Abstract
Analysis of GLOBOCAN-2012 data shows clearly here that cancer incidence worldwide is highly related with low average annual temperatures and extreme low temperatures. This applies for all cancers together or separately for many frequent or rare cancer types (all cancers P = 9.49×10−18). Supporting fact is that Inuit people, living at extreme low temperatures, have the highest cancer rates today. Hypothesizing an evolutionary explanation, 240 cancer genome-wide association studies, and seven genome-wide association studies for cold and high-altitude adaptation were combined. A list of 1,377 cancer-associated genes was created to initially investigate whether cold selected genes are enriched with cancer-associated genes. Among Native Americans, Inuit and Eskimos, the highest association was observed for Native Americans (P = 6.7×10−5). An overall or a meta-analysis approach confirmed further this result. Similar approach for three populations living at extreme high altitude, revealed high association for Andeans-Tibetans (P = 1.3×10−11). Overall analysis or a meta-analysis was also significant. A separate analysis showed special selection for tumor suppressor genes. These results can be viewed along with those of previous functional studies that showed that reduced apoptosis potential due to specific p53 variants (the most important tumor suppressor gene) is beneficial in high-altitude and cold environments. In conclusion, this study shows that genetic variants selected for adaptation at extreme environmental conditions can increase cancer risk later on age. This is in accordance with antagonistic pleiotropy hypothesis.
Keywords: evolutionary medicine, antagonistic pleiotropy, natural selection, extreme environmental conditions, cancer evolution, tumor suppressor genes, population genetics, meta-analysis, genomics, average annual temperature, cancer epidemiology, high altitude, breast cancer, GWAS
Introduction
Cancer is a leading cause of morbidity and mortality worldwide (Jemal et al. 2005). It is well established that genetic factors have an important role in the etiology of many cancer types (Amundadottir et al. 2004). A major question is why cancer incidence, defined as the number of new cancer cases occurring in a population within a specified period of time, is so increased in human populations. Another major question is why cancer incidence varies a lot among human populations (Ferlay et al. 2015). Analysis of epidemiological data show that human populations in specific geographic areas exhibit very high cancer incidence and mortality rates (Sharma et al. 2015). This is known as spatial or geographic distribution of cancer. There is limited information today about the factors that determine this spatial distribution (Sharma et al. 2015). Environmental and genetic factors are equally suspected, since in multifactorial diseases environmental variables and gene variants shape the risk per population.
It is also well known that genetic variants that predispose to a disease could have been selected by natural selection if offering a survival advantage (Nesse 2011). This phenomenon is known as “antagonistic pleiotropy,” first proposed by the evolutionary biologist George Williams in 1957 as an evolutionary explanation for senescence (Williams 1957). This is a longstanding hypothesis for cancer, but to date not any reliable evidence has been presented yet (Crespi and Summers 2005; Vittecoq et al. 2013). Some scientists believe that this phenomenon is not so rare and might be a fundamental mechanism for the survival of deleterious alleles predisposing for multifactorial diseases (Carter and Nguyen 2011). On the other site, due to factual difficulties, very few studies exist investigating antagonistic pleiotropy in certain diseases.
In this study, an evolutionary relationship is hypothesized between adaptation at extreme environmental conditions and increased cancer risk in humans. In order to prove this, reliable and accurate data on cancer incidence worldwide were needed. The most accurate and complete survey for global cancer incidence, being public available, is GLOBOCAN-2012 (Ferlay et al. 2015). GLOBOCAN-2012 permits a variety of incidence/prevalence analysis per country or per cancer type (http://globocan.iarc.fr/). Additionally, bibliographic cancer incidence and genetic data for human populations living at extreme cold and extreme high-altitude plus 240 cancer genome-wide association studies (GWAS) were analyzed as well. Evidence was found that cancer rates have been increased in those populations through natural selection procedures. This is the first study that provides evidence that high cancer risk may be a result of evolutionary adaptation in certain environmental conditions.
Results and Discussion
GLOBOCAN-2012, Average Annual Temperature, and Extreme Low Temperature Analyses
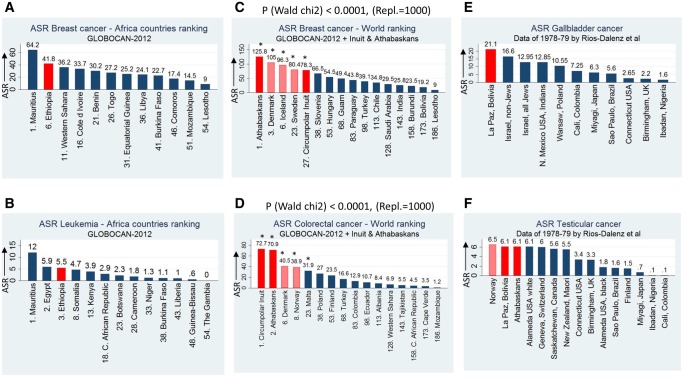
A number of different analyses were performed with GLOBOCAN-2012 data, according the research hypothesis of this study. Interestingly, at “all cancers” analysis (excl. nonmelanoma skin cancer) at a descending age-standardized rate (ASR) order, number two in ranking is Norway and number eight is Denmark. Indeed, Scandinavian countries, being among the coldest countries in Earth, rank very high in ASR rates for many frequent and rare cancer types, like breast and colorectal cancer (fig. 1 and supplementary table S1, Supplementary Material online). This can be attributed to genetic background of Scandinavian populations. Hypothesizing here an evolutionary relationship between adaptation at very cold environments and high cancer incidence, published data for Inuit (Canadian and Greenlandic native populations) and Athabascans (Native Americans living mainly in Alaska), that live at extreme cold, were investigated. These data show that these populations exhibit extreme high cancer incidence, especially for lung, breast, and colorectal cancer (Friborg and Melbye 2008; Lemrow et al. 2008; Day et al. 2010; Moore et al. 2014; Perdue et al. 2014; Foote et al. 2016; Young et al. 2016) (fig. 1C and D). Lung cancer can be attributed to high rates of smoking in these populations (Young et al. 2016). These populations are not part of GLOBOCAN-2012 project.
Fig. 1.
Cancer age-standardized (ASR) data by country and by cancer type. ASR data of 184 populations can be found in supplementary table S1, Supplementary Material online. Bars of populations of main interest are in red color. Bars showing noticeable data of other populations are in pink color. For (A), (B), and (C), (D), ASR ranking is indicated by the ranking number before each country/population name. ASR values are according GLOBOCAN-2012 and Young et al. (2016) (2004–2008 ASR data). Bootstrapping for 1,000 random replications in (C) and (D) shows that the association of temperature with ASR is highly significant under a Wald χ2 statistic. Additionally, in (C) and (D), statistical analysis showed that when countries/populations were separated in six quartiles according ASR ranking, the first quartile (31 populations in total, asterisk symbolled, 5 of them are shown) had a significantly lower mean of average annual temperatures when compared with the mean of average annual temperatures of the other five quartiles (P=5.09×10−8, P=3.01×10−9, respectively). For (E) and (F), ASR 1978–1979 data were taken from Rios-Dalenz et al. (1981). In (F), Athabascans’ ASR of testicular cancer is included in the plot according Young et al. (2016), since data show that ASR values are stable long-term (1989–2008). ASR is termed as “new cases/time period/100,000 individuals.”
