Abstract
Most banana cultivars are triploid seedless parthenocarpic clones derived from hybridization between Musa acuminata subspecies and sometimes M. balbisiana. M. acuminata subspecies were suggested to differ by a few large chromosomal rearrangements based on chromosome pairing configurations in intersubspecies hybrids. We searched for large chromosomal rearrangements in a seedy M. acuminata ssp. malaccensis banana accession through mate-pair sequencing, BAC-FISH, targeted PCR and marker (DArTseq) segregation in its progeny. We identified a heterozygous reciprocal translocation involving two distal 3 and 10 Mb segments from chromosomes 01 and 04, respectively, and showed that it generated high segregation distortion, reduced recombination and linkage between chromosomes 01 and 04 in its progeny. The two chromosome structures were found to be mutually exclusive in gametes and the rearranged structure was preferentially transmitted to the progeny. The rearranged chromosome structure was frequently found in triploid cultivars but present only in wild malaccensis ssp. accessions, thus suggesting that this rearrangement occurred in M. acuminata ssp. malaccensis. We propose a mechanism for the spread of this rearrangement in Musa diversity and suggest that this rearrangement could have played a role in the emergence of triploid cultivars.
Keywords: Musa, chromosome, translocation, segregation distortion, mate-pair sequencing, genotyping by sequencing
Introduction
Speciation is considered to be the consequence of population divergence due to halted gene flow (Dobzhansky 1937; Mayr 1942). Mechanisms that limit gene flow could involve prezygotic reproductive isolation, which prevents hybrid zygote formation, or postzygotic isolation, which occurs after mating and results in decreased hybrid fitness (Ramsey et al. 2003). Postzygotic reproductive isolation is often associated with speciation genes and/or chromosomal rearrangements (see review by Rieseberg and Blackman 2010). The causal or incidental accumulation of structural variations in relation to speciation events is a matter of debate, with Rieseberg (2001) arguing that large structural variations reduce gene flow more by suppressing recombination and extending the effect of linked isolation genes than by reducing fitness. Regardless of the mechanism, large chromosome structural variations generally cause chromosomal segregation distortion and/or recombination reduction in hybrids (e.g., Tadmor et al. 1987; Quillet et al. 1995; Jáuregui et al. 2001; Ostberg et al. 2013), therefore reducing fertility and gene flow. Mechanisms limiting gene flow can be a constraint for breeding programs that exploit genetic resources with the aim of enhancing crop diversity. Conversely, mechanisms reducing fertility have been important factors in the domestication of plants such as bananas with seedless edible fruit.
The Musa genus generates bananas, a major starchy staple food and cash crop in tropical and subtropical regions (Lescot 2014), while also providing a valuable model for studying chromosomal rearrangements. Most banana cultivars are derived from Musa acuminata (2n = 2× = 22, A genome), sometimes combined with Musa balbisiana (2n = 2× = 22, B genome). M. acuminata is divided into six to nine subspecies (banksii, burmannica, malaccensis, microcarpa, zebrina, burmannicoïdes, truncata, siamea, and errans) which diverged following geographical isolation in distinct Southeast Asian continental regions and islands (Daniells 2001; Perrier et al. 2009). The currently accepted domestication scenario suggests that human migrations, probably during the Holocene, led to contacts between these subspecies through the transport of plant material (Perrier et al. 2011). This resulted in the emergence of intersubspecific hybrids with reduced fertility (Dodds and Simmonds 1948; Fauré et al. 1993a; Shepherd 1999). Early farmers would then have selected parthenocarpic diploid and triploid hybrids producing fruit with high flesh and low seed content.
Cytogenetic studies have shown that chromosomal pairing at meiosis in Musa acuminata is generally regular in bivalents within subspecies, but irregular with some multivalents and univalents in hybrids between subspecies (Dodds 1943; Dodds and Simmonds 1948; Dessauw 1987; Fauré et al. 1993a; Shepherd 1999). Chromosomal structural variations between subspecies have been put forward to explain those irregularities. Based on pairing configurations in intersubspecific hybrids, Shepherd (1999) suggested the presence of seven translocation groups, differing from each other by 1–4 translocations. These groups only partly overlap with subspecies delimitation. The Standard group (ST) is the largest one, consisting of banksii, microcarpa, and malaccensis spp. accessions. The other groups were named according to the geographic origins of their representatives. The Northern Malayan group (NM) includes some malaccensis accessions, the Northern 1 group includes some burmannicoïdes and siamea accessions, the Northern 2 group includes different burmannica and siamea accessions, the Malayan Highland group is based on one truncata accession, the Javanese group is based on two zebrina accession, whereas the East African group is based on one unclassified accession. Overall, only a limited number of accessions have been studied by cytogenetics and hence little is currently known about the distribution and exact nature of these translocations in Musa germplasm.
Genetic mapping studies involving Musa acuminata highlighted substantial segregation distortions involving a few linkage groups (Fauré et al. 1993b; Hippolyte et al. 2010; D’Hont et al. 2012; Mbanjo et al. 2012). Chromosomal pairing at meiosis observed in some of the parents has always been irregular, suggesting the presence of chromosomal structural heterozygosity associated with these segregation distortions (Fauré et al. 1993b; Hippolyte et al. 2010). However, no direct links between segregation distortions and the nature of the structural heterozygosity have been established and no large structural variations have been precisely characterized so far in Musa.
The aim of the present study was to characterize large structural variations in Musa and their impact on chromosome segregation and Musa evolution based on the recent availability of a reference genome sequence assembly for M. acuminata (D’Hont et al. 2012), while taking advantage of new sequencing potential offered by next-generation sequencing (NGS) technology. We thus focused on a M. acuminata ssp. malaccensis accession (PT-BA-00267) originally used to produce a genetic map to anchor the M. acuminata reference sequence assembly to Musa chromosomes and that displayed 17% skewed markers with a high concentration in linkage groups corresponding to chromosomes 01 and 04 (D’Hont et al. 2012).
We sequenced the M. a. ssp. malaccensis accession PT-BA-00267 through mate-pair sequencing and developed bioinformatics tools to interpret the detected discordant mapping sequences relative to the M. acuminata reference sequence. In addition, we refined the analysis of chromosome segregation in the self-progeny of this accession through genotyping by sequencing (DArTseq) and in a biparental cross using SSR markers. A large reciprocal translocation at a heterozygous state was identified in this accession, validated and accurately characterized through PCR and BAC-FISH experiments. The impact of this translocation on chromosome recombination and transmission was also measured and its distribution in accessions representative of Musa acuminata germplasm was investigated, enabling us to put forward hypotheses regarding its origin.
Results
Evidence for a Large Translocation In the PT-BA-00267 Accession
Marker Segregation Analysis Revealed High Distortion Involving Two Regions On Chromosomes 1 and 4
A total of 180 self-progeny individuals of the PT-BA-00267 accession were genotyped using DArTseq technology generating 9,968 SNPs, of which 7,417 were kept after the filtering steps. The markers were well distributed along the 11 reference chromosomes of the M. acuminata reference genome assembly, with an average of 1.9 markers per 100 kb (supplementary table S1, Supplementary Material online) and, as expected, a much higher marker density in gene-rich regions than in repeat-rich pericentromeric regions (supplementary fig. S1, Supplementary Material online).
The average recombination rate was 0.045 recombinations per Mb (supplementary table S1, Supplementary Material online), representing around one recombination event per chromosome arm per meiosis. The recombination rate was in general positively correlated with the gene density, with the exception of two gene-rich regions that showed a very limited recombination rate: a 4 Mb region in the median part of metacentric reference chromosome 04 (24.5–28.5 Mb) and a 7.5 Mb distal region of the acrocentric reference chromosome 01 (0–7.5 Mb) (supplementary fig. S1, Supplementary Material online). No recombination events were observed between 0 and 2.9 Mb for reference chromosome 01.
