Abstract
Objective
The mechanisms that determine the efficacy or inefficacy of MTX in JIA are ill-defined. The objective of this study was to identify a gene expression transcriptional signature associated with poor response to MTX in patients with JIA.
Methods
RNA sequencing was used to measure gene expression in peripheral blood mononuclear cells collected from 47 patients with JIA prior to MTX treatment and 14 age-matched controls. Differentially expressed baseline genes between responders and non-responders were evaluated. Biological differences between all JIA patients and controls were explored by constructing a signature of differentially expressed genes. Unsupervised clustering and pathway analysis was performed.
Results
A signature of 99 differentially expressed genes (Bonferroni-corrected P < 0.05) capturing the biological differences between all JIA patients and controls was identified. Unsupervised clustering of samples based on this list of 99 genes produced subgroups enriched for MTX response status. Comparing this gene signature with reference signatures from sorted cell populations revealed high concordance between the expression signatures of monocytes and of MTX non-responders. CXCL8 (IL-8) was the most significantly differentially expressed gene transcript comparing all JIA patients with controls (Bonferroni-corrected P = 4.12 × 10−10).
Conclusion
Variability in clinical response to MTX in JIA patients is associated with differences in gene transcripts modulated in monocytes. These gene expression profiles may provide a basis for biomarkers predictive of treatment response.
Keywords: juvenile idiopathic arthritis, methotrexate, gene expression
Rheumatology key messages
Pre-methotrexate JIA disease signature can distinguish between subsequent poor responses to methotrexate.
The mean profile signature of methotrexate non-responders in JIA is concordant with monocyte gene expression.
Introduction
JIA is the most common rheumatic disease of childhood with 16–150 cases per 100 000 population. It is defined as joint inflammation of unknown aetiology, with duration of at least 6 weeks beginning before the age of 16 years [1, 2]. Oligoarticular and polyarticular JIA subtypes are the most common forms of JIA. They share a number of clinical and genetic similarities [2, 3] and are distinguished by the total number of involved joints. Oligoarticular arthritis is defined as four or fewer involved joints whereas patients with polyarthritis have five or more affected joints. In an era of increasing treatment options for patients with inflammatory arthritis, MTX remains a first choice DMARD. MTX is a commonly used, safe and inexpensive treatment with an acceptable safety profile established over 25 years [4]. Approximately 70% of patients will respond to MTX but with varying levels of success. Delays in identifying those patients that will have an inadequate response to MTX can result in delayed introduction of other treatments and as a result these children may have reduced quality of life, greater risk of structural damage to joints and more long-term disability.
Other studies have sought to identify biological and genetic predictors of response to MTX to create an evidence-based selection tool for timely introduction of alternative treatment such as biologic drugs to minimize the risk of disease progression. Genetic associations have been reported for MTX response status in JIA [5–7]. Higher MRP8/14 (S100A8/A9) serum levels prior to treatment may predict poor MTX responses [8]. The presence of long-chain MTX polyglutamates is also a putative indicator of favourable MTX response [9]. An algorithm incorporating genetic polymorphisms in transporter genes multi-drug resistance 1/ABCB1, multi-drug resistance protein 1 (MRP-1/ABCC1) and proton-coupled folate transporter with the clinical variable ESR has been reported to aid in the identification of MTX non-responders [10]. Despite these advances, there are no validated clinical tools available for decision making in the treatment of JIA.
The goal of the current study was to characterize differences in the peripheral blood mononuclear cell (PBMC) gene expression profiles of patients with JIA prior to MTX treatment in comparison with age-matched controls without autoimmune disease, and then to assess JIA disease heterogeneity with respect to responsiveness to MTX treatment.
Methods
More detailed methods are available in supplementary Methods, available at Rheumatology Online (see the following sections: ‘Patients and samples’, ‘RNA sequencing’, ‘Gene expression analysis’, ‘Identifying the cellular origins of gene expression signatures’).
Patients and samples
Patients for this study were selected from a cohort of patients enrolled in a multicentre JIA gene expression study. This study was approved by the institutional review board at all centres. Written informed parental consent was obtained for each subject prior to participation, and child assent was obtained where appropriate.
Patients were included if they had appropriate peripheral blood mononuclear cells collected prior to beginning treatment with MTX, adequate RNA for sequencing, and clinical data to define response based on two time points: prior to MTX treatment and at least 8 weeks into therapy. Suitable RNA was available from a total of 47 children who met the ILAR criteria [1] for oligoarticular (both persistent and extended) or polyarticular (RF negative and positive) JIA (38 female, 9 male). Fourteen age-matched controls (10 female, 4 male) were included for comparison. Six age-matched controls were from the Cincinnati Children's Hospital Test Referral Center, and their primary diagnoses are listed in supplementary Table S1, available at Rheumatology Online.
Clinical data, were recorded at baseline and after at least 2 months of MTX treatment (median 5.2 months). Of the 47 patients with JIA, 4 individuals had <3 months of MTX treatment: 3 non-responders and 1 responder. MTX was given at a dose of 6–21 mg/m2/week (median dose 11.3 mg/m2). There was no significant difference in MTX dose between responders and non-responders. The JIA core set variables used to define improvement were the following: physician’s and parent’s global assessment of disease activity, number of joints with active arthritis and loss of motion, functional capability as measured by the Childhood Health Assessment Questionnaire and ESR [11]. A response meeting ACR-Ped50 criteria was defined by improvement of at least 50% from baseline in at least three of the six core variables with no more than one variable worsening by > 30% [11]. Here responders are defined as subjects who meet the ACR-Ped50 response criteria and conversely, the non-responder category was defined as those subjects who failed to meet the ACR-Ped50 response criteria as previously described [8].