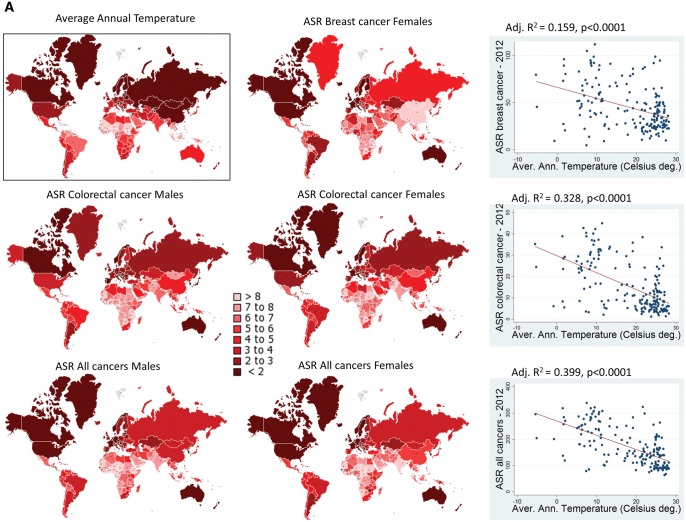
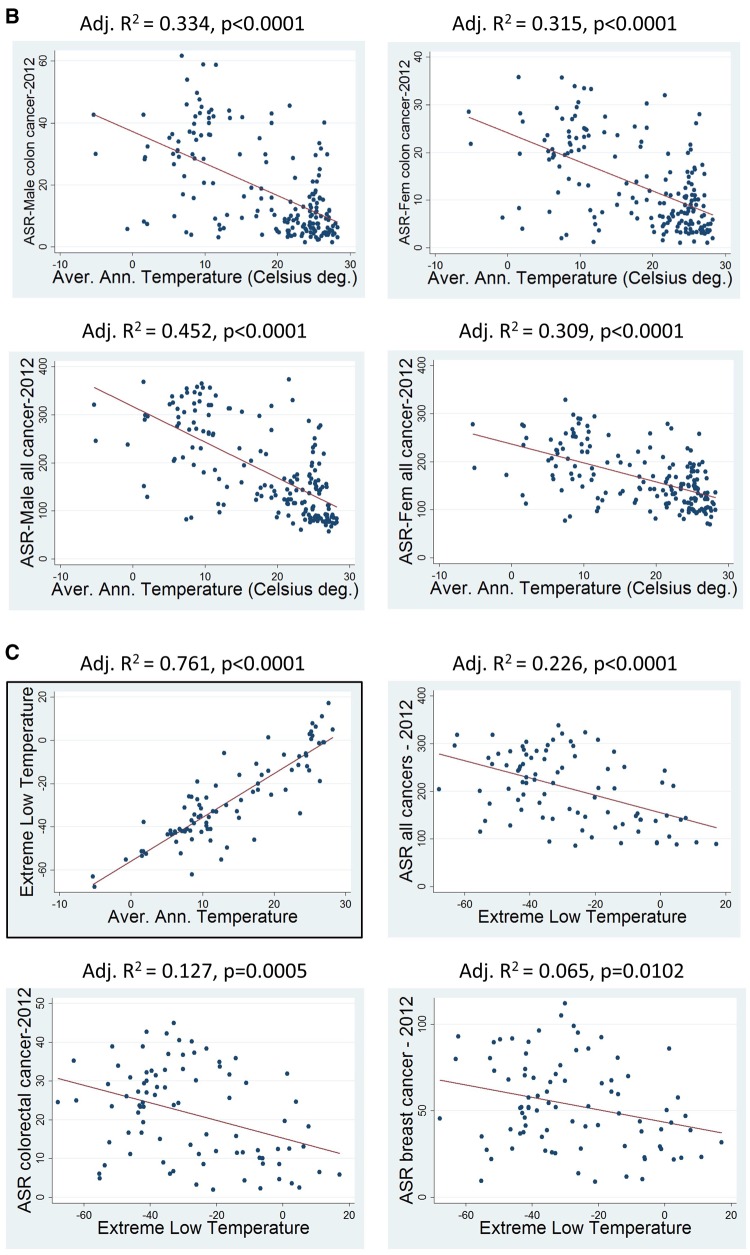
In order to investigate if temperature–cancer incidence association is an extended phenomenon in human populations, linear regression analysis was performed of average annual temperatures (AAT) per country with cancer ASR per country (184 countries included). Results are shown in figure 2A and B. ASRs of “all-cancers” (excl. nonmelanoma skin cancer), breast cancer, and colorectal cancer were found to be linearly related with AAT. Prediction power of the model is highly significant (P < 0.0001). In order to visualize these results, ASRs and AATs were distributed in eight quartiles and then plotted in global maps, presenting each gender separately (fig. 2A). It is obvious that the ASR pattern follows the AAT one. Since here it is speculated an extreme environment—extreme cancer rate relationship, a further statistical analysis was attempted by comparing ASR means of the countries belonging to the two 25% AAT extremes (table 1). ASR means of the ten most frequent cancer types were compared between 45 countries (AAT: −5.1 to 10.6 °C) and 43 countries (AAT: 25.1–28.3 °C) by independent t-test. All means differ significantly (highest association: lung cancer P = 8.80×10−18, bladder cancer P = 3.54×10−14, colorectal cancer P = 7.28×10−13), except ones of liver cancer. On the contrast, comparison for cervix cancer was significant for the opposite effect (high AAT—high ASR). Results for cervix cancer is quite logical, since this cancer is highly related with infectious factors (HPV viruses) that thrive in warm countries and especially in Africa (de Martel et al. 2017).
Fig. 2.
(A) ASR values of certain cancers and average annual temperatures plotted in Earth maps, aside to linear regression plots. (B) Linear regression plots, separately for each gender, for colon cancer and for all cancers. (C) Linear regression plots showing i) the high linearity between average annual temperatures (Celsius degrees) and extreme low temperatures (Celsius degrees) and ii) significant correlation between cancer ASR values and extreme low temperatures. ASR values were plotted by ascending order of eight quartiles (the darker the color the higher the incidence, quartiles can be found in supplementary table S1, Supplementary Material online). Greenland was included in the maps according cancer ASRs reported by Young et al. (2016). Average annual temperatures were plotted according an inverse ascending order (the darker the color the lower the temperature) of eight quartiles. Only countries reported by GLOBOCAN-2012 were included to linear regression analyses. Adjusted R2 is above of each regression plot (both gender included for “all cancers” and “colorectal cancer” plots). Prediction probability is highly significant for all regression analyses.
Table 1.
Age-Standardized Rate Mean Comparison (independent t-test, two-tailed) between Cold and Warm Countries, for the Ten Most Frequent Cancer Types Worldwide, Plus Gallbladder and Testis Cancera.
| Cancer Site | Countries Number (AAT range in °C) | Mean ASR | SD | [95% CI] | t Value | P Value |
|---|---|---|---|---|---|---|
| All cancers | 45 (−5.1 to 10.6) | 240.3644 | 61.9635 | 221.7485 258.9803 | 10.8334 | 9.49×10−18 |
| 43 (25.1 to 28.3) | 118.2488 | 41.21378 | 105.5651 130.9326 | |||
| Lung | 45 (−5.1 to 10.6) | 28.65778 | 9.817505 | 25.70827 31.60728 | 10.8496 | 8.80×10−18 |
| 43 (25.1 to 28.3) | 7.890698 | 7.998982 | 5.428974 10.35242 | |||
| Breast | 45 (−5.1 to 10.6) | 61.27778 | 26.92922 | 53.18735 69.36821 | 5.3538 | 7.06×10−7 |
| 43 (25.1 to 28.3) | 36.22326 | 15.04381 | 31.59346 40.85306 | |||
| Colorectum | 45 (−5.1 to 10.6) | 25.96 | 10.58173 | 22.7809 29.1391 | 8.4250 | 7.28×10−13 |
| 43 (25.1 to 28.3) | 9.402326 | 7.523186 | 7.087031 11.71762 | |||
| Prostate | 45 (−5.1 to 10.6) | 54.39333 | 38.2683 | 42.89626 65.8904 | 4.2772 | 4.88×10−5 |
| 43 (25.1 to 28.3) | 24.4814 | 25.84096 | 16.52872 32.43407 | |||
| Stomach | 45 (−5.1 to 10.6) | 11.00444 | 6.338876 | 9.100035 12.90885 | 6.2743 | 1.37×10−8 |
| 43 (25.1 to 28.3) | 4.52093 | 2.445848 | 3.768209 5.273651 | |||
| Liver | 45 (−5.1 to 10.6) | 6.624444 | 11.51497 | 3.164963 10.08393 | −1.1090 | 0.2705 |
| 43 (25.1 to 28.3) | 8.823256 | 6.17016 | 6.924361 10.72215 | |||
| Cervix | 45 (−5.1 to 10.6) | 13.74889 | 6.854846 | 11.68947 15.80831 | −4.1028 | 9.24×10−5 |
| 43 (25.1 to 28.3) | 22.62791 | 12.71393 | 18.71514 26.54068 | |||
| Esophagus | 45 (−5.1 to 10.6) | 4.097778 | 3.434153 | 3.066044 5.129511 | 3.3467 | 1.21×10−3 |
| 43 (25.1 to 28.3) | 2.023256 | 2.223772 | 1.33888 2.707632 | |||
| Bladder | 45 (−5.1 to 10.6) | 8.473333 | 3.937027 | 7.290519 9.656147 | 9.0700 | 3.54×10−14 |
| 43 (25.1 to 28.3) | 2.660465 | 1.501609 | 2.198338 3.122592 | |||
| Non-Hodgkin lymphoma | 45 (−5.1 to 10.6) | 6.2 | 3.370932 | 5.18726 7.21274 | 3.8072 | 2.63×10−4 |
| 43 (25.1 to 28.3) | 4.027907 | 1.658646 | 3.517451 4.538363 | |||
| Gallbladder | 45 (−5.1 to 10.6) | 1.935556 | 1.546279 | 1.471002 2.400109 | 4.7545 | 7.92×10−6 |
| 43 (25.1 to 28.3) | 0.7186047 | 0.666996 | 0.513334 0.923876 | |||
| Testis | 45 (−5.1 to 10.6) | 5.293333 | 3.571631 | 4.220297 6.36637 | 8.8007 | 1.25×10−13 |
| 43 (25.1 to 28.3) | 0.465116 | 0.433086 | 0.331832 0.598401 |
Note.—Italic P values indicate statistical significance. Degrees of freedom: 86.
