Abstract
Novel adaptations are generally assembled by co-opting pre-existing genetic components, but the factors dictating the suitability of genes for new functions remain poorly known. In this work, we used comparative transcriptomics to determine the attributes that increased the likelihood of some genes being co-opted for C4 photosynthesis, a convergent complex trait that boosts productivity in tropical conditions. We show that independent lineages of grasses repeatedly co-opted the gene lineages that were the most highly expressed in non-C4 ancestors to produce their C4 pathway. Although ancestral abundance in leaves explains which genes were used for the emergence of a C4 pathway, the tissue specificity has surprisingly no effect. Our results suggest that levels of key genes were elevated during the early diversification of grasses and subsequently repeatedly used to trigger a weak C4 cycle via relatively few mutations. The abundance of C4-suitable transcripts therefore facilitated physiological innovation, but the transition to a strong C4 pathway still involved consequent changes in expression levels, leaf specificity, and coding sequences. The direction and amount of changes required for the strong C4 pathway depended on the identity of the genes co-opted, so that ancestral gene expression both facilitates adaptive transitions and constrains subsequent evolutionary trajectories.
Keywords: C4 photosynthesis, evolvability, grasses, phylogenetics, transcriptomics, gene co-option
Introduction
The evolution of novel physiological adaptations occasionally requires the development of new biochemical cascades, which are generally achieved via the co-option of pre-existing genes into new functions (Duboule and Wilkins 1998; True and Carroll 2002; Monson 2003; Monteiro and Podlaha 2009). Rewiring of biochemical pathways can require both modifications of spatial and temporal gene expression patterns and alterations of the coding sequences (CDSs) to adapt the encoded enzymes to the new catalytic context (Duret and Mouchiroud 2000; Carroll 2008; Aubry et al. 2014). In cases where numerous modifications are needed, the novel pathways can be assembled by natural selection only if a functional version can emerge through relatively few changes, allowing subsequent selection to fix mutations that increase the efficiency of the pathway. Genomic factors that reduce the phenotypic distance between ancestral and novel physiologies, thereby enabling the emergence of novel cascades via few mutations, would consequently be expected to increase accessibility to novel phenotypes. However, in most cases these factors remain poorly understood.
The ability of given genes or genomic features to trigger evolutionary innovation can be investigated via experimental evolution (e.g. Weinreich et al. 2006; Blount et al. 2012), but such studies are restricted to short-lived organisms that do not encapsulate the existing diversity of phyla. For larger organisms with long generation times, a historical approach is the most appropriate. Indeed, phylogenetic inference allows explicit tests of how specific features affect the accessibility of new phenotypes (e.g. Marazzi et al. 2012). Conversely, genomic features that have recurrently contributed to independent origins of a given phenotype can be safely assumed to be suitable for the trait of interest, and their origin can be regarded as potentially facilitating later adaptive transitions (Huang, O’Donnell, et al. 2016). For example, the same autosome pairs were repeatedly co-opted to evolve sex chromosomes in turtles (Montiel et al. 2017), the same gene families encoding crystallins were used to evolve camera eyes in cephalopods and vertebrates (Zinovieva et al. 1999; Yoshida et al. 2014), and homologous genes recurrently contributed to the diversification of coloration patterns in butterflies (Jiggins et al. 2017). Although such evidence indicates that some genomic regions or genes preferentially contribute to specific evolutionary transitions (Tenaillon et al. 2012), multiple factors might increase the adaptive potential, and their identification requires the comparison of the ancestral condition of genes or genomic regions that were recurrently co-opted, to those that were not.
