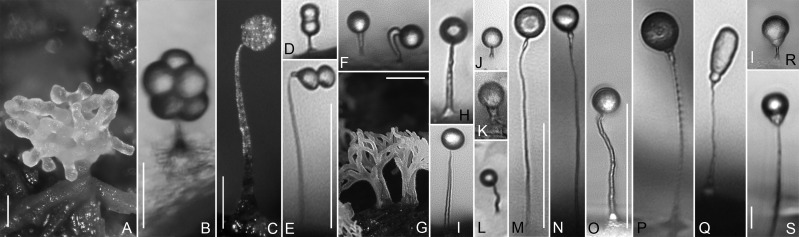
Fig. 2.
Representative fruiting amoebozoans sampled. (A) Sorocarp of Copromyxa protea, scale bar = 100 µm. (B–S) sporocarps of sporocarpic amoebozoans. (B) Echinosteliopsis oligospora, scale bar = 25 µm. (C) Echinostelium minutum, scale bar = 100 µm. (D) Echinostelium bisporum. (E) Protosproangium articulatum, scale bar = 50 µm. (F) Clastostelium recurvatum. (G) Ceratiomyxa fruticulosa, scale bar = 1 mm. (H) Schizoplasmodiopsis vulgaris. (I) Tychosporium acutostipes. (J) Schizoplasmodiopsis pseudoendospora. (K) Cavostelium apophysatum. (L) Protostelium nocturnum. (M) Soliformovum irregularis, scale bar = 50 µm. (N) Nematostelium gracile, same sporocarp morphology as Ceratiomyxella tahitiensis. (O) Vannella fimicola, scale bar = 50 µm. (P) Endostelium zonatum. (Q) Luapelamoeba arachisporum. (R) Luapelamoeba hula, scale bar = 10 µm. (S) Acanthamoeba pyriformis, scale bar = 10 µm. Images D–F, H–K, O–S are to scale with each other. Images M and N are to scale.
Fruiting bodies among amoebozoans produce dormant, walled propagules (spores, see further definition in supplementary table S1, Supplementary Material online) that are derived through the development of these structures. Sorocarps like those in the well-studied Dictyostelia are formed by aggregation of individual cells that work in concert to form an emergent, aerial multicellular structure (fig. 2A) with many spores (Brown and Silberman 2013). This trait evolved at least seven times independently in eukaryotes (Brown and Silberman 2013). In contrast, sporocarps are formed by a single cell that produces an elevated extracellular, subaerial stalk upon which it develops into a spore or cleaves into several spores (fig. 2B–S; Olive 1975; Shadwick et al. 2009; Tice et al. 2016a). Unlike sorocarps, sporocarpic development is unique to Amoebozoa, where it is found in the microscopic protosteloid amoebae as well as in the mostly macroscopic myxogastrid plasmodial (i.e., large multinucleate cell) slime molds (see Olive 1975; Shadwick et al. 2009).
It is has not been well established if the various types of amoeboid cells among Amoebozoa are homologs of each other (Spiegel and Feldman 1985; Spiegel et al. 1995). In addition to being trophozoites (feeding cells), some amoeboid cells may function as gametes or zygotes or they may become committed to differentiate into fruiting bodies or resting stages (Spiegel and Feldman 1985).
Amoebozoans evolved from their last common ancestor around 1.2 billion years ago (Eme et al. 2014), and as such their evolutionary relationships are deep and are currently not well understood. Without such understanding, it is difficult to discern the macroevolutionary patterns responsible for the extreme diversity in cell types and life cycles within this supergroup. The morphological characters, alone, are considered too few and ambiguous to accurately resolve a phylogeny over the evolutionary distances in Amoebozoa (Page 1988; Patterson 1999). However, early molecular phylogenetic analyses, based mostly on nuclear small subunit ribosomal RNA gene sequences (SSU), yielded no better insights than morphology because they suffered from very poor taxon sampling and more issues (i.e., extreme rate differences and compositional bias). With increasing taxon sampling and/or number of genes analyzed, the monophyly of what we now call Amoebozoa began to emerge (Bolivar et al. 2001; Fahrni et al. 2003; Cavalier-Smith 1998; Pawlowski 2008; Shadwick et al. 2009; Lahr et al. 2011a), and many, robust lower-level taxa that mostly correspond with classical orders have been established (Amaral Zettler et al. 2000; Tekle et al. 2008; Lahr et al. 2011a; Smirnov et al. 2011). These early analyses hinted at the existence of several higher, supra-ordinal taxa (Cavalier-Smith 1998; Cavalier-Smith et al. 2004; Smirnov et al. 2005), although only one of these (Tubulinea) appears to be well supported on a consistent basis (Cavalier-Smith et al. 2004; Lahr et al. 2011a; Smirnov et al. 2011; Berney et al. 2015). The remaining deepest branches in the Tree of Amoebozoa have remained very difficult to resolve (Cavalier-Smith et al. 2016; Tekle et al. 2016).
Recent phylogenomic studies strongly support the monophyly of Amoebozoa (Brown et al. 2013), but robust inferences on the deepest relationships and even the composition of proposed major lineages are likely compromised by a severe undersampling of the known amoebozoan diversity (Cavalier-Smith et al. 2015, 2016; Tekle et al. 2016). Taxa underrepresented in these analyses include those that have 1) complex life cycles involving the sequential development of multiple amoeboid morphologies, 2) flagellated cells, 3) sporocarps, and 4) tests. Here, we present data derived from a set of 325 protein-coding genes drawn from a sampling of 86 amoebozoans that represent the known morphological diversity of the supergroup in its entirety (figs. 1,2). We include taxa from previous phylogenomic studies that focused on Amoebozoa (Cavalier-Smith et al. 2015, 2016; Tekle et al. 2016; Tice et al. 2016a), and collected new transcriptomic data for 61 additional species (figs. 1–3, and robust site sampling per taxon (fig. 3, histogram, see supplementary table S2, Supplementary Material online). Our transcriptomic data include the archetypal amoebozoan, A. proteus, nearly all genera for which a flagellate state is known, most taxa that are known or suspected to be sexual (Lahr et al. 2011b), several sporocarpic myxogastrids, and almost all genera that are known to produce unicellular stalked, protosteloid fruiting bodies via sporocarpy (Spiegel 1990; Shadwick et al. 2009). We also include representatives of both clades where sorocarpic fruiting is known (Brown et al. 2011; Romeralo et al. 2013) and both known types of testate amoebozoans, the leathery shelled amoebae (herein named corycid amoebae) and the rigid-shelled arcellinids. To a great extent, our capacity to sample such taxonomic depth and diversity is due to our utilization of single cell transcriptomic methods. This allowed us to include species in our analyses that have not been cultivated or are predators of other eukaryotes (see supplementary table S3, Supplementary Material online).
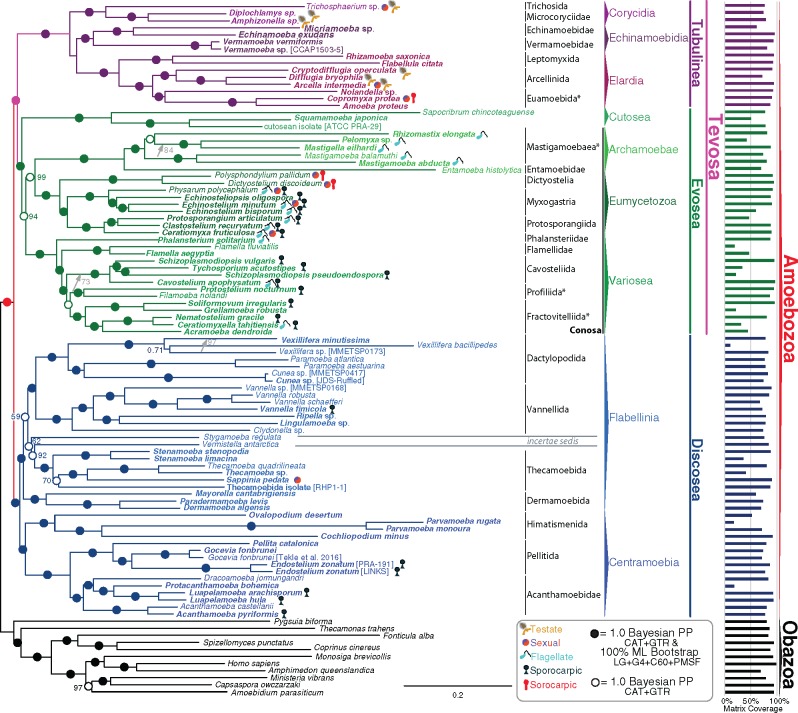
Fig. 3.