Overall, 24% of the markers deviated from the expected Mendelian ratio (0.25: 0.5: 0.25) (χ2 test, P < 0.005) (supplementary table S1, Supplementary Material online). These markers were mainly located on chromosomes 01 and 04, which each exhibited a large region with very high segregation distortion (fig. 1, supplementary fig. S1, Supplementary Material online). Reference chromosomes 01 and 04 displayed 100% and 63% of distorted markers, respectively. Regions of chromosomes 01 and 04 with reduced recombination showed the highest distortion. The segregation bias consisted in an excess of one homozygous genotype (53% observed vs. 25% expected) at the expense of the alternative homozygous (8% vs. 25%) and heterozygous genotypes (39% vs. 50%).
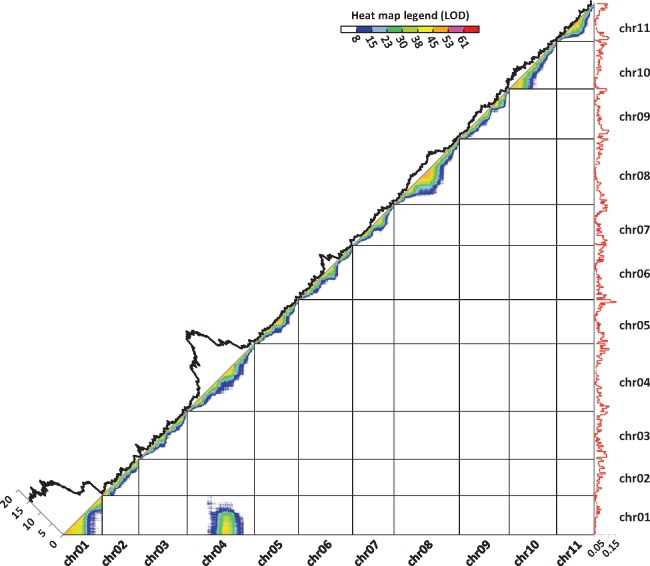
Fig. 1.
Representation of marker linkage, recombination rates, and segregation distortion in PT-BA-00267 self-progeny along the 11 Musa acuminata chromosomes. Each dot represents linkage between two markers. Marker linkage is represented by a color gradient from red to dark blue for strong and weak linkages, respectively. The black curve represents marker segregation distortions calculated as -log10 (P value of the chi-square test testing the deviation of the expected Mendelian segregation ratio). The red curve represents the recombination rate.
Several clustered markers from reference chromosomes 01 and 04 appeared to be highly linked, with a linkage intensity similar to that observed for physically close markers belonging to one reference chromosome (fig. 1). These markers belonged to the distal region of acrocentric chromosome 01 and a pericentromeric region of chromosome 04 in the reference assembly. These two regions corresponded to those showing both high segregation distortion and a low recombination rate (fig. 1).
5 kb Mate-Pair Analysis Suggested the Presence of a Heterozygous Reciprocal Translocation
Evidence of a structural variation involving chromosomes 01 and 04 was sought based on mapping of 5 kb paired-end reads of the PT-BA-00267 accession on the M. acuminata reference sequence assembly and analysis of discordant paired reads. A significant cluster of discordant paired reads was found linking the regions of reference chromosomes 01 and 04 that displayed high distortion and low recombination (fig. 2A). This linkage, supported by 150 read pairs, suggested a translocation in the PT-BA-00267 accession linking a distal region of reference chromosome 04 (at position 26.7 Mb) to a distal region of reference chromosome 01 (at position 2.9 Mb). Concordant mapped paired reads overlapping the two translocation breakpoints were also observed (fig. 2B), indicating that the structural variation was at a heterozygous state in the PT-BA-00267 accession.
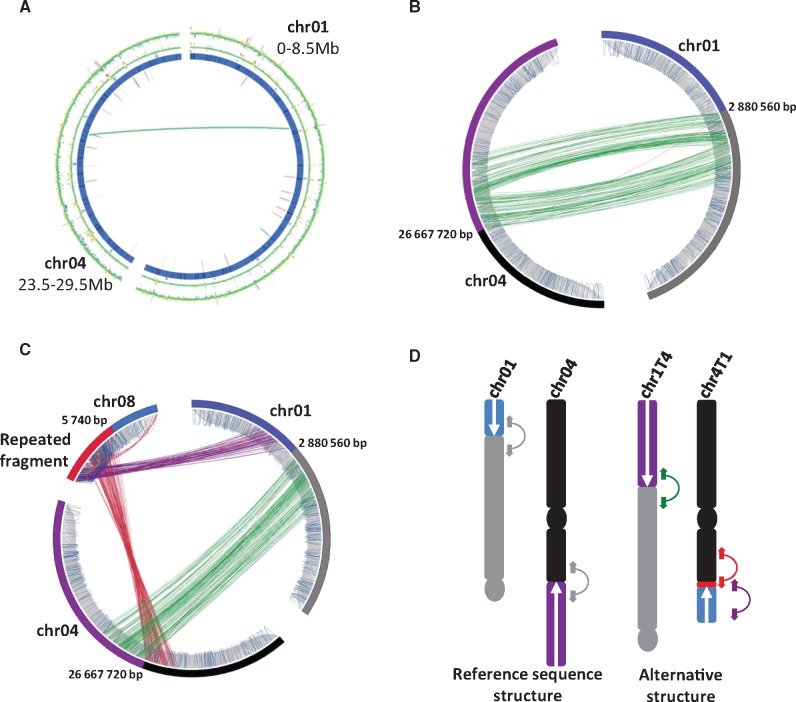
Fig. 2.
Paired read evidence for a reciprocal translocation involving chromosomes 01 and 04. (A) Circos representation of significant discordant read clusters from PT-BA-00267 identified in the targeted regions of reference chromosomes 01 (0–8.5 Mb) and 04 (23.5–29.5 Mb). (B) Circos with focus on paired reads in a 25 kb region around the discordant read cluster detected in chromosomes 01 and 04 in (A). Grey lines correspond to concordant pairs (correct orientation and insert size), orange and red lines correspond to discordant pairs with smaller and greater insert sizes, respectively. Purple lines correspond to pairs showing a reverse–reverse orientation, green lines a forward–forward orientation, and blue lines correspond to pairs with a complete reverse orientation relative to the paired library construction. (C) Circos with focus on paired read clusters detected in the targeted region of chromosomes 01 and 04 and with chromosome 08. (D) Hypothesized chromosome structures for PT-BA-00267 based on the paired read mapping interpretation. Linked colored arrows correspond to the read pairs shown in (C). Centromeres are indicated by circles.
Analysis of discordant paired reads revealed only one discordant cluster linking reference chromosomes 01 and 04, although the presence of a translocation should generate at least two distinct discordant paired read clusters. Among potential translocation configurations, several required additional clusters of discordant paired reads contiguous to at least one extremity of the detected discordant cluster. We carefully searched for additional links involving these regions and found a cluster (64 paired reads) linking reference chromosome 01 to the extremity of reference chromosome 08 and a second cluster (104 paired reads) that linked the same region of reference chromosome 08 to reference chromosome 04 (fig. 2C). Coverage of this chromosome 08 region appeared to be excessive (38×) relative to the average coverage on chromosome 08 (14×), thus showing its repeated nature.