RNA sequencing
Patients had blood sampled at time of clinical care, prior to starting MTX. Briefly, PBMCs were purified and RNA rapidly stabilized. All resulting RNA samples were of high quality as indicated by an RNA integrity number of ⩾9. Genomic libraries were created using TruSeq Stranded mRNA Sample Prep Kits (Illumina, San Diego, CA, USA) and next generation sequencing was performed using an Illumina HiSeq2500 to generate an average of over 40 million raw, single-end reads per sample.
Gene expression analysis
Analysis was performed on RNA sequencing data from two independent cohorts of JIA patients and controls collected over different periods of time. RNA-seq reads were aligned to the reference genome (hg19) and the number of reads aligning to each gene were counted following the RNA-seq protocol described in Anders et al. [12]. Statistical analysis to identify genes differentially expressed between all JIA patients and age-matched controls was performed using the edgeR Bioconductor package [13]. Bonferroni-corrected P < 0.05 was used as evidence of statistical significance. Differences between the two sequencing cohorts were adjusted for batch effects using ComBat [14]. Unsupervised clustering of genes and samples were performed using Bayesian infinite mixture model-based clustering of the normalized log-2 reads per kilobase per million mapped reads (rpkm) gene expression profiles [15]. The statistical significance of the association between the clustering and the MTX response status was assessed by computing the P-values of Fisher’s exact test on the contingency table of association between clustered groups and MTX response status (control, responder or non-responder). Since the response status information has not been used in the process of selecting the genes and clustering samples, this procedure correctly calculates the probability of observing such an association under the null hypothesis of no difference between the MTX responders and non-responders. This point was further demonstrated by calculating empirical P-values under random permutation of MTX response labels in JIA samples. Detailed explanations are presented in supplementary Methods (see ‘Gene expression analysis’), available at Rheumatology Online.
Ingenuity pathway analysis (IPA, Qiagen, Redwood City, CA, USA; www.qiagen.com/ingenuity) was used to identify canonical pathways reflected by genes showing differences between all JIA patients and controls. Upstream regulators and their predicted activation state were characterized using IPA. Gene function was summarized using information available in PubMed, and other NCBI databases including OMIM and Gene. The background reference set for IPA were the 18079 genes expressed in our PBMC dataset.
Identifying the cellular origins of gene expression signatures
Cellular origins of the JIA gene signature were derived using gene expression profiles of purified cell populations in the Gene Atlas dataset [16], as previously described in studies of autoimmune disease [17, 18]. Cell populations of interest included CD14+ monocytes, CD4+ T cells, CD8+ T cells and CD19+ B cells. Briefly, reference profiles were constructed for cell populations of interest (CD14+ monocytes, CD4+ T cells, CD8+ T cells and CD19+ B cells) and correlated with average expression profiles of MTX responder, MTX non-responder and control samples. Detailed explanations are presented in supplementary Methods (see ‘Identifying the cellular origins of gene expression signatures’), available at Rheumatology Online.
Results
This prospective study enrolled patients who were due to start MTX to control joint inflammation. Blood specimens were collected for RNA sequencing and clinical variables recorded to determine MTX response status. Both oligoarticular and polyarticular JIA subtypes were enrolled in this study, as patients with these subtypes have clinical similarities and comprise the majority of JIA cases. The demographics of the JIA patient cohort and age-matched controls are summarized in Table 1. Control blood samples from healthy children were matched by age and gender. As expected for these JIA subtypes, females were the majority of cases at nearly 81% of the patient cohort. The disease duration prior to starting MTX was relatively short (median = 0.6 years). Patients were classified by MTX response status using the ACR-Ped criteria. Patients that met ACR-Ped50 criteria or better were defined as the responder category whereas those patients that failed to meet this ACR-Ped50 level of response were categorized as non-responders, consistent with previous definitions [8]. The responder and non-responder baseline demographics were similar prior to starting MTX with no significant differences apart from ESR (Table 1). This clinical variable differed significantly between the two patient groups (Wilcoxon rank-sum test P = 0.01) at a median (interquartile range) of 12.5 (5.75–30) mm/h compared with 33 (28–49) mm/h, but alone would not have classified patients into subsequent responder or non-responder status, respectively. Notably, MTX dose, duration and route of administration did not differ between the responder and non-responder patient groups. Concomitant medications were permitted in this non-interventional study. A similar percentage of responders and non-responders had concomitant NSAIDS (83.3 and 86.2%, respectively) at the start and during the treatment study period. Only 2 of 29 patients (6.9%) who would respond to MTX had concomitant steroids (prednisone/prednisolone) compared with 5 of 18 (27.8%) MTX non-responders. No biologic therapies were permitted.
Table 1.
