Abstract
Reproductive isolation is an intrinsic aspect of species formation. For that reason, the identification of the precise isolating traits, and the rates at which they evolve, is crucial to understanding how species originate and persist. Previous work has measured the rates of evolution of prezygotic and postzygotic barriers to gene flow, yet no systematic analysis has studied the rates of evolution of postmating-prezygotic (PMPZ) barriers. We measured the magnitude of two barriers to gene flow that act after mating occurs but before fertilization. We also measured the magnitude of a premating barrier (female mating rate in nonchoice experiments) and two postzygotic barriers (hybrid inviability and hybrid sterility) for all pairwise crosses of all nine known extant species within the melanogaster subgroup. Our results indicate that PMPZ isolation evolves faster than hybrid inviability but slower than premating isolation. Next, we partition postzygotic isolation into different components and find that, as expected, hybrid sterility evolves faster than hybrid inviability. These results lend support for the hypothesis that, in Drosophila, reproductive isolation mechanisms (RIMs) that act early in reproduction (or in development) tend to evolve faster than those that act later in the reproductive cycle. Finally, we tested whether there was evidence for reinforcing selection at any RIM. We found no evidence for generalized evolution of reproductive isolation via reinforcement which indicates that there is no pervasive evidence of this evolutionary process. Our results indicate that PMPZ RIMs might have important evolutionary consequences in initiating speciation and in the persistence of new species.
Keywords: premating isolation, postmating-prezygotic isolation, conspecific sperm precedence, postzygotic isolation, hybrids, Drosophila, genomic alignment
Introduction
Barriers to gene flow, or reproductive isolating mechanisms (RIMs), evolve as a byproduct of divergence between populations that accrue genetic differences over time (Coyne and Orr 1989, 1997). The process of speciation is thus the accumulation of RIMs. The strength of reproductive isolation (RI) dictates whether nascent species persist or whether they merge into a single lineage once they come into contact with each other. In cases where RI is absolute and no intermixing through hybridization is possible, speciation is complete. In other cases, RIMs are weak and can be overcome by gene flow, thus merging nascent species into a single lineage (Comeault et al. 2015; Cenzer 2016). There is an intermediate scenario, which is likely to be common, in which hybridization—and admixture—occurs but species persist (Rosenblum et al. 2012). Therefore, the nature and magnitude of RIMs that evolve between groups and the rate at which they evolve are key factors influencing the origin of new species. The systematic identification of these barriers in a phylogenetic context (to infer their rates of evolution) is a prerequisite for understanding which barriers are important drivers of speciation and which result from postspeciation divergence (Coyne and Orr 2004).
Depending on when they occur in the reproductive cycle, RIMs may be classified as premating, postmating-prezygotic, or postzygotic (Dobzhansky 1937; Coyne and Orr 2004). Premating RIMs encompass all the biological traits that preclude populations from encountering or mating with each other. Niche specificity, habitat preferences, reproductive timing, and mate choice are all examples of premating barriers. A second type of barrier that acts after mating but before a zygote is formed (i.e., postmating-prezygotic [PMPZ] barriers) involves discordant interactions between gametes or between the female reproductive tract and components of the male seminal fluid. Gametic interactions include the physical and chemical cues that allow for mutual gametic recognition and eventual formation of a zygote. Gametic incompatibilities may arise if these cues are incompatible between gametes from different species, thereby restricting gene flow. Finally, postzygotic isolation includes all defects that lead to fitness reductions in interspecific hybrid individuals and not in the pure species and includes developmental (Orr and Presgraves 2000; Presgraves 2010) and behavioral defects (e.g., McBride and Singer 2010; Turissini et al. 2017) in hybrids.
Several meta-analyses have inferred the rate at which RIMs evolve over time and most have found a positive relationship between their strength and genetic distance (reviewed in Edmands 2002). Hybrid sterility and hybrid inviability both scale with genetic distance, with the former seemingly evolving before the latter (Wu 1992; Coyne and Orr 1997; Sasa et al. 1998; Presgraves 2002; Price and Bouvier 2002; Scopece et al. 2008; De Vienne et al. 2009; Tubaro and Lijtmaer 2002). Premating isolation usually evolves before postzygotic isolation in Drosophila, amphibians, and certain groups of plants and fish (e.g., Bolnick and Near 2005 reviewed in Coyne and Orr 2004). This body of work has led to the widespread notion that prezygotic isolation is necessary to initiate speciation (e.g., Abbott et al. 2013, Seehausen et al. 2014 among many others). However, premating, and postzygotic isolation have similar rates of evolution in some plant genera (e.g., Bolnick and Near 2005 reviewed in Widmer et al. 2009), and in copepods, postzygotic isolation evolves before prezygotic isolation (Ganz and Burton 1995; Palmer and Edmands 2000; Edmands et al. 2009). Clearly, more comparative work, in terms of traits and taxa, is needed before a conclusion on what RIMs are responsible for setting the process of speciation in motion.
In contrast to studies measuring the magnitude of premating, and postzygotic isolation relative to genetic distance, the rates at which PMPZ isolation is achieved have rarely been investigated, with the notable exception of plant taxa. In orchids and Fragaria, there is no apparent correlation between the magnitude of prezygotic isolation (either premating isolation or postpollination prezygotic, the equivalent of PMPZ) and genetic distance (Scopece et al. 2007, 2008; Nosrati et al. 2011). In Glycine (Fabaceae) and Silene (Caryophyllaceae), postpollination prezygotic and postzygotic isolation both increase monotonically with divergence time and at similar rates (Moyle et al. 2004). In Chilean Bellflowers (Nolana, Solanaceae), postzygotic isolation evolves faster than postpollination prezygotic isolation (Jewell et al. 2012). The results from these five taxa suggest that postpollination prezygotic isolation is important but the extent of importance is heterogeneous across groups.
In animals, less is known about the evolution and prevalence of PMPZ RIMs compared with premating or postzygotic mechanisms (Servedio 2004; Pitnick et al. 2009 but see Markow 1997; Civetta and Clark 2000; Birkhead and Pizzari 2002; Civetta et al. 2008; Sweigart 2010; Jennings et al. 2011; Ahmed-Braimah and McAllister 2012; Larson et al. 2012; Ahmed-Braimah 2016; Ala‐Honkola et al. 2016; Miller and Pitnick 2002, Pitnick et al. 2001, Sagga and Civetta 2011). This is surprising because this type of barrier seems to be common (e.g., Fricke and Arnqvist 2004; Mendelson et al. 2007; Dopman et al. 2010). Few studies have explored the effect of genetic distance on the magnitude of PMPZ isolation. Gametic incompatibilities are crucial in maintaining species boundaries in sea urchins of the genus Echinometra. Qualitative measurements revealed no apparent increase in the magnitude of gametic isolation in two species pairs (Lessios and Cunningham 1990). A second study measured the magnitude of conspecific sperm precedence in two pairs of species of Drosophila and suggested that this type of gametic barrier does scale with genetic distance and evolves after premating isolation (Dixon et al. 2003). Finally, a comparative analysis of in vitro fertilization rates (i.e., percentage of fertilized eggs) in toads revealed that genetic distance between the parental species had no effect on the magnitude of gametic interactions (Malone and Fontenot 2008). These three studies highlight there is no current consensus on whether PMPZ isolation increases with divergence. These disparate conclusions indicate that a more systematic approach is needed to understand the rate of evolution of these traits. More generally, this lack of precise estimates is surprising given that PMPZ isolation has been implicated in the origin of new species (Palumbi 1994; Palumbi 2009; Manier et al. 2013; Jennings et al. 2014) and with the persistence of species in nature in the face of gene flow (Howard et al. 1998; Howard 1999; Wolstenholme 2004; Matute and Coyne 2010; Hart et al. 2014). These types of RIMs have also been shown to be the object of reinforcing selection, the evolutionary process that increments the magnitude of prezygotic isolation (in this case PMPZ) as an indirect response to avoid maladaptive hybridization (Matute 2010a; Davis et al. 2017; Castillo and Moyle 2016).
We measured the rate of evolution of RI in a common environment for all pairwise crosses of all nine known extant species within the Drosophila melanogaster species subgroup (supplementary fig. S1, Supplementary Material online). We provide fine scale measurements of two PMPZ RIMs: noncompetitive gametic isolation (i.e., the number of eggs a female lays after a mating with males from a heterospecific species; NCGI) and conspecific sperm precedence (i.e., the number of individuals a conspecific male sires after mating with a female that also mated with a heterospecific male; CSP). We also improve upon previous summaries of premating, and postzygotic isolation in the melanogaster subgroup by attempting all possible hybridizations in the group, measuring the magnitude of these barriers in a controlled laboratory environment, and incorporating genome-wide information to quantify genetic distance between species. Our results show that PMPZ barriers evolve faster than hybrid inviability but slightly slower than premating RIMs. Using this data set, we also quantify the rate of evolution of different mechanisms of postzygotic isolation and find that hybrid sterility evolves faster than hybrid inviability. Of the different types of hybrid inviability, we find that RI acting early in development evolves faster than RI acting later in the life cycle. In general, our results provide support for the idea that reproductive isolating barriers that act earlier evolve more rapidly.
Results
Our goal was to quantify the magnitude of four mechanisms of RI—premating, NCGI, CSP, and hybrid inviability—in a controlled laboratory environment for all possible crosses between species of the Drosophila melanogaster species subgroup (supplementary fig. S1, Supplementary Material online). The indexes we used to measure the magnitude of each RIM are shown in table 1. Indexes equal to 1 indicate complete isolation. We report our results for each barrier in the Supplementary Material (supplementary text, figs. S2–S5, and tables S1–S6, Supplementary Material online). Next, we compared the rates of evolution of these four RIMs using the melanogaster subgroup data set and then did similar comparisons performing phylogenetic corrections. Even though our main goal was to assess the rate of evolution of PMPZ RIMs, our data set also allowed us to quantify the rate of evolution of different types of postzygotic isolation. Finally, we also assessed whether PMPZ RIMs were stronger in sympatric populations than in allopatric populations, a test of the influence of reinforcement.
Table 1.
Reproductive Isolation Barriers Studied in This Report.
| Mechanism | Success | Failure | Index of isolation |
|---|---|---|---|
| Premating | Mated | Unmated | 1—mated/total paired |
| Postmating-prezygotic—NCGI | Hybrid eggs | Conspecific female eggs—hybrid eggs | 1—hybrid eggs/conspecifically mated female eggs |
| Postmating-prezygotic—CSP | NA | NA | See text for description |
| Postzygotic—hybrid inviability | Hybrid adults | Hybrid eggs - hybrid adults | 1—hybrid adults/hybrid eggs |
| Postzygotic—embryonic lethality | Hybrid egg cases | Dead embryos | Dead hybrid embryos/hybrid eggs |
| Postzygotic—larval lethality | Hybrid pupae | Hybrid egg cases—hybrid pupae | 1—hybrid pupae/hybrid egg cases |
| Postzygotic—pupal lethality | Hybrid adults | Hybrid pupae—hybrid adults | 1—hybrid adults/hybrid pupae |
| Postzygotic—female sterility | Females with ovarioles | Females without ovarioles | 1—females with ovarioles/total females |
| Postzygotic—male sterility | Males with motile sperm | Males without motile sperm | 1—males without motile sperm/total males |
NCGI: noncompetitive gametic isolation; CSP: conspecific sperm precedence.
Rate of Evolution of Reproductive Isolating Mechanisms: no Phylogenetic Corrections
We evaluated the rate at which PMPZ isolation (which has rarely been measured in animals) evolves compared with premating, and postzygotic isolation. To do this, we tested whether the genetic distance between the parental species influenced the magnitude of RI between them. Ks, the number of per site synonymous substitutions between a pair of species was used as a proxy for genetic distance (and therefore divergence time; supplementary table S7, Supplementary Material online), and πs, the per synonymous site nucleotide diversity was used as the average genetic distance between individuals of the same species (table 2). It is worth noting that genetic divergence can be underestimated by Ks between divergent species because of mutational saturation at synonymous sites (Gojobori 1983; Smith and Smith 1996) and reference genome bias (i.e., reduced ability to map reads from a divergent species).
Table 2.
Within Species Nucleotide Diversity.
| Species | N | πs | Genes |
|---|---|---|---|
| D. melanogaster | 599 | 0.013 | 10,499 |
| D. simulans | 29 | 0.0329 | 8,975 |
| D. sechellia | 41 | 0.0018 | 9,157 |
| D. mauritiana | 13 | 0.0201 | 9,097 |
| D. yakuba | 56 | 0.0243 | 8,598 |
| D. santomea | 11 | 0.0172 | 8,952 |
| D. teissieri | 13 | 0.0367 | 8,951 |
Note.—Average heterozygosity values across the whole genome based on synonymous sites (πs) values. N represents the number of sequenced lines per species. Ks, the genetic distance between species, was calculated with 8,923 genes (supplementary table S7, Supplementary Material online).
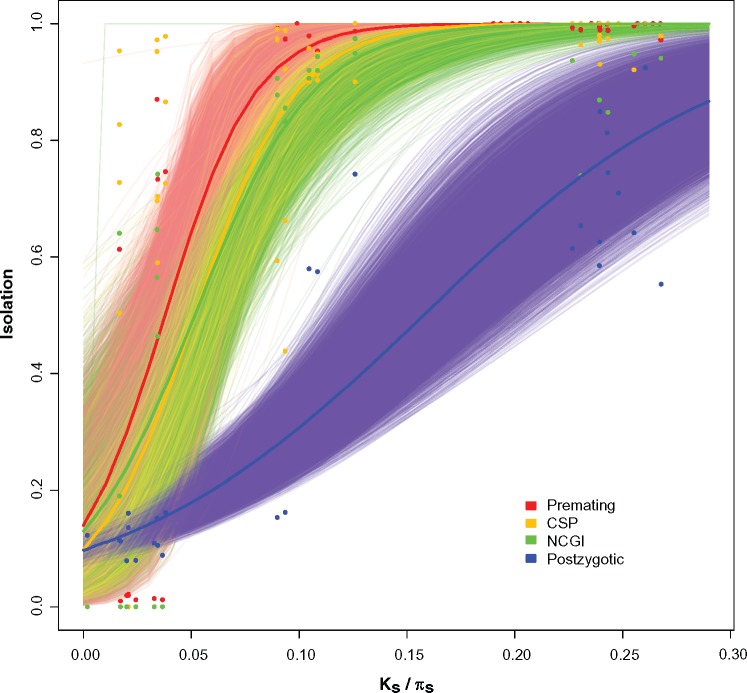
As expected, the magnitude of all types of RI scales positively with divergence time. A logistic regression for each of the RIMs showed a strong positive relationship between the magnitude of RI and the genetic distance between the parentals (fig. 1). Linear regressions show the same pattern but we restrict our analyses to the logistic regressions because of the bounded nature of the traits (i.e., RI cannot be higher than 1). These regressions are approximations and are likely to not encompass the whole level of variability in the traits (see points around the regressions in fig. 1). The fit of each of these regressions is shown in supplementary table S8, Supplementary Material online. The increase in premating isolation (fig. 1, red lines) is rapid and almost complete at Ks ≥ 10% between the hybridizing species. The two mechanisms of PMPZ also follow a similar pattern. The magnitude of both NCGI and CSP is almost complete between species with Ks ≥ 12%. This is in contrast to hybrid inviability, which also scales positively with divergence but evolves more slowly; hybrid inviability is complete in only one of the possible crosses in the melanogaster species subgroup (♀D. santomea × ♂D. sechellia; fig. 1, blue lines). Analyses with phylogenetically independent pairs yielded similar results (see below).