The Tree of Amoebozoa. 325 gene (63,157 sites) phylogeny of Amoebozoa rooted with Obazoa. The tree was built using PhyloBayes-MPI v1.6j under the CAT + GTR model of protein evolution, with two converged independent chains, burnin 1,850 generations, with a postburnin sampling of ∼2,800 generations. Values at nodes are posterior probability and ML bootstrap (MLBS) (1,000 ultrafast BS reps, IQ-Tree LG + Γ4 + FMIX(emprical,C60) + PMSF values respectively. Filled in circles at nodes represents full support in both analyses (1.0/100). Open circles represent full Bayesian support without full MLBS support. Colors of dots represent taxa and branches are colored according to their respective taxonomy (i.e., red is corresponding to the deepest branches of Amoebozoa). Nodes not recovered in the corresponding ML analysis are represented by an asterisk and the differences between the Bayesian and ML tree topologies are depicted with gray arrows. The length of the Entamoeba branch has been reduced by 50%. Bars along the right side of the figure show the percent of the total data set available for each taxon. Novel data were generated in this study for taxa whose names are bold.
We robustly recover for the first time major lineages of amoebozoans (Evosea, Tubulinea, and Discosea) and a highly supported dichotomy between Tubulinea + Evosea (Tevosa) and Discosea. We can now infer that the last common ancestor of the group most likely had a multistate life cycle with sex, flagella, and probably sporocarps. From these data and the mapping of these phenotypic traits, we are able to develop several testable hypotheses concerning the macroevolutionary patterns of morphology in Amoebozoa, which will enable subsequent studies to test our hypotheses.
Results
A Resolved Tree of Amoebozoa
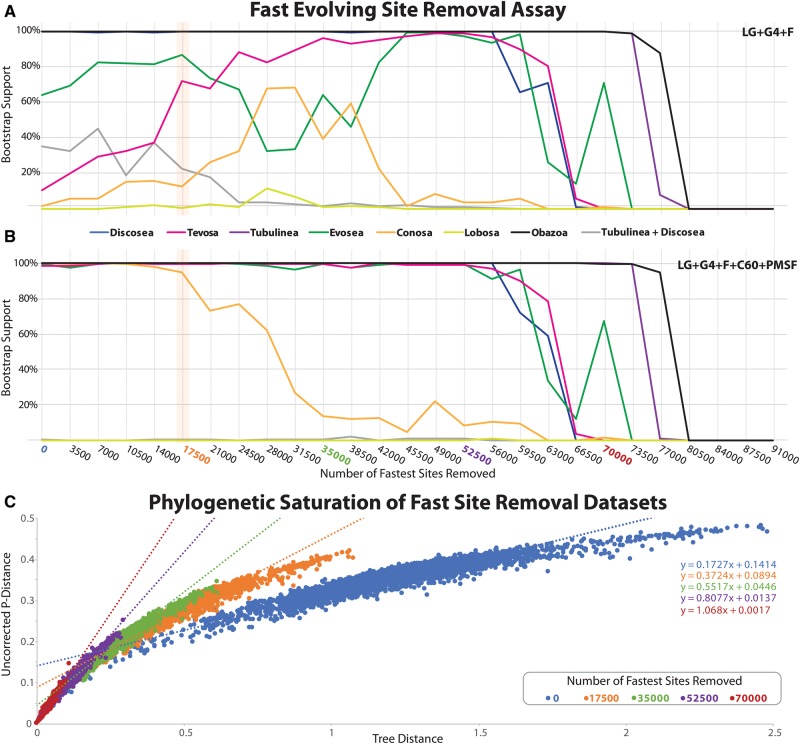
For our phylogenomic results presented herein, a supermatrix composed of 325 proteins from 98 taxa was constructed. Initial analyses of this full supermatrix with simple site-homogeneous (LG + Γ4 + F; see supplementary fig. S1, Supplementary Material online) and more sophisticated site-heterogeneous (LG + Γ4 + F + C60 + PMSF; see supplementary fig. S2, Supplementary Material online) models (detailed below) conflicted regarding several deep nodes of the Amoebozoa tree. For example, the Tevosa clade was recovered by the site-heterogeneous model (see supplementary fig. S2, Supplementary Material online), but not by the site-homogeneous model (see supplementary fig. S1, Supplementary Material online). To examine this conflict, we estimated the rate of evolution at sites in the supermatrix and progressively removed them in a stepwise fashion, plotting bootstrap values for nodes of interest in the tree per site deletion step under the models LG + Γ4 + F and LG + Γ4 + F + C60 + PMSF models estimated in IQ-Tree (fig. 4A, B). The rationale for this procedure is that the fastest evolving sites within a deep phylogenetic analysis are often saturated with multiple substitutions and, as a result of model-misspecification can manifest nonphylogenetic signal especially when overly simplistic models are used (Jeffroy et al. 2006; Olsen 1987; Lartillot and Philippe 2008; Brown et al. 2013). Analysis of the numbers of substitutions per sites in the various deletion data sets revealed that the full data set displayed substitutional saturation and removal of fastest evolving sites did ameliorate the problem (fig. 4C). With the progressive removal of the fastest sites, the site homogeneous analyses yielded increasing support for the same major groups as the initial site-heterogeneous analyses. In contrast, the topology estimated by site-heterogeneous model and its support values were unaffected by fast-site removal until well over half the data set was removed (fig. 4B). To avoid problems associated with model misspecification in fast-evolving sites, all of our subsequent phylogenomic analyses were based on a data set that excludes fastest evolving sites as well as the removal of uninformative constant sites (fig. 4, the 17,500 fast site removal data set, less constant sites). This noise reduction step also had the benefit of reducing the data set size, lessening the extreme computational burden of phylogenomic tree inference under Phylobayes, which is a particularly important consideration when working with data sets of this size. For example, our Phylobayes analyses presented herein required ∼979,200 CPU hours.
Fig. 4.
Effects of fast evolving sites on our phylogenomic analyses. (A, B) Sites were sorted based on their rates of evolution estimated under LG + Γ4 and removed from the data set from highest to lowest rate. Each step has 3,500 of the fastest evolving sites removed. The bootstrap values for each bipartition of interest are plotted under the LG + Γ4 + F (A) and LG + Γ4 + C60 + F + PMSF (B). The data set with 17,500 sites removed (orange bar) was the basis of the main phylogenomic analyses shown in figure 3. This data set showed the most drastic changes in MLBS values in the LG + Γ4 + F MLBS assay (A). (C) Phylogenetic signal saturation assay of five data sets within the fast site removal assay shown in A. Signal saturation was assessed by plotting uncorrected pairwise distance to the per taxon tip to base tree distance the best scoring ML tree inferred under LG + Γ4 + C60 + F + PMSF. Data sets plotted correspond to the whole data set and a subset of fast site deletion data sets (17,500, 35,000, 52,500, and 70,000 sites removed) shown in A. The linear equation (y = mx + b) for each plot is shown on right of the figure and is color coded to each examined data set. The orange data set (17,500 sites removed) was used for subsequent analyses as it shows limited phylogenetic saturation to phylogenetic signal.