Cold countries are termed these of the first 25% percentile of average annual temperature (AAT) and the warm ones those of the last 25% percentile of AAT distribution.
An additional linear regression analysis was performed, using this time extreme low temperatures (ELT) data. Since such data do not exist for all countries (data were found for 86 countries), a linear regression was performed for AAT and ELT values (fig. 2C). The model is highly linear and predictive, this showing that ELT values follow closely the AAT ones. That means that AAT data can be used safely for extracting conclusions for extreme environmental conditions. Additionally, linear regression analyses were performed for ELT versus ASR/all cancers, ASR/colorectal cancer, and ASR/breast cancer. Adjusted R2 were smaller than the AAT ones (this is because less countries were included), but the models were significant.
GWAS and Populations in Extreme Cold Environments
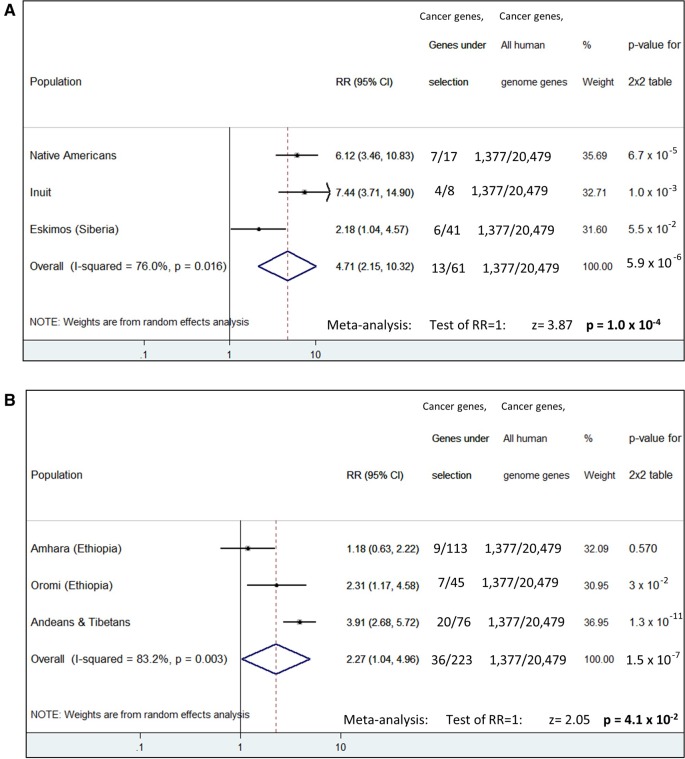
Speculating an evolutionary mechanism, genetic results for population living in extreme cold conditions were collected. Three lists of genes under selection were adopted from three evolutionary GWAS, Native Americans (Amorim et al. 2017), Inuit (Fumagalli et al. 2015), and Siberian Eskimos (Cardona et al. 2014), all abbreviated as “COLD.” Additionally, 240 cancer GWAS (see Materials and Methods for details) were combined to create a reliable list of cancer-associated genes (CAG). The main aim here was to investigate if genes found to be under selection for cold environments, predispose for cancer too. This is coming out by comparing the list of genes under selection with the CAG one, for any genes in common. Results are shown in figure 3. Population analysis, one by one, gives statistically significant results (P = 6.7×10−5, P = 6.7×10−3, P = 0.055, respectively). By adding all genes together from all three populations (overall analysis), significance is P = 5.9×10−6. By a meta-analysis approach, significance is P = 1.0×10−4. Part of the significance for Native Americans and Inuit is derived from a positive signal on chromosomal region 11q12.2, where the genes FADS1 and FADS2 are located. These genes encode for fatty acid desaturases implicated to lipids metabolism, which is considered a highly important pathway for northern populations (Fumagalli et al. 2015; Amorim et al. 2017). On the other hand, certain alleles on these genes have been found to predispose for colorectal cancer in East Asians (Zhang et al. 2014). This is in accordance to the fact that Native Americans (Athabascans) and Inuit have the highest incidence of colorectal cancer in the world (Young et al. 2016) (fig. 1D).
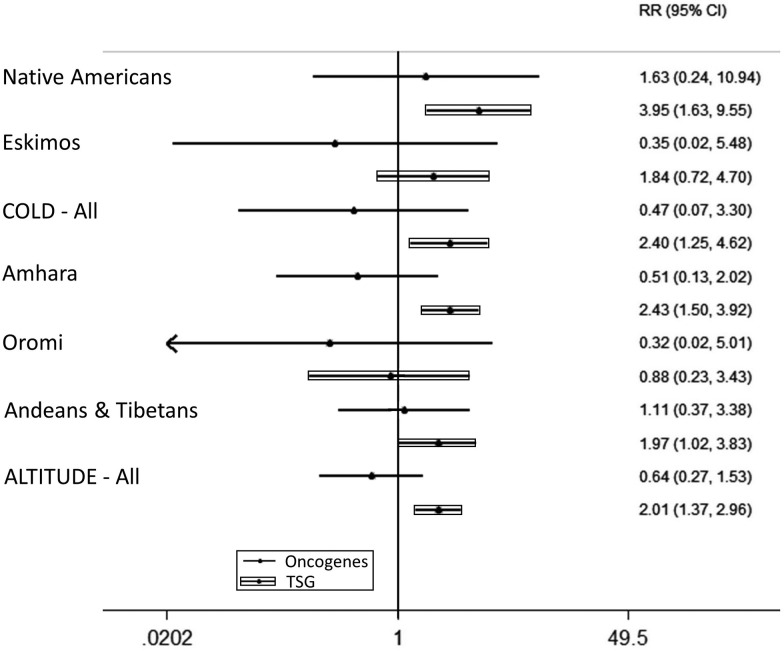
Fig. 3.
Forest plot showing statistical analysis for enrichment of genes under selection with cancer associated genes. Results are presented for each population separately, for an overall analysis and for a meta-analysis. (A) COLD populations and (B) ALTITUDE populations. Analysis was based on list comparisons that are included in supplementary table S2, Supplementary Material online. The highest significances are observed for Native Americans, Tibetans-Andeans, and for overall COLD. RR, risk ratio.
CAG and Populations in Extreme High-Altitude Environments
Under the concept of extreme environment—extreme cancer risk, human populations adapted at extreme high altitude were investigated using the same approach as above. These populations are Amhara and Oromi, the major part of population of Ethiopia, Aymara Indians in Andes (Andeans), and Tibetans (all three abbreviated as “ALTITUDE”). Are these populations at extreme cancer risk? Unfortunately not any analytical cancer incidence data are public available for Tibet (part of China). Regarding Aymara, the only detailed published cancer study is for La-Paz, the highest altitude town in the world. La-Paz is inhabited mainly by Aymara Indians. Data published on 1981 show that Aymara are at extreme risk for gallbladder and testicular cancer (Rios-Dalenz et al. 1981) (fig. 1E and F), cancers with significant genetic component (Mhatre et al. 2017; Wang et al. 2017), and cancers found to be highly associated with low AAT (table 1). High-altitude environments are also considered as extreme cold environments. A recent archeological study revealed that Andes mountains were inhabited by people much more longer than previously thought, ∼8,000 years ago (Randall et al. 2017). It is worthy to underline here that Athabascans and Norwegians are also at extreme risk for testicular cancer (Norway ranks presently first for testicular cancer according GLOBOCAN-2012) (fig. 1F). Ethiopia cancer ASR data are in the same line of research hypothesis. All cancer cases of Ethiopia that are registered in GLOBOCAN-2012, are coming from the only cancer center of the capital town, Addis-Abeba. The major part of patients of this center is of Amhara or Oromi ethnicity (Abate et al. 2016). Analysis showed that Ethiopia ranks 6th for breast cancer (fig. 1A) and 12th for colorectal cancer (data not shown), out of 54 Africa countries.