Interpretation of these discordant relationships suggested the presence, in PT-BA-00267, of a reciprocal translocation in a heterozygous state involving chromosomes 01 and 04 (fig. 2D). For convenience, we will refer to chromosomes 01 and 04 for chromosomal structures corresponding to the reference genome sequence and chromosomes 1T4 and 4T1 for chromosomal structures resulting from the reciprocal translocation compared with this reference (fig. 2D). The presence of a segment of repeated sequences larger than the paired read insert size at the breakpoint of chromosome 4T1 could explain why we did not directly detect discordant reads between the involved regions of chromosomes 01 and 04 (fig. 2D). This repeated sequence was also present on chromosome 08, but not on chromosomes 01 and 04 in the reference assembly. This explained why an indirect link between chromosomes 01 and 04 through chromosome 08 was detected (fig. 2D).
PCR and BAC-FISH Validation of the Heterozygous Reciprocal Translocation
PCR primers were designed to assess the possible presence of a breakpoint on chromosomes 01, 04, and 1T4 resulting from a translocation, as suggested based on paired read mapping (fig. 3A and B). However, chromosome 4T1 could not be tested by PCR due to the presence of repeated sequences at the translocation breakpoint, which rendered potential PCR products too long for amplification. The PCR product sizes confirmed the presence of these three chromosome structures in PT-BA-00267. PCR product sequences confirmed that the amplified region corresponded to the expected one.
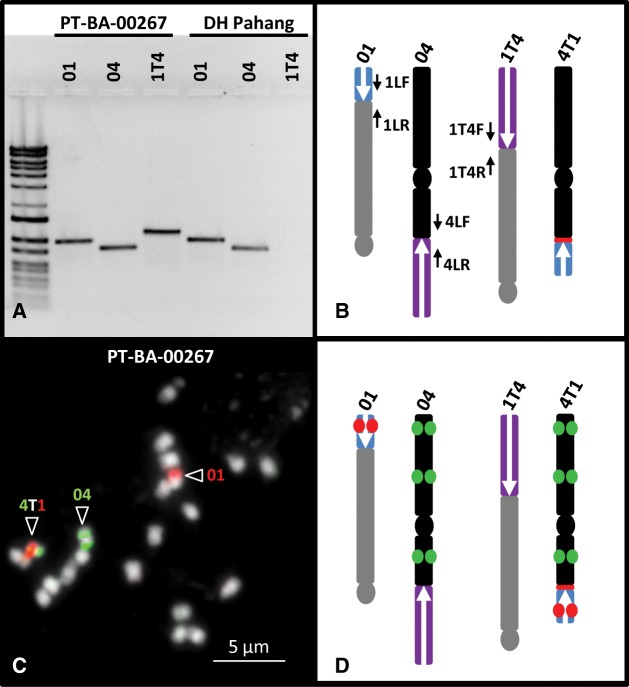
Fig. 3.
Validation of PT-BA-00267 structural heterozygosity through PCR and BAC-FISH. (A) PCR amplification of breakpoints using primers located along the reference and hypothesized chromosome structures (B). (C) BAC-FISH on a PT-BA-00267 chromosome preparation using BACs MAMB_34N11 (red) and MAMB_51M04 + MAMH_47D06 + MAMB_01M16 (green). (D) Location of BACs along reference and hypothesized chromosome structures.
BAC-FISH experiments were performed to check the presence of a reciprocal translocation involving chromosomes 01 and 04 and thus the presence of the four chromosome structures. BAC clones from both sides of the breakpoints were selected (supplementary table S2, Supplementary Material online) and used for BAC-FISH on PT-BA-00267 metaphase chromosome preparations. The results confirmed the presence of the four chromosome structures, reference chromosomes 01 and 04 and also chromosomes 1T4 and 4T1 resulting from reciprocal translocation (fig. 3C and D and supplementary fig. S2, Supplementary Material online).
Structural Heterozygosity Consequences on Genotype Representation In Progeny and Gamete Viability
In the breakpoint regions of chromosomes 01 and 04 involved in the structural heterozygosity, marker segregation was biased in favor of an excess of one homozygous genotype at the expense of the other. PCR tests were performed on selected PT-BA-00267 self-progeny individuals (five homozygotes for each chromosome structure and five heterozygotes). This showed that the underrepresented homozygous genotypes corresponded to chromosomes 01 and 04 structures, whereas the overrepresented homozygous genotypes in the progeny corresponded to chromosomes 1T4 and, by extension, to 4T1 structures (supplementary table S3, Supplementary Material online).
Among the nine possible genotype combinations, only four were found in the most distorted regions in the PT-BA-00267 progeny using DArTseq data (supplementary table S4, Supplementary Material online). Three genotype combinations exhibited a higher than expected frequency in case of Mendelian segregation whereas a last genotype combination had a lower than expected frequency. The three genotype combinations exceeding expectations were a heterozygous genotype for both chromosome structures (37.78% vs. 25% expected) and the two genotype combinations homozygous for both chromosome structures. Among these homozygous combinations, the 1T4-1T4-4T1-4T1 chromosome combination was highly overrepresented (51.67% vs. 6.25% expected) whereas the 01-01-04-04 chromosome combination was only slightly overrepresented (8.33% vs. 6.25% expected). The sole genotype combinations found with underrepresentation (2.22% vs. 12.5% expected) corresponded to 01-1T4-4T1-4T1 chromosome structures.
To investigate whether these proportions were due to gametic or zygotic selection, we analyzed a second population involving PT-BA-00267 (female parent) and “ChicameT” (male parent), which gave access to gamete transmission ratios. This F1 population was genotyped with 35 SSR markers located on chromosomes 01 and 04. Analysis of PT-BA-00267 alleles in this population revealed high segregation distortion and linkage involving chromosomes 01 and 04, consistent with those observed for the PT-BA-00267 self-progeny population (supplementary fig. S3, Supplementary Material online). These results suggested differential gamete viability rather than zygotic selection. Moreover, the F1 population generated information on the allelic composition of maternal gametes transmitted to the offspring. Only PT-BA-00267 maternal gametes having chromosomes 1T4 and 4T1 or gametes having the chromosomes 01 and 04 were transmitted (supplementary table S5, Supplementary Material online). Although gamete viability can be estimated at 100% (best transmission) for gametes with chromosomes 1T4 and 4T1, gamete viability for those with chromosomes 01 and 04 was 28%.
Paternal gamete viability was then estimated based on the PT-BA-00267 selfing population segregations (proportions of genotypes and genotype combinations between chromosomes 01 and 04) using the maternal gamete viability value calculated from the PT-BA-00267 × “ChicameT” population. The estimated paternal gamete viability in PT-BA-00267 was close to the observed maternal gamete viability (supplementary table S5, Supplementary Material online).
Distribution of Chromosomes 01, 04, 1T4, and 4T1 In Musa acuminata Germplasm
Mate-pair resequencing data were used to search for the presence of chromosomes 01, 04, 1T4, and 4T1 in 14 M. acuminata accessions by inspecting paired reads mapped on the reference sequence at the identified translocation breakpoints (supplementary fig. S4 and table S6, Supplementary Material online). Out of the 14 resequenced accessions, eight were found to be homozygous for chromosomes 01 and 04 (“Galeo”, “Manang”, “Pisang Madu”, “Banksii”, “Calcutta4”, “DH-Pahang”, “Bornéo,” and “Maia Oa”), five accessions were found heterozygous with a copy of each chromosome (01, 1T4, 04, and 4T1) (“IDN110”, “Pisang Lilin”, “Akondro Mainty,” and “Paka”) and one accession (“Malaccensis Nain”) was found to be homozygous for chromosomes 1T4 and 4T1. The triploid “Grande Naine” accession had at least one copy of each chromosome structure.