Fig. 1.
Premating, conspecific sperm precedence, noncompetitive gametic isolation, and postzygotic isolation increase with pairwise genetic distance between species. Proxies of premating isolation (red), conspecific sperm precedence (CSP, yellow), noncompetitive gametic isolation (NCGI, green), and postzygotic isolation (blue) were regressed against phylogenetic distance (Ks between species and πs within species). The four types of isolation increase with genetic distance, and premating isolation evolves faster than hybrid inviability. The thick red, yellow, green, and blue lines represent fitted logistic regressions for the premating, and postzygotic data, respectively. The thinner lines of each of the four colors are the regressions for each of 10,000 bootstrap resamplings of the data.
We tested whether any of the four RIMs evolved more quickly than others. Due to the perfect separation of values along Ks in hybrid sterility, we analyzed this trait separately (see below). The rate at which the magnitude of RI increments (shown in fig. 1) is different among RIMs (logistic regression, test of rate of increment with a common intercept: F4,1115= 325.02, P < 1 × 10−15; results with other regressions are shown in supplementary tables S9 and S10, Supplementary Material online). Pairwise comparisons of the rate of increment indicate that of all four types of RI, premating isolation seems to evolve quickest followed by the two types of premating-postzygotic isolation (supplementary table S10, Supplementary Material online). We found that of the two PMPZ barriers, CSP evolves faster than noncompetitive gametic isolation (supplementary table S10, Supplementary Material online). All prezygotic barriers evolve quicker than postzygotic isolation (supplementary table S10, Supplementary Material online). Note that in one analysis—linear regression with a common intercept—both types of premating-postzygotic isolation show similar rates of increase to premating isolation (supplementary table S10, Supplementary Material online).
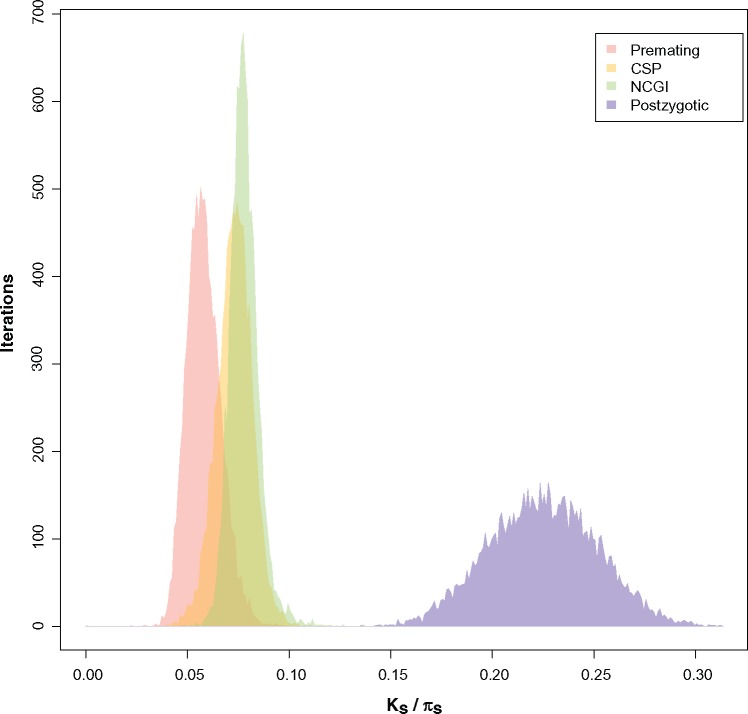
To illustrate how fast each RIM achieves completion, we calculated Threshold_Ks, the genetic distance at which 95% of RI is achieved (i.e., any of the four indexes of RI equals 0.95; fig. 2). This quantity shows how quickly the logistic regression approaches 1, and constitutes a proxy of how fast a RIM evolves. These distributions show the same results from the regression coefficients. Collectively, the results from the linear models and the bootstrapped distributions indicate that earlier acting RIMs evolve faster than those that evolve later in the reproductive cycle.
Fig. 2.
Premating, NCGI, CSP, and postzygotic isolation evolve at different rates in the melanogaster species subgroup. Threshold_Ks illustrates the Ks value for which a given RI barrier achieves a value of 0.95. Premating isolation values are red, conspecific sperm precedence (CSP) are yellow, noncompetitive gametic isolation (NCGI) are green, and postzygotic values are blue. Regression coefficients (supplementary table S9, Supplementary Material online) show that the rates at which each RIM increases differ from each other.
Rate of Evolution of Reproductive Isolating Mechanisms: Phylogenetic Corrections
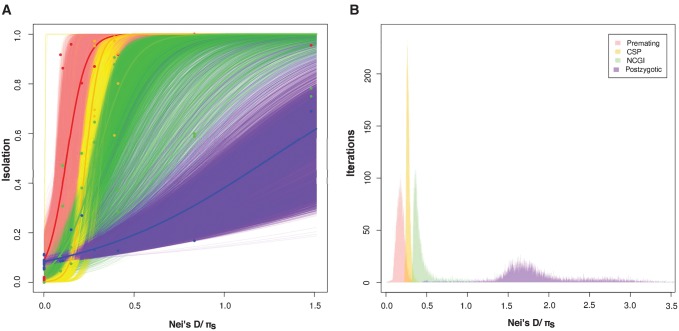
We collected additional data for four more species pairs from other Drosophila subgroups different from melanogaster to address two potential issues. First, we needed to assess whether our measurements of the rate of evolution of different RIMs were robust to phylogenetic nonindependence (i.e., multiple overlapping branches when only studying the melanogaster species subgroup). Second, when premating isolation is complete, the number of measurements of estimates of any type of postmating isolation (either PMPZ or postzygotic) is reduced. This will inflate the estimates of the rate of evolution of premating isolation (Wu 1992). To address these two potential issues, we identified species for which we could measure the magnitude of all the four types of RI. We added these four species pairs to the data from our original study for three melanogaster species pairs that are phylogenetically independent (the most possible hybridizations without overlapping branches; supplementary fig. S1, Supplementary Material online). In total, this gave us seven independent species pairs to compare. Results did not change when measuring isolation between any random two overlapping branches in the melanogaster subgroup—accounting for the D. simulans, D. sechellia, D. mauritiana polytomy—for a total of seven species pairs, or when excluding these species in this polytomy altogether—for a total of six species pairs. As observed before, the rate of evolution of these traits differs (logistic regression, test of rate of increment with a common intercept: F4,433 = 115.77, P < 1 × 10−15; results with other regressions are shown in supplementary tables S11 and S12, Supplementary Material online). Similar to what we observed in the melanogaster subgroup-only analysis, premating isolation is the fastest RIM to evolve, followed by conspecific sperm precedence, noncompetitive gametic isolation, and finally postzygotic isolation (fig. 3A; pairwise comparisons in supplementary table S12, Supplementary Material online). Bootstrap Threshold_Ks values illustrate this difference (fig. 3B). These results indicate that the ranking of the rates of evolution of the four RIMs is not exclusive to the melanogaster species subgroup of Drosophila, and instead is a more general pattern that might pertain to the whole Sophophora genus. The extent which this generalization can be made for the whole Drosophila genus and other clades will require additional data.
Fig. 3.
Premating, CSP, NCGI, and postzygotic isolation increase with pairwise genetic distance between species across the Sophophora subgenus. To account for the possibility of phylogenetic nonindependence in the melanogaster species subgroup, we subsampled phylogenetically independent crosses and added a pair of species from other four Drosophila clades (all within the Sophophora genus). Proxies of RI are similar to the ones shown in figure 1. Premating isolation (red), conspecific sperm precedence (CSP, yellow), noncompetitive gametic isolation (NCGI, green), and postzygotic isolation (blue) were regressed against phylogenetic distance (Nei’s distance between species and πs within species). (A) As observed in the melanogaster species subgroup, the four types of isolation increase with genetic distance, and premating isolation evolves faster than hybrid inviability. The thick red, yellow, green, and blue lines represent fitted logistic regressions for the premating and postzygotic data, respectively. The thinner lines of each of the four colors are the regressions for each of 10,000 bootstrap resamplings of the data. (B) Premating, NCGI, CSP, and postzygotic isolation (hybrid inviability) evolve at different rates in the Drosophila genus in a set of phylogenetically independent species pairs (supplementary table S10, Supplementary Material online). Bootstrapped distributions of Threshold_Ks show the Ks value for which a given RI barrier achieves a value of 0.95 and illustrate that all RIMs evolve at a different rate.
Rate of Evolution of Different Types of Postzygotic Isolation
Postzygotic isolation includes all developmental defects that might reduce the fitness of interspecific hybrids. By performing all the possible pairwise crosses among species of the melanogaster subgroup, we could measure the rate of evolution of hybrid sterility and of hybrid inviability in the melanogaster species subgroup. The results from this section are threefold. First we report all the possible hybridizations in the species group. Second, we compare the rate of evolution of hybrid inviability with that of hybrid sterility. Finally, we compare the rate of evolution of hybrid inviability at three developmental stages.
Pairwise Hybridizations in the melanogaster Species Group
We set up interspecific crosses that lasted for over 4 weeks. Thirty-three crosses produce progeny (table 3). Of these, 32 produce viable adult hybrids of at least one sex. Among these 33 crosses, we find seven previously undescribed hybridizations, mostly between highly divergent species of the yakuba species complex and the melanogaster/simulans clade. The list of hybridizations that produced progeny is shown in table 3. We found that hybrid inviability is rare in the D. melanogaster subgroup, even after 15 million years of divergence. Of all crosses, only ♀ D. santomea × ♂ D. sechellia showed complete hybrid inviability (table 3). In this cross, half of the progeny died as embryos, and the other half died as pupae. Dissection of these pupae revealed that all individuals had testes (N = 34) suggesting (but not confirming) that females died at an earlier developmental stage, possibly as embryos.
Table 3.
Postzygotic Isolation in the melanogaster Species Group.
| Male | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Female | melanogaster | simulans | sechellia | mauritiana | orena | erecta | yakuba | santomea | teissieri |
| melanogaster | Sterile ♀, dead ♂ | Sterile ♀, dead ♂ | Sterile ♀, dead ♂ | NA | NA | NA | Sterile ♀, dead ♂ | Sterile ♀, dead ♂ | |
| simulans | Dead ♀, Sterile ♂ | Fertile ♀, sterile ♂ | Fertile ♀, sterile ♂ | NA | NA | NA | Sterile ♀, dead ♂ | Sterile ♀, dead ♂ | |
| sechellia | Dead ♀, Sterile ♂ | Fertile ♀, sterile ♂ | Fertile ♀, sterile ♂ | NA | NA | NA | Sterile ♀, dead ♂ | Sterile ♀, dead ♂ | |
| mauritiana | Dead ♀, Sterile ♂ | Fertile ♀, sterile ♂ | Fertile ♀, sterile ♂ | NA | NA | Sterile ♀, dead ♂ | Sterile ♀, dead ♂ | Sterile ♀, dead ♂ | |
| orena | NA | NA | NA | Sterile ♀, dead ♂ | NA | NA | NA | NA | |
| erecta | NA | NA | NA | Sterile ♀, dead ♂ | NA | NA | NA | NA | |
| yakuba | NA | NA | NA | Sterile ♀, sterile ♂ | NA | NA | Fertile ♀, sterile ♂ | Fertile ♀, sterile ♂ | |
| santomea | NA | NA | dead ♀, dead ♂ | Sterile ♀, sterile ♂ | NA | NA | Fertile ♀, sterile ♂ | Fertile ♀, sterile ♂ | |
| teissieri | NA | NA | NA | Sterile ♀, sterile ♂ | NA | NA | Fertile ♀, sterile ♂ | Fertile ♀, sterile ♂ |
Note.—In the majority of crosses for which we observed interspecific matings (i.e., inseminated females), we obtained viable interspecific hybrids. Black cells mark conspecific crosses which produce fertile progeny of both sexes. NA indicates crosses for which we obtained no progeny (i.e., behavioral isolation was complete). Thirty-three hybridizations produced viable progeny out of 72 possible interspecific crosses in the melanogaster species subgroup in long term interspecific matings. Only one cross (♀D. santomea × ♂D. sechellia) showed complete hybrid inviability. We failed to obtain inseminated females from other 39 crosses. Note that the cross ♀D. melanogaster × ♂D. orena produces sterile female progeny in rare cases (Comeault et al. 2017).
Hybrid Sterility versus Hybrid Inviability
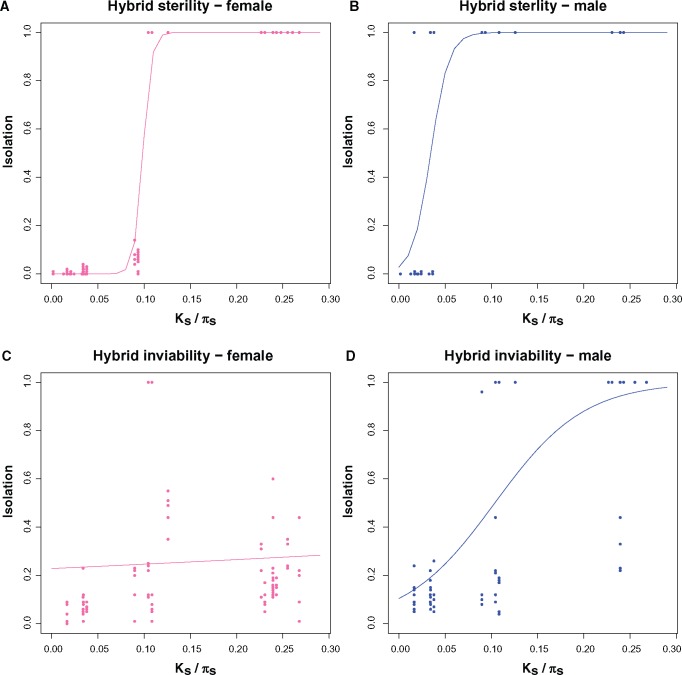
We compared the rates of evolution of hybrid sterility (fig. 4A and B) and of hybrid inviability (fig. 4C and D). We analyzed the two sexes separately. First, we compared the rate of evolution of male inviability with that of female inviability. Clearly, hybrid male inviability evolves faster than hybrid female inviability (logistic regression, test of rate of increment with a common intercept: F2,191 = 94.013, P < 1 × 10−15; results are identical with other regressions which are shown in supplementary tables S13 and S14, Supplementary Material online). This is not surprising since hybrid female inviability is not present in most crosses (at least when Ks ≤ 0.25; (fig. 4C). On the other hand, hybrid male inviability (95% complete) occurs at Ks ∼0.15.