A number of evolutionary GWAS studies exist for ALTITUDE populations (Bigham et al. 2010; Simonson et al. 2010; Peng et al. 2011; Xu et al. 2011; Alkorta-Aranburu et al. 2012; Scheinfeldt et al. 2012; Huerta-Sanchez et al. 2013; Tekola-Ayele et al. 2015; Valverde et al. 2015). All available GWAS for Amhara and Oromi were combined (Alkorta-Aranburu et al. 2012; Scheinfeldt et al. 2012; Huerta-Sanchez et al. 2013). A single study was taken into account for Andeans-Tibetans (Foll et al. 2014), due to the highly reliable “convergent evolution” method that was used by the authors for identifying genes under selection. As in COLD populations, list of genes under selection were compared for any common genes with CAG list. This analysis gave a significant result for Oromi (P = 0.03), a highly significant result for Andeans-Tibetans (P = 1.3×10−11) and not significance at all for Amhara (fig. 3). Due to the nonsignificance of Amhara, meta-analysis of all three ALTITUDE populations was marginally significant (fig. 3). On the other hand, an overall analysis was highly significant (P = 1.5×10−7). A possible cause for the nonsignificance for Amhara is that Ethiopians are rarely part of cancer GWAS.
Housekeeping Genes versus CAG
In order to increase reliability of the above results, a separate control experiment was done. A complete list of housekeeping genes was used (Eisenberg and Lavanon 2013) for performing similar comparisons as described earlier for CAG. This list (3,800 genes) was first compared for common genes with CAG. In total, 175 genes out of the 1,377 CAG were found to be housekeeping genes. This is just a percentage of 12.7% in comparison with the percentage (18.6%) of all housekeeping genes in the human genome. That means that housekeeping genes are underrepresented (P = 5.19×10−8) in the list of CAG, so they can serve as a separate independent and reliable comparison with genes under selection for cold and high-altitude. Analysis was performed under the same logic as it was described earlier. This can be found in table 2. Not any statistical significance came out after this comparison, that meaning that genes under selection are not significantly enriched with housekeeping genes. This result gives an increased confidence to the result that a significant number of CAG have undergone selection procedures in the studied populations.
Table 2.
GUS in COLD and ALTITUDE Populations Were Statistically Tested for Containing HG or CAG.
| Population | HG/HGG | HG/GUS | RR | P value | CAG/HGG | CAG/GUS | RR | P value |
|---|---|---|---|---|---|---|---|---|
| COLD | ||||||||
| Native Americans | 3,800/20,479 | 3/17 | 0.77 | 1.00 | 1,377/20,479 | 7/17 | 6.12 | 6.7×10−5 |
| Inuit | 3,800/20,479 | 1/8 | 0.55 | 1.00 | 1,377/20,479 | 4/8 | 7.44 | 1.0×10−3 |
| Eskimos (Siberia) | 3,800/20,479 | 7/41 | 0.75 | 1.00 | 1,377/20,479 | 6/41 | 2.18 | 5.5×10−2 |
| All | 3,800/20,479 | 10/61 | 0.72 | 0.664 | 1,377/20,479 | 13/61 | 4.71 | 5.9×10−6 |
| ALTITUDE | ||||||||
| Amhara | 3,800/20,479 | 16/113 | 0.62 | 0.297 | 1,377/20,479 | 9/113 | 1.18 | 0.570 |
| Oromi | 3,800/20,479 | 7/45 | 0.68 | 0.704 | 1,377/20,479 | 7/45 | 2.31 | 3.0×10−2 |
| Andeans-Tibetans | 3,800/20,479 | 18/76 | 1.04 | 0.251 | 1,377/20,479 | 20/76 | 3.91 | 1.3×10−11 |
| All | 3,800/20,479 | 41/223 | 0.81 | 0.948 | 1,377/20,479 | 36/223 | 2.27 | 1.5×10−7 |
Note.—Results show significant enrichment of genes under selection with cancer associated genes but no with housekeeping genes (Fisher’s exact test was used for counts <10). HG, housekeeping genes; GUS, genes under selection; CAG, cancer-associated genes; HGG, human genome genes; RR, risk ratio.
Italic P values indicate statistical significance.
All related gene lists and analyses can be found in supplementary tables S2 and S3, Supplementary Material online.
Oncogenes and Tumor Suppressor Genes
In order to investigate which cancer pathways have been favored by evolution in COLD and ALTITUDE populations, a separate analysis was performed for oncogenes and tumor suppressor genes. A number of 724 protein coding oncogenes were downloaded from the oncogene database http://ongene.bioinfo-minzhao.org/ (Liu et al. 2017) and 1,038 protein coding tumor suppressor genes were downloaded from the tumor suppressor gene database https://bioinfo.uth.edu/TSGene/ (Zhao et al. 2016). Analysis was performed under the same way with housekeeping genes and CAG (supplementary table S4, Supplementary Material online). Interestingly, only tumor suppressor genes were found to be under selection in COLD (P = 1.2×10−2) and ALTITUDE (P = 1.3×10−5) populations (table 3). Significance is somewhat lower than the CAG one, since now the analysis was split in two cancer genes categories and due to the fact that the CAG list contains genes that have not yet been categorized as oncogenes or tumor suppressor genes. On the other hand, risk ratios show clearly the overrepresentation of tumor suppressor genes (compared with oncogenes) in genes under selection (fig. 4). The fact that there is a preferential selection for tumor suppressor genes is very important, showing a potential survival advantage for organisms living in very cold and high-altitude environments. Such an advantage can be apoptosis resistance, to escape cell death under extreme environmental conditions. In addition, this mechanism has been proved through functional studies by other research teams (see below).
Table 3.
GUS in COLD and ALTITUDE Populations Were Statistically Tested for Containing ONC or TSG.
| Population | ONC/HGG | ONC/GUS | RR | P value | TSG/HGG | TSG/GUS | RR | P value |
|---|---|---|---|---|---|---|---|---|
| COLD | ||||||||
| Native Americans | 724/20,479 | 1/17 | 1.63 | 0.458 | 1,038/20,479 | 4/17 | 3.95 | 9.0×10−3 |
| Inuit | 724/20,479 | 0/8 | 1.62 | 1.00 | 1,038/20,479 | 0/8 | 1.15 | 1.00 |
| Eskimos (Siberia) | 724/20,479 | 0/41 | 0.35 | 1.00 | 1,038/20,479 | 4/41 | 1.84 | 0.153 |
| All | 724/20,479 | 1/61 | 0.47 | 0.726 | 1,038/20,479 | 8/61 | 2.40 | 1.2×10−2 |
| ALTITUDE | ||||||||
| Amhara | 724/20,479 | 2/113 | 0.51 | 0.443 | 1,038/20,479 | 15/113 | 2.43 | 7.8×10−5 |
| Oromi | 724/20,479 | 0/45 | 0.32 | 1.00 | 1,038/20,479 | 2/45 | 0.88 | 1.00 |
| Andeans-Tibetans | 724/20,479 | 3/76 | 1.11 | 0.752 | 1,038/20,479 | 8/76 | 1.97 | 0.059 |
| All | 724/20,479 | 5/223 | 0.64 | 0.364 | 1,038/20,479 | 24/223 | 2.01 | 1.3×10−5 |
Note.—Results show preferential selection of tumor suppressor genes and not oncogenes (Fisher’s exact test was used for counts <10). ONC, oncogenes; GUS, genes under selection; TSG, tumor suppressor genes; HGG, human genome genes; RR, risk ratio.
Italic P values indicate statistical significance.
Fig. 4.