The presence of chromosomes 01, 04, and 1T4 was also tested by PCR on 169 M. acuminata banana accessions (including the 14 mentioned above) (supplementary table S6, Supplementary Material online). A total of 119 accessions amplified only chromosomes 01 and 04, ten accessions amplified only chromosome 1T4, 34 amplified all tested chromosome structures (01, 04, and 1T4), one accession amplified only chromosome 04 and 1T4 and five accessions amplified only chromosome 01. The PCR results were in agreement with the paired read mapping data for the resequenced accessions, with the exception of “IDN110” which did not amplify the chromosome 01 structure, whereas paired read mapping identified it as structurally heterozygous. This discrepancy may have been due to local micro-rearrangements or sequence divergence preventing PCR primer hybridization. Chromosome 4T1 could not be tested by PCR, but since nearly all of the observed gametes in progenies were 01-04 or 1T4-4T1 and since the resequenced accessions bearing chromosome 1T4 also had chromosome 4T1, we hypothesize that 4T1 was present when chromosome 1T4 was detected.
The vast majority (45 out of 48) of the diploid wild accession representatives of M. acuminata subspecies amplified reference chromosomes 01 and 04 only (supplementary table S6, Supplementary Material online). Among the three exceptions, “Malaccensis nain” and “Pa songkla” amplified only the 1T4 chromosome, whereas the PT-BA-00267 accession amplified all of the tested chromosomes (01, 04, and 1T4). Among the 73 diploid cultivated accessions tested, 48 amplified only chromosomes 01 and 04, eight amplified only chromosome 1T4 and 17 amplified chromosomes 01, 04, and 1T4 (supplementary table S6, Supplementary Material online).
Among the 31 triploid cultivated accessions tested, around half amplified only chromosomes 01 and 04, including the AAA accessions from the Red, Orotava, and Lujujira/Mutika subgroups, AAB accessions from the Popoulou/Maia Maoli, Mysore, Pisang Kelat, and Plantain subgroups, and ABB accessions from the Bluggoe, Pelipita, Peyan, and Saba subgroups (supplementary table S6, Supplementary Material online). The other half amplified chromosomes 01, 04, and 1T4, including AAA accessions from to the Ambon, Rio, Ibota, Cavendish, and Gros Michel subgroups, AAB accessions from the Silk, Nendra Padaththi, and Pome subgroups and ABB accessions from the Pisang Awak subgroup. The only tetraploid accession tested, i.e., “Yawa2” (ABBT), also amplified chromosomes 01, 04, and 1T4.
In addition to Musa acuminata accessions, nine Musa species were tested for the various structures, including Musa acuminata close relatives M. laterita, M. ornata, M. rosea, and M. velutina, along with more divergent species M. sanguinea, M. balbisiana, M. maclayi, and M. textilis, as well as the Fe’i type. In all cases, 01 and 04 structures were found but not the 1T4 structure.
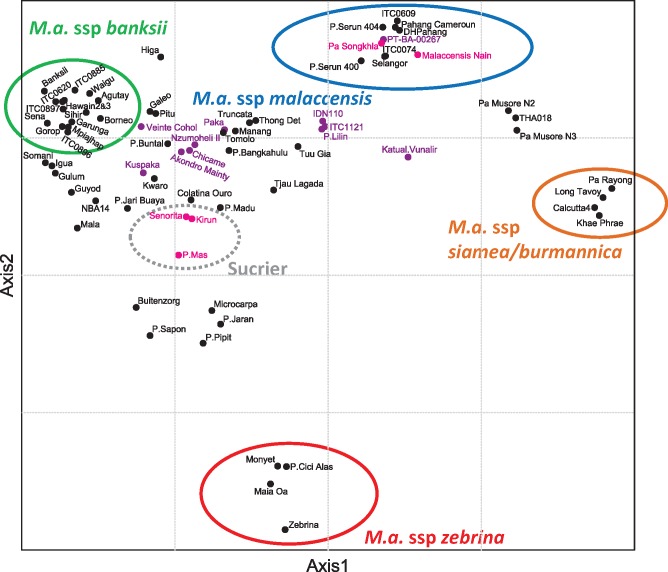
Finally, to refine the position in the Musa acuminata classification of key accessions, in particular the two wild diploid accessions homozygous for 1T4 and 4T1 chromosomes, we performed a factorial analysis with Musa acuminata diploid accessions for which genotyping by sequencing information was available for 3043 SNP markers. The first two axes that, respectively, explained 28% and 22% of the diversity allowed us to differentiate the four main Musa acuminata subspecies (i.e., banksii, zebrina, malaccensis, and siamea/burmannica) (fig. 4). The accession distribution along these axes showed that homozygotes for chromosome 1T4 and 4T1 were present in two distinct genetic groups corresponding to ssp. malaccensis and the Sucrier cultivar subgroup. Heterozygous accessions were mostly located in a triangle between M. acuminata ssp. banksii, M. acuminata ssp. Malaccensis, and the Sucrier cultivar subgroup.
Fig. 4.
Factorial analysis performed on 35 wild Musa acuminata accessions with projection of 40 cultivars along the synthetic axes. The dissimilarity matrix was based on genotyping by sequencing data. Pink dots indicate accessions homozygous for chromosomes 1T4 and 4T1, black dots indicate accessions homozygous for chromosomes 01 and 04 and purple dots indicate structurally heterozygote accessions.
Discussion
Characterization of a Reciprocal Translocation in Musa acuminata
The presence of large chromosome structural variations within M. acuminata was proposed by cytogeneticists on the basis of the observation of chromosome pairing irregularities at meiosis in hybrids between M. acuminata accessions (Dodds 1943; Dodds and Simmonds 1948; Dessauw 1987; Fauré et al. 1993a; Shepherd 1999). Seven Musa acuminata translocation groups, within which accessions were structurally homogeneous, were proposed by Shepherd (1999), with a supposedly ancestral Standard group (ST) and six groups suggested to differ by 1–4 translocations (Northern Malayan (NM), Malayan Highland, Northern 1, Northern 2, Javanese, and East Africa). These groups only partially corresponded to the Musa acuminata subspecies classification. Here, for the first time, we used resequencing approaches and were able to characterize one of these structural variations in the PT-BA-00267 M. acuminata ssp. malaccensis accession in the form of a reciprocal translocation involving 3 Mb of the distal region of reference chromosome 01 and 10 Mb of one distal region of reference chromosome 04.
A few of the accessions studied by Shepherd (1999) were included in our study. Among them, we characterized “P. Lilin” and “Paka” accessions as structurally heterozygous with chromosomes 01, 04, 1T4, and 4T1 whereas “Pahang”, “Selangor”, “Madang,” and “Borneo” accessions were characterized as homozygous for chromosomes 01 and 04. The first two accessions belonged to the NM translocation group and the last four to the ST group. These two groups were proposed to differ by one translocation event (Shepherd 1999) and our results revealed that this event was a reciprocal translocation involving reference chromosomes 01 and 04.
Molecular marker segregation analysis in an F1 population involving “P. Lilin” as male parent revealed distorted segregation and, based on the pattern of these distortions, Hippolyte et al. (2010) suggested the presence of a duplication of the chromosome 01 distal region in chromosome 04. In the present study, thanks to the availability of a reference genome sequence for Musa acuminata, we were able to reinterpret the data and refute the duplication hypothesis but showed that “P. Lilin” presented a heterozygous reciprocal translocation of distal regions of chromosomes 01 and 04, identical to that identified in PT-BA-00267.