Baseline demographic data of patients with JIA at MTX start and age-matched controls
| Characteristic | JIA patients (all) | JIA R | JIA NR | Controls |
|---|---|---|---|---|
| Number of cases | 47 | 29 | 18 | 14 |
| Gender, female, n (%) | 38 (81) | 25 (86) | 13 (72) | 10 (71) |
| JIA subtype, n (%) | ||||
| Oligoarticular persistent | 3 (6) | 2 (7) | 1 (6) | |
| Oligoarticular extended | 3 (6) | 3 (10) | 0 (0) | |
| Polyarticular RF− | 32 (68) | 17 (59) | 15 (83) | |
| Polyarticular RF+ | 9 (19) | 7 (24) | 2 (11) | |
| Clinical variables | ||||
| Disease duration at starting MTX, decimal years | 0.6 (0.3–1.6) | 0.6 (0.3–1.2) | 0.6 (0.3–1.6) | |
| Age at starting MTX, decimal years | 11.8 (7.2–15.4) | 13.0 (8.8–15.3) | 9.7 (5.7–14.7) | |
| Physician’s VAS | 5 (4–7) | 5 (4–7) | 5 (4–6.8) | |
| Active joints | 9 (5–17.5) | 9 (5–12) | 13 (4.5–19.5) | |
| Restricted joints | 6 (2–12) | 4.5 (2–9.3) | 7 (4.3–21.3) | |
| Parent’s VAS | 3 (1.5–6.7) | 3 (1.4–6.3) | 4 (1.5–7.9) | |
| CHAQ | 0.4 (0–1.13) | 0.4 (0.03–0.96) | 0.7 (0.03–1.41) | |
| ESR, mm/h | 25 (8–42) | 12.5 (5.75–30) | 33 (28–49) | |
| MTX dose, mg/m2 | 11.4 (9.1–13.3) | 10.6 (8.3–13.0) | 12.3 (10.2–13.7) | |
Data are the median and interquartile range, unless otherwise indicated. CHAQ: childhood health assessment questionnaire; NR: non-responder; R: responder; VAS: visual analogue scale.
RNA sequencing was performed to characterize molecular heterogeneity of JIA and then relate this heterogeneity to MTX responsiveness. A summary of quality control information reported by RNA-SeQC [19] (supplementary Table S2, available at Rheumatology Online) demonstrated that all the RNA-seq data is of high quality. At least 24 million RNA sequence reads (mean 43 million reads) were achieved per sample.
Six significantly differentially expressed genes were identified comparing MTX responders directly with MTX non-responders: ADAMTS2, FOSL1, F3, NR4A3, NFKBID and CD83 had Bonferroni-adjusted P < 0.05 (Table 2). This gene signature was not predictive of overall response status, but it distinguished a subgroup of six non-responders from the remaining 41 JIA samples.
Table 2.
Differentially expressed genes between JIA patients classified as MTX responders and non-responders
| Gene | Bonferroni corrected P-value | log-2 fold change |
|---|---|---|
| ADAMTS2 | 2.43 × 10−7 | 5.29 |
| FOSL1 | 1.63 × 10−3 | 1.82 |
| F3 | 3.45 × 10−3 | 2.21 |
| NR4A3 | 3.52 × 10−3 | 1.79 |
| NFKBID | 5.78 × 10−3 | 0.83 |
| CD83 | 3.02 × 10−2 | 1.91 |
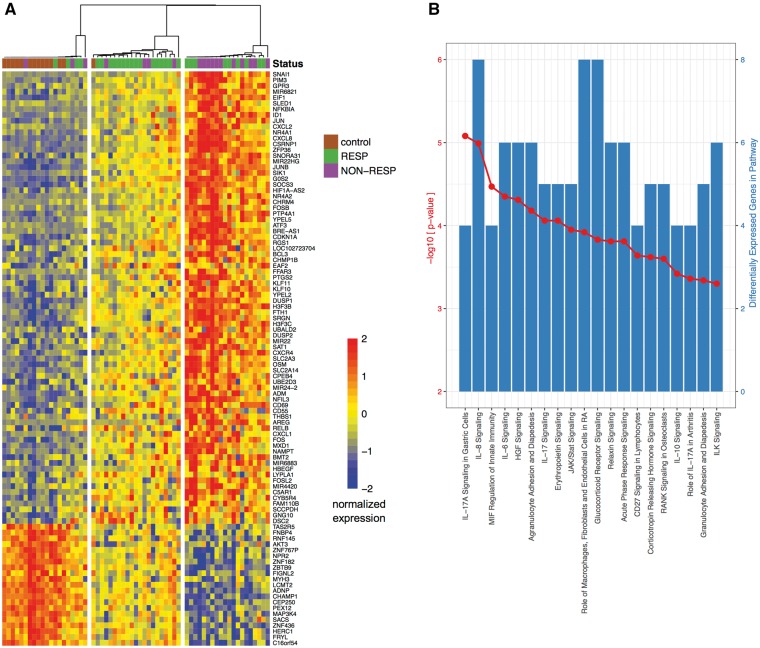
When all JIA patient samples pre-treatment were compared with controls, significant differences in gene expression levels were detected in the peripheral blood. Specifically, 99 genes passed a stringent filter of Bonferroni-corrected P < 0.05 (supplementary Table S3, available at Rheumatology Online). Remarkably, unsupervised clustering of all samples using these 99 genes that were differentially expressed comparing JIA patients to age-matched controls revealed patients groups that reflect MTX response status (Fig. 1A). Three distinct clusters emerged: cluster I included predominantly control samples (13/20) along with a few JIA patient samples (four responders, three non-responders), while clusters II and III, with the exception of one control were limited to JIA patients only. Unsupervised clustering uses no information about MTX response status, yet the contingency table (supplementary Table S4, available at Rheumatology Online) showed statistically significant association between responders and non-responders with clusters II and III (Fisher’s exact test P < 0.05 and empirical odds ratio P < 0.05). Cluster II was highly enriched for responders (1 control, 4 non-responders, 16 responders). In contrast, cluster III was enriched for non-responders (11 non-responders, 9 responders). A gradient of expression levels is visible in the heatmap shown in Fig. 1A: the MTX responder-enriched cluster II appears midway in intensity between cluster I, enriched for controls, and cluster III, enriched for MTX non-responders.