Forest plot showing content of genes under selection with oncogenes and tumor suppressor genes (TSG). It is obvious that in all populations there is a positive trend over TSG in comparison with oncogenes (Inuit were not included on this plot since zero genes were found in both gene categories). RR, risk ratio.
Analysis by DAVIDv.6.8 Software
Aiming at further confirmation of the described results, the online software DAVIDv.6.8 was used to analyze gene lists under selection of COLD and ALTITUDE populations. DAVIDv.6.8 is connected with Genetic Association Database (GAD) and P values are provided for genes that are associated with specific diseases of the database. Analytical disease output is found in table 4 and supplementary table S5, Supplementary Material online. Cancer findings are in accordance with the previously presented results: Colorectal cancer for Natives Americans, Colorectal cancer/Esophageal cancer/Lung cancer for Siberian Eskimos, not any association for Amhara (as before), leukemia for Oromi, and a variety of cancers for Andeans–Tibetans. Justification of these results: 1) Natives Americans rank first together with Inuit for colorectal cancer incidence (fig. 1D); 2) esophageal cancer and Lung cancer are indeed major cancer types for Siberian Eskimos (Zaridze et al. 1993); 3) regarding Amhara and Oromi, Ethiopia ranks third in Africa for leukemia, according GLOBOCAN-2012 (fig. 1B); and 4) due to limited data for cancer incidence in Andeans–Tibetans, results for this data set are difficult to be interpreted.
Table 4.
Cancer Types Found to be Associated (Fisher’s exact test) with Genes under Selection in COLD and ALTITUDE Populations.
| Population | Cancers Found to be Significant | P Values (range) |
|---|---|---|
| Native Americansa (Amorim et al. 2017) | Colorectal cancer | 9.8×10−3 |
| Inuit—Greenland (Fumagalli et al. 2015) | Not any | – |
| Note: The most frequent BRCA1 mutation worldwide (founder mutation) was found in Inuit (Harboe et al. 2009) | – | |
| Eskimos—Siberiab (Cardona et al. 2014) | Colorectal cancer, esophageal cancer, lung cancer, head and neck cancer, breast cancer, bladder cancer, hepatocellular carcinoma, lymphoma | 1.9×10−4–3.9×10−2 |
| Amhara—Ethiopia ( Alkorta-Aranburu et al. 2012; Scheinfeldt et al. 2012; Huerta-Sanchez et al. 2013) | Not any | – |
| Oromi—Ethiopiac (Alkorta-Aranburu et al. 2012; Huerta-Sanchez et al. 2013) | Chronic myelogenous leukemia, acute lymphoblastic leukemia, myeloid leukemia | 1.5×10−4–1.5×10−3 |
| Andeans—Tibetans (Foll et al. 2014) | Esophageal cancer, head and neck cancer, squamous cell carcinoma, stomach cancer, breast cancer, melanoma, Hodgkin disease, mouth neoplasms, pharyngeal neoplasms, nasopharyngeal neoplasms, precursor cell lymphoblastic leukemia, adenoma, colorectal cancer, uterine cervical neoplasms, mesothelioma, pleural neoplasms, laryngeal neoplasm, prostate cancer, lung cancer | 2.4×10−7–2.4×10−2 |
| Note: Variants in EGLN1 (gene under selection in high altitude) predispose for lung cancer in Tibetans (Lanikova et al. 2017) | – |
Note.—Data retrieved from Genetic Association Database (GAD) through DAVIDv.6.8 software (analysis details in supplementary table S5, Supplementary Material online).
Native Americans (Athabascans) rank at the first place worldwide (together with Inuit) for colorectal cancer incidence (fig. 1).
Siberian Eskimos have very high rates of lung and esophagus cancer (Zaridze et al. 1993).
Ethiopians rank at the third place in Africa countries for leukemia incidence (fig. 1).
Italic P values indicate statistical significance.
Role of Apoptosis Genes and DNA Repair Genes at Extreme Environmental Conditions
It is useful to discuss here that the most frequent BRCA1 mutation worldwide (founder mutation) has been found in Greenlandic Inuit population (Harboe et al. 2009). This maybe a coincidence, but evolutionary forces could be hypothesized as well. Additionally, variants in EGLN1 gene, one of the most important genes under selection in high altitude (Bigham et al. 2010; Simonson et al. 2010; Yi et al. 2010; Lorenzo et al. 2014; Bigham 2016; Peng et al. 2017; Tashi et al. 2017), have been recently associated with lung cancer in Tibetans (Lanikova et al. 2017). The last study is a direct connection of a gene under selection with cancer susceptibility, in a high-altitude population.
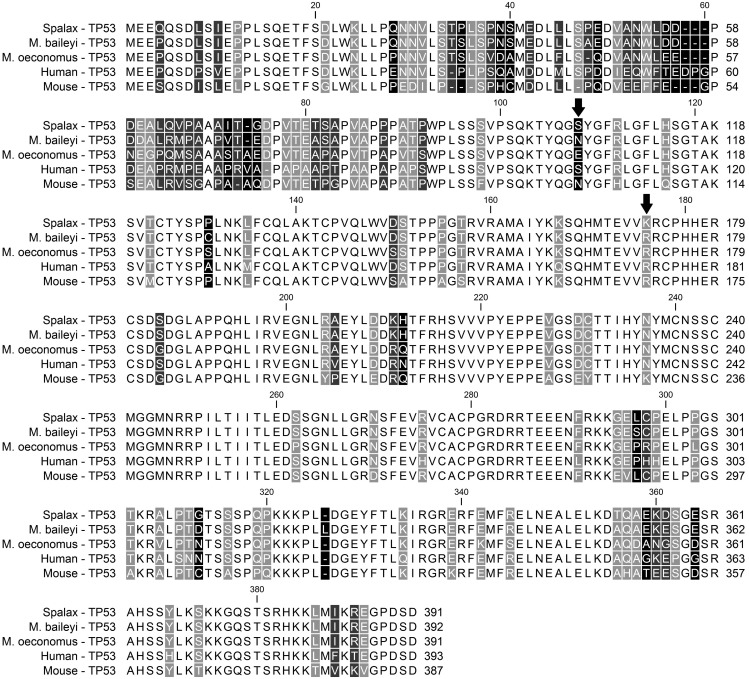
Colorectal cancer, breast cancer, and leukemia, are cancers that are closely related with apoptosis and DNA repair procedures (Jeggo et al. 2016). It is well known for unicellular organisms (bacteria, protozoans, and fungi), that randomly appeared mutations in DNA repair genes can convert them in “mutator” strains (LeClerc et al. 1996; Notley-McRobb et al. 2002; Lujan et al. 2011; Byrne et al. 2014; Bui et al. 2015; Grazielle-Silva et al. 2015; Healey et al. 2016). These strains are resistant in a variety of adverse environments. Same genes cause colorectal or breast cancer in humans (Jeggo et al. 2016) (table 5). In addition, published evidence shows that reduced apoptosis potential (certain alleles in TP53 gene) helps mammals to survive in cold and high-altitude environments (Ashur-Fabian et al. 2004; Zhao et al. 2013) (table 5). Interestingly, two mammals living at the Tibet plateau, Myospalax baileyi and Microtus oeconomus, have a certain amino acid in their p53 protein at the same position where a germ-line mutation was reported in a patient with multiple primary cancers. The authors (Zhao et al. 2013) found evidence that this amino acid in those two mammals is needed for divergent responses of IGFBP3 and Apaf1 to hypoxia and cold stresses. Ashur-Fabian et al. (2004) found similar evidence for a certain amino acid position in Spalax, contributing to hypoxia adaptation of this mammal. This amino acid is found in a highly conserved DNA-binding domain of TP53 (fig. 5) that it is a hotspot in cancer mutagenesis. This amino acid is reported to be affected in 57 different human tumors of various types (Ashur-Fabian et al. 2004). Results of those two studies were combined and are presented in figure 5. It is obvious, that these studies can be considered as direct functional justification of the results presented here. Additionally, these data are in perfect match with the finding that that tumor suppressor genes are under selection in COLD and ALTITUDE human populations. It is useful to remind that many cancers that rank high in COLD and ALTITUDE populations, are related with tumor suppressor genes. Gene Ontology analysis with DAVIDv.6.8 for genes under selection in COLD and ALTITUDE populations (presented in supplementary table S5, Supplementary Material online), revealed certain cell cycle and apoptosis biological procedures (BP) in Native Americans, Amhara, and Andeans-Tibetans.