Fig. 4.
Hybrid sterility evolves faster than hybrid inviability. Values of female and male inviability and female and male sterility were regressed against phylogenetic distance (Ks between species and πs within species). The four types of isolation increase with genetic distance. In both sexes, fertility evolves faster than hybrid inviability. Given the perfect separation of values along the x-axis (Ks/πs), hybrid sterility was not directly compared with other RIMs.
We observed a similar pattern in the evolution of hybrid sterility. Hybrid male sterility evolves faster than hybrid female sterility (logistic regression, test of rate of increment with a common intercept: F2,211= 47.67, P < 1 × 10−15; results with other regressions are shown in supplementary tables S15 and S16, Supplementary Material online). All interspecific male hybrids were sterile while only hybrid females from parents with a Ks ∼ 0.10 were sterile. This pattern holds regardless of how hybrid sterility is measured.
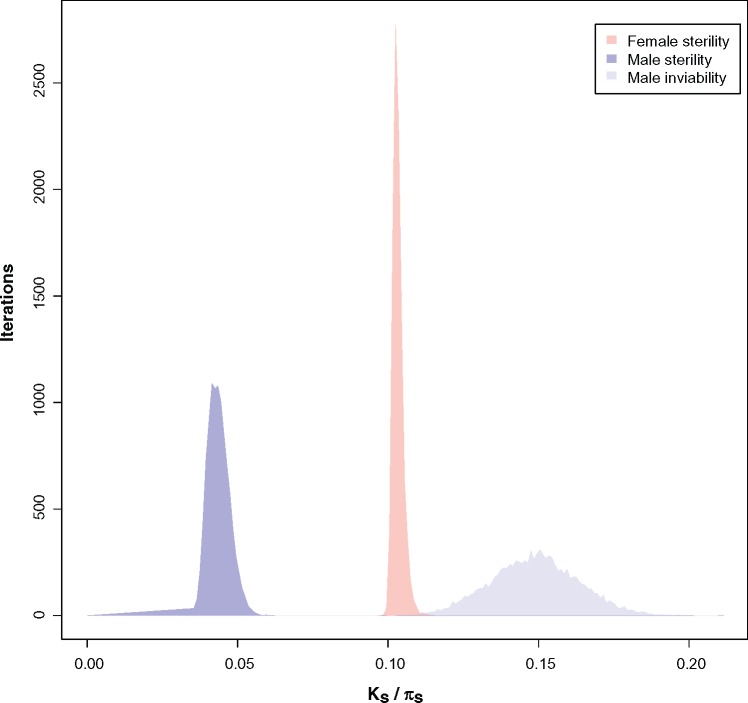
Third, we compared the rates of evolution of hybrid sterility and of hybrid inviability for each sex. The rate at which these two RIMs increment is different for both, males (logistic regression, test of rate of increment with a common intercept: F2,179 = 21.643, P = 3.82 × 10−9; supplementary tables S17 and S18, Supplementary Material online) and females (logistic regression, test of rate of increment with a common intercept: F2,223 = 179.5, P < 1 × 10−15; supplementary tables S19 and S20, Supplementary Material online). Not surprisingly, we found that hybrid male sterility is achieved much earlier than hybrid male inviability (pairwise comparisons in supplementary table S18, Supplementary Material online). Hybrid male sterility (95% complete) evolves at Ks ∼ 0.05. An equivalent strength of hybrid male inviability takes more divergence to occur (Ks ∼ 0.15). A similar comparison in females revealed that hybrid female sterility evolves faster than female inviability (pairwise comparisons in supplementary table S20, Supplementary Material online). Hybrid female sterility (95% complete) evolves at Ks ∼ 0.10. An equivalent strength of hybrid female inviability must necessarily take a Ks value higher than 0.25 (the maximum level of pairwise genetic divergence in this study). Bootstrap distributions of Threshold_Ks illustrate the difference between hybrid sterility and hybrid inviability (fig. 5). These results confirm the largely accepted, but untested, hypothesis that hybrid sterility evolves faster than hybrid inviability in both sexes.
Fig. 5.
Female sterility, male sterility, and female sterility evolve at different rates in the melanogaster species subgroup. Threshold_Ks indicates the Ks value for which a given RI barrier achieves a value of 0.95 (similar to the distributions shown in fig. 2). Even though these distributions of bootstrapped values are not a statistical test, Threshold_Ks illustrates how quickly isolation approaches 1. Female sterility values are red, male sterility are purple, and male inviability are blue. Female viability did not reach (or approached) an asymptote in our study and for that reason there is no distribution of bootstrapped values for this RIM.
Hybrid Inviability by Developmental Stage
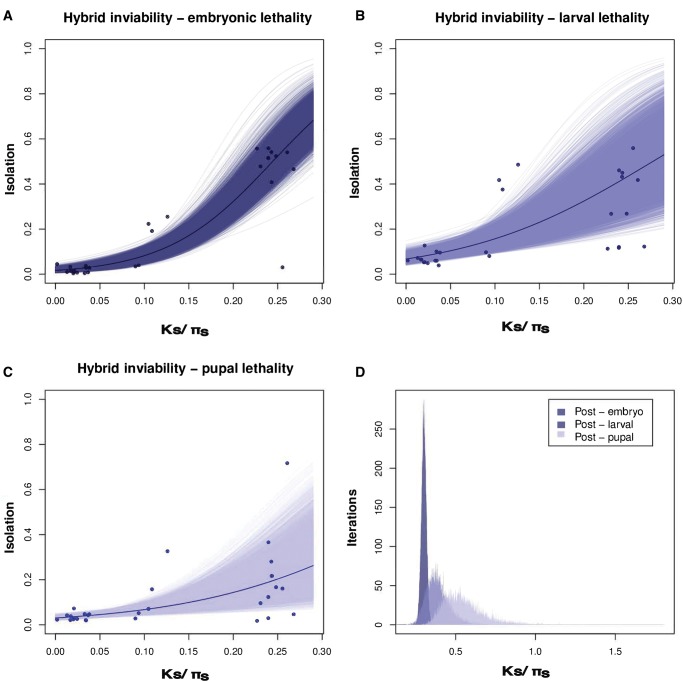
Hybrid inviability can manifest within three discrete developmental stages in holometabolan insects: the larvae, the pupae, or the adults. We assessed if hybrid inviability evolved faster at any of these three developmental stages. We quantified developmental stage specific rates of inviability by scoring the number of individuals that die at each of three crucial developmental transitions: embryo-to-L1 larva (embryonic lethality), L1 larvae-to-pupa (larval lethality), and pupa-to-adult (pupal lethality). As expected, the strength of all three types of postzygotic isolation increased with divergence time (fig. 6A–C), and in general, the lowest viabilities were observed for the crosses between the most distantly related species (supplementary table S5, Supplementary Material online). We next compared the rate at which hybrid inviability increased with genetic distance. We find significant differences in the rate of increase of each type of hybrid inviability (logistic regression, test of rate of increment with a common intercept: F3,491 = 14.621, P = 3.904 × 10−9; supplementary tables S21 and S22, Supplementary Material online). Pairwise comparisons (supplementary table S22, Supplementary Material online) and Threshold_Ks (fig. 6D) illustrate that complete embryonic lethality is reached before larval lethality, but complete larval lethality and pupal lethality are reached at similar rates.
Fig. 6.
Rates at which inviability increases with genetic distance for three developmental stages. Measures of inviability at each developmental stage were regressed against phylogenetic distance (Ks between species and πs within species). The three types of postzygotic isolation (i.e., death at a particular developmental stage) increase with genetic distance. (A) Embryonic lethality. (B) Larval lethality. (C) Pupal lethality. The thick lines represent fitted logistic regressions for each developmental stage. The thinner lines are the regressions for each of 10,000 bootstrap resamplings of the data. (D) Distributions of bootstrapped values of Threshold_Ks, a parameter that determines how quickly isolation approaches 1, are largely nonoverlapping. Regression coefficients (supplementary table S15, Supplementary Material online) show that early inviability (hybrid embryonic lethality) evolves faster than later inviability (hybrid pupal lethality).
Detection of Reinforcement Using Comparative Analyses
We evaluated the possibility of different mechanisms of RI evolving through reinforcing selection. Linear models revealed that the geographic origin effect, whether a line was sympatric or not, did not significantly predict the strength of any RIM (table 4). Threshold_Ks also suggests that there is no difference between the rates at which allopatric and sympatric lines in the magnitude of premating, NCGI, CSP, or postzygotic isolation approach completion (supplementary fig. S6, Supplementary Material online). We thus find no support for the hypothesis that reinforcing selection consistently acts on any one of the RIMs. This does not mean that reinforcement has not played a role in the evolution of RI in some of these pairs (e.g., Matute 2010a); rather, the influence of reinforcing selection is likely to be idiosyncratic and does not always influence the same RIM.
Table 4.
Linear Regression of PMPZ RIMs in Sympatric and Allopatric Lines.
| Sympatric Lines |
Allopatric Lines |
Origin Effect |
|||||
|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Degrees of Freedom (numerator, denominator) | F Value | P Value | |
| Premating isolation | 0.702 | 0.580 | 0.703 | 0.584 | 17,306 | 1.1462 | 0.3089 |
| NCGI | 0.8938 | 0.0751 | 0.8644 | 0.0966 | 11,141 | 6.9568 | 2.561 × 10−9 |
| CSP | 0.1488 | 1.5807 | 0.1434 | 1.7568 | 18,88 | 0.4602 | 0.9679 |
| Postzygotic isolation | 0.5036 | 0.2687 | 0.5142 | 0.2824 | 12,203 | 2.2074 | 0.01262 |
| Postzygotic isolation—embryonic lethality | 0.0905 | 0.1909 | 0.0905 | 0.1900 | 12,203 | 1.6188 | 0.08846 |
| Postzygotic isolation—larval lethality | 0.2280 | 0.2231 | 0.2224 | 0.2388 | 12,203 | 1.3817 | 0.1767 |
| Postzygotic isolation—pupal lethality | 0.2048 | 0.1923 | 0.2282 | 0.2058 | 12,203 | 0.6689 | 0.7801 |
Note.—Linear models show no difference at the strength of most types of RI between sympatric and allopatric pairs of lines. The only RIM that shows an origin effect is NCGI, whose significance is exclusively driven by the cross ♀D. yakuba × ♂D. santomea (Matute 2010). When this cross is excluded, the origin effect is not significant anymore (F10,123 = 1.7319, P = 0.0808).
Finally, we compared the magnitude of RI in two species triads characterized by an allopatric pair and a sympatric pair. The comparisons for each RIM for each triad are shown in supplementary table S23, Supplementary Material online. The majority of RIMs show no difference in magnitude between allopatric and sympatric pairs in any of the two triads. There are two notable exceptions. Behavioral isolation is stronger between D. yakuba and D. teissieri (a sympatric pair) than between D. santomea and D. teissieri (an allopatric pair). This observation has been reported before and is consistent with the action of reinforcement (Turissini et al. 2015). Second, and contrary to the expectations of reinforcing selection, D. sechellia and D. melanogaster (a mostly allopatric pair) show stronger hybrid inviability than D. simulans and D. melanogaster (a mostly sympatric pair). In particular, hybrid inviability is stronger in crosses where D. melanogaster is the female (D. melanogaster × D. simulans—mean = 0.593—vs. D. melanogaster × D. sechellia—mean = 0.833—Welch’s two sample t-test: t7.8 = 6.5561, P = 1.984 ×10−4). These results are opposite to the expectations if hybrid inviability evolved through reinforcement in this species pair. Regardless of how it was measured, our analyses indicate that reinforcement has indeed occurred in the melanogaster species subgroup but does not leave a consistent signature on any one RIM.
Discussion
Little is known about the rate of evolution of postmating-prezygotic (PMPZ) isolation in animals. Studies on plants have found that PMPZ and postzygotic isolation evolve at a similar rate in at least three plant genera. Unlike plant studies, most studies evaluating the rate of accumulation of reproductive isolation in animals have a common limitation: they have not looked at the rate of evolution of PMPZ barriers. We thus measured the rate of evolution of such barriers in Drosophila species pairs while assessing the magnitude of premating, and postzygotic isolation in the same crosses. This makes our study the first to measure the magnitude of PMPZ isolation, compare it with other RIMs, and explicitly test its rates of evolution in animals. Our results have implications for our understanding of three large topics in speciation: 1) the evolution of PMPZ barriers, 2) the evolution of postzygotic isolation, and 3) the role of PMPZ isolation and other RIMs on speciation via reinforcement. We discuss each of these topics as follows. We also present a series of caveats and general conclusions of our analyses.
The Evolution of PMPZ Barriers
PMPZ RIMs, both noncompetitive and CSP have been hypothesized to evolve through the influence of postcopulatory sexual selection (Birkhead and Pizzari 2002) and natural selection (Knowles et al. 2004). The female × male interactions that underlie NCGI can be interpreted as discrimination against heterospecific sperm by an inseminated female and are thus sexually selected (Price et al. 2001; Birkhead and Pizzari 2002; Fricke and Arnqvist 2004). Sperm morphology and traits associated with fertilization success show fast rates of change across species (Pitnick et al. 1999; Presgraves et al. 1999; Byrne et al. 2003; Manier et al. 2013). These phenotypes also evolve rapidly within and between species due to the constant influence of antagonistic sexual conflict (e.g., Knowles and Markow 2001; Comeault et al. 2016; reviewed in Pizzari and Snook 2003). Proteins involved in fertilization, gametic fusion, and stimulation of oviposition show signatures of positive selection across all taxa for which this signature has been systematically sought (e.g., Lee et al. 1995; Swanson et al. 2001; Galindo et al. 2003; Marshall et al. 2011; Harrison et al. 2015). This accelerated evolution at the phenotypic and molecular level is consistent with evolution of these interactions via either sexual or natural selection.
The second type of PMPZ barrier we examined, CSP, is also affected by sexual selection. CSP and its plant equivalent, conspecific pollen precedence, is ubiquitous and has been uncovered in a variety of organisms (references in Yeates et al. 2013). CSP is the aggregate of the three possible interactions between the female reproductive tract, conspecific sperm, and heterospecific sperm. These interactions create the grounds for reproductive incompatibilities due to sexual antagonism between sexes of different species, sexual antagonism between sexes of the same species, and sexual competition between sperm of different species (Howard 1999; Simmons 2005). Our results indicate that at least one of these interactions scales up with genetic divergence which leads to stronger CSP in divergent species.