Impact of NM Translocation on Chromosomal Segregation
SNP segregation in PT-BA-00267 self-progeny showed that the reciprocal translocation of distal regions of chromosomes 01 and 04 at a heterozygous state highly distorted chromosome segregation. It also induced a reduction of recombination in regions around breakpoints and generated genetic linkage between the reference chromosome 01 and 04 regions involved. Less expected, we observed no recombination in the 3 Mb translocated region of chromosome 01. In his review, and based on the observation of an anaphase bridge and a minute chromosome fragment, Shepherd (1999) proposed the presence of a small subterminal inversion in one of the translocated fragments between the ST and NM groups (supplementary fig. S5, Supplementary Material online). This inversion could explain the lack of crossover in this region. Careful searches for inversion between chromosomes 1 and 4T1 in this region in the PT-BA-00267 accession using resequencing data did not reveal such a structure. However, if this inversion is terminal and due to the fact that telomeric regions are missing in the assembly such evidence may not yet be detectable with paired read data. Finally, the relatively small size of the translocated fragment (3 Mb, 3-fold smaller than for chromosome 04) combined with the acrocentric nature of chromosome 01 could explain this lack of recombination.
The consequences of this structural heterozygosity on gamete transmission to PT-BA-00267 progeny were found to be similar in paternal and maternal gametes and resulted in the quasi-absence of gametes displaying a combination of Standard and Northern Malayan groups. The lethality of such gametes could be explained by the lack of either of the translocated regions of chromosomes 01 or 04 in these cells. This observation is in agreement with the report on STxNM hybrids by Shepherd (1999), based on chromosome pairing analysis in hybrid progenies.
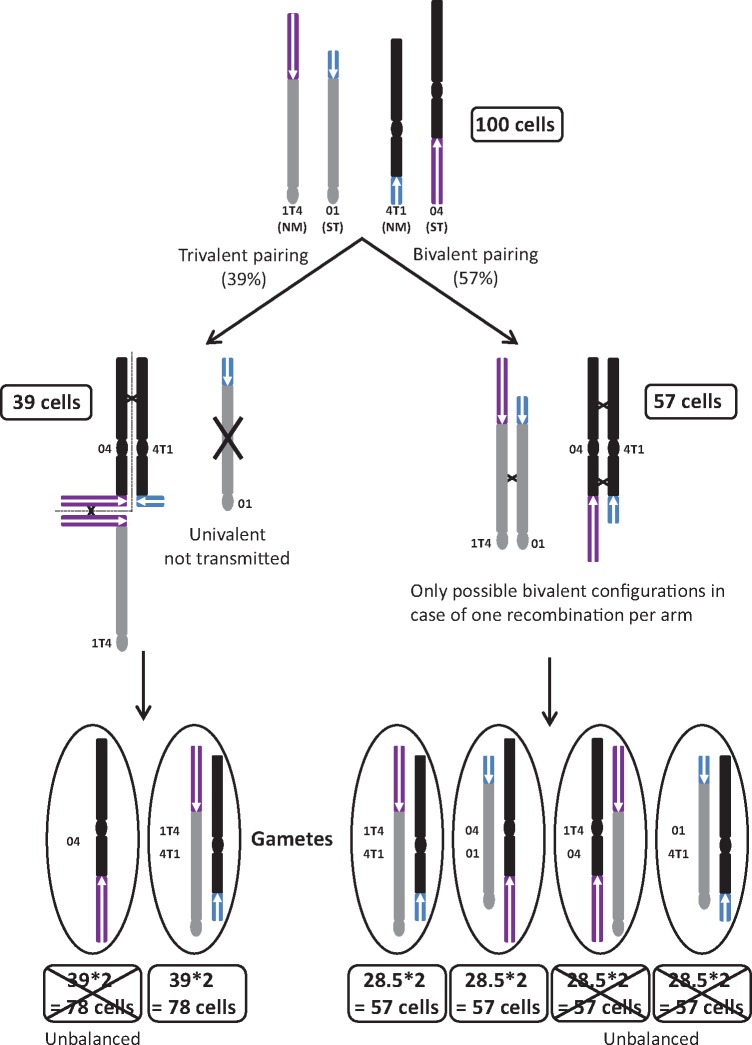
The observation of differential gamete transmission of the two remaining gamete types with a biased proportion in favor of increased transmission of chromosomes 1T4 and 4T1 compared with chromosomes 01 and 04 was more intriguing. Such bias has also been reported in STxNM hybrids by Dodds and Simmonds (1948), reviewed in Shepherd (1999), based on chromosome pairing observations. Interestingly, in these hybrids, the authors observed a ratio of 57% of pollen mother cells (PMC) displaying 11 bivalents and 39% PMC displaying 9 bivalents, 1 V-shaped trivalent and 1 univalent. Based on these proportions and on the hypothesis that unbalanced gametes are not viable, among 100 PMC, 57 PMC should generate 57 gametes with an ST structure and 57 gametes with an NM structure (fig. 5). Among the 100 PMCs, 39 showed a tetravalent V-shape and a univalent. Based on the absence of recombination in the translocated region of chromosome 01 associated with its acrocentric nature and the hypothesis of no more than one chiasma per chromosome arm, chromosome 01 was strongly favored as being the univalent observed by Shepherd (1999) (fig. 5). Note that Shepherd (1999) reached the same conclusion on the nature of the univalent based on the fact that chromosome bridges and univalents were mutually exclusive events. Therefore, the trivalent should involve the two NM chromosomes (1T4 and 4T1) separated by chromosome 04. Based on the V-shape, it could be assumed that during anaphase 1 the two NM chromosomes segregated to the same pole and chromosome 04 segregated to the other. Regarding the fate of the univalent (chromosome 01), it could be expected that it segregated randomly (half with chromosome 04 and half with NM chromosomes), leading to an equal proportion of ST and NM gametes plus unbalanced gametes. However, a few studies have suggested that univalents could be transmitted in a lower proportion than expected. In Brassica addition lines, supernumerary chromosomes (which formed univalent at meiosis) were found at a rate of 1.3–30% in gametes, while 50% was expected (Chèvre et al. 1997). Similar results have also been found in wheat addition lines with an average transmission ratio of 25% (Morrison 1953). In the extreme case of no univalent transmission to gametes, the 39 PMCs will give 78 NM gametes and 78 nonviable unbalanced gametes with only chromosome 04. In this context, a total of 57 (30%) ST and 137 (70%) NM gametes would be obtained from 100 PMCs (fig. 5). Interestingly, these proportions corresponded to that we deduced from DArTseq analysis in PT-BA-00267 self-progeny. This meiosis mechanistic hypothesis could explain the observed segregation. However, other mechanisms involving incompatible gene combinations in hybrids such as those reviewed in Maheshwari and Barbash (2011), Larracuente and Presgraves (2012), and Sweigart and Willis (2012) cannot be excluded.
Fig. 5.
Schematic representation of chromosomal pairing within a heterozygous accession for chromosomes 01, 04, 1T4, and 4TI (ST × NM hybrids) and the hypothesis that led to the observed gamete frequencies.
The Northern Malayan Translocation May Have Emerged in M. acuminata ssp. malaccensis
All wild diploid M. acuminata accessions tested, except three M. a. malaccensis accessions, showed the ST structure only. In addition, the ST structure was found in all tested Musa species, suggesting that this structure occurred in the M. acuminata lineage after its divergence from the other Musa species. The NM structure at the homozygous state was only found in a few M. acuminata ssp. malaccensis wild accessions and in the Sucrier cultivar subgroup. The Sucrier subgroup is composed of diploid AA clones with very low fertility, thus reducing the likelihood that this new structure emerge in the Sucrier subgroup and then spread within Musa. Therefore, it is likely that this structure emerged in spp. malaccensis, the only other group showing the NM structure in a homozygous state. However, due to its rare occurrence within the M. a. spp. malaccensis sample tested here, it cannot be completely excluded that this structure could have been present in an untested Musa species and was then incorporated into M. a. ssp. malaccensis. Further phylogenetic analyses are needed to confirm its origin in ssp. malaccensis.