The resulting Tree of Amoebozoa, in figure 3 is based on the Bayesian site-heterogeneous mixture model, CAT-GTR, the most realistic phylogenetic model available (Lartillot et al. 2013). Although the CAT-GTR model cannot be used in the ML framework, the LG + Γ4 + F + C60 + PMSF model that similarly estimates site-specific amino acid (AA) profiles for phylogenomic analyses based on the C60 empirical frequency profiles (Le and Gascuel 2008) was used for ML analyses in IQ-Tree (Nguyen et al. 2014). The Bayesian analyses recovered a highly resolved, rooted Tree of Amoebozoa in which all nodes are fully supported by Bayesian posterior probabilities (BPP) of the two converged Phylobayes chains except for one node within a terminal genus, Vexillifera (fig. 3). With a few exceptions (illustrated with arrows in fig. 3), Maximum Likelihood (ML) recovers these nodes with high support values (ML bootstrap, MLBS, >95%; fig. 3, see supplementary fig. S3, Supplementary Material online). However, we believe caution is warranted in interpreting branches that are not fully supported by both BPP and MLBS methods.
As with many previous molecular phylogenetic analyses (Cavalier-Smith 1998; Pawlowski 2008; Shadwick et al. 2009; Lahr et al. 2011a), we find that Amoebozoa is a fully supported clade. Armed with strong statistical support for the deepest nodes within the Tree of Amoebozoa we show that there are three major lineages. The lineages Tubulinea and Discosea continue to be recovered as previously suggested (Cavalier-Smith et al. 2015, 2016; Tice et al. 2016a). However, we do not recover Lobosa sensuSmirnov et al. (2011), that is, a clade comprising Discosea + Tubulinea as sister lineages or Lobosa sensuCavalier-Smith et al. (2016) (Discosea + Tubulinea + Cutosea, listed as Lobosa in fig. 4 and table 1). Additionally, constraining the tree with Lobosa (Discosea + Tubulinea, with or without Cutosea) can be rejected under approximately unbiased (AU) tests (Shimodaira, 2002) at a confidence interval of 95% using our data set (P-value = 0.0059 and 0.0014, respectively, table 1). Instead Tubulinea is sister to a major monophyletic lineage we call Evosea (named herein composed of Eumycetozoa, Variosea, Archamoebae, and Cutosea) (BPP = 1.0, MLBS = 99%, fig. 3, see supplementary fig. S3, Supplementary Material online). We propose the name Tevosa for the clade Tubulinea + Evosea.
Table 1.
Approximately Unbiased Topological Test Constraints
| Constrained Tree | P-Value AU | log L | Δlog L |
|---|---|---|---|
| Best ML tree | 0.9018 | −3291377.510 | 0.000 |
| Phylobayes CAT-GTR tree | 0.2747 | −3291408.504 | 30.995 |
| (Tubulinea + Discosea) | 0.0059 | −3291481.495 | 103.985 |
| (Evosea + Discosea) | 0.0011 | −3291482.573 | 105.063 |
| (Cutosea + Tubulinea + Discosea) | 0.0014 | −3291580.832 | 203.323 |
| ((Cutosea + Archamoebae) + (Variosea + Eumycetozoa)) | 0.0621 | −3291477.789 | 100.280 |
| (Archamoebae + (Cutosea + (Variosea + Eumycetozoa))) | 0.0046 | −3291526.597 | 149.088 |
| ((Cutosea + Variosea) + (Archamoebae + Eumycetozoa)) | 0.1995 | −3291407.372 | 29.863 |
| (Variosea + (Cutosea + (Archamoebae + Eumycetozoa))) | 0.0377 | −3291429.080 | 51.571 |
| (Eumycetozoa + (Cutosea + (Archamoebae + Variosea))) | 0.0000 | −3291664.894 | 287.384 |
| (Eumycetozoa + (Archamoebae + (Cutosea + Variosea))) | 0.0325 | −3291639.763 | 262.253 |
| ((Cutosea + Eumycetozoa) + (Archamoebae + Variosea)) | 0.0027 | −3291688.843 | 311.334 |
Each tree was loosely constrained with the hypothetical groupings and optimized under LG + Γ4 + F + C60 + PSMF in IQ-tree using the 325 gene (63,157 sites) data set as presented in figure 3. The optimized trees were compared using the approximately unbiased test with 10,000 RELL bootstrap replicates. Maximum log likelihoods of each constraint and their differences from the optimal ML tree are listed. The hypotheses within the 95% confidence interval that could not be rejected are where P ≥ 0.05.
These backbone clades of Amoebozoa are in concordance with Tekle et al. (2016), but not with Cavalier-Smith et al. (2016). The deepest clades we confidently accept in Amoebozoa are Tevosa and Discosea. The group names we are using and have named herein are mapped on the tree in figure 3 and are listed with a novel clade in the supplementary text, Supplementary Material online. We have modified the composition and definitions of all major lineages with respect to our findings. On the basis of the phylogenomic evidence, the deepest node in Amoebozoa appears to be between Discosea and Tevosa. Our results lead us to reject the concepts of Lobosa sensuSmirnov et al. (2011) (Discosea + Tubulinea) and Lobosa sensuCavalier-Smith et al. (2016) (Discosea + Tubulinea + Cutosea) (fig. 3, table 1). Because the clade Tevosa has no obvious unifying morphological traits, we will discuss Tubulinea and Evosea separately.
Tubulinea
Most members of Tubulinea have an amoeboid state in their life histories that is tubular in cross section and lacks subpseudopodia, though a few have a flat cross section and subpseudopodia (Smirnov et al. 2011; fig. 1A–O, see supplementary table S1, Supplementary Material online). We recover Tubulinea, a group that often shows strong statistical support even when few genes are analyzed (Lahr et al. 2011a, 2013; Cavalier-Smith et al. 2015, 2016). The subclades of Tubulinea are all strongly supported by both BPP and MLBS. With our broad sampling of Tubulinea taxa, we provide deep resolution and reveal a novel subclade, which we name Corycidia (fig. 3, see supplementary text, Supplementary Material online). Corycidia is the strongly supported sister lineage to the rest of the Tubulinea (includes Echinamoebidia and Elardia [see supplementary taxonomic summary, Supplementary Material online]). Corycidian taxa are characterized by having a leathery flexible tests (fig. 1A–C), distinct from the rigid tests found within Arcellinida (fig. 1J–L, see supplementary table S1, Supplementary Material online). Corycidia includes Diplochlamys sp. (fig. 1B), Amphizonella sp. (fig. 1C), and Trichosphaerium sp. (fig. 1A), which were difficult to place within Amoebozoa in previous phylogenomic studies due to poor taxon sampling, specifically from within Tubulinea (Tekle et al. 2008; Cavalier-Smith et al. 2016; Tekle et al. 2016). In previous, less comprehensive studies, the only member of Corycidia included was Trichosphaerium sp.; it was placed either within Tubulinea (Cavalier-Smith et al. 2016) or sister to Tubulinea (Tekle et al. 2016). This particular isolate of Trichosphaerium was renamed “Atrichosa algivora” in (Cavalier-Smith et al. 2016); however, given our observations of this isolate (fig. 1A) it likely represents a spicule-less stage (gamont) of Trichosphaerium the details of which are discussed in (Page 1983). A spicule-bearing Trichosphaerium must be examined further to confirm this.