Since premating and PMPZ traits usually cooccur in organisms with internal fertilization (such as Drosophila), the rapid evolution of PMPZ traits poses a conundrum: how can sexual selection or reinforcing selection influence the evolution of PMPZ barriers in the presence of premating isolation? First, premating isolation is often not complete, giving natural and sexual selection the opportunity to drive the evolution of PMPZ traits. Second, in organisms with internal fertilization, the evolution of PMPZ traits might be accelerated in instances where “the wallflower effect” applies (Kokko and Mappes 2005); females might adaptively lower their sexual preferences for males of high condition (conspecifics) if they are only exposed to males of low condition (heterospecifics). Similar models (Wilson and Hedrick 1982) and experimental measurements (Matute 2014) have shown that females might mate with heterospecifics if mates are rare. These instances where premating isolation is not an effective RIM might favor the accelerated evolution of PMPZ. The formal test of this hypothesis will require measuring PMPZ isolation in sister populations (or species) that differ in their strength of premating isolation.
The Evolution of Postzygotic Isolation
Our measurements also allowed us to discern which developmental stage was most affected by hybrid defects. Of the three possible transitions (embryo to larva, larva to pupa, and pupa to adult), we found that embryonic lethality arises first and evolves faster than larval or pupal lethality. A possible explanation for this result is that more genes are involved in embryogenesis than in other developmental stages, which seems to be the case from multispecies analyses of gene expression (Graveley et al. 2011). Hybrid incompatibilities would, therefore, have more potential targets at the embryonic stage. The GO category “embryo development” indeed contains more genes (740) than “larvae and prepupal development” (177), which in turn contains more genes than “pupal development” (17; supplementary table S24, Supplementary Material online). The identification of alleles involved in hybrid inviability has yielded mixed support for this hypothesis. Mapping of X-linked dominant factors in three Drosophila interspecific hybrids revealed that the majority of X-linked alleles in mel/san hybrids cause embryonic inviability. In the other two hybrids (mel/sim and mel/mau), however, no embryonic lethality alleles were found (Matute and Gavin-Smyth 2014). Similarly, in mel/sim hybrid males the vast majority of alleles involved in hybrid inviability cause postembryonic and not embryonic lethality (Presgraves 2003). We found that genes associated to all RIMs (clustered by GO terms and including different developmental transitions) show average rates of molecular evolution (as determined by their ratio of nonsynonymous to synonymous substitutions; KA/KS) comparable with the rest of the genome (supplementary text and in tables S24 and S25, Supplementary Material online). This is important because a nontrivial fraction of genes found to cause hybrid inviability and hybrid sterility have signatures of positive selection (e.g., Presgraves et al. 2003; Tang and Presgraves 2009 reviewed in Coyne and Orr 2004 and Nosil and Schluter 2011). Yet, we find no evidence for a consistent signature of positive selection at a particular RIM. Even though KA/KS and GO analyses have important caveats (reviewed in Rhee et al. 2008 and Venkat et al. 2017), and we cannot rule out that strong selection has occurred at cis-regulatory elements, our results generally refute the idea that genes involved in RIMs that act early in reproduction evolve faster than genes involved in RIMs that act later on.
Finally, we also saw complete hybrid male sterility for all interspecific crosses where males were viable (Ks higher than ∼0.05), and hybrid females were consistently sterile when Ks exceeded ∼0.10. It is worth noting how dramatically full hybrid sterility can arise when compared with the other forms of isolation we investigated. Since hybrid inviability is the slowest barrier to evolve, our results are consistent with the idea that hybrid sterility evolves faster than hybrid inviability in both sexes (Wu 1992), an idea that had remained formally untested because indexes of postzygotic isolation usually conflate sterility and inviability.
The Role of PMPZ Isolation on Speciation via Reinforcement
Our results are also important in the context of reinforcement. Two different approaches have been historically used to detect reinforcement: detecting reproductive character displacement in areas of secondary contact, and detecting the phylogenetic signal in sympatric species pairs. We used a modified version of the former approach. Comparing allopatric and sympatric lines from the same species can reveal reinforcement at recent scales (after secondary contact). On the other hand, phylogenetic comparison of the magnitude of RIMs detects reinforcement at deeper scales of divergence (Hopkins 2013, Hudson and Price 2014). We found no new evidence for cases of reinforcement besides the already reported influence of reinforcing selection in NCGI in the D. yakuba/D. santomea hybrid zone (Matute 2010a), and the phylogenetic signature of reinforcement at behavioral isolation in the D. teissieri/D. yakuba species pair (Turissini et al. 2015). It is possible that our experiment has little power to detect differences because all RIMs are already strong and the influence of sympatry is minimal compared with the amount of divergence that has already occurred between species.
A surprising result comes from the comparisons between the pairs D. simulans/D. melanogaster and D. sechellia/D. melanogaster. The latter pair shows extremely high hybrid inviability compared with the former pair. Given that D. melanogaster and D. sechellia are largely allopatric while D. melanogaster and D. simulans are largely sympatric, this pattern goes against the expectation of evolution of RI by reinforcing selection. The reasons behind such stark difference in the magnitude of RI remain unknown but we can formulate two possibilities. First, D. sechellia may have accumulated more hybrid incompatibilities due to extreme bottlenecks during its evolutionary history. Second, D. melanogaster and D. simulans may have had more opportunities to interbreed in the distant past, thus purging hybrid incompatibilities that still separate D. sechellia and D. melanogaster. More research on the demographic history of these species, as well as the effect of different demographic events on the accumulation of incompatibilities is needed before addressing the reasons for this difference.
The influence of PMPZ isolation on speciation by reinforcement remains largely unstudied. Reinforcement is traditionally viewed as the process of strengthening premating isolation driven by selection against unfit hybrids. However, PMPZ acts earlier in the reproductive cycle and has a faster rate of evolution compared with hybrid inviability and hybrid female sterility (shown here). Therefore, deleterious and costly PMPZ incompatibilities, such as reduced female fertility after interspecific crosses, might lead to the evolution of behavioral barriers in the same manner that postzygotic costs lead to premating isolation during conventional reinforcement (Harrison 1993; Servedio 2001). Even though we did not perform a formal comparison between the rates of evolution of PMPZ barriers and hybrid male sterility, the two types of RIMs seem to evolve at roughly the same rate. Thus, both PMPZ and hybrid male sterility might be equally important in inducing the evolution of premating isolation via reinforcement. Currently, the evidence that reproductive interference (excluding the production of unfit hybrids) might be costly is scattered and has been circumscribed to premating interactions (e.g., reproductive character displacement caused by noisy neighbors, Mullen and Andrés 2007 but see Matute 2015).
Theoretical arguments have also suggested that CSP can hamper the evolution of premating isolation by reinforcement because if CSP is complete, and a female has the chance to mate with multiple males, then no hybrids are likely to be produced if one of those males is a conspecific (Lorch and Servedio 2007). A similar argument can also be made about noncompetitive gametic isolation. If NCGI is strong, then no hybrids will be produced after interspecific crosses and if a female remates with a conspecific, then most of her progeny will be pure species and fit. In both of these cases, there will be no cost to hybridization and no incentive to strengthen premating isolation. This hypothesis yields a clear prediction: reinforcement of premating isolation should be rarer in clades where PMPZ isolation is strong. In spite of its straightforwardness, it might be premature to test this hypothesis because bona fide cases of reinforcement and of gametic isolation are still rare.
Conversely, postzygotic isolation might also lead to the evolution of PMPZ traits in the same manner that it leads to the evolution of premating isolation. This is obvious in aquatic organisms that spawn in open waters but it has been more controversial in animals with internal fertilization. Overall, reinforcement should affect the evolution of any trait that minimizes parental investment on an unfit hybrid (Coyne 1974; Servedio and Noor 2003). Two examples show that reinforcement can indeed lead to the evolution of PMPZ traits. In the case of Drosophila yakuba, females from the hybrid zone with D. santomea show stronger noncompetitive gametic isolation than females from areas where D. yakuba is not present (Matute 2010b; Comeault et al. 2016). Similarly, CSP in D. pseudoobscura is stronger in areas of sympatry with D. persimilis (Castillo and Moyle 2016). Both patterns of reproductive character displacement are highly suggestive of reinforcement and indicate that reinforcement of PMPZ barriers might not be a rare instance even in animals with internal fertilization.
Caveats
Our study is not devoid of caveats. The first one pertains to how much reinforcement can affect different types of reproductive barriers. Since reinforcement is thought to affect prezygotic isolation more commonly than other RIMs (Servedio and Noor 2003; but see Coyne 1974), then it is possible that reinforcing selection has led to an increase in the rate of evolution in premating isolation, NCGI, an CSP. This in turn would lead to an inflation of our estimated rate of evolution of these three RI barriers. Even though we compared allopatric and sympatric populations from eight species pairs, reinforcement might act at deeper levels of divergence that do not involve contemporary coexistence. An obvious research avenue is to test whether sympatric species evolve PMPZ mechanisms faster than allopatric pairs. This approach is not trivial as the range of species contracts and expands along their history making the distinction between allopatric and sympatric a gray area. Our data set does not allow us to split between currently allopatric and currently sympatric pairs because species from the melanogaster subgroup are largely sympatric (Lachaise et al. 1988) and an expanded data set will be necessary to address the importance of reinforcement.
A second bias is that when an early acting barrier is complete, we cannot measure the magnitude of later acting barriers. This introduces a bias that might inflate the rates of evolution of premating isolation because there will be more measurements of strong premating isolation than of postmating isolation. However, we attempted to minimize this bias by trying all possible crosses and, for one analysis, only including species for which we had measured the magnitude of the four types of RI barriers.
Conclusions
In general, we find that in Drosophila (subgenus Sophophora) both PMPZ barriers evolve faster than postzygotic isolation, but slower than premating behavioral mechanisms. These results indicate that there is a qualitative difference between the rate of evolution of PMPZ barriers in Drosophila and plants; in the latter PMPZ and postzygotic barriers evolve at similar rates (Moyle et al. 2004; Jewell et al. 2012). A possible explanation for this dichotomy is that in Drosophila, mate choice is a primary source of intrinsic isolation, whereas in plants the main source of intrinsic isolation might occur as pollen reaches the stigma. More research in different plant and animal taxa is needed to establish whether this difference is real or whether it is the byproduct of sparse taxonomic sampling. Similarly, and even though there is evidence for the existence of PMPZ in fungi (Turner et al. 2010, 2011) the rate of evolution of premating, and postmating isolation in this group and other eukaryotes remains largely unexplored (but see Gourbière and Mallet 2010; Giraud and Gourbiere 2012). These estimates across currently understudied taxa are sorely needed to understand what biological features drive the origin of new species.
Across metazoans, Drosophila has been one of the premier model systems for studying the evolution of reproductive isolation which in turn has provided support for several hypotheses such as the existence of reinforcement (Coyne and Orr 1989; Coyne and Orr 1997; Nosil 2013), the relative rate of evolution of RI (Yukilevich 2012; Rabosky and Matute 2013), and the role of ecology in speciation (Funk et al. 2006; Turelli et al. 2014). Overall, the fast accumulation of PMPZ isolation indicates that these RIMs are likely to be driven by selection, either sexual or natural. The integration of our results show that the earlier a barrier acted during the reproductive/developmental process, the faster its rate of accumulation over time. PMPZ isolation accumulates quickly in Drosophila thus indicating that this type or RI might be an important source of isolation in promoting the evolution of new species and maintaining species barriers.
Materials and Methods
Drosophila m elanogaster Subgroup: Species and Stocks
All wild-type stocks are described in supplementary table S26, Supplementary Material online. Briefly, for all genetic crosses we used synthetic stocks (i.e., outbred stocks derived from a combination of isofemale lines) with the exception of Drosophila erecta. Stocks from D. santomea, D. yakuba, D. teisseiri, D. orena, D. sechellia, D. simulans, and D. melanogaster were collected by D.R. Matute (supplementary table S26, Supplementary Material online Matute and Harris 2013). Stocks from these species were kept in large numbers (>200 flies) since their creation. Drosophila erecta was purchased at the San Diego Stock Center (Stock number: 14021-0224.00). All lines were reared on standard cornmeal/Karo/agar medium at 24 °C under a 12 h light/dark cycle in 100 ml bottles. Adults were allowed to oviposit for 1 week and after that time they were cleared from the bottles. We added 1 ml of propionic acid (0.5% V/V solution to the vials and provided a pupation substrate to the vial (Kimberly Clark, Kimwipes Delicate Task; Irving, TX). At least ten bottles of each species were kept in parallel to guarantee the collection of large numbers of virgins.
To measure conspecific sperm precedence, we also used mutants from each of eight of the species (with the exception of D. orena, see below). All mutants were raised in identical conditions to the wild-type stocks.
Virgin Collection
Pure species males and females of each species were collected as virgins within 8 h of eclosion under CO2 anesthesia and kept for 3 days in single-sex groups of 20 flies in 30 ml, corn meal food-containing vials. Flies were kept at 24 °C under a 12 h light/dark cycle. On day four, we assessed whether there were larvae in the media. If the inspection revealed any progeny, the vial was discarded.
Premating Isolation: Insemination Rates
We measured premating isolation as the number of females that did not accept heterospecific males when housed together in no-choice experiments for 24 h. Two hundred females (i.e., individuals pooled from ten virgin vials) were housed with 200 males either from the same species or from a different species. Females and males were housed together for 24 h. After that time, females were anesthetized with CO2 and males were discarded. We dissected all the females and extracted their reproductive tract (spermathecae, seminal receptacles, and uterus) and placed it in chilled (4 °C) Ringer’s solution. We assessed whether the female carried any sperm, either dead or alive anywhere in their reproductive tract. The vials where the crosses occurred were kept to study the magnitude of hybrid inviability (see below “Postzygotic isolation: Hybrid inviability”). We used the proportion of females inseminated in the en masse matings in each bottle (see “Insemination rates”) and calculated a proxy of the strength of premating isolation:
Five batches (bottles) per cross (each with 100 females) were counted.
We assessed whether there was heterogeneity in insemination rates among conspecific matings. We counted how many females were inseminated in five replicates. To detect heterogeneity, we fit a linear model in which the proportion of inseminated females in these conspecific crosses was the response and the cross (i.e., species) was the only factor.
Next, we studied whether there was heterogeneity in premating isolation among interspecific crosses. We fit two linear regressions to analyze the data. First, to assess if any particular combination of species was more prone to mating than others, we fit a linear regression in which in each bottle was the response, and the identity of the cross was the only fixed effect. There were five replicates per species for a total of 360 bottles. Second, we analyzed whether any type of female (and male) were more prone to hybridize with other species. To do so, we used the same data set but fit a factorial model in which in each bottle was the response and the identity of the female and that of the male were fixed effects. We also included an interaction term. All statistical analyses were carried out using the package “stats” in R (function: lm; R Core Team 2016).