Interestingly, the NM structure was found to be overrepresented in the heterozygous PT-BA-00267 self-population (70% of haplotypes) as well as in other populations from heterozygous parents (Shepherd 1999). These results suggested that the new emerging structure may progressively replace the ST structure. However, in natural populations, the distinct fertility pattern of homozygotes compared with heterozygotes for these chromosome structures may affect spreading of the new structure.
The Northern Malayan Translocation May Have Favored the Emergence of Triploid Cultivars
The NM structure was found in 39% of the tested banana cultivars (seedless parthenocarpic accessions), mainly in a heterozygous state. Interestingly, this structure was found to be scattered among diverse cultivar groups. These results suggest that the new NM structure could have been incorporated via hybridization and backcrosses in distinct genetic backgrounds. This would imply that there were more hybridization steps in the formation of current cultivars than currently assumed (Perrier et al. 2011), as was already suggested by De Langhe et al. (2010).
The diversity pattern of diploid cultivars with structural heterozygosity in the factorial analysis was interesting as most of them were located between M. a. ssp. malaccensis, Sucrier and M. a. sp. banksii, likely as a result of secondary hybridization between these gene pools. This pattern is consistent with the hypothesis of separate South-East Asia and New Guinea domestication events proposed by Sardos et al. (2016).
The NM structure was found in half of the triploid cultivar subgroups tested, all of them being of the dessert type, highlighting a substantial contribution of this new chromosome structure to polyploid cultivars, while suggesting a role of this structure in dessert banana domestication. In particular, this structure was found in the Cavendish subgroup of dessert bananas, which represents above half of global banana production and also in its suggested 2n gamete donors belonging to the Mlali subgroup (Raboin et al. 2005; Perrier et al. 2009). The reasons underlying the success of this spreading remain to be clarified. The hypotheses include overrepresentation of the NM structure in gametes from STxNM hybrids, reduced fertility of heterozygous genotypes (important for fruit edibility) or some other important agronomical traits associated with this structure that have been selected by farmers. In addition, as structural heterozygosity perturbs meiosis, it could have favored the production of unreduced gametes (Ramsey and Schemske 1998) and may thus have been an important factor in the formation of triploid cultivars and thus in banana domestication. This hypothesis remains to be tested on larger progenies since no triploids were observed in the selfed PT-BA-00267 progeny analyzed (D’Hont et al. 2012). Triploidy is the most efficient ploidy level for agronomic performance in banana (Bakry et al. 2009). These characteristics have generated more vigorous plants, larger fruits, and higher sterility, resulting in a complete absence of seeds in the fruits.
The presence of NM structures in the heterozygous state in accessions used in breeding programs could have important breeding implications. This knowledge could be exploited to either foster recombination or fix allele combinations in the rearranged regions by choosing adequate parental combinations.This applies also to genetic studies involving QTLs or GWAS, etc., aimed at identifying chromosome regions involved in agronomic traits since recombination reduction and biased gamete transmission would hamper and reduce the resolution in the vicinity of rearranged regions.
Finally, Musa with its combination of vegetative propagation with occasional sexual reproduction, is likely to display ongoing slow-motion genetic differentiation and represents a valuable model for unravelling various facets of the speciation process in plants.
Materials and Methods
Plant Material
A total of 169 Musa accessions, wild and cultivated, representative of known Musa acuminata diversity, were analyzed (see supplementary table S6, Supplementary Material online).
In addition, two populations, i.e., a self-progeny population of 180 PT-BA-00267 diploid individuals (D’Hont et al. 2012, Supplementary Material online) and a biparental progeny population of 57 triploid individuals from a cross between PT-BA-00267 and a tetraploid derived from chromosome doubling of the parthenocarpic AA cultivar “Chicame” (PT-BA-00056; “ChicameT”) obtained at the CIRAD research station in Guadeloupe were also analyzed.
Methods
DArTseq Genotyping, Segregation Distortion Analysis and Recombination Rate Estimation
PT-BA-00267 self-progeny was genotyped using the DArTseq technology (Cruz et al. 2013) as described in Martin et al. (2016). Only codominant DArTseq markers were used for this analysis. DArTseq markers were filtered to reduce the technical genotyping error rate using a similar approach to that described in Spindel et al. (2013) for genotyping by sequencing data. This step was based on the assumption that: i) recombination could not occur several times in a small window of contiguous markers, ii) only markers that could be located on the 11 M. acuminata pseudomolecules were preserved, and iii) markers accounting for multiple recombination breakpoints in more than 10% of the progeny were removed. The filtration step was automatically performed with locOnRef and GBS_corrector tools available on the South Green platform https://github.com/SouthGreenPlatform (last accessed May 29, 2017) in the Scaffremodler toolbox (Martin et al. 2016).
Pairwise marker linkage LOD was calculated using JoinMap4.1 and represented along the chromosomes using the pwd2figure tool in the Scaffhunter toolbox (Martin et al. 2016) available on the South Green platform https://github.com/SouthGreenPlatform (last accessed May 29, 2017)
Recombination rates were estimated on sliding windows of 500 kb along the 11 chromosomes. Genotyping error should be close to 0% when calculating recombination rates because genotyping errors lead to artefactual recombination breakpoints. Genotyping data were thus corrected based on the same principle applied to marker filtering (i.e., if the genotype of an individual for a marker differed from the strict consensus of the six surrounding markers, the genotype of this individual was converted to the consensus genotype).
Mate-Pair Sequencing and Structural Variation Detection
Five kilobase insert mate-pair libraries were constructed for 15 Musa accessions. Libraries were sequenced using the Illumina Hiseq platform at GENOSCOPE http://www.genoscope.cns.fr (last accessed May 29, 2017) and BGI http://www.genomics.cn/en (last accessed May 29, 2017).
Paired reads from the 15 sequenced accessions were aligned against the Musa acuminata DH Pahang reference genome sequence (D’Hont et al. 2012) using bowtie2 in very-sensitive mode. Only single hit paired reads were conserved, and redundant paired reads were removed using the MarkDuplicates tool of the Picard toolkit (http://broadinstitute.github.io/picard/; last accessed May 29, 2017). Filtered paired reads were then used to identify discordant read clusters. Resequencing data from the reference genome accession “DH Pahang,” was also used. Discordant read clusters detected using paired reads from “DH Pahang,” were considered as resulting from assembly errors and were thus removed in the analysis of other accessions. Discordant reads were searched and interpreted using the Scaffremodler tools previously developed and available on the South Green platform https://github.com/SouthGreenPlatform (last accessed May 29, 2017) in the Scaffremodler toolbox (Martin et al. 2016). Discordant read clusters were visualized using CIRCOS software (Krzywinski et al. 2009).
Targeted-PCR Validation
Primer pairs were designed at the boundaries of the identified rearrangement breakpoint in the PT-BA-00267 accession. Primers 1LF (5′-TGGAGTTGGCCTGTTAAACC-3′) and 1LR (5′-ACTTGCCGTTTGAACCATC-3′) on chromosome 1 and 4LF (5′-TGGTGAAAGCATTATCTCTTGG-3′) and 4LR (5′-AGACGCAGCATTTGGATG-3′) on chromosome 4 were used to validate the reference genome structure. Primers 1T4F (5′-CGCACTTGGAGCTTGTTCTT-3′) and 1T4R (5′-AACTTGCCGTTTGAACCATC-3′) were used to validate the alternative structure 1T4. The alternative 4T1 structure could not be tested by PCR due to the presence of repeated sequences at the translocation breakpoint, which rendered potential PCR products too long for amplification.