Fig. 1.
Unsupervised clustering between JIA patients and controls reveals patient groups that reflect MTX response status
(A) Heatmap of normalized log-2 rpkm RNA-seq data from PBMC samples collected prior to MTX treatment. Response status of JIA patients to MTX indicated by top annotation bar (RESP: ACR50 or better; NON-RESP: less than ACR50). Unsupervised clustering of rows (genes) and columns (individual persons) produces clusters of individuals enriched for controls, MTX responders or MTX non-responders. (B) Top 20 most significant canonical pathways predicted from the differentially expressed genes in (A) identified with ingenuity pathway analysis. Left axis, red: –log-10 P-value; right axis, blue: number of differentially expressed genes in pathway.
Ingenuity pathway analysis based on the list of 99 genes differentially expressed between all JIA patients and controls found the list is enriched for genes involved in cytokine signalling pathways, including IL-1, IL-6 and IL-17 (Fig. 1B). Other fundamental immunomodulatory pathways include Janus kinase (JAK)–signal transducer and activator of transcription (STAT) signalling, peroxisome proliferator-activated receptor (PPAR) signalling and CD40 signalling, which have key roles in mediating the immune response. Table 3 summarizes the functions of the top 10 differentially expressed genes between JIA patients and age-matched controls. CXCL8 (IL-8), a major mediator of the inflammatory response, was upregulated 69-fold in JIA compared with controls (P = 4.12 × 10−10). As expected, genes involved in inflammation were also upregulated: oncostatin M belongs to the IL-6 family and contributes to matrix metalloprotease (MMP) expression, and junB proto-oncogene leads to cytokine activation. A pathway analysis to identify molecules that are active upstream of the 99 gene transcripts differentially expressed between JIA patients and controls confirmed key cytokines such as IL-1β, TNF and the growth factor TGF-β1 are activated in our JIA patient blood samples (supplementary Table S5, available at Rheumatology Online).
Table 3.
Characteristics of the top 10 genes that distinguish JIA patients and controls
| Gene | Bonferroni corrected P-value | log-2 fold change | Function of gene product |
|---|---|---|---|
| CXCL8 (IL8) | 4.12 × 10−10 | 6.11 | CXCL8 mediates the activation and migration of neutrophils into tissue from peripheral blood. IL-6 treatment of PBMCs or RA synoviocytes leads to increased CXCL8 production [20] |
| SOCS3 | 4.21 × 10−10 | 2.98 | Suppressor of cytokine signalling 3 (SOCS3) inhibits multiple pathways including IL-6 and NF-κB signalling. Expression induced by various cytokines, including IL-6, IL-10 and IFN-γ. Protein binds to and inhibits JAK kinase. SOCS3 has been shown to limit severe joint inflammation in a mouse model of arthritis. SOCS3 is a major negative regulator of STAT3 [21, 22] |
| G0S2 | 5.59 × 10−10 | 5.56 | G0/G1 SWITCH GENE 2 is highly conserved between vertebrates and was identified during cell cycle progression. Roles in lipid metabolism, cell proliferation. Expression of G0S2 was significantly upregulated in primary human fibroblasts treated with the proapoptotic factor TNF-α [23, 24] |
| JUNB | 1.49 × 10−8 | 1.82 | JunB is a member of the activator protein-1 family and regulates T cell function. JunB activates several cytokine genes including IL-2, IL-4 and IL-10. TNF-α and IL-17 induce nuclear translocation of JunB in RA synoviocytes [25, 26] |
| OSM | 1.62 × 10−7 | 2.55 | Oncostatin M (OSM) is an IL-6 cytokine family member. It contributes to MMP expression and human cartilage destruction. Anti-OSM mAbs have been assessed in phase II clinical trials for RA and suggest a need for an anti-OSM mAb with high affinity for greater efficacy [27] |
| DUSP1 | 4.51 × 10−7 | 2.45 | Dual-specificity phosphatase 1 (DUSP1) inactivates members of the MAPK family and controls mediators of inflammation including TNF. In an arthritis model, Dusp1−/− mice had earlier disease onset and increased disease severity [28] |
| NR4A2 | 2.50 × 10−6 | 3.58 | Orphan nuclear receptor NR4A2 is a key regulator of inflammation. NR4A2 induces synoviocyte proliferation and invasion, and MMP13 transcription [29] |
| MIR22HG | 6.71 × 10−6 | 1.37 | MIR22HG encodes a long ncRNA that is 2699 nucleotides in length. Chemical stressors including cycloheximide have been shown to prolong the decay rate of MIR22HG in human induced pluripotent stem cells [30] |
| AREG | 1.10 × 10−5 | 3.82 | Amphiregulin (AREG) is a growth factor shown to be expressed by several types including activated Th2 cells. AREG enhances Treg suppressive function Treg function at the site of inflammation via EGFR [31] |
| RGS1 | 1.19 × 10−5 | 2.21 | Regulator of G-protein signalling 1 (RGS1) reduces macrophage chemotaxis and dampens chemokine receptor signalling. RGS1 is expressed at low basal levels in healthy PBMCs, with constitutive expression reported to be limited to monocytes. TNF-α and IL-17 have been shown to upregulate RGS1 expression in a monocyte-derived human cell line (U937) [32, 33] |
EGFR: epidermal growth factor receptor; MAPK: mitogen-activated protein kinase; MMP: matrix metalloprotease; ncRNA: non-coding RNA; NF-κB: nuclear factor-κB; PBMC: peripheral blood mononuclear cell; STAT3: signal transducer and activator of transcription 3.