PMPZ Isolation: noncompetitive Gametic Isolation (NCGI)
We next measured gametic incompatibilities between females and males from different species in single matings, namely, the inability of a male to induce a female to lay eggs (Price et al. 2001; Matute 2010b; Marshall and DiRienzo 2012). We watched single heterospecific and conspecific pairs for 8 h and kept the females that mated successfully for each of the 81 possible hybrid crosses (72 heterospecific + 9 conspecific). We repeated this approach until we collected at least five females from each of the interspecific and conspecific crosses. We kept all females who mated (either to con- or heterospecific males) to measure gametic isolation. To prevent females from remating, males were removed from the vial by aspiration after mating. Each mated female was allowed to oviposit for 24 h in a vial. The female was then transferred to a fresh vial, and the total number of eggs were subsequently counted daily for 10 days. At least five females were scored for each cross.
Ig, an index of PMPZ isolation which was calculated as:
Ig values were compared across crosses using a linear model in which cross was the only fixed factor. This index is not devoid of caveats. We list two of them. First, some species may differ in the number of eggs a sexually mature virgin female will lay. This will lower the confidence level for the estimate of NCGI because some of the laid eggs will be noninseminated. Second, it is also possible that laid eggs are dying before embryogenesis begins and thus conflates early embryonic lethality with NCGI. This latter concern is further addressed below (section “Postzygotic isolation: Hybrid inviability”).
PMPZ Isolation: Competitive Gametic Isolation
CSP Indexes
We also scored how much hybrid progeny a female produces after mating with two males: a heterospecific, and a conspecific male. To do so, we used a combination of mutants to differentiate between hybrid and pure species progeny in crosses that involved more than one male. Traditionally, CSP is measured as P2, the proportion of progeny sired by the second male in double matings (Boorman and Parker 1976; Chang 2004). Nonetheless, this measurement conflates two important biological aspects. First, second males have an advantage over first males (regardless of their genotype) and in conspecific crosses, they invariably sire more progeny than first males. Second, conspecific sperm might indeed have an advantage over heterospecific sperm (true conspecific sperm precedence). We propose to quantify CSP as the proportion of progeny sired by a heterospecific male by a doubly mated female respective to a conspecific mating that mated to two conspecific males. To account by the fact that the second male usually has an advantage over the first male, we propose to do normalizations taking into account the order of mating. Matings with two males can occur in two different orders. In an interspecific/conspecific mating, we counted the number of hybrid progeny (HCH) and the number of conspecific progeny (HCc). In crosses where the heterospecific male was mated first followed by a conspecific male, the indexes took the form HCH1 and HCC2. In crosses where the conspecific male was first and was followed by a heterospecific male, the indexes took the form HCC1 (i.e., the progeny sired by the heterospecific male in females mated to a heterospecific male and then to a conspecific male) and HCH2 (i.e., the progeny sired by the heterospecific male in females mated to a conspecific male and then to a heterospecific male). In conspecific/conspecific matings, we counted the progeny sired by a wildtype and a mutant stock of the same species. This yielded two quantities: the progeny sired by the first male, CCC1 (i.e., the progeny sired by the first male in females mated to two conspecific males) and the progeny sired by the second male, CCC2 (i.e., the progeny sired by the second male in females mated to two conspecific males).
These quantities were then incorporated into two indexes of conspecific sperm precedence, one for crosses when heterospecific males were the first to mate, and one for crosses where conspecific males were first. The two indexes followed the form:
and
There was only true CSP if both and indexes were low.
Mutant Stocks
In order to quantify CCC1and CCC2 we needed to obtain females that mated to conspecific males twice and be able to distinguish between the progeny sired by each father. To do so, we needed mutant stocks that could be visually recognized from the wild type. We described the mutants for eight species in the melanogaster species subgroup (no mutants were available for D. orena).
Drosophila melanogaster: D. melanogaster yellow white (mely1 w1) males and females were derived from a stock purchased at the Bloomington Stock Center (Stock number: 1,495).
Drosophila mauritiana: D. mauritiana yellow (mauy1 w1 f1) males and females were derived from a stock purchased at the San Diego Stock Center (Stock number: 14021-0241.55).
Drosophila yakuba: D. yakuba yellow (yaky) males and females were derived from a stock originally collected in the Täi Forest (Liberia) in 1998.
Drosophila teissieri: D. teissieri yellow (teiy)was isolated from a line collected in Bioko (2009). Both, yakyand teiy, are a fully recessive body color mutation identical to that on the D. melanogaster X chromosome (Llopart et al. 2002).
Drosophila erecta: D. erecta yellow (erey) also has a body color mutation. Whether this yellow mutation complements mely remains unknown.
Drosophila santomea: D. santomea white (sanw) males and females were derived from the STO.4 isofemale line, originally collected on São Tomé in 1998. All the white-eyed mutations described in this contribution are fully recessive eye color mutations orthologous (i.e., fail to complement) to white eyed mutations on the D. yakuba and/or D. melanogaster X chromosomes.
Drosophila sechellia: D. sechellia white (sechw) was isolated from a recently collected line in Denis island.
Drosophila simulans: D. simulans white (simw) was donated by D.C. Presgraves.
Measuring CSP
First matings were watched as described above (Section “Premating isolation: Insemination rates”). Mated females were then separated from the males and housed in groups of 1–5 females. On the morning of day 4, females were individually transferred to a new vial with cornmeal food. The male to be mated was also transferred to the vial by aspiration. We observed up to 300 individual matings at the same time. Second matings were allowed to proceed for 16 h. To identify true CSP, we attempted to measure the magnitude of sperm precedence in two orders of the cross. Crosses that involved a conspecific male first and an interspecific male second are challenging and in some cases estimates involved only a few measurements. The sample sizes of each mating are shown in supplementary table S3, Supplementary Material online. Once doubly mated females were obtained, we removed the male from the vial, kept the females and tended their progeny. Females were transferred to a new vial every 7 days until they died. Vial tending and fly husbandry were done as described immediately below. ICSP were compared using a linear model with the identity of the cross and the order (i.e., what male was mated first) as the two fixed factors.
Postzygotic Isolation: Hybrid Inviability
Finally, we measured viability in F1 hybrids. For each interspecific and conspecific species pair, we calculated overall F1 inviability and three components of inviability: embryonic lethality (death during the embryo-to-L1 transition), larval lethality (death during the L1 larvae-to-pupa transition), and pupal lethality (death during the pupa-to-adult transition). We collected virgin males and females as described above (See “Virgin collection”). We studied hybrid inviability following two approaches: by letting crosses occur over a period longer than 4 weeks, and by following the development of the embryos deposited by females over a period of 24 h (obtaining roughly staged hybrid individuals). The procedure for the former approach is described somewhere else (Turissini et al. 2015). Briefly, on the morning of day four after collection, we placed 40 males and 20 females together at room temperature (21°–23 °C) to mate en masse on corn meal media. We set up 50 crosses per species pair for a total of 4,050 crosses (81 crosses × 50 replicates). Vials were inspected every 5 days to assess the presence of larvae and/or dead embryos. We transferred all the pure species adults to a new vial (without anesthesia) every ten days. This procedure was repeated until the cross stopped producing progeny (all females died, over 5 weeks). L1 larvae were allowed to feed on an apple-agar plate and were tended daily. Once L2 larvae were observed, we added a solution of 0.05% propionic acid and a KimWipe (Kimberly Clark, Kimwipes Delicate Task, Roswell, GA) to the vial. Adult hybrids were collected and counted using CO2 anesthesia. In order to maximize the lifespan of the parents, we kept all the vials lying on their sides. We repeated this procedure until we obtained five cages that produced hybrid progeny for the crosses for which we could obtain inseminated females.
To follow the development of hybrid embryos, we collected mated females as described above (See “Premating isolation: Insemination rates” and “PMPZ isolation: noncompetitive gametic isolation (NCGI)”). The development of the eggs laid by these females and the rates of transition to the next developmental stage (egg to larvae, larvae to pupae, pupae to adult) were recorded daily. We partitioned overall inviability into three components by comparing the number of individuals that entered a developmental stage (Total) to the number that survived it (Successes) using the equations shown in table 1. The proportions were then transformed to a logistic index following the form:
Overall inviability was based on the number of individuals that died during development (from embryo to adulthood). For a global estimate of inviability, we counted the total number of viable eggs and the number of those that developed into adults. To quantify embryonic inviability, we counted the total number of embryos defined as the total number of egg cases (successes) plus the number of dead embryos (brown eggs). This procedure was applied to 69 interspecific crosses (the exception being crosses between females from the simulans clade and D. melanogaster males; See below). To quantify larval lethality, we counted the number of egg cases (Total) and pupae (Successes) in each vial. If larvae pupated on the food media, the vial was discarded. Finally, to quantify pupal viability we counted the number of pupae (Total) and adults (Successes). We quantified lethality for at least five replicates per cross and summed the results. The number of replicates per cross is listed in supplementary table S27, Supplementary Material online.
For embryonic lethality, we assessed the robustness of our two proxies: egg cases for viable eggs, and brown eggs for dead embryos. First, we studied how robust was the proxy of empty cases for the number of live embryos. As larvae feed, they churn the food and egg cases can disappear from the surface. As a result, counts of the number of egg cases + brown eggs (dead embryos) were sometimes less than the initial egg count. To account for this missing data, we inferred the number of missing dead embryos from the consolidated data from the replicates using the formula: missing dead embryos = (dead embryos/eggs) × missing embryos. This estimate was rounded to the nearest integer and added to the number of dead embryos. The number of egg cases was likewise adjusted by adding the difference between missing and missing dead embryos. Accounting for the missing data does not affect our results (supplementary fig. S7, Supplementary Material online).
Second, we adjusted the estimates of dead embryos. Three crosses have shown extensive embryonic mortality before the zygote stage is achieved: ♀ D. simulans × ♂D. melanogaster, ♀ D. sechellia × ♂D. melanogaster, and ♀ D. mauritiana × ♂D. melanogaster (Sawamura et al. 1993). We focused on these three crosses because no other cross in the melanogaster species group exhibit the same phenomenon. In these three interspecific crosses, brown eggs are rare, as the female diploid embryos that do not develop do not achieve the status of zygote. To measure the magnitude of female embryonic lethality in these three crosses, we collected one-hundred 72-h embryos from each cross. From each of these embryos, we extracted DNA using the QIAamp DNA Micro Kit (Qiagen, Chatsworth, CA, USA) following the manufacturer’s instructions and amplified the yellow locus of D. melanogaster by PCR using the primers mel_y_F: 5′ CGGCTCCCTTGGCCACTTTA3′ and mel_y_R: 5′ CGGGCATTCACATAAGTTTTTAACC 3′. Both primers include sites private to D. melanogaster in positions 20 and 25, respectively, and do not amplify in D. simulans. The primers amplify a 412 bp fragment (Tm = 59.4 °C). The presence of this locus meant an embryo had been fertilized, and its absence meant the embryo was unfertilized. We used the primers control_y_F: 5′ CTGACTTGGATTATTCAGATACTAATTTC3′ and control_y_R: 5′ CTACATTGCCTGAATTGGCG3′ as a positive amplification control. These primers amplify a PCR product of 267 bp (Tm = 56.0 °C). PCR conditions were identical to those described elsewhere (Matute and Ayroles 2014). Amplicons were run in a 2% agarose gel and visualized using ethidium bromide and UV. None of the newly described hybrids produces only adult males, so there were no additional cases of female early embryonic lethality and thus no need to correct these estimates.
The proportion of dead embryos in an oviposition cage was calculated by multiplying the total number of eggs in the oviposition cage with the average proportion of embryos that were diploid (i.e., carried the D. melanogaster yellow locus) and did not hatch. To detect heterogeneity among crosses, we used the ‘lm’ function in the “stats” package in R to fit linear model where the strength of hybrid inviability was the response and the identity of the cross was the only fixed effect. Since there were three different types of hybrid inviability (one for each developmental transition) and a life-long estimate of inviability, there were four linear models.
Correlations between viability at different development transitions were calculated using a Pearson’s product-moment correlation (R package “Stats”: function “cor.test”; ρ). Critical P-value for significance was 0.01 to account for multiple comparisons (three comparisons).
To measure sex-specific hybrid viability we took advantage of the existence of yellow mutant stocks, mutant stocks for which we could differentiate between males and females early in development. In crosses between a yellow-null carrying mother and a wild-type father, female progeny is heterozygote (y+y−) and larvae have black mouthparts. Male progeny will be hemizygous for the y-null and their mouthparts will be brown. Four of five yellow− used in this experiment stocks are described in above (i, ii, iii, iv in section “PMPZ isolation: competitive gametic isolation, Mutant stocks”). For these experiments, we also used an additional stock, Drosophila simulans yellow−, which was donated by J.A. Coyne. This yellow mutation does not complement mely.
Postzygotic Isolation: Fertility Assessments
For all pure-species and interspecific crosses, we assessed whether their progeny were fertile or sterile. The protocol was similar for both sexes: we extracted and dissected their gonads to look for the production of gametes. In the case of female hybrids, we looked at the presence of ovarioles in the ovaries; females with ovarioles were classified as fertile. We used this binary scale to avoid the significant effects that environment has on ovariole number (Wayne and Mackay 1998; Wayne et al. 2006). In the case of male hybrids, testes were dissected, mounted in Ringer’s solution and squashed to assess for the presence of motile sperm. We scored 100 individuals per cross per sex. We measured isolation separately for each sex as:
Hybrids that were thought to be sterile were then housed with pure species individuals from the opposite sex (both parentals) to make sure our assessment of fertility was qualitatively adequate. Obviously, individuals that had been dissected and scored for fertility could not be mated; from a single cross we dissected half of the progeny and kept at least 50 of them to do en masse matings. Hybrid females were housed with males of the two species as described in (Turissini et al. 2015); hybrid males were housed with virgin females from both species. For both sexes, we assessed whether the crosses produced progeny until hybrid individuals were dead. Male sterility, and to a lesser extent hybrid female inviability were binomial traits that showed separation (i.e., all crosses above a particular Ks produce only sterile progeny) and for that reason these two traits were not directly compared with premating, NCGI, CSP or hybrid inviability which showed a continuous trait distribution.
Genome Sequencing
DNA Extraction
DNA was extracted from single female flies using the QIAamp DNA Micro Kit (Qiagen, Chatsworth, CA, USA). We followed the manufacturer’s instruction using cut pipette tips to avoid shearing the DNA. This protocol can yield up to ∼50 ng of DNA per fly per extraction.
Library Construction
For short read sequencing, we constructed libraries following two options. 54 libraries were built using the Kappa protocol for TrueSeq at the sequencing facility of the University of North Carolina, Chapel Hill. For these libraries, DNA was sheared by sonication. Briefly ∼10 µg of DNA were sonicated with a Covaris S220 to 160 bp mean size (120–200 bp range) with the program: 10% duty cycle; intensity 5; 100 cycles per burst; six cycles of 60 s in frequency sweeping mode The second type of libraries were Nextera libraries which were built at the sequencing facility of the University of Illinois, Urbana-Champaign. For this type of library, DNA was segmented using Nextera kits which uses proprietary transposases to fragment DNA. Libraries were built following standard protocols.