Evosea
The clade we name Evosea contains the well-supported subclades Cutosea, Archamoebae, Eumycetozoa, and Variosea (fig. 3). However, we are hesitant to state categorically the exact branching orders among them because of the lack of MLBS support for the deepest nodes within the group (fig. 3, see supplementary fig. S3, Supplementary Material online) and AU test results show no significant resolution of the branching order (table 1). We demonstrate that Cutosea (Cavalier-Smith et al. 2016) belongs within this group and not within Lobosa (i.e., Tubulinea + Discosea) as argued in (Cavalier-Smith et al. 2016), in part because the proposed group Lobosa is not recovered in our analyses. Evosea corresponds, more or less, with the traditional Conosa (Cavalier-Smith 1998) plus the Cutosea. However, Conosa is morphologically defined as having taxa with flagella associated with a radiating cone of microtubules emerging from either their anteriorly directed basal body or a microtubule organizing center associated with this basal body. As far as currently known, Cutosea is devoid of flagellated taxa (Cavalier-Smith et al. 2016). We feel that the most robust result and conclusion should be to provide a new name for this clade and not to subsume Cutosea into Conosa. We feel that leaving Conosa as a valid and nonsynonymous clade to Evosea is a more reliable and stable taxonomic option. More work should be focused on this clade, particularly, isolation and transcriptomic sequencing efforts to collect more data from cutosean taxa.
Cutosea is represented by Sapocribrum, Squamamoeba, and American Type Culture Collection (ATCC) strain PRA-29 (deposited as “Pessonella”). The former two taxa are small amoebae covered with very small scales (Pussard 1973; Tekle et al. 2008; Kudryavtsev and Pawlowski 2013; Lahr et al. 2015). ATCC PRA-29 was misidentified as Pessonella sensu (Pussard 1973; Tekle et al. 2008), and further work should be done to examine this strain in order to formally describe it.
Cutosea sensuCavalier-Smith et al. (2016) was originally placed as sister to the rest of Lobosa. However, since their Amoebozoa-only trees (Cavalier-Smith et al. 2016) are unrooted, a topology where Cutosea is sister to Conosa (Archamoebae, Eumycetozoa, and Variosea) cannot be ruled out. In Tekle et al. (2016) the authors treat Cutosea as sister to Himatismenida, which, together, are sister to Tubulinea in their analyses. This result of Tekle et al. (2016) is probably due to a long branch attraction artifact that was not remedied by a more realistic evolutionary model such as the site heterogeneous model used in our analyses. Here, Cutosea appears to be sister to the rest of Evosea, that is, the traditional Conosa (fig. 3). In the full data set, there is full MLBS support for Conosa, but the order among Archamoebae, Eumycetozoa, and Variosea, with or without Cutosea branching within, is ambiguous when fast evolving sites are removed under the LG + Γ4 + F + C60 + PMSF model (fig. 4B).
We rename the well-supported group that contains myxogastrids, dictyostelids, and protosporangiids (represented here by Ceratiomyxa, Clastostelium, and Protosporangium) Eumycetozoa rather than the recently coined term Macromycetozoa (Fiore-Donno et al. 2010) for two major reasons. First, Eumycetozoa is the older name such that it even has priority over the entirety of Amoebozoa (see Shadwick et al. 2009; Adl et al. 2012). However, the conservation of Amoebozoa was argued for because of its literature familiarity in Adl et al. (2012). Secondly, Eumycetozoa is a group that should include the myxogastrids and our usage corresponds to the Eumycetozoa hypothesis (Olive 1975) that posits a monophyletic group of exclusively fruiting protists that includes myxogastrids, dictyostelids, and some protosteloid amoebae, in this case, the protosporangiids (fig. 3). The previous incertae sedis protosteloid amoeba, Echinosteliopsis oligospora (Spiegel 1990), a species that lacks a flagellate state in its life cycle and has an unusual multinucleolate nucleus (Reinhardt 1968) is a myxogastrid slime mold (fig. 3). As previously suggested (Whitney et al. 1982), we confirm that the protosteloid amoeba Echinostelium bisporum (figs. 1W, 2C), which lacks a plasmodial state (Spiegel and Feldman 1989), is also a myxogastrid and is sister to the more typical Echinostelium minutum (figs. 1V,2B).
Our recovery of Variosea conforms with the hypothesis of Berney et al. (2015), although they were unable to show deep resolution with their 18S rRNA gene-based trees. Notably, we still lack the key multiflagellate variosean, Multicilia (Nikolaev et al. 2006); however, we conclusively demonstrate the monophyly of Variosea. Although some ambiguities exist between ML and Bayesian results (fig. 3, see supplementary fig. S1, Supplementary Material online) the backbone topology of Archamoebae corresponds well with the hypotheses about the relationships within this exclusively anaerobic clade, with a dichotomy yielding Pelobionta and Entamoebida (Pánek et al. 2016).
Evosea includes amoebae that are either tubular or flat in cross section (see supplementary table S1, Supplementary Material online). Archamoebae, Eumycetozoa, and Variosea each contain some species that are flagellated (usually amoeboflagellates sensuSpiegel 1990), and at least some members of all three groups (Adl et al. 2012) have flagella that contain an electron-dense element in their transition zone, a character not found in any other eukaryotes (see Spiegel 1990, 1991; Walker et al. 2001). Sex has been demonstrated in Eumycetozoa (in both Dictyostelia and Myxogastria) and life cycles consistent with sex (plasmogamy or uninucleate cells with two divisions similar to meiosis and/or obligate amoebae sensuSpiegel and Feldman (1985) arising from amoeboflagellates or genetic evidence of possible recombination) are present in several varioseans (e.g., Cavostelium aphophysatum, Ceratiomyxella tahitiensis; Spiegel 1990) as well as in Archamoebae (i.e., Entamoeba histolytica; Lahr et al. 2011b). In protosteloid evoseans, sex is sometimes thought to be associated with life cycles that include amoeboflagellates that germinate from spores and precede the development of nonflagellate obligate amoebae that subsequently produce the sporocarps (Spiegel and Feldman 1985; Spiegel et al. 1995; Spiegel 2011; see supplementary table S1, Supplementary Material online). However, additional work on sex in Evosea should be conducted to examine the true nature of these life cycles.
Discosea
Discosean amoebozoans are relatively flat in cross section, though some can be somewhat dome shaped, and they may or may not exhibit subpseudopodia (Brown et al. 2007; Smirnov et al. 2011; Shadwick et al. 2016; Tice et al. 2016a). Many of the flabellinids are fan-shaped and mostly uniaxial, though some can be multiaxial. Protosteloid, sporocarpic fruiting is found in both Flabellinia and Centramoebida (fig. 3 and see supplementary table S1, Supplementary Material online). Sex is suspected in the flabellinid Sappinia (Brown et al. 2007; fig. 3 and see supplementary table S1, Supplementary Material online). No sorocarpic or flagellated taxa are found in Discosea (fig. 3 and see supplementary table S1, Supplementary Material online).
Discosea is fully supported in our analyses and is divided into two fully supported groups, Flabellinia and Centramoebida sensuTekle et al. (2016). Even with their paucity of taxon sampling, the analyses of Cavalier-Smith et al. (2016) correspond with this dichotomy. In Cavalier-Smith et al. (2016) they recover Centramoebida and Himatismenida in their Centramoebia, but they did not sample any taxa in Pellitida (here represented by the genera Pellita, Gocevia, and Endostelium). All the species in their Flabellinia that correspond with species in our Flabellinia occur within the taxon, but the poorly supported relationships they inferred within the group are quite different than those we recover. Although the analyses of (Tekle et al. 2016) focused on Discosea, they did not recover the group as monophyletic. Their interpretation of results was plagued by incorrectly identified taxa, “Mayorella sp.” (strain BSH, MMETSP0417), which is a Cunea sp. (Cavalier-Smith et al. 2015), and “Pessonella sp.” (strain PRA-29, MMETSP0420), which is not true Pessonella. An assemblage that they called Eudiscosea contains the Centramoebida (including one pellitid, Gocevia fonbrunei) as the sister to a clade that corresponds very closely to our interpretation of Flabellinia. However, they placed the Himatismenida as sister to the Cutosea, which together group as sister to a severely undersampled Tubulinea.