To identify the possible cellular origins contributing to expression signature driving the clustering that correlated with patient MTX response outcome in Fig. 1A, transcriptional profiles from the human U133A/GNF1H Gene Atlas dataset [16] for purified B cells, CD4 T cells, CD8 T cells and monocytes were used for comparison. Seventy-one of the 99 genes that comprise the JIA signature were available in the Human Gene Atlas reference dataset. For each of these genes, the mean normalized log-2 rpkm expression for samples in clusters I, II and III was calculated to produce an averaged expression profile corresponding to each cluster. In a similar manner, mean expression values were also calculated after separating samples by MTX response status to produce averaged profiles for all healthy controls, responders and non-responders. These mean expression profiles were also compared with the Human Gene Atlas profiles for the cell types listed above. As shown in Fig. 2, the mean profiles of clusters I, II and III, respectively, enriched for controls, responders and non-responders, were similar to the corresponding mean profiles of only controls, responders or non-responders. Cluster III, which is enriched for non-responders, is correlated to the monocyte profile (Pearson’s correlation coefficient, r = 0.55), while cluster II, which is enriched for responders, is not (r = −0.008). Complete blood count data revealed no difference in monocyte percentage either between responders and non-responders (Wilcoxon rank-sum test P = 0.93) or between clusters II and III (Wilcoxon rank-sum test P = 0.42). In contrast, the cluster I profile and control profile were most similar to the T cell profiles (CD8+, r = 0.36; CD4+, r = 0.48).
Fig. 2.
The mean profile signature of MTX non-responders is concordant with the monocyte gene expression signature (Human Gene Atlas)
Genes that were differentially expressed between JIA patients and controls and also present in the Human Gene Atlas dataset [16] are shown in rows. The columns represent the mean normalized expression profile for controls, MTX responders and MTX non-responders (left), the mean normalized expression profile for samples in clusters I, II and III in Fig. 1A (centre) and the mean normalized expression signatures of different cell types from the Human Gene Atlas: CD14+ monocytes, CD4+ T cells, CD8+ T cells and B cells (right). Control and cluster I profiles are most strongly correlated with the T cell signatures, while non-responder and cluster III profiles are correlated with the monocyte signature.
The primary analysis presented relied on RNA sequencing data from two JIA cohorts that were subsequently combined for greater statistical power. We assessed reproducibility between the two cohorts by analysing each independently. Supplementary Fig. S1, available at Rheumatology Online, displays the log-2 fold change between JIA patients and controls in each cohort for all genes. There is a positive correlation of fold change between the two cohorts (Pearson’s correlation coefficient r = 0.46, P < 2 × 10−16). The overlap and direction of change of differentially expressed genes were highly consistent between both cohorts (supplementary Fig. S2, available at Rheumatology Online). All genes that were among the 800 most significant in both cohorts had a consistent direction of change (binomial test, P < 2 × 10−16). Furthermore, we assessed the residuals from the gene-specific multivariate generalized linear models used to assess the differential expression between control and JIA samples. A batch covariate was added to account for differences between the two JIA cohorts. Principal component and multidimensional scaling analysis was performed on log-2 residuals for models with and without a batch covariate. No significant difference in residuals between cohorts remained following the addition of a batch covariate to the model (supplementary Fig. S3, available at Rheumatology Online).
Discussion
MTX remains the first line DMARD therapy for patients with JIA, and a response is seen in < 70% of the patients. The first signs of a good response are not usually seen until at least 6–8 weeks into the treatment, and it may take several months to achieve the full benefit of this treatment. Importantly, there was no significant difference in the dose, route or duration of MTX between responders and non-responders. Furthermore, the use of NSAIDS was largely equal between MTX responders and non-responders. Notably, concomitant steroid usage was lower in MTX responders, which is consistent with the response measured being due to MTX rather than steroids. In patients who eventually fail MTX, the introduction of alternative treatments is often delayed by months waiting for improvement on MTX, and this delay may even decrease the chance of achieving clinically inactive disease with any treatment and increase the risk for irreversible joint damage in the long term [34]. Therefore, defining with precision if MTX is the appropriate treatment will advance clinical management of JIA [35]. Consensus treatment plans for patients with new-onset polyarticular JIA demonstrate the continued importance of MTX therapy in JIA [36]. Long-term safety data for two commonly used biologic treatments for JIA, etanercept and adalimumab, show that at 4.5–4.7 events per 100 exposure years there is a 2.2-fold relative risk of having a serious adverse event using these biologics to treat JIA compared with MTX [37]. Thus there remains a pressing need to understand the mechanisms of MTX action and develop biomarkers to predict response in JIA.