BAC-FISH Validation
Chromosome preparations were performed as described in D’Hont et al. (2000). Seven BAC clones (MAMB_34N11, MAMB_17B03, MAMB_51M04, MAMH_47D06, MAMB_01M16, MAMB_51J24, and MAMH_66D03) from both sides of the breakpoints were selected from a BamH1 and HindIII BAC libraries of accession DH-Pahang (D’Hont et al. 2012; http://banana-genome.cirad.fr/; last accessed May 29, 2017). BAC clones were labeled by random priming with biotin-14-dUTP (Invitrogen, Life Technologies) or Alexa 488-5-dUTP (Invitrogen, Life Technologies). In situ hybridization was performed as described in D’Hont et al. (1996) with the following modifications. Chromosome preparations were incubated in RNAse A (100 ng/µl), pepsin (100 mg/ml) in 0.01M HCl and fixed with paraformaldehyde (4%). Biotinylated probes were immunodetected by Texas Red avidin DCS (Vector Laboratories) and the signal was amplified with biotinylated antiavidin D (Vector Laboratories). Fluorescence images were captured using a CoolSnap HQ camera (Photometrics, Tucson, Ariz) via an Axioplan 2 microscope (Zeiss, Oberkochen, Germany) and analyzed using MetaVueTM (Universal Imaging Corporation, Downington, PA).
Estimation of PT-BA-00267 Maternal and Paternal Gamete Transmission Ratios
PT-BA-00267 × “ChicameT” biparental progeny was genotyped with 35 SSR markers located along chromosomes 01 and 04. SSR genotyping was performed with the Applied Biosystems® 35006L Genetic Analyzer. PT-BA-00267 maternal gamete transmission ratios were then estimated in the most distorted region of chromosomes 01 and 04.
PT-BA-00267 selfing progeny could not directly give access to the gamete transmission ratios. However, for this type of cross, the expected genotype combination proportions for the two chromosomal structures in case of a reciprocal translocation can be expressed as a function of parental gamete transmission ratios for the 01 and 04 chromosome regions around the translocation breakpoints (supplementary fig. S6, Supplementary Material online). As maternal gametes can be fixed using the biparental cross, paternal gamete transmission ratios were estimated by searching quadruplets (a value for each chromosome 01 and 04 combination) complying with both observed genotype proportions and genotype combination proportions in the population. A quadruplet was kept if, for each genotype proportion, the deviation from the observed values was <0.02. A total of 10,000 quadruplets were searched and the most probable quadruplets were then identified based on their distribution and mean value (supplementary fig. S7, Supplementary Material online).
Diversity Analysis and Genotyping by Sequencing
To investigate the occurrence of the two chromosomal structures in a panel of Musa diversity, PCR amplification was performed on 169 Musa accessions using the 1LF/1LR, 4LF/4LR, and 1T4F/1T4R PCR primers pairs (supplementary table S6, Supplementary Material online).
For a part of the studied accessions, Illumina sequencing data (RNAseq and DNAseq) were available through various ongoing projects (supplementary table S6, Supplementary Material online). RNAseq and DNAseq reads were aligned against version 2 of the “DH Pahang” Musa acuminata reference genome sequence (Martin et al. 2016) using STAR (Dobin et al. 2012) and BWA (Li and Durbin 2010), respectively. Reads were locally realigned around indels using the IndelRealigner tool of GATK software, version 3.3 (McKenna et al. 2010). For each accession, at each covered position, all mapping bases that had a mapping quality equal to or greater than ten were counted with the bam-readcount program (https://github.com/genome/bam-readcount; last accessed May 29, 2017). For each accession and at each variant site, a genotype was called based on the maximum likelihood of the genotype, calculated based on a binomial distribution assuming a sequencing error rate of 0.005. The variant calling file was formatted in VCF format. The VCF file was then filtered according to the following criteria: i) data points covered by less than ten reads were converted to missing data, ii) data points with a minor allele frequency inferior to three reads were converted to missing data. Accessions and sites (available at http://banana-genome-hub.southgreen.fr/download; last accessed May 29, 2017) were then selected to have no more than 50% missing sites per accession. The final VCF file, composed of 3,043 polymorphous sites for 75 Musa acuminata wild and cultivar accessions, was used to calculate a dissimilarity matrix using custom python scripts. The dissimilarity index between two accessions was calculated as the proportion of unmatching alleles. The dissimilarities matrix was used to perform a factorial analysis using R (v3.2.4) software (http://www.r-project.org; last accessed May 29, 2017). Considering that cultivar accessions originated from the wild banana genepools, the factorial analysis was performed with the 35 wild accessions. The 40 cultivar accessions were then projected along the synthetic axes.
Availability of Supporting Data
Illumina 5 kb reads mapping in the breakpoint regions of chromosomes 01, 04, and 08, the VCF file comprising 3043 high-quality polymorphic sites for 75 diploid Musa acuminata accessions, the PT-BA-00267 selfing population genotyping matrix and the PT-BA-00267 × “ChicameT” genotyping matrix are available in the download section of the Banana Genome Hub (http://banana-genome-hub.southgreen.fr/download; last accessed May 29, 2017) under Transloc_1-4_reads.tar.gz, Transloc_1-4_vcf.tar.gz, AF-Pahang marker matrix file, and PT-BA-00267_x_ChicameT.txt names, respectively.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
G.M., F.C., F.C.B., and A.D.H. designed the study. G.M. performed bioinformatics and genetic analyses. P.D. designed some bioinformatics tools. O.C performed BAC FISH analyses. F.S and D.R. provided the genetic material. C.H. and C.C. performed PCR analyses. K.L., J.S., and M.R. produced or provided part of the sequencing data. F.C. and F.C.B. contributed to the analysis and edited the manuscript. G.M. and A.D.H wrote the manuscript. ADH: coordinated the study.
Supplementary Material
Acknowledgements
This work was supported by the Centre de cooperation Internationale en Recherche Agronomique pour le Développement (CIRAD). The authors thank for financial support the CGIAR Research Program on Roots, Tubers and Bananas (RTB) and Agropolis Fondation under the reference ID 1504-006 through the Investissements d'avenir programme Labex Agro:ANR-10-LABX-0001-01 (ARCAD project, GenomeHarvest project). We thank the South Green Bioinformatics Platform (http://www.southgreen.fr; last accessed May 29, 2017) for providing us with computational resources. We thank Christophe Jenny (CIRAD research station, French West Indies) for providing the PT-BA-00267 self-progeny population, Lionel Toubi (CRB Plantes tropicales, French West Indies) for providing roots and leaves samples and Jeff Daniells (Department of Agriculture, Fisheries and Forestery, Australia) for providing some leaf samples.
References
- Bakry F, Carreel F, Jenny C, Horry J-P.. 2009. Genetic improvement of banana In: Jain SM, Priyadarshan PM, editors. Breeding plantation tree crops: tropical species. New York: Springer; p. 3–50. [Google Scholar]
- Chèvre AM, Eber F, Barret P, Dupuy P, Brace J.. 1997. Identification of the different Brassica nigra chromosomes from both sets of B. oleracea–B. nigra and B. napus–B. nigra addition lines with a special emphasis on chromosome transmission and self-incompatibility. Theor Appl Genet. 94:603–611. [Google Scholar]
- Cruz VM, Kilian A, Dierig DA.. 2013. Development of DArT marker platforms and genetic diversity assessment of the U.S. collection of the new oilseed crop lesquerella and related species. PLoS One 85:e64062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniells J. 2001. Musalogue : a catalogue of Musa germplasm: diversity in the genus Musa. Biover Int. 213. [Google Scholar]
- De Langhe E, Hřibová E, Carpentier S, Doležel J, Swennen R.. 2010. Did backcrossing contribute to the origin of hybrid edible bananas? Ann Bot. 106:849–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessauw D. 1987. Etude des facteurs de la stérilité du bananier (Musa spp.) et des relations cytotaxinomiques entre M. acuminata Colla et M. balbisiana Colla. PhD thesis. University of Paris-Sud Centre d’Orsay, France.