Discussion
A well-resolved rooted tree allows us to construct testable hypotheses concerning the macroevolutionary patterns of morphology that characterize Amoebozoa. Here, we have mapped life cycle characters onto the Tree of Amoebozoa (fig. 3) and list these characters by species in supplementary table S1, Supplementary Material online. On the basis of parsimony principles we can assess the macroevolutionary trends across the Tree of Amoebozoa, as well as the nature of the last common ancestor of Amoebozoa (LCAA; fig. 3, table S1). For instance, it is possible to conclude that LCAA had a flagellate state in its life history (Spiegel 1991, 2011; Spiegel et al. 1995; Lahr et al. 2011b; Adl et al. 2012; Yubuki and Leander 2013) as this character must have been present in the Last Eukaryote Common Ancestor (LECA; Goodenough and Heitman 2014). Relatively few amoebozoan lineages have a flagellate state, and all confirmed flagellate taxa are found in Evosea (fig. 3, see supplementary table S1, Supplementary Material online; Spiegel 1991; Spiegel et al. 1995; Mikrjukov and Mylnikov 1998; Smirnov et al. 2011; Adl et al. 2012; Ptackova et al. 2013; Berney et al. 2015; Zadrobilkova et al. 2015; Pánek et al. 2016).
Although some exceptions exist (e.g., Multicilia [Nikolaev et al. 2006] and some species of Phalansterium [Smirnov et al. 2011]), almost all amoebozoans have an amoeboid state in their life history. It is likely LCAA had an amoeboid state, but the evolution of particular types of amoebae could have been quite complex (Spiegel et al. 1995). Alternatively, if the ancestor had a complex life cycle with one amoeboid state that alternated with another, as in myxogastrids and several protosteloid amoebae (Spiegel and Feldman 1985), an interesting hypothesis presents itself. For example in Amoebozoa, amoebae can be divided into two major morphological categories, tubular in cross section with axial cytoplasmic flow, and flattened in cross section with more irregular cytoplasmic flow (Smirnov et al. 2005, 2011; see supplementary table S1, Supplementary Material online). Several amoebozoans have life cycles with amoeboid cells that can assume both morphologies at alternate stages (see supplementary table S1, Supplementary Material online), for example, the myxogastrids and some protosteloid amoebae (Olive 1975; Adl et al. 2012). Other amoebozoans, such as some tubulinean taxa (e.g., the Leptomyxida—Rhizamoeba, Leptomyxa, and Flabellula; Page 1988), can transition back and forth in the same cell. It is clear from our results that the myxogastrids contain examples of the loss of the tubular plasmodial state seen in E. bisporum (Spiegel and Feldman 1989) and the loss of the alternate flagellate states found in E. oligospora (Reinhardt 1968).
Outside of Evosea, Trichosphaerium (here shown to be in Corycidia in Tubulinea), reportedly has a flagellate state (Schmarda 1871), which should be further investigated. Nonetheless, most flagellated amoebozoans, barring a few derived taxa (e.g., Pelomyxa [Seravin and Goodkov 1987]), have typical eukaryotic axonemes (a 9 × 2 + 2 microtubule configuration) and basal bodies (a 9 × 3 microtubule configuration). These nearly universal features of flagella are consistent with their shared evolutionary history with LECA; thus, the presence of a flagellate state in LCAA. Flagellates in Archamoebae have a less complex flagellar apparatus compared with those within Variosea and Eumycetozoa (Ptackova et al. 2013; Zadrobilkova et al. 2015; Pánek et al. 2016), and careful work has yet to conclude what homologies exist between the flagellar rootlets in Archamoebae and those of the more complex flagellar apparatuses in Variosea and Eumycetozoa. Some superficial comparisons have been made (Cavalier-Smith 1998; Cavalier-Smith et al. 2004, 2015, 2016). Since most rootlet elements have apparent homologs to those outside Amoebozoa (Spiegel 1991; Yubuki and Leander 2013), none is considered synapomorphies of the group, though their overall conformation might be synapomorphic. However, one potential synapomorphy of Amoebozoa, or at least in examined Evosea, is the presence of an electron dense plug in the flagellar transition zone of many Eumycetozoa, Variosea, and Archamoebae (Spiegel 1990, 1991; Ptackova et al. 2013). The presence of this character outside of Evosea can only be evaluated if flagellates of Trichosphaerium are rediscovered and examined.
There are members within each of the deep amoebozoan clades that have life cycles indicative of sex (fig. 3, see supplementary table S1, Supplementary Material online; Lahr et al. 2011b) and sex is relatively common outside of Amoebozoa. Therefore, LCAA must have been sexual (Spiegel 2011; Tekle et al. 2017). Since sex does not appear necessary for reproduction in any amoebozoan, it was likely facultative in this ancestor. It is interesting to note that the stages of sex, or suspected sex, are often associated with transitions from one somatic state to another, for example, the alternation between amoeboflagellates and plasmodia, a form of obligate amoeba, in the myxogastrid slime molds and between amoeboflagellates and obligate amoebae in protosteloid varioseans and eumycetozoans (Martin and Alexopoulos 1969; Spiegel and Feldman 1985; Spiegel et al. 1995).
Most amoebozoans, but not all (e.g., A. proteus [Page 1988]), make resting cysts that are different from spores in that they are sessile (i.e., not elevated above the substratum via a stalk as in protosteloid and myxogastrid amoebae), walled, dormant cells (see supplementary table S1, Supplementary Material online). This is also true of most protistan groups. Moreover, most sporocarpic amoebae are also capable of producing sessile cysts that are morphologically distinct from spores (Tice et al. 2016a). We however do not yet know the genetic basis for encystment across the breadth of Amoebozoa, which leaves us ill-informed about whether all cysts are homologous throughout the group as well as to outgroup protistan taxa. Future developmental studies should be undertaken to examine this more fully. Nevertheless, we hypothesize that LCAA was capable of encystment.
Sporocarpy, where a single cell develops into a subaerial, stalked fruiting structure that bears spores, is unique to Amoebozoa (Shadwick et al. 2009). Other than amoebae and cysts, both of whose genetic basis need to be worked out to confirm if the various types of amoebae and the types of cysts are homologs throughout the group, the most widespread developmental state found across Amoebozoa is sporocarpy (figs. 2,3, see supplementary table S1, Supplementary Material online; Olive 1975; Spiegel 1990; Shadwick et al. 2009; Kudryavtsev et al. 2014; Berney et al. 2015; Tice et al. 2016a). The most evolutionarily common type of sporocarpy across the Tree of Amoebozoa is protosteloid sporocarpy, where the cell that develops into a sporocarp and the sporocarp itself are microscopic and contain only one to a few spores (fig. 2B,D–S; Shadwick et al. 2009). Variosea and Eumycetozoa contain members that have sporocarpic fruiting (protosteloid and myxogastrid; fig. 2B–S, see supplementary table S1, Supplementary Material online). Protosteloid amoebae are present in both Evosea (Variosea and Eumycetozoa) and Discosea (Centramoebia and Flabellinia; fig. 3). If the root of Amoebozoa is between Discosea and the rest of the amoebozoans as suggested in our results, we may hypothesize that the last common ancestor of the whole group was sporocarpic, a possibility previously suggested (Shadwick et al. 2009). This hypothesis was recently rejected in Cavalier-Smith et al. (2016), where the authors suggested that protosteloid sporocarpy is simply the result of the addition of stalks to already existing cyst stages and could easily have evolved independently several times. However, in most protosteloid amoebae, cysts, when present, are morphologically distinct from spores (Olive 1975; Spiegel and Feldman 1993; Tice et al. 2016a). Also, fruiting body ultrastructure is remarkably similar in protosteloid amoebae in both Evosea and Discosea (Spiegel et al. 1979; Olive et al. 1984; Spiegel and Feldman 1993). Our hypothesis of the origin sporocarpy is quite testable. Future work using comparative developmental transcriptomics will help us to determine if the molecular mechanisms contributing to sporocarpy have a common evolutionary basis. Should sporocarpy in Evosea and Discosea prove homologous, that would support the hypothesis that it was present in their common ancestor, and thus, given our phylogeny, LCAA.