Peripheral blood is routinely available as part of clinical care, compared with samples from the inflamed joint. In this study, initial analysis of two independent cohorts of patients and controls separated by time showed significant overlap in their gene expression profiles and also a consistent direction of change. Unsupervised clustering of pre-treatment peripheral blood samples using gene expression signatures distinguishing all JIA patients from age-matched controls yielded a monocyte-correlated gene expression signature that reflects subsequent MTX response status. This was not explained by monocyte numbers as there was no difference in complete blood count monocyte percentages between responders and non-responders. The mRNA of the most significant upregulated gene, CXCL8 (IL8), has high stability and abundance in primary human monocytes [38]. This chemokine results in neutrophil activation and migration from peripheral blood to tissues. The upregulated genes in this profile (supplementary Table S3, available at Rheumatology Online) included putative therapeutic targets that had previously been identified for arthritis including PTGS2 and FOS. PTGS2, also known as COX2, is an inducible mediator that contributes to inflammatory response and pain. Selective inhibition of FOS protein (c-fos) protects from collagen-induced arthritis in mice [39]. The PPAR pathways differentially expressed between JIA cases and controls modulate monocyte/macrophage immune function and affect expression of cytokines including TNF-α, IL-1β and IL-6 [40]—which were predicted to be activated in this study and are also current major therapeutic targets in JIA. Our findings are consistent with previously reported data showing a monocyte signature in patients with polyarticular JIA [41]. This is consistent with a role for monocyte activation in a subset of patients with oligoarticular and polyarticular JIA.
NFKBIA, a differentially expressed gene between all JIA patients and controls, is a member of the nuclear factor-κB (NF-κB) inhibitor family. NFKBIA gene expression aided the molecular discrimination of RA anti-TNFα therapy responders and non-responders [42]. Another study also identified the upregulation of genes such as NFKB1 and NR4A1 in an oligoarticular JIA cohort when compared with controls [43]. In the Trial of Early Aggressive Therapy study, whole blood RNA analysis showed higher NFKB1 levels in polyarticular JIA patients before treatment compared with healthy child controls [44]. Differences in the inflammatory profile between JIA cases and controls were identified but not further resolved by cell population. In our study, we correlate gene expression profiles with those of purified cell populations and so advance beyond the whole blood analysis in the Trial of Early Aggressive Therapy study to reveal new insights into the cell types that drive JIA inflammation and MTX response.
There are some limitations to our study. JIA is a heterogeneous disease and this study was not powered to analyse individual ILAR subtypes. Collecting large numbers of PBMC samples from JIA patients pre-treatment is not trivial. This study leveraged a multi-site cohort collected over 11 years. The power to detect clinically relevant signatures would improve with a larger sample size. We considered gender, route, age at onset, disease duration, as well as lymphocyte, monocyte and neutrophil percentage, but these parameters do not provide an explanation for the imperfect stratification and therefore other uncharacterized disease heterogeneity is a likely explanation. We use ACR50 as the primary outcome in this study. This is a more stringent definition than ACR30, which is accepted by the US Food and Drug Administration as defining a clinical meaningful response for clinical trials in JIA. However, while ACR50 identifies a relative improvement in disease symptoms, considerable disease burden can remain, particularly in patients who started with more severe disease. The limitations on blood volume collection on paediatric samples from sick children meant it was not feasible to test sorted cell populations. The monocyte-enriched signature is limited to a subgroup of non-responders suggesting that there might be more than one mechanism leading to poor MTX response. This apparent heterogeneity may explain why the direct comparison of non-responders and responders identified only six genes and driven by a subset of patients. Our data provide a basis for study of new prospective cohorts and focus on purified monocytes as a hypothesis-driven test to further validate the findings demonstrated here. In this discovery phase of profiling non-responders, large-scale transcriptional profiling has been performed. For future validation studies, determining the feasibility of protein assays, such as ELISA, on a subset of proteins would be beneficial for clinical utility.
The gene expression profile that distinguishes a subgroup of non-responders strongly correlates with a monocyte signature, suggesting a prominent role of innate immunity in the pathogenesis of the disease in these patients. To understand the relevance of this potentially distinct subgroup of MTX non-responders, a larger number of patient samples is needed. This feature is reminiscent of gene expression profiles in systemic JIA, a subtype that has been linked to the abnormalities in innate rather than adaptive immunity and consistent with this, systemic JIA patients, as a group, show poor responses to MTX in general [45].
To our knowledge, this is the first demonstration that suggests a cell type-specific signature associated with MTX non-response in patients with JIA. We also observe that responders display a gene expression signature prior to MTX treatment that is distinct from, but more similar to, age-matched controls than the MTX non-responders. We confirmed that RNA-sequencing is of sufficient sensitivity to detect the presence of key inflammatory pathways active in PBMC samples from JIA patients. This method of identifying gene expression patterns specific to cell populations may be a valuable tool in understanding the complex mechanisms of drug response and enable greater precision in treatment selection for patients with JIA.
Supplementary Material
Acknowledgements
We thank the patients, families, physicians and staff at of the participating hospitals including Cincinnati Children’s Hospital Medical Center, Cincinnati, OH; Levine Children’s Hospital, Charlotte, NC; Medical College of Wisconsin, Milwaukee, WI; Emory University, Atlanta, GA; Arkansas Children’s Hospital Research Institute, Little Rock, AR; and Boston Children’s Hospital, Boston, MA. We also acknowledge and appreciate the contributions of Kimberly Solomon, Allen Watts, Judyann C. Olson and Paula Morris. Data deposition: the datasets supporting the conclusions of this article are available in the Gene Expression Omnibus repository [Series GSE81259, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=sxgjoeawjredlon&acc=GSE81259].
Funding: This work was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health [Grant Award Numbers P01AR048929, P30AR47363, P30AR070549, and P30AR070253] and in part by the Cincinnati Children’s Research Foundation and its Cincinnati Genomic Control Cohort, as well as Fundación Bechara. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Disclosure statement: J.D. has received research support from AbbVie, AstraZeneca, Bristol-Myers Squibb, Horizon Pharma, Medac, Pfizer, Roche and UCB. P.A.N. has received consulting fees from Sobi, Novartis, UCB, Genentech, Alkermes and Casebia, and has ongoing investigator-initiated research support from Sobi, Novartis and Genentech. A.A.G. has received consulting fees from Novartis, NovImmune and Juno. S.P. has served on advisory board to Novartis, UCB and Medac pharmaceuticals; this relationship had no bearing on the work reported in this paper. The other authors have declared no conflicts of interest.