- D’Hont A, Grivet L, Feldmann P, Glaszmann JC, Rao S, Berding N.. 1996. Characterisation of the double genome structure of modern sugarcane cultivars (Saccharum spp.) by molecular cytogenetics. Mol Gen Genet. 250:405–413. [DOI] [PubMed] [Google Scholar]
- D’Hont A, Paget-Goy A, Escoute J, Carreel F.. 2000. The interspecific genome structure of cultivated banana, Musa spp. revealed by genomic DNA in situ hybridization. Theor Appl Genet. 100:177–183. [Google Scholar]
- D’Hont A, Denoeud F, Aury J-M, Baurens F-C, Carreel F, Garsmeur O, Noel B, Bocs S, Droc G, Rouard M, et al. 2012. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488:213–217. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR.. 2012. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 291:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. 1937. Genetics and the origin of species. New York: Columbia University Press; p. 364. [Google Scholar]
- Dodds K, Simmonds N.. 1948. Sterility and parthenocarpy in diploid hybrids of Musa. Heredity 2:101–117. [DOI] [PubMed] [Google Scholar]
- Dodds KS. 1943. Genetical and cytological studies of Musa. V. Certain edible diploids. J Genet. 45:113–138. [Google Scholar]
- Fauré S, Bakry F, González de Leon D.. 1993a. Cytogenetic studies of diploid bananas In: Breeding banana and plantain for resistance to diseases and pests. Montpellier: CIRAD-FLHOR; p. 77–92. [Google Scholar]
- Fauré S, Noyer JL, Horry JP, Bakry F, Lanaud C, de León DG.. 1993b. A molecular marker-based linkage map of diploid bananas (Musa acuminata). Theor Appl Genet. 87:517–526. [DOI] [PubMed] [Google Scholar]
- Hippolyte I, Bakry F, Seguin M, Gardes L, Rivallan R, Risterucci A-M, Jenny C, Perrier X, Carreel F, Argout X, et al. 2010. A saturated SSR/DArT linkage map of Musa acuminata addressing genome rearrangements among bananas. BMC Plant Biol. 10:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jáuregui B, de Vicente MC, Messeguer R, Felipe A, Bonnet A, Salesses G, Arús P.. 2001. A reciprocal translocation between ‘Garfi’ almond and ‘Nemared’ peach. Theor Appl Genet. 102:1169–1176. [Google Scholar]
- Krzywinski M, Schein J, Birol İ, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA.. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larracuente AM, Presgraves DC.. 2012. The selfish segregation distorter gene complex of Drosophila melanogaster. Genetics 192:33–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot T. 2014. La diversité génétique des bananiers. FruiTrop 221:98. [Google Scholar]
- Li H, Durbin R.. 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari S, Barbash DA.. 2011. The genetics of hybrid incompatibilities. Annu Rev Genet. 45:331–355. [DOI] [PubMed] [Google Scholar]
- Martin G, Baurens F-C, Droc G, Rouard M, Cenci A, Kilian A, Hastie A, Doležel J, Aury J-M, Alberti A, et al. 2016. Improvement of the banana “Musa acuminata” reference sequence using NGS data and semi-automated bioinformatics methods. BMC Genomics 17:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E. 1942. Systematics and the origin of species, from the viewpoint of a zoologist. Cambridge (MA: ): Harvard University Press. [Google Scholar]
- Mbanjo E, Tchoumbougnang F, Mouelle A, Oben J, Nyine M, Dochez C, Ferguson M, Lorenzen J.. 2012. Molecular marker-based genetic linkage map of a diploid banana population (Musa acuminata Colla). Euphytica 188:369–386. [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison J. 1953. Heredity—Abstract of article: chromosome behaviour in wheat monosomics. Heredity 7:203–217. [Google Scholar]
- Ostberg CO, Hauser L, Pritchard VL, Garza JC, Naish KA.. 2013. Chromosome rearrangements, recombination suppression, and limited segregation distortion in hybrids between Yellowstone cutthroat trout (Oncorhynchus clarkii bouvieri) and rainbow trout (O. mykiss). BMC Genomics 14:570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrier X, Bakry F, Carreel F, Jenny C, Horry J-P, Lebot V, Hippolyte I.. 2009. Combining biological approaches to shed light on the evolution of edible bananas. Ethnobot Res Appl. 7:199–216. [Google Scholar]
- Perrier X, De Langhe E, Donohue M, Lentfer C, Vrydaghs L, Bakry F, Carreel F, Hippolyte I, Horry J-P, Jenny C, et al. 2011. Multidisciplinary perspectives on banana (Musa spp.) domestication. Proc Natl Acad Sci U S A. 108:11311–11318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillet MC, Madjidian N, Griveau Y, Serieys H, Tersac M, Lorieux M, Bervillé A.. 1995. Mapping genetic factors controlling pollen viability in an interspecific cross in Helianthus sect. Helianthus. Theor Appl Genet. 91:1195–1202. [DOI] [PubMed] [Google Scholar]
- Raboin LM, Carreel F, Noyer J-L, Baurens F-C, Horry JP, Bakry F, Tézenas du Montcel H, Ganry J, Lanaud C, Lagoda PJL.. 2005. Diploid ancestors of triploid export banana cultivars: molecular identification of 2n restitution gamete donors and n gamete donors. Mol Breed. 16:333–341. [Google Scholar]
- Ramsey J, Schemske DW.. 1998. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu Rev Ecol Syst. 29:467–501. [Google Scholar]
- Ramsey J, Bradshaw HD, Schemske DW.. 2003. Components of reproductive isolation between the monkeyflowers Mimulus lewisii and M. cardinalis (Phrymaceae). Evolution 57: 1520–1534. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH. 2001. Chromosomal rearrangements and speciation. Trends Ecol Evol. 16:351–358. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Blackman BK.. 2010. Speciation genes in plants. Ann. Bot. 1063:439–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd K. 1999. Cytogenetics of the genus Musa. Montpellier, France: IPGRI. [Google Scholar]
- Sardos J, Perrier X, Doležel J, Hřibová E, Christelová P, Houwe IV, den Kilian A, Roux N.. 2016. DArT whole genome profiling provides insights on the evolution and taxonomy of edible Banana (Musa spp.). Ann Bot. 1187: 1269–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spindel J, Wright M, Chen C, Cobb J, Gage J, Harrington S, Lorieux M, Ahmadi N, McCouch S.. 2013. Bridging the genotyping gap: using genotyping by sequencing (GBS) to add high-density SNP markers and new value to traditional bi-parental mapping and breeding populations. Theor Appl Genet. 126: 2699–2716. [DOI] [PubMed] [Google Scholar]
- Sweigart AL, Willis JH.. 2012. Molecular evolution and genetics of postzygotic reproductive isolation in plants. F1000 Biol Rep. 4:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadmor Y, Zamir D, Ladizinsky G.. 1987. Genetic mapping of an ancient translocation in the genus Lens. Theor Appl Genet. 73:883–892. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.