Sequencing
Lines were sequenced in a HiSeq 2000 machine and were a mixture between single end and paired end sequencing. Supplementary table S28, Supplementary Material online indicates the sequencing type and coverage for each line. Six individuals were sequenced per lane. The HiSeq 2000 machine was run with chemistry v3.0 and using the 2 × 100 bp paired-end read mode and original chemistry from Illumina following the manufacturer’s instructions. To assess the read quality, we performed initial image analyses and base calling using HiSeq Control Software 2.0.5 and RTA 1.17.20.0 (real time analysis). CASAVA-1.8.2 generated and reported run statistics of each of the final FASTQ files. Resulting reads ranged from 100 or 150 bp and the target average coverage for each line was 30×. The actual coverage for each line is shown in supplementary table S28, Supplementary Material online.
Public Data
We accessed and used two additional sources of genomic data. We downloaded available raw reads (FASTQ files) from NCBI and mapped them to the corresponding reference genome (Mackay et al. 2012, Rogers et al. 2014, Rogers et al. 2015; see below). Additionally, we downloaded D. melanogaster sequences from the NEXUS sequencing project (Lack et al. 2015).
Read Mapping and Variant Calling
Reads were mapped using bwa version 0.7.12 (Li and Durbin 2010). Drosophila yakuba, D. teissieri, and D. santomea reads were mapped to the D. yakuba genome version 1.04 (Clark et al. 2007), D. simulans, D. sechellia, and D. mauritiana reads were mapped to the D. simulans w501 genome (Hu et al. 2013), and D. orena reads were mapped to the D. erecta genome (Drosophila 12 Genomes Consortium et al. 2007). Bam files were merged using Samtools version 0.1.19 (Li et al. 2009). Indels were identified and reads were locally remapped in the merged bam files using the GATK version 3.2-2 RealignerTargetCreator and IndelRealigner functions (McKenna et al. 2010; DePristo et al. 2011). SNP genotyping was done independently for the D. yakuba clade, D. simulans clade, and D. orena using GATK UnifiedGenotyper with the parameter het = 0.01. The following filters were applied to the resulting vcf file: QD = 2.0, FS_filter = 60.0, MQ_filter = 30.0, MQ_Rank_Sum_filter = −12.5, and Read_Pos_Rank_Sum_filter = −8.0. Sequences were created for individual lines with perl scripts using the GATK genotype calls and coverage information obtained from pileup files generated using the samtools’ mpileup function. Ambiguous nucleotide characters were used to identify the two alleles at heterozygous sites. Sites were replaced with an “N” if the coverage was <5 or greater than the 99th quantile of the genomic coverage distribution for the given line or if the SNP failed to pass one of the GATK filters.
Genomic Alignments
Alignments were made based on the dmel6.01 annotation downloaded from Flybase: ftp.flybase.net/genomes/Drosophila_melanogaster/dmel_r6.01_FB2014_04/gff/ dmel-all-r6.01.gff.gz (Santos dos et al. 2015). The D. yakuba, D. simulans, and D. erecta reference genomes were separately aligned to the D. melanogaster genome using nucmer version 3.23 with parameters –r and –q. A custom perl script then combined the nucmer genomic alignment coordinates and individual line sequences to create genomic sequences for each line that were syntenic to the D. melanogaster reference genome. A perl script then called a consensus sequence for each species using these D. melanogaster syntenic genome sequences.
Between Species Genetic Distance
The number of synonymous substitutions in protein coding genes (Ks) was used as a measure of genetic distance between species pairs in the melanogaster species subgroup. A perl script generated a CDS alignment for each gene using the consensus D. melanogaster syntenic genomic sequences for D. simulans, D. sechellia, D. mauritiana, D. yakuba, D. santomea, D. teissieri, and D. orena; the reference genome sequences for D. melanogaster; and the melanogaster syntenic genome for D. erecta. For genes with multiple annotated transcripts in dmel6.01, we used the longest transcript. We excluded codons that had an N in any of the aligned species. We also excluded genes with either a premature stop codon in any species or whose length was less than 100 bases. We ran PAML version 4.8 (Yang 1997; Yang 2007) to calculate Ks individually for 8,923 genes using the basic model (model=0). PAML was also run with additional models: free ratios (model=1), 3 ratios (model=2, tree = ((mel, (sim, sech, mau)2)1, (((yak, san), tei), (ore, ere))3)), and 2 ratios (model=2, tree = ((mel, (sim, sech, mau))1, (((yak, san), tei), (ore, ere))2)). The super-indices indicate the branches that were allowed to vary in their KA/KS. Pairwise Ks divergences were obtained by taking the average over all genes.
This genome wide data set was also used to test whether genes involved in a particular RIM showed evidence for positive selection. We used KA, the number of synonymous substitutions in coding genes and Ks (described above). The ratio KA/KS is a proxy of whether a gene has undergone rapid evolution in its coding sequence. We selected genes annotated for 11 GO terms that were related to the nature of each RIM included in this study and calculated their average KA/KS. The list of relevant GO terms is shown in supplementary table S24, Supplementary Material online. We assumed a constant KA/KS across the tree (model = 0, described immediately above). We could not compare the mean KA/KS value for each GO term with that of the rest of the genome because the sample sizes for each GO term were rather small (mean=7.5 genes per GO term), and there was extensive overlap across GO terms. In lieu of the comparisons, we present all the raw data for each GO term in supplementary table S25, Supplementary Material online.
Within Species Neutral Variation
We also calculated the level of genetic variation within species as a proxy of genetic distance between individuals of the same species. πs was used as a measure of the average genetic distance between individuals of the same species. We calculated πs and the number of synonymous sites in each gene for each species using Polymorphorama (Andolfatto 2007; Haddrill et al. 2008). A perl script generated a CDS alignment for each species for each gene using the D. melanogaster syntenic genome sequences and the dmel 6.01 gene annotations. Since the NEXUS D. melanogaster sequences were mapped to dmel5 (Lack et al. 2015), we used the dmel5.10 gene annotations for that species. As was the case for interspecific alignments, we only used the longest transcript for genes with multiple transcripts. We only used sequences that were less than 5% Ns and required that at least five individuals met this criterion. We also excluded genes if a premature stop codon was encountered in any individual. A measure of within species variation for each species was obtained by averaging πs over all genes weighted by the number of synonymous sites.
Rate of Evolution of Reproductive Isolating Mechanisms: No Phylogenetic Corrections
Finally, we used a logistic regression with complete taxon sampling to analyze whether the distance between potentially hybridizing species influence the magnitude of reproductive isolation. Each index of RI was independently regressed against the divergence between the parental species using the glm function with a logit link function with binomial errors in R (“stats” package, R Core Team 2016). Ks was used as a measure of neutral species divergence, and πs was used as a proxy of neutral within species variation.
The logistic function is given by
where the parameter β1 determines how quickly the function approaches 1 with larger values producing steeper slopes and x represents an independent variable. The fit of each function was determined with a McFadden’s pseudo-r2 calculated with the R package “pscl” (function “pR2”, Jackman et al. 2007).
To compare the rate of evolution of different RIMs we used three different analyses. First, we fitted a logistic regression to the data where the magnitude of RI depended on the two fixed effects (type of RIM and genetic distance) and their interaction. The regression followed the form:
Second, we also fitted a logistic regression where RI depended exclusively on the magnitude of the interaction between the fixed effects (i.e., assumed a common intercept, ). The regression followed the form:
Finally, we fitted a linear regression where the magnitude of RI depended only on the interaction between RIM type and genetic distance (also assuming a common intercept, ). The regression followed the form:
To determine which RIM evolved faster, we compared the magnitude of the least squares mean value for each interaction category on each of the three regressions listed above. We used Tukey’s Honest difference pairwise comparisons as implemented in the package lsmeans (function “lsmeans”) with a P value adjustment for each regression (four estimates per regression).
This approach was used to compare the rates of evolution in six instances. First, to compare the rate of evolution of premating isolation, NCGI, CSP, and hybrid inviability (with and without phylogenetic corrections). Second, to compare the rate of evolution of hybrid female inviability and hybrid male inviability. Third, to compare the rate of evolution of hybrid female sterility and hybrid male sterility. Fourth, to compare the rate of evolution of hybrid male sterility and hybrid male inviability. Fifth, to compare the rate of evolution of hybrid female sterility with hybrid female inviability. Finally, to compare the rate of evolution of different types of hybrid inviability.
To illustrate the differences in the rates of evolution of different types of RI, we calculated the level of genetic divergence where the magnitude of RI crossed a 0.95 threshold. This is not formally a statistical test (to that end see the regression coefficients) but the quantity illustrates how fast a RIM approaches completion. For the melanogaster subgroup, genetic divergence was measured as Ks (Threshold_Ks). For the Sophophora subgenus, genetic distance was measured as Nei’s D (Threshold_D; See below “Rate of evolution of reproductive isolating mechanisms: phylogenetic corrections”). The same approach was applied to illustrate the differences between evolutionary rates of hybrid sterility and hybrid inviability, and between embryo, larval, and pupal stage inviability.
Rate of Evolution of Reproductive Isolating Mechanisms: Phylogenetic Corrections
Our measurements of the rate of evolution of RI on the melanogaster subgroup have an important caveat: since all the species are closely related and our design involved measuring all possible pairwise interactions, we could not apply phylogenetic corrections. This is an important limitation because if one species is more likely than others to be reproductively isolated and the branch leading to that species is used more than once, it might inflate the rate of evolution of a particular RI. Several approaches have been proposed to correct nonindependent measurements of RI (i.e., those that include a species or a branch more than once). Nonetheless, reconstructing levels of ancestral RI at a node might be problematic as RI does not follow the regular assumptions of quantitative traits (e.g., Moyle et al. 2004). We opted for a more conservative approach in which we performed regression using only strictly independent species pairs (i.e., nonoverlapping branches; supplementary fig. S1, Supplementary Material online). We thus evaluated whether the relative ranking of the rates of evolution of the four types of RIMs obtained in the melanogaster comparisons also held when we did a similar analysis with phylogenetically independent points. We evaluated our hypothesis in a phylogenetically independent subset of species from the melanogaster subgroup. In this case, three species pairs is the maximum number of independent species pairs (supplementary fig. S1, Supplementary Material online). We also included measurements for hybridizations of the willinstoni (D. paulistorum Centroamerica, D. paulistorum Interior), pseudoobscura (D. pseudoobscura, D. persimilis, D. bogotana), virilis (D. virilis, D. lummei, D. americana, D. novamexicana), and mojavensis (movavensis baja, mojavensis sonora) group. All these species belong to the Sophophora subgenus. Nei’s D distance between these species was obtained from Yukilevich (2012). The choice of groups and species was dictated by the existence of phenotypic mutants and limited by the ability to measure sperm precedence in conspecific crosses; we needed mutant stocks to be able to quantify CCC1 and CCC2, two required components of the ICSP indexes (see above). To this end, only four species satisfied the requirement: D. paulistorum Centroamerica white (14030-0771.04), D. virilis eGFP (15010-1051.108), D. pseudoobscura GFP (10411-0121.201), and D. mojavensis w (15081-1352.05). The performed crosses and sample sizes are shown in supplementary table S29, Supplementary Material online. We next estimated the rate of evolution of premating, noncompetitive gametic isolation, competitive gametic isolation, and hybrid inviability in crosses for each group as described above.
Detection of Reinforcement Using Comparative Analyses
The magnitude on RI can be affected by the influence of reinforcement, a type of natural selection that strengthens prezygotic isolation as a byproduct to reduce maladaptive hybridization (Servedio and Noor 2003; but see Coyne 1974 for an argument of reinforcement of postzygotic isolation). To detect whether reinforcement has played a widespread role on the evolution of any particular of RI, we followed two approaches. First, we compared the magnitude of all the above mentioned types of RI in eight pairs of species for which we had sympatric and allopatric species (N = 8 species, 1 sympatric line pair and 1 allopatric line pair per species pair). In both sympatric and allopatric crosses, we measured RI as described above. We compared the magnitude of each RIM (three types of prezygotic isolation, hybrid inviability as a whole, and the three developmental components of inviability) using two methods. First we quantified the amount of genetic divergence required to achieve 95% of the maximum value of RI in sympatric and allopatric populations independently. The expectation of this comparison is that if a RIM is evolving through reinforcement, sympatric lines should show a stronger RI than allopatric lines from the same species for that RIM. Second, we assessed whether the effect of geographic overlap (i.e., whether lines were sympatric or allopatric) influenced the magnitude of RI (while controlling by cross) by fitting linear models where each type of RI (7 linear models excluding female and male sterility) was the response and depended on the identity of the cross, the geographic origin (whether a line was sympatric or allopatric), and the interaction between these two main effects.
The second approach aimed to detect the phylogenetic signature of reinforcement (Noor 1997). We compared the magnitude of all RIMs in two species triads: (D. teissieri, (D. yakuba, D. santomea)) and (D. melanogaster, (D. sechellia, D. simulans)). Notably these triads include a pair of species that is sympatric (D. teissieri, D. yakuba; and D. melanogaster, D. simulans) and one that is allopatric (D. teissieri, D. santomea; and D. melanogaster, D. sechellia). If reinforcing selection has acted, then the magnitude of RI should be stronger in the sympatric pairs than in the allopatric pairs. We pooled the two directions of the cross for each pair and compared the mean strength of each RIM using permutation tests (function “oneway_test” with and 9,999 Monte Carlo iterations; R package “coin”). All raw data and analytical code have been deposited to Dryad (doi:10.5061/dryad.6v774).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We would like to thank C. H. Martin, C.J. Jones, A.A. Comeault, V. Courtier-Orgogozo, Y. Brandvain, R. Marquez, K. L. Gordon and the members of the Matute lab for helpful scientific discussions and comments. We would also like to thank the Bioko Biodiversity Protection Program, and the Ministry of Environment, Republic of São Tomé and Príncipe for permission to collect and export specimens for study. D.R.M. was supported in part by NIH award R01GM121750. The authors have no conflicts of interest.