Table 5.
Human Homologue Cancer Genes That Confer Adaptive Advantage to Certain Organisms.
| Organism That the Cancer Gene Was Found Mutated | Human Homologous Gene | Gene Function | Survival Advantage (selected variants) | Study |
|---|---|---|---|---|
| Prokaryotic | ||||
| Escherichia coli, Salmonella enterica | MSH2 | DNA Mismatch Repair | Antibiotic resistance | LeClerc et al. (1996), and many other studies |
| Pseudomonas aeruginosa | MSH2 | DNA Mismatch Repair | Increased adaptation in biofilms | Lujan et al. (2011), and other studies |
| Escherichia coli | RAD51 | DNA homologous recombination | Ionizing radiation resistance | Byrne et al. (2014) |
| Escherichia coli | MUTYH | DNA glycosylase (DNA repair) | Survival at nutrient limitation | Notley-McRobb et al. (2002) |
| Eucaryotic—fungi and protozoans | ||||
| Candida glabrata | MSH2 | DNA Mismatch Repair | Drug resistance | Healey et al. (2016) |
| Trypanosoma brucei | MSH2 | DNA Mismatch Repair | Adaptation to oxidative stress | Grazielle-Silva et al. (2015) |
| Sacharomyces cerevisiae | MSH2, PMS1 | DNA Mismatch Repair | Adaptation to stress conditions | Bui et al. (2015) |
| Eucaryotic—mammals | ||||
| Spalax ehrenbergi | TP53 | Stress response (DNA damage, hypoxia) leading to growth arrest and apoptosis | Hypoxia stress tolerance | Ashur-Fabian et al. (2004) |
| Myospalax baileyi, Microtus oeconomus | TP53 | Stress response (DNA damage, hypoxia) leading to growth arrest and apoptosis | Survival to hypoxia and cold (Tibet) | Zhao et al. (2013) |
Fig. 5.
Multiple alignment for TP53 protein for five species. The two critical amino-acid positions contributing to extreme environmental adaptation (Ashur-Fabian et al. 2004; Zhao et al. 2013) are shown with black arrows (lysine-176 for Spalax, asparagine-108, and glutamic acid-108 for Myospalax baileyi and Microtus oeconomus, respectively).
Conclusion
Concluding, findings of this study provide evidence that genetic variants found to be beneficial in extreme environments, can also predispose for cancer. This can be considered as an antagonistic pleiotropy phenomenon. Cell resistance at low temperatures and high altitude, it probably increases probability for malignancy. This effect hardly could be filtered out by natural selection since most cancers appear late on age, after most people have their children. It is also tempting to hypothesize that similar events of the past, contributed to rise of cancer rates in all human populations today. Future research may clear this out.
Materials and Methods
Worldwide Cancer Incidence and Temperature Data
Cancer incidence data (ASR) for 184 countries and for different cancer types were adapted from GLOBOCAN-2012 (Ferlay et al. 2015) (http://globocan.iarc.fr). Cancer incidence for populations living at extreme cold environments (Athabascans and Circumpolar Inuit) was adapted by Young et al. (2016). Unfortunately, recent cancer incidence data do not exist for Andeans and Tibetans living at extreme high-altitude environments. The only available study is by Rios-Dalenz et al. (1981) referring analytical ASR cancer data for Aymara Indians living in La Paz, Bolivia (highest altitude town in the world, in Andes, 4,000 meters above the sea level).
AAT by country (years 1961–1990) are based on gridded climatologies from the Climatic Research Unit (http://www.cru.uea.ac.uk/). Extreme low-temperature data for 86 countries were retrieved from Arizona State University climate extremes archive (https://wmo.asu.edu/) and from reliable sources that are referred in List of weather records (https://en.wikipedia.org/).
GLOBOCAN-2012, AAT, and ELT data are listed in supplementary table S1, Supplementary Material online. ASR is termed “new cases/time period/100,000 individuals.” ASR data of GLOBOCAN-2012 are recorded for year 2012.
Cancer Genes’ Data Set
NHGRI-EBI GWAS catalog (MacArthur et al. 2017) (https://www.ebi.ac.uk/gwas/) was used for downloading all genes that have been associated with cancer through GWAS studies (supplementary table S2, Supplementary Material online) since May of 2017. NHGRI-EBI GWAS catalog is a continuously updating GWAS database. Keyword “cancer” and genetic association with P ≤ 5×10−8 were used as filtering criteria for retrieving all GWAS cancer association studies that are archived in NHGRI-EBI GWAS catalog. Totally 240 GWAS studies were included for analysis (supplementary table S2, Supplementary Material online). All cancer association studies were downloaded except cervix cancer, due to its high correlation with infectious factors. GWAS for drug response and disease progression were excluded too. In order to create a “CAG” list, genes termed as “mapped” or “reported” by the NHGRI-EBI GWAS catalog, were included for listing. Genes in duplicate (same in different studies) were excluded in order to have only unique gene names. CAG list can be found to supplementary table S2, Supplementary Material online. Nonprotein coding genes “MIR” and “LINC” and genes of uncertain importance or existence (“LOC”) were not taken into account for the statistical analysis. Despite this, readers can find them listed in supplementary table S2, Supplementary Material online.
A separate analysis was performed for protein coding oncogenes and tumor suppressor genes. Oncogenes list was downloaded from oncogene database (Liu et al. 2017), http://ongene.bioinfo-minzhao.org/, and tumor suppressor genes were downloaded from tumor suppressor gene database (Zhao et al. 2016), https://bioinfo.uth.edu/TSGene/. All related analyses can be found in supplementary table S4, Supplementary Material online.
Housekeeping Genes
A separate analysis was performed with housekeeping genes, serving as a control experiment. Housekeeping genes’ list was adapted by Eisenberg and Lavanon (2013). This list (3,800 genes) is based on analysis of next-generation sequencing (RNA-seq) data. As the authors state, at least one variant of these genes is expressed in all tissues uniformly. Analysis was performed by comparing this list for any common genes with genes under selection, under the same logic that it is described below. The housekeeping genes’ list and all comparisons performed can be found in supplementary table S3, Supplementary Material online.
Genes under Selection in Extreme Cold and High-Altitude Environments
A number of GWAS exist, indicating genes under selection for populations living at extreme cold and high-altitude environments. These populations are: Inuit (cold), Eskimos (cold), Native Americans (cold), Tibetans (high-altitude), Andeans (high-altitude), and Ethiopians (high-altitude). For creating lists of genes under selection (supplementary table S2, Supplementary Material online) for extreme cold and high-altitude populations, seven GWAS studies were taken into account, as justified below.
Data and studies that were analyzed for “COLD” group of populations are: 1) First Americans (Native American populations living or their close ancestors used to live at extreme cold environments): Table S2 of genes of Amorim et al. (2017), 2) Greenlandic Inuit: Table S2 of genes of Fumagalli et al. (2015), and 3) Siberian Eskimos: Table S6 of genes (only those being in 0.1% of PBS values) of Cardona et al. (2014).
Data and studies that were analyzed for the “ALTITUDE” group of populations are: 1) Amhara population (Ethiopians living in high altitude): Table S7 of genes of Scheinfeldt et al. (2012), Table S23 of genes of Alkorta-Aranburu et al. (2012), and Table S4 of genes of Huerta-Sanchez et al. (2013), 2) Oromi population (Ethiopians living in high altitude): Table S24 of genes of Alkorta-Aranburu et al. (2012), and Table S5 of genes of Huerta-Sanchez et al. (2013), and 3) Andeans and Tibetans: Table S1 of genes of Foll et al. (2014). The last study was considered as the most reliable for Andeans and Tibetans since a convergent evolution model of analysis was taken into account, giving results of high confidence.
Every excel sheet of supplementary table S2, Supplementary Material online, contains the CAG list, created by combining all GWAS studies, the list of genes under selection of each study or population and the ENSG code of each gene under selection. CAG list contains 1,377 genes. Comparison of the CAG list with each list of selected genes was performed though the “duplicate” function of Microsoft Excel 2016 and genes in common are highlighted. The same approach has been followed for housekeeping genes, oncogenes, and tumor suppressor genes.
Statistical approach is a 2×2 table: CAG (1,377) out of all genes (Howe et al. 2013) of human genome (20,479) versus (n) cancer genes found in (z) genes under selection.