Supplementary data
Supplementary data are available at Rheumatology Online.
References
- 1. Petty RE,, Southwood TR,, Manners P. et al. International League of Associations for Rheumatology classification of juvenile idiopathic arthritis: second revision, Edmonton, 2001. J Rheumatol 2004;31:390–2. [PubMed] [Google Scholar]
- 2. Prakken B, Albani S, Martini A.. Juvenile idiopathic arthritis. Lancet 2011;377:2138–49. [DOI] [PubMed] [Google Scholar]
- 3. Hinks A,, Cobb J,, Marion MC. et al. Dense genotyping of immune-related disease regions identifies 14 new susceptibility loci for juvenile idiopathic arthritis. Nat Genet 2013;45:664–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rose CD,, Singsen BH,, Eichenfield AH. et al. Safety and efficacy of methotrexate therapy for juvenile rheumatoid arthritis. J Pediatr 1990;117:653–9. [DOI] [PubMed] [Google Scholar]
- 5. Cobb J,, Cule E,, Moncrieffe H. et al. Genome-wide data reveal novel genes for methotrexate response in a large cohort of juvenile idiopathic arthritis cases. Pharmacogenomics J 2014;14:356–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Rotte MC,, Bulatovic M,, Heijstek MW. et al. ABCB1 and ABCC3 gene polymorphisms are associated with first-year response to methotrexate in juvenile idiopathic arthritis. J Rheumatol 2012;39:2032–40. [DOI] [PubMed] [Google Scholar]
- 7. Hinks A,, Moncrieffe H,, Martin P. et al. Association of the 5-aminoimidazole-4-carboxamide ribonucleotide transformylase gene with response to methotrexate in juvenile idiopathic arthritis. Ann Rheum Dis 2011;70:1395–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moncrieffe H,, Ursu S,, Holzinger D. et al. A subgroup of juvenile idiopathic arthritis patients who respond well to methotrexate are identified by the serum biomarker MRP8/14 protein. Rheumatology 2013;52:1467–76. [DOI] [PubMed] [Google Scholar]
- 9. Ćalasan MB,, den Boer E,, de Rotte MC. et al. Methotrexate polyglutamates in erythrocytes are associated with lower disease activity in juvenile idiopathic arthritis patients. Ann Rheum Dis 2015;74:402–7. [DOI] [PubMed] [Google Scholar]
- 10. Bulatovic M,, Heijstek MW,, Van Dijkhuizen EH. et al. Prediction of clinical non-response to methotrexate treatment in juvenile idiopathic arthritis. Ann Rheum Dis 2012;71:1484–9. [DOI] [PubMed] [Google Scholar]
- 11. Giannini EH,, Ruperto N,, Ravelli A. et al. Preliminary definition of improvement in juvenile arthritis. Arthritis Rheum 1997;40:1202–9. [DOI] [PubMed] [Google Scholar]
- 12. Anders S,, McCarthy DJ,, Chen Y. et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc 2013;8:1765–86. [DOI] [PubMed] [Google Scholar]
- 13. Robinson MD, McCarthy DJ, Smyth GK.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johnson WE, Li C, Rabinovic A.. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007;8:118–27. [DOI] [PubMed] [Google Scholar]
- 15. Freudenberg JM,, Sivaganesan S,, Wagner M. et al. A semi-parametric Bayesian model for unsupervised differential co-expression analysis. BMC Bioinformatics 2010;11:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Su AI,, Wiltshire T,, Batalov S. et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A 2004;101:6062–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Banchereau R,, Hong S,, Cantarel B. et al. Personalized immunomonitoring uncovers molecular networks that stratify lupus patients. Cell 2016;165:1548–50. [DOI] [PubMed] [Google Scholar]
- 18. Grayson PC,, Carmona-Rivera C,, Xu L. et al. Neutrophil-related gene expression and low-density granulocytes associated with disease activity and response to treatment in antineutrophil cytoplasmic antibody-associated vasculitis. Arthritis Rheumatol 2015;67:1922–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. DeLuca DS,, Levin JZ,, Sivachenko A. et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 2012;28:1530–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Caiello I,, Minnone G,, Holzinger D. et al. IL-6 amplifies TLR mediated cytokine and chemokine production: implications for the pathogenesis of rheumatic inflammatory diseases. PLoS One 2014;9:e107886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wong PK,, Egan PJ,, Croker BA. et al. SOCS-3 negatively regulates innate and adaptive immune mechanisms in acute IL-1-dependent inflammatory arthritis. J Clin Invest 2006;116:1571–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liang Y,, Xu WD,, Peng H,, Pan HF,, Ye DQ.. SOCS signaling in autoimmune diseases: molecular mechanisms and therapeutic implications. Eur J Immunol 2014;44:1265–75. [DOI] [PubMed] [Google Scholar]
- 23. Heckmann BL,, Zhang X,, Xie X,, Liu J.. The G0/G1 switch gene 2 (G0S2): regulating metabolism and beyond. Biochim Biophys Acta 2013;1831:276–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim TH,, Choi SJ,, Lee YH,, Song GG,, Ji JD.. Gene expression profile predicting the response to anti-TNF treatment in patients with rheumatoid arthritis; analysis of GEO datasets. Joint Bone Spine 2014;81:325–30. [DOI] [PubMed] [Google Scholar]