Our data clearly demonstrate that it is reasonable to assume that most macroevolutionary patterns in Amoebozoa are likely the result of the loss of characters that were present in a complex last common ancestor that was sexual and flagellate and perhaps had more than one somatic state and the ability to disperse propagules using sporocarpy. Nonetheless, there has also been the evolution of novel traits in certain lineages of amoebozoans. Some of these clearly need to be evaluated with more in-depth study, but two clearly appear to be the evolution of tests (herein) and sorocarpy (Brown et al. 2011), the formation of fruiting bodies via aggregation into multicellular assemblages. Tests appear to have originated twice within Tubulinea: in the arcellinids in Elardia and in the Corycidia (fig. 3). Sorocarpy has not only evolved many times outside of Amoebozoa (see Brown and Silberman 2013) it has independently emerged twice (fig. 3) within the group: once in Copromyxa (Brown et al. 2011; fig. 2A, see supplementary table S1, Supplementary Material online) in Elardia (Tubulinea) and also in the developmentally distinct dictyostelids in Eumycetozoa (Romeralo et al. 2013; Evosea; fig. 3; see supplementary table S1, Supplementary Material online). It is interesting to note that both tests and sorocarpy are not unique to Amoebozoa and have convergently evolved in other major lineages of amoeboid eukaryotes. Sorocarpy is known in Heterolobosea (Acrasis), Opisthokonta (Fonticula), Rhizaria (Guttulinopsis), Stramenopiles (Sorodiplophrys; Brown and Silberman 2013, Tice et al. 2016b). Testate amoebae are also present in Stramenopiles and widely distributed in Rhizaria (Pawlowski 2008).
With this well-supported phylogenetic hypothesis based on many genes and covering the taxonomic and developmental breadth of Amoebozoa, it has finally become possible to begin to address the macroevolutionary patterns of amoebozoans in an objective and nonspeculative manner. Robust studies of this nature provide a starting point for evolutionary developmental approaches to systematically examine the homology of the proposed characters. Additionally, with the data presented here, we can begin to identify convergent and homologous characters and character-states, not only morphological but also genomic, that have shaped and consequently led to the evolution of this vastly diverse and ecologically important supergroup of eukaryotes.
Materials and Methods
Details of experimental methods for isolation, identification, culturing, microscopic methods, nucleic acid extraction, cDNA construction, Illumina sequencing, cluster assembly, phylogenomic matrix assembly, and fast evolving site removal from the phylogenomic matrix were performed as in Tice et al. (2016a) and are described in the supplementary text 1, Supplementary Material online. Methods associated with our AU tests are also described in the supplementary text 1, Supplementary Material online.
Phylogenomic Tree Inference
Concatenated matrices compiled of 325 genes from 98 taxa resulted in an alignment of 93,478 amino acid (AA) sites. From this data set a subset of the fastest evolving sites (17,500 AA sites; fig. 3, discussed below and in supplementary text, Supplementary Material online) and constant sites (12,821 AA sites), which do not contribute to phylogenetic signal, were removed, resulting in our primary matrix of 63,157 AA sites. Bayesian inferences were performed in Phylobayes-MPI v1.6j (Lartillot et al. 2013). To account for site heterogeneous amino acid substitutions, we used CAT-GTR (Lartillot et al. 2013). For Bayesian analyses we ran four independent Markov chain Monte Carlo chains for ∼4,500 generations. Two of the chains converged at 1,850 generations with the largest discrepancy in posterior probabilities (PPs) (maxdiff) <0.069. The consensus of the two converged chains is presented in figure 3. While the other three chains did not converge, their overall topology was largely congruent with our converged chains and a consensus tree of all four chains is presented in supplementary figure S4, Supplementary Material online. ML trees were inferred in IQ-Tree v. 1.5.0 (Nguyen et al. 2014). IQ-Tree is currently the only high-performance ML program capable of implementing C-series models, which offer more realistic phylogenetic site-heterogeneous models, that are not options in other ML programs. The best-fitting available model for ML analyses was LG + Γ4 + C60 + F with class weights optimized from the data set. We used this model to estimate the “posterior mean site frequencies” using the Phylobayes tree as a guide tree (using the exchangeabilities from the LG matrix; Wang et al. 2014; Pánek et al. 2016) followed by tree-searching and bootstrapping (http://www.iqtree.org/doc/Complex-Models/; last accessed December 20, 2016). Topological support for trees of the supermatrix was conducted using 1,000 MLBS pseudoreplicates (fig. 3, see supplementary fig. S1, Supplementary Material online).
Data Availability
All transcriptomic data generated in this manuscript have been deposited with National Center for Biotechnology Information under the BioProject PRJNA380424 as detailed in supplementary table S2, Supplementary Material online. All single gene alignments, masked and unmasked, and phylogenomic matrices are available in the supplementary file Kang_etal.2017.tar.gz.
Supplementary Material
Acknowledgments
Mississippi State University’s High Performance Computing Collaboratory provided some computational resources. We wish to thank the anonymous reviewers of this manuscript who provided detailed constructive comments. We wish to thank Dr Franck Gael Carbonero at the University of Arkansas for the MiSeq sequencing. Additionally, we thank Génome Québec Innovation Centre for HiSeq sequencing. This project was supported by the National Science Foundation (NSF) Division of Environmental Biology (DEB) grant 1456054 (http://www.nsf.gov), awarded to M.W.B., F.W.S., and A.K., the São Paulo Research Foundation (FAPESP) Young Investigator Award to D.J.G.L. (#2013/04585), and the Henry Hunter Family Research Foundation at Mississippi State University Initiation Award to M.W.B. A.J.R. thanks the Canada Research Chairs Program for support.
Author Contributions
M.W.B., F.W.S., A.K., and D.J.G.L. conceived this project and experiments. S.K., M.W.B., A.K.T., J.D.S., I.C., T.P., D.M.C.A, A.K., D.J.G.L., M.K., L.L.S., and F.W.S. collected, isolated, and maintained cultures of the amoebozoans sampled within. S.K., A.K.T., D.J.G.L., I.C., T.P., and M.W.B. collected transcriptomic data. M.W.B. and A.J.R. conceived phylogenomic experiments. S.K., A.K.T., and M.W.B. conducted the phylogenomic experiments. A.S. and A.K. helped with in depth interpretation of results. S.K. composed the first draft of this manuscript. All authors contributed to and agree with the text within the manuscript.