References
- Abbott R, Albach D, Ansell S, Arntzen JW, Baird SJE, Bierne N, Boughman J, Brelsford A, Buerkle CA, Buggs R, et al. 2013. Hybridization and speciation. J Evol Biol. 262: 229–246. [DOI] [PubMed] [Google Scholar]
- Ahmed-Braimah YH. 2016. Multiple genes cause postmating prezygotic reproductive isolation in the Drosophila virilis group. G3 6:4067–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed-Braimah YH, McAllister BF.. 2012. Rapid evolution of assortative fertilization between recently allopatric species of Drosophila. Int J Evol Biol. 2012:285468.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ala‐Honkola O, Ritchie MG, Veltsos P.. 2016. Postmating–prezygotic isolation between two allopatric populations of Drosophila montana: fertilisation success differs under sperm competition. Ecol Evol. 66: 1679–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P. 2007. Hitchhiking effects of recurrent beneficial amino acid substitutions in the Drosophila melanogaster genome. Genome Res. 1712: 1755–1762.http://dx.doi.org/10.1101/gr.6691007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkhead TR, Pizzari T.. 2002. Postcopulatory sexual selection. Nat Rev Genet. 34: 262–273.http://dx.doi.org/10.1038/nrg774 [DOI] [PubMed] [Google Scholar]
- Bolnick DI, Near TJ.. 2005. Tempo of hybrid inviability in centrarchid fishes (Teleostei: Centrarchidae). Evolution 598: 1754–1767.http://dx.doi.org/10.1111/j.0014-3820.2005.tb01824.x [PubMed] [Google Scholar]
- Boorman E, Parker GA.. 1976. Sperm (ejaculate) competition in Drosophila melanogaster, and the reproductive value of females to males in relation to female age and mating status. Ecol Entomol. 13: 145–155.http://dx.doi.org/10.1111/j.1365-2311.1976.tb01217.x [Google Scholar]
- Byrne PG, Simmons LW, Roberts JD.. 2003. Sperm competition and the evolution of gamete morphology in frogs. Proc R Soc Lond B Biol Sci. 2701528: 2079–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo DM, Moyle LC.. 2016. Reinforcement of conspecific sperm precedence weakens sexual selection in sympatric populations of Drosophila bioRxiv Doi: 10.1101/071886. [DOI] [PMC free article] [PubMed]
- Cenzer ML. 2016. Adaptation to an invasive host is driving the loss of a native ecotype. Evolution 7010: 2296–2307.http://dx.doi.org/10.1111/evo.13023 [DOI] [PubMed] [Google Scholar]
- Chang AS. 2004. Conspecific sperm precedence in sister species of Drosophila with overlapping ranges. Evolution 584: 781–789.http://dx.doi.org/10.1111/j.0014-3820.2004.tb00411.x [DOI] [PubMed] [Google Scholar]
- Civetta A, Clark AG.. 2000. Correlated effects of sperm competition and postmating female mortality. Proc Natl Acad Sci U S A. 9724: 13162–13165.http://dx.doi.org/10.1073/pnas.230305397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civetta A, Rosing KR, Fisher JH.. 2008. Differences in sperm competition and sperm competition avoidance in Drosophila melanogaster. Anim Behav. 755: 1739–1746.http://dx.doi.org/10.1016/j.anbehav.2007.10.031 [Google Scholar]
- Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, and the Drosophila 12 Genomes Consortium, et al. 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 4507167: 203–218. [DOI] [PubMed] [Google Scholar]
- Comeault AA, Flaxman SM, Riesch R, Curran E, Soria-Carrasco V, Gompert Z, Farkas TE, Muschick M, Parchman TL, Schwander T, et al. 2015. Selection on a genetic polymorphism counteracts ecological speciation in a stick insect. Curr Biol. 2515: 1975–1981. [DOI] [PubMed] [Google Scholar]
- Comeault AA, Serrato‐Capuchina A, Turissini DA, McLaughlin PJ, David JR, Matute DR.. 2017. A nonrandom subset of olfactory genes is associated with host preference in the fruit fly Drosophila orena. Evol Lett. 12: 73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeault AA, Venkat A, Matute DR.. 2016. Correlated evolution of male and female reproductive traits drive a cascading effect of reinforcement in Drosophila yakuba. Proc R Soc B 283:20160730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA. 1974. The evolutionary origin of hybrid inviability. Evolution 28:505–506 [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA.. 1989. Patterns of speciation in Drosophila. Evolution 43:362–381 [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA.. 1997. “Patterns of speciation in Drosophila” revisited. Evolution 511: 295–303. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA.. 2004. Speciation. Sunderland (MA): Sinauer Associates. [Google Scholar]
- Davis JS, Castillo DM, Moyle LC.. 2017. 2016. Remating responses are consistent with male post-copulatory manipulation but not reinforcement in D. pseudoobscura. Ecol Evol. 72: 507–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 435: 491–498.http://dx.doi.org/10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vienne DM, Refrégier G, Hood ME, Guigue A, Devier B, Vercken E, Smadja C, Deseille A, Giraud T.. 2009. Hybrid sterility and inviability in the parasitic fungal species complex Microbotryum. J Evol Biol. 224: 683–698. [DOI] [PubMed] [Google Scholar]
- Dixon SM, Coyne JA, Noor MAF.. 2003. The evolution of conspecific sperm precedence in Drosophila. Mol Ecol. 125: 1179–1184.http://dx.doi.org/10.1046/j.1365-294X.2003.01742.x [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. 1937. Genetics and the origin of species. New York: Columbia University Press. [Google Scholar]
- Dopman EB, Robbins PS, Seaman A.. 2010. Components of reproductive isolation between North American pheromone strains of the European corn borer. Evolution 644: 881–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmands S. 2002. Does parental divergence predict reproductive compatibility?. Trends Ecol Evol. 1711: 520–527.http://dx.doi.org/10.1016/S0169-5347(02)02585-5 [Google Scholar]
- Edmands S, Northrup SL, Hwang AS.. 2009. Maladapted gene complexes within populations of the intertidal copepod Tigriopus californicus?. Evolution 638: 2184–2192.http://dx.doi.org/10.1111/j.1558-5646.2009.00689.x [DOI] [PubMed] [Google Scholar]
- Fricke C, Arnqvist G.. 2004. Divergence in replicated phylogenies: the evolution of partial post‐mating prezygotic isolation in bean weevils. J Evol Biol. 176: 1345–1354. [DOI] [PubMed] [Google Scholar]
- Fricke C, Arnqvist G.. 2004. Conspecific sperm precedence in flour beetles. Anim Behav. 674: 729–732.http://dx.doi.org/10.1016/j.anbehav.2003.08.014 [Google Scholar]
- Funk DJ, Nosil P, Etges WJ.. 2006. Ecological divergence exhibits consistently positive associations with reproductive isolation across disparate taxa. Proc Natl Acad Sci U S A. 1039: 3209–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo BE, Vacquier VD, Swanson WJ.. 2003. Positive selection in the egg receptor for abalone sperm lysin. Proc Natl Acad Sci U S A. 1008: 4639–4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz HH, Burton RS.. 1995. Genetic differentiation and reproductive incompatibility among Baja California populations of the copepod Tigriopus californicus. Mar Biol. 1234: 821–827. [Google Scholar]
- Giraud T, Gourbiere S.. 2012. The tempo and modes of evolution of reproductive isolation in fungi. Heredity (Edinb) 1094: 204–214.http://dx.doi.org/10.1038/hdy.2012.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gojobori T. 1983. Codon substitution in evolution and the “saturation” of synonymous changes. Genetics 1054: 1011–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourbière S, Mallet J.. 2010. Are species real? The shape of the species boundary with exponential failure, reinforcement, and the “missing snowball.” Evolution 641: 1–24. [DOI] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. 2011. The developmental transcriptome of Drosophila melanogaster. Nature 4717339: 473–479.http://dx.doi.org/10.1038/nature09715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill PR, Bachtrog D, Andolfatto P.. 2008. Positive and negative selection on noncoding DNA in Drosophila simulans. Mol Biol Evol. 259: 1825–1834.http://dx.doi.org/10.1093/molbev/msn125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison RG. 1993. Hybrid zones and the evolutionary process. New York (NY) Oxford University Press. [Google Scholar]
- Harrison PW, Wright AE, Zimmer F, Dean R, Montgomery SH, Pointer MA, Mank JE.. 2015. Sexual selection drives evolution and rapid turnover of male gene expression. Proc Natl Acad Sci U S A. 11214: 4393–4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart MW, Sunday JM, Popovic I, Learning KJ, Konrad CM.. 2014. Incipient speciation of sea star populations by adaptive gamete recognition coevolution. Evolution 685: 1294–1305. [DOI] [PubMed] [Google Scholar]
- Hopkins R. 2013. Reinforcement in plants. New Phytol. 1974: 1095–1103. [DOI] [PubMed] [Google Scholar]
- Howard DJ. 1999. Conspecific sperm and pollen precedence and speciation. Annu Rev Ecol Evol Syst. 301: 109–132.http://dx.doi.org/10.1146/annurev.ecolsys.30.1.109 [Google Scholar]
- Howard DJ, Gregory PG, Chu J, Cain ML.. 1998. Conspecific sperm precedence is an effective barrier to hybridization between closely related species. Evolution 522: 511–516.http://dx.doi.org/10.1111/j.1558-5646.1998.tb01650.x [DOI] [PubMed] [Google Scholar]
- Hu TT, Eisen MB, Thornton KR, Andolfatto P.. 2013. A second-generation assembly of the Drosophila simulans genome provides new insights into patterns of lineage-specific divergence. Genome Res. 231: 89–98.http://dx.doi.org/10.1101/gr.141689.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson EJ, Price TD.. 2014. Pervasive reinforcement and the role of sexual selection in biological speciation. J Hered. 105(S1): 821–833. [DOI] [PubMed] [Google Scholar]
- Jackman S, Fearon J, Jackman MS, MCMCpack S.. 2007. The pscl package. Software. Available from: http://cran.rproject.org/src/contrib/Descriptions/pscl.html. May 16, 2016.
- Jennings JH, Mazzi D, Ritchie MG, Hoikkala A.. 2011. Sexual and postmating reproductive isolation between allopatric Drosophila montana populations suggest speciation potential. BMC Evol Biol. 14: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings JH, Snook RR, Hoikkala A.. 2014. Reproductive isolation among allopatric Drosophila montana populations. Evolution 6811: 3095–3108.http://dx.doi.org/10.1111/evo.12535 [DOI] [PubMed] [Google Scholar]
- Jewell C, Papineau AD, Freyre R, Moyle LC.. 2012. Patterns of reproductive isolation in Nolana (Chilean Bellflower). Evolution 668: 2628–2636.http://dx.doi.org/10.1111/j.1558-5646.2012.01607.x [DOI] [PubMed] [Google Scholar]
- Knowles LL, Markow TA.. 2001. Sexually antagonistic coevolution of a postmating-prezygotic reproductive character in desert Drosophila. Proc Natl Acad Sci U S A. 9815: 8692–8696.http://dx.doi.org/10.1073/pnas.151123998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles LL, Hernandez BB, Markow TA.. 2004. Exploring the consequences of postmating–prezygotic interactions between the sexes. Proc R Soc Lond B Biol Sci. 271:S357–S359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokko H, Mappes J.. 2005. Sexual selection when fertilization is not guaranteed. Evolution 599: 1876–1885.http://dx.doi.org/10.1111/j.0014-3820.2005.tb01058.x [PubMed] [Google Scholar]
- Lachaise D, Cariou M-L, David JR, Lemeunier F, Tsacas L, Ashburner M.. 1988. Historical biogeography of the Drosophila melanogaster species subgroup In: Hecht MK, Wallace B, Prance GT, editors. Evolutionary biology. Boston (MA): Springer US; p. 159–225. [Google Scholar]
- Lack JB, Cardeno CM, Crepeau MW, Taylor W, Corbett-Detig RB, Stevens KA, Langley CH, Pool JE.. 2015. The Drosophila genome NEXUS: a population genomic resource of 623 Drosophila melanogaster genomes, including 197 from a single ancestral range population. Genetics 1994: 1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson EL, Hume GL, Andrés JA, Harrison RG.. 2012. Post-mating prezygotic barriers to gene exchange between hybridizing field crickets. J Evol Biol. 251: 174–186. [DOI] [PubMed] [Google Scholar]
- Lee YH, Ota T, Vacquier VD.. 1995. Positive selection is a general phenomenon in the evolution of abalone sperm lysin. Mol Biol Evol. 122: 231–238. [DOI] [PubMed] [Google Scholar]
- Lessios HA, Cunningham CW.. 1990. Gametic incompatibility between species of the sea urchin Echinometra on the two sides of the Isthmus of Panama. Evolution 444: 933..http://dx.doi.org/10.1111/j.1558-5646.1990.tb03815.x [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R.. 1000 Genome Project Data Processing Subgroup 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2516: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lijtmaer DA, Mahler B, Tubaro PL.. 2003. Hybridization and postzygotic isolation patterns in pigeons and doves. Evolution 57: 1411–1418. [DOI] [PubMed] [Google Scholar]
- Llopart A, Elwyn S, Lachaise D, Coyne JA.. 2002. Genetics of a difference in pigmentation between Drosophila yakuba and Drosophila santomea. Evolution 5611: 2262–2277.http://dx.doi.org/10.1111/j.0014-3820.2002.tb00150.x [DOI] [PubMed] [Google Scholar]
- Lorch PD, Servedio MR.. 2007. The evolution of conspecific gamete precedence and its effect on reinforcement. J Evol Biol. 203: 937–949.http://dx.doi.org/10.1111/j.1420-9101.2007.01306.x [DOI] [PubMed] [Google Scholar]
- Mackay TFC, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han Y, Magwire MM, Cridland JM, et al. 2012. The Drosophila melanogaster genetic reference panel. Nature 4827384: 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone JH, Fontenot BE.. 2008. Patterns of reproductive isolation in toads. PLoS One 312: e3900.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manier MK, Lüpold S, Belote JM, Starmer WT, Berben KS, Ala-Honkola O, Collins WF, Pitnick S.. 2013. Postcopulatory sexual selection generates speciation phenotypes in Drosophila. Curr Biol. 2319: 1853–1862. [DOI] [PubMed] [Google Scholar]
- Markow TA. 1997. Assortative fertilization in Drosophila. Proc Natl Acad Sci U S A. 9415: 7756–7760.http://dx.doi.org/10.1073/pnas.94.15.7756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall JL, DiRienzo N.. 2012. Noncompetitive gametic isolation between sibling species of cricket: a hypothesized link between within-population incompatibility and reproductive isolation between species. Int J Evol Biol. 2012:1–7.http://dx.doi.org/10.1155/2012/593438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall JL, Huestis DL, Garcia C, Hiromasa Y, Wheeler S, Noh S, Tomich JM, Howard DJ.. 2011. Comparative proteomics uncovers the signature of natural selection acting on the ejaculate proteomes of two cricket species isolated by postmating, prezygotic phenotypes. Mol Biol Evol. 281: 423–435. [DOI] [PubMed] [Google Scholar]