- 25. Granet C, Maslinski W, Miossec P.. Increased AP-1 and NF-kappaB activation and recruitment with the combination of the proinflammatory cytokines IL-1beta, tumor necrosis factor alpha and IL-17 in rheumatoid synoviocytes. Arthritis Res Ther 2004;6:R190–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Garaude J,, Farrás R,, Bossis G. et al. SUMOylation regulates the transcriptional activity of JunB in T lymphocytes. J Immunol 2008;180:5983–90. [DOI] [PubMed] [Google Scholar]
- 27. Choy EH,, Bendit M,, McAleer D. et al. Safety, tolerability, pharmacokinetics and pharmacodynamics of an anti-oncostatin M monoclonal antibody in rheumatoid arthritis: results from phase II randomized, placebo-controlled trials. Arthritis Res Ther 2013;15:R132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vattakuzhi Y,, Abraham SM,, Freidin A,, Clark AR,, Horwood NJ. et al. Dual-specificity phosphatase 1-null mice exhibit spontaneous osteolytic disease and enhanced inflammatory osteolysis in experimental arthritis. Arthritis Rheum 2012;64:2201–10. [DOI] [PubMed] [Google Scholar]
- 29. Mix KS,, McMahon K,, McMorrow JP. et al. Orphan nuclear receptor NR4A2 induces synoviocyte proliferation, invasion, and matrix metalloproteinase 13 transcription. Arthritis Rheum 2012;64:2126–36. [DOI] [PubMed] [Google Scholar]
- 30. Tani H,, Onuma Y,, Ito Y,, Torimura M.. Long non-coding RNAs as surrogate indicators for chemical stress responses in human-induced pluripotent stem cells. PLoS One 2014;9:e106282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zaiss DM,, van Loosdregt J,, Gorlani A. et al. Amphiregulin enhances regulatory T cell-suppressive function via the epidermal growth factor receptor. Immunity 2013;38:275–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Denecke B, Meyerdierks A, Bottger EC.. RGS1 is expressed in monocytes and acts as a GTPase-activating protein for G-protein-coupled chemoattractant receptors. J Biol Chem 1999;274:26860–8. [DOI] [PubMed] [Google Scholar]
- 33. Patel J,, McNeill E,, Douglas G. et al. RGS1 regulates myeloid cell accumulation in atherosclerosis and aortic aneurysm rupture through altered chemokine signalling. Nat Commun 2015;6:6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wallace CA,, Giannini EH,, Spalding SJ. et al. Clinically inactive disease in a cohort of children with new-onset polyarticular juvenile idiopathic arthritis treated with early aggressive therapy: time to achievement, total duration, and predictors. J Rheumatol 2014;41:1163–70. [DOI] [PubMed] [Google Scholar]
- 35. Calasan MB, Wulffraat NM.. Methotrexate in juvenile idiopathic arthritis: towards tailor-made treatment. Expert Rev Clin Immunol 2014;10:843–54. [DOI] [PubMed] [Google Scholar]
- 36. Ringold S,, Weiss PF,, Colbert RA. et al. Childhood Arthritis and Rheumatology Research Alliance consensus treatment plans for new-onset polyarticular juvenile idiopathic arthritis. Arthritis Care Res 2014;66:1063–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Klotsche J,, Niewerth M,, Haas JP. et al. Long-term safety of etanercept and adalimumab compared to methotrexate in patients with juvenile idiopathic arthritis (JIA). Ann Rheum Dis 2016;75:855–61. [DOI] [PubMed] [Google Scholar]
- 38. Mahmoud L,, Al-Enezi F,, Al-Saif M. et al. Sustained stabilization of Interleukin-8 mRNA in human macrophages. RNA Biol 2014;11:124–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Aikawa Y,, Morimoto K,, Yamamoto T. et al. Treatment of arthritis with a selective inhibitor of c-Fos/activator protein-1. Nat Biotechnol 2008;26:817–23. [DOI] [PubMed] [Google Scholar]
- 40. Kiss M, Czimmerer Z, Nagy L.. The role of lipid-activated nuclear receptors in shaping macrophage and dendritic cell function: From physiology to pathology. J Allergy Clin Immunol 2013;132:264–86. [DOI] [PubMed] [Google Scholar]
- 41. Griffin TA,, Barnes MG,, Ilowite NT. et al. Gene expression signatures in polyarticular juvenile idiopathic arthritis demonstrate disease heterogeneity and offer a molecular classification of disease subsets. Arthritis Rheum 2009;60:2113–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Koczan D,, Drynda S,, Hecker M. et al. Molecular discrimination of responders and nonresponders to anti-TNF alpha therapy in rheumatoid arthritis by etanercept. Arthritis Res Ther 2008;10:R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stevens A,, Meyer S,, Hanson D,, Clayton P,, Donn RP.. Network analysis identifies protein clusters of functional importance in juvenile idiopathic arthritis. Arthritis Res Ther 2014;16:R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jiang K,, Sawle AD,, Frank MB. et al. Whole blood gene expression profiling predicts therapeutic response at six months in patients with polyarticular juvenile idiopathic arthritis. Arthritis Rheumatol 2014;66:1363–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fall N,, Barnes M,, Thornton S. et al. Gene expression profiling of peripheral blood from patients with untreated new-onset systemic juvenile idiopathic arthritis reveals molecular heterogeneity that may predict macrophage activation syndrome. Arthritis Rheum 2007;56:3793–804. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.