References
- Adl SM, Simpson AG, Lane CE, Lukes J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, et al. 2012. The revised classification of eukaryotes. J Eukaryot Microbiol. 59:429–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral Zettler LA, Nerad TA, O’Kelly CJ, Peglar MT, Gillevet PM, Silberman JD, Sogin ML.. 2000. A molecular reassessment of the Leptomyxid amoebae. Protist 151:275–282. [DOI] [PubMed] [Google Scholar]
- Berney C, Geisen S, Van Wichelen J, Nitsche F, Vanormelingen P, Bonkowski M, Bass D.. 2015. Expansion of the ‘reticulosphere’: diversity of novel branching and network-forming amoebae helps to define Variosea (Amoebozoa). Protist 166:271–295. [DOI] [PubMed] [Google Scholar]
- Bolivar I, Fahrni JF, Smirnov AV, Pawlowski J.. 2001. SSU rRNA-based phylogenetic position of the genera Amoeba and Chaos (Lobosea, Gymnamoebia): the origin of gymnamoebae revisited. Mol Biol Evol. 18:2306–2314. [DOI] [PubMed] [Google Scholar]
- Brown MW, Sharpe SC, Silberman JD, Heiss AA, Lang BF, Simpson AGB, Roger AJ.. 2013. Phylogenomics demonstrates that breviate flagellates are related to opisthokonts and apusomonads. Proc R Soc B: Biol Sci. 280:20131755–20131755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MW, Silberman JD.. 2013. The non-dictyostelid sorocarpic amoebae. In Romeralo, Escalante, Baldauf, editors. Dictyostelids – evolution, genomics and cell biology. Heidelberg (Germany: ): Springer; pp 219–242. [Google Scholar]
- Brown MW, Silberman JD, Spiegel FW.. 2011. “Slime molds” among the Tubulinea (Amoebozoa): molecular systematics and taxonomy of Copromyxa. Protist 162:277–287. [DOI] [PubMed] [Google Scholar]
- Brown MW, Spiegel FW, Silberman JD.. 2007. Amoeba at attention: phylogenetic affinity of Sappinia pedata. J Eukaryot Microbiol. 54:511–519. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. 1998. A revised six-kingdom system of life. Biol Rev Camb Philos Soc. 73:203–266. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Chao EE, Lewis R.. 2016. 187-Gene phylogeny of protozoan phylum Amoebozoa reveals a new class (Cutosea) of deep-branching, ultrastructurally unique, enveloped marine Lobosa and clarifies amoeba evolution. Mol Phylogenet Evol. 99:275–296. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Chao EEY, Oates B.. 2004. Molecular phylogeny of Amoebozoa and the evolutionary significance of the unikont Phalansterium. Eur J Protistol. 40:21–48. [Google Scholar]
- Cavalier-Smith T, Fiore-Donno AM, Chao E, Kudryavtsev A, Berney C, Snell EA, Lewis R.. 2015. Multigene phylogeny resolves deep branching of Amoebozoa. Mol Phylogenet Evol. 83:293–304. [DOI] [PubMed] [Google Scholar]
- Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, et al. 2005. The genome of the social amoeba Dictyostelium discoideum. Nature 435:43–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eme L, Sharpe SC, Brown MW, Roger AJ.. 2014. On the age of eukaryotes: evaluating evidence from fossils and molecular clocks. Cold Spring Harb Perspect Biol. 6: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrni JF, Bolivar I, Berney C, Nassanova E, Smirnov AV, Pawlowski J.. 2003. Phylogeny of lobose amoebae based on actin and small-subunit ribosomal RNA genes. Mol Biol Evol. 20:1881–1886. [DOI] [PubMed] [Google Scholar]
- Fiore-Donno AM, Nikolaev SI, Nelson M, Pawlowski J, Cavalier-Smith T, Baldauf SL.. 2010. Deep phylogeny and evolution of slime moulds (mycetozoa). Protist 161:55–70. [DOI] [PubMed] [Google Scholar]
- Goodenough U, Heitman J.. 2014. Origins of eukaryotic sexual reproduction. Cold Spring Harb Perspect Biol. 6:a016154–a016154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffroy O, Brinkmann H, Delsuc F, Philippe H.. 2006. Phylogenomics: the beginning of incongruence? Trends Genet. 22:225–231. [DOI] [PubMed] [Google Scholar]
- Kudryavtsev A, Brown MW, Tice A, Spiegel FW, Pawlowski J, Anderson OR.. 2014. A revision of the order pellitida Smirnov et al., 2011 (Amoebozoa, Discosea) based on ultrastructural and molecular evidence, with description of Endostelium crystalliferum n. sp. Protist 165:208–229. [DOI] [PubMed] [Google Scholar]
- Kudryavtsev A, Pawlowski J.. 2013. Squamamoeba japonica n. g. n. sp. (Amoebozoa): a deep-sea amoeba from the Sea of Japan with a novel cell coat structure. Protist 164:13–23. [DOI] [PubMed] [Google Scholar]
- Lahr DJ, Grant J, Molestina R, Katz LA, Anderson OR.. 2015. Sapocribrum chincoteaguense n. gen. n. sp.: a small, scale-bearing Amoebozoan with Flabellinid affinities. J Eukaryot Microbiol. 62:444–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahr DJ, Grant J, Nguyen T, Lin JH, Katz LA.. 2011a. Comprehensive phylogenetic reconstruction of amoebozoa based on concatenated analyses of SSU-rDNA and actin genes. PLoS ONE. 6:e22780.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahr DJG, Parfrey LW, Mitchell EAD, Katz LA, Lara E.. 2011b. The chastity of amoebae: re-evaluating evidence for sex in amoeboid organisms. Proc R Soc B: Biol Sci. 278:2081–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahr DJ, Grant JR, Katz LA.. 2013. Multigene phylogenetic reconstruction of the Tubulinea (Amoebozoa) corroborates four of the six major lineages, while additionally revealing that shell composition does not predict phylogeny in the Arcellinida. Protist 164:323–339. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Philippe H.. 2008. Improvement of molecular phylogenetic inference and the phylogeny of Bilateria. Philos Trans R Soc Lond B Biol Sci. 363:1463–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Rodrigue N, Stubbs D, Richer J.. 2013. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol. 62:611–615. [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel O.. 2008. An improved general amino acid replacement matrix. Mol Biol Evol. 25:1307–1320. [DOI] [PubMed] [Google Scholar]
- Martin GW, Alexopoulos CJ.. 1969. Monograph of the Myxomycetes. Iowa City (IA: ): University of Iowa Press. [Google Scholar]
- Mikrjukov KA, Mylnikov AP.. 1998. The fine structure of a carnivorous multiflagellar protist Multicilia marina Cienkowski, 1881 (flagellata incertae sedis). Eur J Protistol. 34:391–401. [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ.. 2014. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev SI, Berney C, Petrov NB, Mylnikov AP, Fahrni JF, Pawlowski J.. 2006. Phylogenetic position of Multicilia marina and the evolution of Amoebozoa. Int J Syst Evol Microbiol. 56:1449–1458. [DOI] [PubMed] [Google Scholar]
- Olive LS. 1975. The mycetozoans. New York, NY: Academic Press. [Google Scholar]
- Olive LS, Bennett WE, Deasey MC.. 1984. The new protostelid genus Endostelium. Mycologia 76:884. [Google Scholar]
- Olsen GJ. 1987. Earliest phylogenetic branchings: comparing rRNA-based evolutionary trees inferred with various techniques. Cold Spring Harb Symp Quant Biol. 52:825–837. [DOI] [PubMed] [Google Scholar]
- Page FC. 1976. A revised classification of the Gymnamoebia (Protozoa: Sarcodina). Zool J Linnean Soc. 58:61–77. [Google Scholar]
- Page FC. 1983. Marine gymnamoebae. Cambridge: Institute of Terrestrial Ecology, Culture Centre of Algae and Protozoa. [Google Scholar]
- Page FC. 1988. A new key to freshwater and soil gymnamoebae. Ambleside, Cumbria (United Kingdom: ): Freshwater Biological Association. [Google Scholar]