Statistical Analysis and Multiple Alignment
All statistical analysis needed for this work was performed though the statistical package STATAv.13 (StataCorp LLC, TX). Basic statistical analysis included univariate linear regression, Pearson χ2 for 2×2 tables, Fisher’s exact test for 2×2 tables including counts <10, independent t-test (two-tailed), quartile analysis, and bar plots for cancer ASR presentation of various populations. Significant level alpha was set to 0.05.
Meta-analysis was performed through the DerSimonian and Laird random effects method in STATAv.13, using the metan algorithm according to Harris et al. (2008).
Software DAVIDv6.8 (Huang da et al. 2009a, b) was applied for analyzing gene lists under selection (supplementary table S2, Supplementary Material online). Analysis done by DAVIDv6.8: 1) association with diseases registered in GAD-Genetic Association Database (supplementary table S5, Supplementary Material online) and 2) analysis for any significance with biological procedures (BP) according Gene Ontology (GO) mappings (directly annotated by the source database) (supplementary table S5, Supplementary Material online).
Paintmaps, a free online map generating tool (http://www.paintmaps.com/) was used for creating global grading Earth maps according AAT and ASR quartile data.
Multiple alignment for TP53 was performed through the CLC Main Workbench 7 software (Qiagen, Aarchus, Denmark).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgment
I would like to thank Dr George Nikolopoulos for advising on epidemiology matters.
References
- Abate Y, Yilma Z, Assefa M, Tigeneh W.. 2016. Trends of breast cancer in Ethiopia. Int J Cancer Res Mol Mech. 2:1–5. [Google Scholar]
- Alkorta-Aranburu G, Beall CM, Witonsky DB, Gebremedhin A, Pritchard JK, Di Rienzo A.. 2012. The genetic architecture of adaptations to high altitude in Ethiopia. PLoS Genet. 812:e1003110.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amorim CE, Nunes K, Meyer D, Comas D, Bortolini MC, Salzano FM, Hunemeier T.. 2017. Genetic signature of natural selection in first Americans. Proc Natl Acad Sci U S A. 1149:2195–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amundadottir LT, Thorvaldsson S, Gudbjartsson DF, Sulem P, Kristjansson K, Arnason S, Gulcher JR, Bjornsson J, Kong A, Thorsteinsdottir U, et al. 2004. Cancer as a complex phenotype: pattern of cancer distribution within and beyond the nuclear family. PLoS Med. 13:e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashur-Fabian O, Avivi A, Trakhtenbrot L, Adamsky K, Cohen M, Kajakaro G, Joel A, Amariglio N, Nevo E, Rechavi G.. 2004. Evolution of p53 in hypoxia-stressed Spalax mimics human tumor mutation. Proc Natl Acad Sci U S A. 10133:12236–12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigham A, Bauchet M, Pinto D, Mao X, Akey JM, Mei R, Scherer SW, Julian CG, Wilson MJ, Lopez HD, et al. 2010. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 69:e1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigham AW. 2016. Genetics of human origin and evolution: high-altitude adaptations. Curr Opin Genet Dev. 41:8–13.http://dx.doi.org/10.1016/j.gde.2016.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui DT, Dine E, Anderson JB, Aquadro CF, Alani EE.. 2015. A genetic incompatibility accelerates adaptation in yeast. PLoS Genet. 117:e1005407.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne RT, Klingele AJ, Cabot EL, Schackwitz WS, Martin JA, Martin J, Wang Z, Wood EA, Pennacchio C, Pennacchio LA, et al. 2014. Evolution of extreme resistance to ionizing radiation via genetic adaptation of DNA repair. Elife 3:e01322.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona A, Pagani L, Antao T, Lawson DJ, Eichstaedt CA, Yngvadottir B, Shwe MT, Wee J, Romero IG, Raj S, et al. 2014. Genome-wide analysis of cold adaptation in indigenous Siberian populations. PLoS One 95:e98076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AJ, Nguyen AQ.. 2011. Antagonistic pleiotropy as a widespread mechanism for the maintenance of polymorphic disease alleles. BMC Med Genet. 12:160.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi B, Summers K.. 2005. Evolutionary biology of cancer. Trends Ecol Evol. 2010:545–552.http://dx.doi.org/10.1016/j.tree.2005.07.007 [DOI] [PubMed] [Google Scholar]
- Day GE, Lanier AP, Bulkow L, Kelly JJ, Murphy N.. 2010. Cancers of the breast, uterus, ovary and cervix among Alaska Native women, 1974-2003. Int J Circumpolar Health. 691:72–86.http://dx.doi.org/10.3402/ijch.v69i1.17388 [DOI] [PubMed] [Google Scholar]
- de Martel C, Plummer M, Vignat J, Franceschi S.. 2017. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int J Cancer. 1414:664–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg E, Levanon EY.. 2013. Human housekeeping genes, revisited. Trends Genet. 2910:569–574.http://dx.doi.org/10.1016/j.tig.2013.05.010 [DOI] [PubMed] [Google Scholar]
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F.. 2015. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 1365:E359–E386. [DOI] [PubMed] [Google Scholar]
- Foll M, Gaggiotti OE, Daub JT, Vatsiou A, Excoffier L.. 2014. Widespread signals of convergent adaptation to high altitude in Asia and america. Am J Hum Genet. 954:394–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote M, Strickland R, Lucas-Pipkorn S, Williamson A, Lamers L.. 2016. The high burden of cancer among American Indians/Alaska natives in Wisconsin. WMJ 1151:11–16. [PubMed] [Google Scholar]
- Friborg JT, Melbye M.. 2008. Cancer patterns in Inuit populations. Lancet Oncol. 99:892–900.http://dx.doi.org/10.1016/S1470-2045(08)70231-6 [DOI] [PubMed] [Google Scholar]
- Fumagalli M, Moltke I, Grarup N, Racimo F, Bjerregaard P, Jorgensen ME, Korneliussen TS, Gerbault P, Skotte L, Linneberg A, et al. 2015. Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science 3496254:1343–1347. [DOI] [PubMed] [Google Scholar]
- Grazielle-Silva V, Zeb TF, Bolderson J, Campos PC, Miranda JB, Alves CL, Machado CR, McCulloch R, Teixeira SM.. 2015. Distinct phenotypes caused by mutation of MSH2 in trypanosome insect and mammalian life cycle forms are associated with parasite adaptation to oxidative stress. PLoS Negl Trop Dis. 96:e0003870.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harboe TL, Eiberg H, Kern P, Ejlertsen B, Nedergaard L, Timmermans-Wielenga V, Nielsen IM, Bisgaard ML.. 2009. A high frequent BRCA1 founder mutation identified in the Greenlandic population. Fam Cancer. 84:413–419. [DOI] [PubMed] [Google Scholar]
- Harris RJ, Deeks JJ, Altman DG, Bradburn MJ, Harbord RM, Sterne JAC.. 2008. metan: fixed- and random-effects meta-analysis. Stata J. 8:3–28. [Google Scholar]
- Healey KR, Zhao Y, Perez WB, Lockhart SR, Sobel JD, Farmakiotis D, Kontoyiannis DP, Sanglard D, Taj-Aldeen SJ, Alexander BD, et al. 2016. Prevalent mutator genotype identified in fungal pathogen Candida glabrata promotes multi-drug resistance. Nat Commun. 7:11128.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, et al. 2013. The zebrafish reference genome sequence and its relationship to the human genome. Nature 4967446:498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA.. 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA.. 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4:44–57. [DOI] [PubMed] [Google Scholar]
- Huerta-Sanchez E, Degiorgio M, Pagani L, Tarekegn A, Ekong R, Antao T, Cardona A, Montgomery HE, Cavalleri GL, Robbins PA, et al. 2013. Genetic signatures reveal high-altitude adaptation in a set of Ethiopian populations. Mol Biol Evol. 308:1877–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggo PA, Pearl LH, Carr AM.. 2016. DNA repair, genome stability and cancer: a historical perspective. Nat Rev Cancer. 161:35–42. [DOI] [PubMed] [Google Scholar]
- Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, Feuer EJ, Thun MJ.. 2005. Cancer statistics, 2005. CA Cancer J Clin. 551:10–30. [DOI] [PubMed] [Google Scholar]
- Lanikova L, Reading NS, Hu H, Tashi T, Burjanivova T, Shestakova A, Siwakoti B, Thakur BK, Pun CB, Sapkota A, et al. 2017. Evolutionary selected Tibetan variants of HIF pathway and risk of lung cancer. Oncotarget 87:11739–11747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClerc JE, Li B, Payne WL, Cebula TA.. 1996. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 2745290:1208–1211.http://dx.doi.org/10.1126/science.274.5290.1208 [DOI] [PubMed] [Google Scholar]
- Lemrow SM, Perdue DG, Stewart SL, Richardson LC, Jim MA, French HT, Swan J, Edwards BK, Wiggins C, Dickie L, et al. 2008. Gallbladder cancer incidence among American Indians and Alaska Natives, US, 1999-2004. Cancer 113(S5):1266–1273. [DOI] [PubMed] [Google Scholar]
- Liu Y, Sun J, Zhao M.. 2017. ONGene: a literature-based database for human oncogenes. J Genet Genomics. 442:119–121.http://dx.doi.org/10.1016/j.jgg.2016.12.004 [DOI] [PubMed] [Google Scholar]
- Lorenzo FR, Huff C, Myllymaki M, Olenchock B, Swierczek S, Tashi T, Gordeuk V, Wuren T, Ri-Li G, McClain DA, et al. 2014. A genetic mechanism for Tibetan high-altitude adaptation. Nat Genet. 469:951–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan AM, Macia MD, Yang L, Molin S, Oliver A, Smania AM.. 2011. Evolution and adaptation in Pseudomonas aeruginosa biofilms driven by mismatch repair system-deficient mutators. PLoS One 611:e27842.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, Junkins H, McMahon A, Milano A, Morales J, et al. 2017. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 45(D1):D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhatre S, Wang Z, Nagrani R, Badwe R, Chiplunkar S, Mittal B, Yadav S, Zhang H, Chung CC, Patil P, et al. 2017. Common genetic variation and risk of gallbladder cancer in India: a case-control genome-wide association study. Lancet Oncol. 184:535–544. [DOI] [PubMed] [Google Scholar]
- Moore SP, Forman D, Pineros M, Fernandez SM, de Oliveira Santos M, Bray F.. 2014. Cancer in indigenous people in Latin America and the Caribbean: a review. Cancer Med. 31:70–80.http://dx.doi.org/10.1002/cam4.134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesse RM. 2011. Ten questions for evolutionary studies of disease vulnerability. Evol Appl. 42:264–277.http://dx.doi.org/10.1111/j.1752-4571.2010.00181.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notley-McRobb L, Pinto R, Seeto S, Ferenci T.. 2002. Regulation of mutY and nature of mutator mutations in Escherichia coli populations under nutrient limitation. J Bacteriol. 1843:739–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Cui C, He Y, Ouzhuluobu, Zhang H, Yang D, Zhang Q, Bianbazhuoma, Yang L, Xiang K, et al. 2017. Down-regulation of EPAS1 transcription and genetic adaptation of Tibetans to high-altitude hypoxia. Mol Biol Evol. 34:818–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Yang Z, Zhang H, Cui C, Qi X, Luo X, Tao X, Wu T, Ouzhuluobu Basang, et al. 2011. Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Mol Biol Evol. 28:1075–1081. [DOI] [PubMed] [Google Scholar]
- Perdue DG, Haverkamp D, Perkins C, Daley CM, Provost E.. 2014. Geographic variation in colorectal cancer incidence and mortality, age of onset, and stage at diagnosis among American Indian and Alaska Native people, 1990-2009. Am J Public Health. 104(S3):S404–S414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall H, Stefanescu IC, Garcia-Putnam A, Aldenderfer MS, Clementz MT, Murphy MS, Llave CV, Watson JT.. 2017. Humans permanently occupied the Andean highlands by at least 7 ka. R Soc Open Sci. 4:170331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios-Dalenz J, Correa P, Haenszel W.. 1981. Morbidity from cancer in La Paz, Bolivia. Int J Cancer. 283:307–314.http://dx.doi.org/10.1002/ijc.2910280309 [DOI] [PubMed] [Google Scholar]
- Scheinfeldt LB, Soi S, Thompson S, Ranciaro A, Woldemeskel D, Beggs W, Lambert C, Jarvis JP, Abate D, Belay G, et al. 2012. Genetic adaptation to high altitude in the Ethiopian highlands. Genome Biol. 131:R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Verma HK, Joshi S, Panwar MS, Mandal CC.. 2015. A link between cold environment and cancer. Tumour Biol. 368:5953–5964. [DOI] [PubMed] [Google Scholar]
- Simonson TS, Yang Y, Huff CD, Yun H, Qin G, Witherspoon DJ, Bai Z, Lorenzo FR, Xing J, Jorde LB, et al. 2010. Genetic evidence for high-altitude adaptation in Tibet. Science 3295987:72–75. [DOI] [PubMed] [Google Scholar]
- Tashi T, Scott Reading N, Wuren T, Zhang X, Moore LG, Hu H, Tang F, Shestakova A, Lorenzo F, Burjanivova T, et al. 2017. Gain-of-function EGLN1 prolyl hydroxylase (PHD2 D4E: C127S) in combination with EPAS1 (HIF-2alpha) polymorphism lowers hemoglobin concentration in Tibetan highlanders. J Mol Med (Berl). 956:665–670. [DOI] [PubMed] [Google Scholar]
- Tekola-Ayele F, Adeyemo A, Chen G, Hailu E, Aseffa A, Davey G, Newport MJ, Rotimi CN.. 2015. Novel genomic signals of recent selection in an Ethiopian population. Eur J Hum Genet. 238:1085–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde G, Zhou H, Lippold S, de Filippo C, Tang K, Lopez Herraez D, Li J, Stoneking M.. 2015. A novel candidate region for genetic adaptation to high altitude in Andean populations. PLoS One 105:e0125444.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vittecoq M, Roche B, Daoust SP, Ducasse H, Misse D, Abadie J, Labrut S, Renaud F, Gauthier-Clerc M, Thomas F.. 2013. Cancer: a missing link in ecosystem functioning? Trends Ecol Evol. 2811:628–635. [DOI] [PubMed] [Google Scholar]
- Wang Z, McGlynn KA, Rajpert-De Meyts E, Bishop DT, Chung CC, Dalgaard MD, Greene MH, Gupta R, Grotmol T, Haugen TB, et al. 2017. Meta-analysis of five genome-wide association studies identifies multiple new loci associated with testicular germ cell tumor. Nat Genet. 497:1141–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G. 1957. Pleiotropy, natural selection, and the evolution of senescence. Evolution 114:398–411.http://dx.doi.org/10.1111/j.1558-5646.1957.tb02911.x [Google Scholar]
- Xu S, Li S, Yang Y, Tan J, Lou H, Jin W, Yang L, Pan X, Wang J, Shen Y, et al. 2011. A genome-wide search for signals of high-altitude adaptation in Tibetans. Mol Biol Evol. 282:1003–1011. [DOI] [PubMed] [Google Scholar]
- Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZX, Pool JE, Xu X, Jiang H, Vinckenbosch N, Korneliussen TS, et al. 2010. Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329:75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TK, Kelly JJ, Friborg J, Soininen L, Wong KO.. 2016. Cancer among circumpolar populations: an emerging public health concern. Int J Circumpolar Health. 75:29787.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaridze DG, Marochko A, Basieva T, Duffy SW.. 1993. Cancer incidence in the native peoples of far eastern Siberia. Int J Cancer 546:889–894.http://dx.doi.org/10.1002/ijc.2910540602 [DOI] [PubMed] [Google Scholar]
- Zhang B, Jia WH, Matsuda K, Kweon SS, Matsuo K, Xiang YB, Shin A, Jee SH, Kim DH, Cai Q, et al. 2014. Large-scale genetic study in East Asians identifies six new loci associated with colorectal cancer risk. Nat Genet. 466:533–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Kim P, Mitra R, Zhao J, Zhao Z.. 2016. TSGene 2.0: an updated literature-based knowledgebase for tumor suppressor genes. Nucleic Acids Res. 44(D1):D1023–D1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Ren JL, Wang MY, Zhang ST, Liu Y, Li M, Cao YB, Zu HY, Chen XC, Wu CI, et al. 2013. Codon 104 variation of p53 gene provides adaptive apoptotic responses to extreme environments in mammals of the Tibet plateau. Proc Natl Acad Sci U S A. 11051:20639–20644. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.