- Matute DR. 2010a. Reinforcement can overcome gene flow during speciation in Drosophila. Curr Biol. 2024: 2229–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute DR. 2010b. Reinforcement of gametic isolation in Drosophila. PLoS Biol. 83: e1000341–e1000311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute DR. 2014. The magnitude of behavioral isolation is affected by characteristics of the mating community. Ecol Evol. 414: 2945–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute DR. 2015. Noisy neighbors can hamper the evolution of reproductive isolation by reinforcing selection. Am Nat. 1852: 253–269. [DOI] [PubMed] [Google Scholar]
- Matute DR, Ayroles JF.. 2014. Hybridization occurs between Drosophila simulans and D. sechellia in the Seychelles archipelago. J Evol Biol. 276: 1057–1068.http://dx.doi.org/10.1111/jeb.12391 [DOI] [PubMed] [Google Scholar]
- Matute DR, Coyne JA.. 2010. Intrinsic reproductive isolation between two sister species of Drosophila. Evolution 644: 903–920.http://dx.doi.org/10.1111/jeb.12391 [DOI] [PubMed] [Google Scholar]
- Matute DR, Gavin-Smyth J.. 2014. Fine mapping of dominant X-linked incompatibility alleles in Drosophila hybrids. PLoS Genet. 104: e1004270.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute DR, Harris A.. 2013. The influence of abdominal pigmentation on desiccation and ultraviolet resistance in two species of Drosophila. Evolution 678: 2451–2460.http://dx.doi.org/10.1111/evo.12122 [DOI] [PubMed] [Google Scholar]
- McBride CS, Singer MC.. 2010. Field studies reveal strong postmating isolation between ecologically divergent butterfly populations. PLoS Biol. 810: e1000529.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA.. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 209: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelson TC, Imhoff VE, Venditti JJ.. 2007. The accumulation of reproductive barriers during speciation: postmating barriers in two behaviorally isolated species of darters (Percidae: Etheostoma). Evolution 6111: 2596–2606.http://dx.doi.org/10.1111/j.1558-5646.2007.00220.x [DOI] [PubMed] [Google Scholar]
- Miller GT, Pitnick S.. 2002. Sperm-female coevolution in Drosophila. Science 2985596: 1230–1233.http://dx.doi.org/10.1126/science.1076968 [DOI] [PubMed] [Google Scholar]
- Moyle LC, Olson MS, Tiffin P.. 2004. Patterns of reproductive isolation in three angiosperm genera. Evolution 586: 1195–1208.http://dx.doi.org/10.1111/j.0014-3820.2004.tb01700.x [DOI] [PubMed] [Google Scholar]
- Mullen SP, Andrés JA.. 2007. Rapid evolution of sexual signals in sympatric Calopteryx damselflies: reinforcement or ‘noisy‐neighbour’ecological character displacement?. J Evol Biol. 204: 1637–1648. [DOI] [PubMed] [Google Scholar]
- Noor MA. 1997. How often does sympatry affect sexual isolation in Drosophila?. Am Nat. 1496: 1156–1163. [DOI] [PubMed] [Google Scholar]
- Nosil P. 2013. Degree of sympatry affects reinforcement in Drosophila. Evolution 673: 868–872.http://dx.doi.org/10.1111/j.1558-5646.2012.01817.x [DOI] [PubMed] [Google Scholar]
- Nosil P, Schluter D.. 2011. The genes underlying the process of speciation. Trends Ecol Evol. 30: 160–167. [DOI] [PubMed] [Google Scholar]
- Nosrati H, Price AH, Wilcock CC.. 2011. Relationship between genetic distances and postzygotic reproductive isolation in diploid Fragaria (Rosaceae). Biol J Linnean Soc. 104: 510–526. [Google Scholar]
- Orr HA, Presgraves DC.. 2000. Speciation by postzygotic isolation: forces, genes and molecules. Bioessays 2212: 1085–1094.http://dx.doi.org/10.1002/1521-1878(200012)22:12 [DOI] [PubMed] [Google Scholar]
- Palumbi SR. 1994. Genetic divergence, reproductive isolation, and marine speciation. Annu Rev Ecol Evol Syst. 251: 547–572.http://dx.doi.org/10.1146/annurev.es.25.110194.002555 [Google Scholar]
- Palumbi SR. 2009. Speciation and the evolution of gamete recognition genes: pattern and process. Heredity (Edinb) 1021: 66–76.http://dx.doi.org/10.1038/hdy.2008.104 [DOI] [PubMed] [Google Scholar]
- Palmer CA, Edmands S.. 2000. Mate choice in the face of both inbreeding and outbreeding depression in the intertidal copepod Tigriopus californicus. Mar Biol. 1364: 693–698. [Google Scholar]
- Pitnick S, Brown WD, Miller GT.. 2001. Evolution of female remating behaviour following experimental removal of sexual selection. Proc R Soc Lond B Biol Sci. 2681467: 557–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitnick S, Marrow T, Spicer GS.. 1999. Evolution of multiple kinds of female sperm-storage organs in Drosophila. Evolution 536: 1804–1822.http://dx.doi.org/10.1111/j.1558-5646.1999.tb04564.x [DOI] [PubMed] [Google Scholar]
- Pitnick S, Wolfner MF, Suarez SS.. 2009. Ejaculate-female and sperm-female interactions In: Birkhead TR, Hosken DJ, Pitnick S. editors. Sperm biology: an evolutionary perspective. Burlington (MA): Academic Press; p. 247–304. [Google Scholar]
- Pizzari T, Snook RR.. 2003. Perspective: sexual conflict and sexual selection: chasing away paradigm shifts. Evolution 576: 1223–1236.http://dx.doi.org/10.1111/j.0014-3820.2003.tb00331.x [DOI] [PubMed] [Google Scholar]
- Presgraves DC. 2002. Patterns of postzygotic isolation in Lepidoptera. Evolution 566: 1168–1183.http://dx.doi.org/10.1111/j.0014-3820.2002.tb01430.x [DOI] [PubMed] [Google Scholar]
- Presgraves DC. 2003. A fine-scale genetic analysis of hybrid incompatibilities in Drosophila. Genetics 1633: 955–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. 2010. The molecular evolutionary basis of species formation. Nat Rev Genet. 113: 175–180.http://dx.doi.org/10.1038/nrg2718 [DOI] [PubMed] [Google Scholar]
- Presgraves DC, Baker RH, Wilkinson GS.. 1999. Coevolution of sperm and female reproductive tract morphology in stalk-eyed flies. Proc R Soc Lond B Biol Sci. 2661423: 1041–1047. [Google Scholar]
- Presgraves DC, Balagopalan L, Abmayr SM, Orr HA.. 2003. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature 4236941: 71571–71579. [DOI] [PubMed] [Google Scholar]
- Price CSC, Kim CH, Gronlund CJ, Coyne JA.. 2001. Cryptic reproductive isolation in the Drosophila simulans species complex. Evolution 551: 81–92.http://dx.doi.org/10.1111/j.0014-3820.2001.tb01274.x [DOI] [PubMed] [Google Scholar]
- Price TD, Bouvier MM.. 2002. The evolution of F1 postzygotic incompatibilities in birds. Evolution 5610: 2083–2089.http://dx.doi.org/10.1111/j.0014-3820.2002.tb00133.x [PubMed] [Google Scholar]
- R Core Team. 2016. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: Available from: https://www.R-project.org/. [Google Scholar]
- Rabosky DL, Matute DR.. 2013. Macroevolutionary speciation rates are decoupled from the evolution of intrinsic reproductive isolation in Drosophila and birds. Proc Natl Acad Sci U S A. 11038: 15354–15359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Wood V, Dolinski K, Draghici S.. 2008. Use and misuse of the gene ontology annotations. Nat Rev Genet. 97: 509–515.http://dx.doi.org/10.1038/nrg2363 [DOI] [PubMed] [Google Scholar]
- Rogers RL, Cridland JM, Shao L, Hu TT, Andolfatto P, Thornton KR.. 2014. Landscape of standing variation for tandem duplications in Drosophila yakuba and Drosophila simulans. Mol Biol Evol. 317: 124–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers RL, Cridland JM, Shao L, Hu TT, Andolfatto P, Thornton KR.. 2015. Tandem duplications and the limits of natural selection in Drosophila yakuba and Drosophila simulans. PLoS One 107: e0132184.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum EB, Sarver BAJ, Brown JW, Roches Des S, Hardwick KM, Hether TD, Eastman JM, Pennell MW, Harmon LJ.. 2012. Goldilocks meets Santa Rosalia: an ephemeral speciation model explains patterns of diversification across time scales. Evol Biol. 392: 255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagga N, Civetta A.. 2011. Male-female interactions and the evolution of postmating prezygotic reproductive isolation among species of the virilis subgroup. Int J Evol Biol. doi: 10.4061/2011/485460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos dos G, Schroeder AJ, Goodman JL, Strelets VB, Crosby MA, Thurmond J, Emmert DB, Gelbart WM.. FlyBase Consortium 2015. FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res. 43:D690–D697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasa MM, Chippindale PT, Johnson NA.. 1998. Patterns of postzygotic isolation in frogs. Evolution 526: 1811..http://dx.doi.org/10.1111/j.1558-5646.1998.tb02258.x [DOI] [PubMed] [Google Scholar]
- Sawamura K, Yamamoto MT, Watanabe TK.. 1993. Hybrid lethal systems in the Drosophila melanogaster species complex. II. The zygotic hybrid rescue (Zhr) gene of D. melanogaster. Genetics 1332: 307–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scopece G, Musacchio A, Widmer A, Cozzolino S.. 2007. Patterns of reproductive isolation in Mediterranean deceptive orchids. Evolution 6111: 2623–2642.http://dx.doi.org/10.1111/j.1558-5646.2007.00231.x [DOI] [PubMed] [Google Scholar]
- Scopece G, Widmer A, Cozzolino S.. 2008. Evolution of postzygotic reproductive isolation in a guild of deceptive orchids. Am Nat. 1713: 315–326. [DOI] [PubMed] [Google Scholar]
- Seehausen O, Butlin RK, Keller I, Wagner CE, Boughman JW, Hohenlohe PA, Peichel CL, Saetre G-P, Bank C, Brännström A, Brelsford A, Clarkson CS.. 2014. Genomics and the origin of species. Nat Rev Genet. 153: 176–192. [DOI] [PubMed] [Google Scholar]
- Servedio MR. 2001. Beyond reinforcement: the evolution of premating isolation by direct selection on preferences and postmating, prezygotic incompatibilities. Evolution 5510: 1909–1920.http://dx.doi.org/10.1111/j.0014-3820.2001.tb01309.x [DOI] [PubMed] [Google Scholar]
- Servedio MR. 2004. The what and why of research on reinforcement. PLoS Biol 212: e420.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servedio MR, Noor M.. 2003. The role of reinforcement in speciation: theory and data. Annu Rev Ecol Evol Syst. 34: 339–364. [Google Scholar]
- Simmons LW. 2005. The evolution of polyandry: sperm competition, sperm selection, and offspring viability. Annu Rev Ecol Evol Syst. 36: 125–146. [Google Scholar]
- Smith JM, Smith NH.. 1996. Synonymous nucleotide divergence: what is “saturation”?. Genetics 1423: 1033–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, Yang Z, Wolfner MF, Aquadro CF.. 2001. Positive Darwinian selection drives the evolution of several female reproductive proteins in mammals. Proc Natl Acad Sci U S A. 985: 2509–2514.http://dx.doi.org/10.1073/pnas.051605998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigart AL. 2010. The genetics of postmating, prezygotic reproductive isolation between Drosophila virilis and D. americana. Genetics 1842: 401–410.http://dx.doi.org/10.1534/genetics.109.111245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Presgraves DC.. 2009. Evolution of the Drosophila nuclear pore complex results in multiple hybrid incompatibilities. Science 3235915: 779–782.http://dx.doi.org/10.1126/science.1169123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubaro PL, Lijtmaer DA.. 2002. Hybridization patterns and the evolution of reproductive isolation in ducks. Biol J Linnean Soc. 772: 193–200.http://dx.doi.org/10.1046/j.1095-8312.2002.00096.x [Google Scholar]
- Turelli M, Lipkowitz JR, Brandvain Y.. 2014. On the Coyne and Orr-igin of species: effects of intrinsic postzygotic isolation, ecological differentiation, X chromosome size, and sympatry on Drosophila speciation. Evolution 684: 1176–1187.http://dx.doi.org/10.1111/evo.12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turissini DA, Liu G, David JR, Matute DR.. 2015. The evolution of reproductive isolation in the Drosophila yakuba complex of species. J Evol Biol. 283: 557–575.http://dx.doi.org/10.1111/jeb.12588 [DOI] [PubMed] [Google Scholar]
- Turissini DA, Comeault AA, Liu G, Lee YC, Matute DR.. 2017. The ability of Drosophila hybrids to locate food declines with parental divergence. Evolution 714: 960–973. 2017 [DOI] [PubMed] [Google Scholar]
- Turner E, Jacobson DJ, Taylor JW.. 2010. Biogeography of postmating reproductive isolation barriers is consistent with reinforcement selection in Neurospora, a model microbial eukaryote. J EvolBiol. 238: 1642–1656. [DOI] [PubMed] [Google Scholar]
- Turner E, Jacobson DJ, Taylor JW.. 2011. Genetic architecture of a reinforced, postmating, reproductive isolation barrier between Neurospora species indicates evolution via natural selection. PLoS Genet. 78: e1002204.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkat AA, Hahn MW, Thornton JW.. 2017. Multinucleotide mutations cause false inferences of positive selection. bioarXiv. doi: 10.1101/165969. [DOI] [PMC free article] [PubMed]
- Wayne ML, Mackay TF.. 1998. Quantitative genetics of ovariole number in Drosophila melanogaster. II. Mutational variation and genotype-environment interaction. Genetics 1481: 201–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne ML, Soundararajan U, Harshman LG.. 2006. Environmental stress and reproduction in Drosophila melanogaster: starvation resistance, ovariole numbers and early age egg production. BMC Evol Biol. 6:57.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmer A, Lexer C, Cozzolino S.. 2009. Evolution of reproductive isolation in plants. Heredity (Edinb) 1021: 31–38.http://dx.doi.org/10.1038/hdy.2008.69 [DOI] [PubMed] [Google Scholar]
- Wilson DS, Hedrick A. 1982. Speciation and the economics of mate choice. Evol Theory 6: 15–24. [Google Scholar]
- Wolstenholme JK. 2004. Temporal reproductive isolation and gametic compatibility are evolutionary mechanisms in the Acropora humilis species group (Cnidaria; Scleractinia). Mar Biol. 1443: 567–582. [Google Scholar]
- Wu C-I. 1992. A Note on Haldane's Rule: Hybrid inviability versus hybrid sterility. Evolution 465: 1584..http://dx.doi.org/10.1111/j.1558-5646.1992.tb01152.x [DOI] [PubMed] [Google Scholar]
- Yang Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 135: 555–556. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 248: 1586–1591.http://dx.doi.org/10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Yeates SE, Diamond SE, Einum S, Emerson BC, Holt WV, Gage MJG.. 2013. Cryptic choice of conspecific sperm controlled by the impact of ovarian fluid on sperm swimming behavior. Evolution 6712: 3523–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukilevich R. 2012. Asymmetrical patterns of speciation uniquely support reinforcement in Drosophila. Evolution 665: 1430–1446.http://dx.doi.org/10.1111/j.1558-5646.2011.01534.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.