- Pánek T, Zadrobílková E, Walker G, Brown MW, Gentekaki E, Hroudová M, Kang S, Roger AJ, Tice AK, Vlček C, et al. 2016. First multigene analysis of Archamoebae (Amoebozoa: Conosa) robustly reveals its phylogeny and shows that Entamoebidae represents a deep lineage of the group. Mol Phylogenet Evol. 98:41–51. [DOI] [PubMed] [Google Scholar]
- Patterson DJ. 1999. The diversity of eukaryotes. Am Nat. 154:S96–S124. [DOI] [PubMed] [Google Scholar]
- Pawlowski J. 2008. The twilight of Sarcodina: a molecular perspective on the polyphyletic origin of amoeboid protists. Protistology 5:281–302. [Google Scholar]
- Ptackova E, Kostygov AY, Chistyakova LV, Falteisek L, Frolov AO, Patterson DJ, Walker G, Cepicka I.. 2013. Evolution of Archamoebae: morphological and molecular evidence for pelobionts including Rhizomastix, Entamoeba, Iodamoeba, and Endolimax. Protist 164:380–410. [DOI] [PubMed] [Google Scholar]
- Pussard M. 1973. Description d'une amibe de type flabellulien, Pessonella marginata n.g. n. sp. (Mayorellidae, amoebaea). Protistologica 9:175–185. [Google Scholar]
- Reinhardt DJ. 1968. Development of the mycetozoan Echinosteliopsis oligospora. J Protozool. 15:480–493. [DOI] [PubMed] [Google Scholar]
- Romeralo M, Skiba A, Gonzalez-Voyer A, Schilde C, Lawal H, Kedziora S, Cavender JC, Glockner G, Urushihara H, Schaap P.. 2013. Analysis of phenotypic evolution in Dictyostelia highlights developmental plasticity as a likely consequence of colonial multicellularity. Proc Biol Sci. 280:20130976.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaap P, Barrantes I, Minx P, Sasaki N, Anderson RW, Benard M, Biggar KK, Buchler NE, Bundschuh R, Chen X, et al. 2015. The Physarum polycephalum genome reveals extensive use of prokaryotic two-component and metazoan-type tyrosine kinase signaling. Genome Biol Evol. 8:109–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmarda LK. 1871. Zoologie. W. Braumüller. Berlin, Germany. [Google Scholar]
- Seravin LN, Goodkov AV.. 1987. The flagella of the freshwater amoeba Pelomyxa palustris. Tsitologiya 29:721–724. [Google Scholar]
- Shadwick LL, Spiegel FW, Shadwick JD, Brown MW, Silberman JD.. 2009. Eumycetozoa = Amoebozoa?: SSUrDNA phylogeny of protosteloid slime molds and its significance for the amoebozoan supergroup. PLoS ONE. 4:e6754.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadwick LL, Brown MW, Tice AK, Spiegel FW.. 2016. A new amoeba with protosteloid fruiting: Luapeleamoeba hula n. g. n. sp. Acta Protozool. 55:123–134. [Google Scholar]
- Shimodaira H. 2002. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 51:492–508. [DOI] [PubMed] [Google Scholar]
- Smirnov AV, Nassonova E, Berney C, Fahrni J, Bolivar I, Pawlowski J.. 2005. Molecular phylogeny and classification of the lobose amoebae. Protist 156:129–142. [DOI] [PubMed] [Google Scholar]
- Smirnov AV, Chao E, Nassonova ES, Cavalier-Smith T.. 2011. A revised classification of naked lobose amoebae (Amoebozoa: obosa). Protist 162:545–570. [DOI] [PubMed] [Google Scholar]
- Spiegel FW. 1990. Phylum plasmodial slime molds, Class Protostelida In: Margulis JOC L., Melkonian M., Chapman D., editors. Handbook of Protoctista. Boston: Jones and Bartlett; pp. 484–497. [Google Scholar]
- Spiegel FW. 1991. A proposed phylogeny of the flagellated protostelids. Biosystems 25:113–120. [DOI] [PubMed] [Google Scholar]
- Spiegel FW. 2011. Commentary on the chastity of amoebae: re-evaluating evidence for sex in amoeboid organisms. Proc R Soc B: Biol Sci. 278:2096–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel FW, Feldman J.. 1985. Obligate amoebae of the protostelids: significance for the concept of Eumycetozoa. Biosystems 18:377–386. [DOI] [PubMed] [Google Scholar]
- Spiegel FW, Feldman J.. 1989. Fruiting body development in the mycetozoan Echinostelium bisporum. Can J Bot. 67:1285–1293. [Google Scholar]
- Spiegel FW, Feldman J.. 1993. Fruiting Body Ultrastructure in the Protostelid Schizoplasmodiopsis vulgare. Mycologia 85:894. [Google Scholar]
- Spiegel FW, Lee SB, Rusk SA.. 1995. Eumycetozoans and molecular systematics. Can J Bot. 73:s738–s746. [Google Scholar]
- Spiegel FW, Olive LS, Brown RM.. 1979. Roles of actin during sporocarp culmination in the simple mycetozoan Planoprotostelium aurantium. Proc Natl Acad Sci U S A. 76:2335–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekle YI, Grant J, Anderson OR, Nerad TA, Cole JC, Patterson DJ, Katz LA.. 2008. Phylogenetic placement of diverse amoebae inferred from multigene analyses and assessment of clade stability within ‘Amoebozoa’ upon removal of varying rate classes of SSU-rDNA. Mol Phylogenet Evol. 47:339–352. [DOI] [PubMed] [Google Scholar]
- Tekle YI, Anderson OR, Katz LA, Maurer-Alcala XX, Romero MA, Molestina R.. 2016. Phylogenomics of ‘Discosea’: a new molecular phylogenetic perspective on Amoebozoa with flat body forms. Mol Phylogenet Evol. 99:144–154. [DOI] [PubMed] [Google Scholar]
- Tekle YI, Wood FC, Katz LA, Ceron-Romero MA, Gorfu LA.. 2017. Amoebozoans are secretly but ancestrally sexual: evidence for sex genes and potential novel crossover pathways in diverse groups of amoebae. Genome Biol Evol doi:10.1093/gbe/evx002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice AK, Shadwick LL, Fiore-Donno AM, Geisen S, Kang S, Schuler GA, Spiegel FW, Wilkinson KA, Bonkowski M, Dumack K, et al. 2016a. Expansion of the molecular and morphological diversity of Acanthamoebidae (Centramoebida, Amoebozoa) and identification of a novel life cycle type within the group. Biol Direct. 11:69.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice AK, Silberman JD, Walthall AC, Le KND, Spiegel FW, Brown MW.. 2016b. Sorodiplophrys stercorea: another novel lineage of sorocarpic multicellularity. J Eukaryot Microbiol. 63:623–628. [DOI] [PubMed] [Google Scholar]
- Visvesvara GS, Moura H, Schuster FL.. 2007. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea. FEMS Immunol Med Microbiol. 50:1–26. [DOI] [PubMed] [Google Scholar]
- Walker G, Simpson AGB, Edgcomb V, Sogin ML, Patterson DJ.. 2001. Ultrastructural identies of Mastigamoeba punctachora, Mastigamoeba simplex and Mastigella commutans and assessment of hypotheses of relatedness of the pelobionts (Protista). Eur J Protistol. 37:25–49. [Google Scholar]
- Wang HC, Susko E, Roger AJ.. 2014. An amino acid substitution-selection model adjusts residue fitness to improve phylogenetic estimation. Mol Biol Evol. 31:779–792. [DOI] [PubMed] [Google Scholar]
- Whitney KD, Bennett WE, Olive LS.. 1982. Observations on Echinostelium bisporum. Mycologia 74:677–680. [Google Scholar]
- Yubuki N, Leander BS.. 2013. Evolution of microtubule organizing centers across the tree of eukaryotes. Plant J. 75:230–244. [DOI] [PubMed] [Google Scholar]
- Zadrobilkova E, Walker G, Cepicka I.. 2015. Morphological and molecular evidence support a close relationship between the free-living archamoebae Mastigella and Pelomyxa. Protist 166: 14–41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All transcriptomic data generated in this manuscript have been deposited with National Center for Biotechnology Information under the BioProject PRJNA380424 as detailed in supplementary table S2, Supplementary Material online. All single gene alignments, masked and unmasked, and phylogenomic matrices are available in the supplementary file Kang_etal.2017.tar.gz.