An excellent system to study the factors that increase gene adaptive potential is C4 photosynthesis. This novel physiology requires a biochemical cascade arising from the high activity of multiple enzymes in specific leaf compartments, and improves autotrophic carbon assimilation in tropical conditions (Pearcy and Ehleringer, 1984; Hatch 1987; Sage et al. 2012, Atkinson et al. 2016). The C4 trait is ecologically and agronomically extremely important (Ehleringer et al., 1997; Still et al., 2003; Byrt et al., 2011). It evolved more than 60 times in independent lineages of flowering plants (Sage et al. 2011), via the co-option of multiple genes that were present in non-C4 ancestors (Hibberd and Quick 2002; Aubry et al. 2011; Brown et al. 2011; Kajala et al. 2012). Most enzymes of the C4 pathway are encoded by multigene families, whose members differed in their expression patterns and catalytic properties of the encoded enzymes before their involvement in C4 photosynthesis (Wang et al. 2009; Hibberd and Covshoff 2010; Aubry et al. 2011; Christin et al. 2013, 2015). Previous comparisons of a handful of C4 species have shown that a subset of gene lineages were recurrently co-opted for C4 evolution, both among grasses and among the distantly related Caryophyllales (Christin et al. 2013, 2015). However, the co-opted genes differed between grasses and Caryophyllales, suggesting that factors predisposing some genes for a C4 function are specific to subgroups of angiosperms (Christin et al. 2015). It has been noted that the co-opted genes appeared to be highly expressed in the non-C4 taxa available at the time for comparison, which might have contributed to their preferential co-option (Christin et al. 2013; Emms et al. 2016). However, systematic tests of the factors underlying the observed co-option bias are still lacking.
In this study, we compare transcriptomes across ten independent C4 origins in grasses, and their non-C4 relatives. Through a combination of phylogeny-based analyses, we test 1) whether a bias in the gene lineages co-opted exists across the whole set of grasses. To determine the causal factors underlying the bias, we then test 2) whether the expression level in leaves and/or 3) whether the tissue specificity in the non-C4 ancestors explain variation in the co-option probability among gene lineages. In addition, we analyze CDSs to test 4) whether adaptive changes in the CDSs occurred during or after the emergence of the C4 physiology. Together, our investigations shed new light on the factors that increase the adaptive potential of some genes, focusing on a complex trait of ecological and agronomical importance.
Results
Sequencing, Read Mapping and Transcriptome Assembly
In total, 74 individually sequenced RNA libraries from 19 species generated over 550 million 100 bp paired-end reads. This represents 98.87 Gb of data, with a mean of 1.34 Gb per library (SD = 0.95 Gb; supplementary table S1, Supplementary Material online). Over 81% of the reads were kept after removing low-quality reads and ribosomal RNA sequences. Transcriptomes were assembled with a mean of 2.23 Gb per species (SD = 1.40 Gb), resulting in a mean of 54,255 Trinity “unigenes” (SD = 17,218.35), 79,566.12 contigs (SD = 23,038.61), and a 1,560.05 bp N50 (SD = 184.95 bp).
The C4-related gene families considered in this study constitute 5.1% (SD = 2.02%) of the reads in the leaf libraries of C4 plants, versus 2.34% in non-C4 plants (SD = 0.75%). On average, 1.05% of the reads from the root libraries mapped to C4-related genes (SD = 0.48%).
Phylogenetic Trees and Identification of Genes Co-Opted for C4 Photosynthesis
A total of 533 nuclear core-orthologs were used to infer the species tree, which was well resolved (fig. 1). The relationships among grass subfamilies mirror those retrieved previously with other data sets (GPWG II 2012). However, relationships within the Paniceae tribe (the group most densely sampled here) differ in several aspects from those based on plastid markers (GPWG II 2012), and were closer to previous analyses that also included nuclear markers (Vicentini et al. 2008). The placement of the different C4 origins within the tree was largely congruent with previous studies, and their non-C4 relatives separated them in the phylogeny as expected (fig. 1).
Fig. 1.
Phylogenetic tree based on nuclear orthologs. C4 taxa are in red, and C4 origins are numbered. One of the tribe and the two main clades of grasses are indicated on the right. The black circle highlights the node representing the common ancestor of the sampled grasses. Bootstrap values are indicated near branches.
For each gene family encoding C4-related enzymes, phylogenetic inference confirmed previous conclusions about orthology (Vilella et al. 2009). The enzyme phosphoenolpyruvate carboxykinase (PCK) and the Na+/H+ antiporter (NHD) are each encoded by a single gene lineage (supplementary fig. S1, Supplementary Material online). The number of grass co-orthologs in other families varies from two (for pyruvate, phosphate dikinase—PPDK) to eight (for triose phosphate–phosphate translocator—TPT; supplementary fig. S1, Supplementary Material online). Groups of co-orthologs were named as in Christin et al. (2015). Phylogenetic relationships inferred in these gene trees were mostly congruent with the species tree. Exceptions include genes for PCK, where Echinochloa stagnina and Alloteropsis semialata grouped with those of Setaria barbata. This pattern has previously been reported for Alloteropsis species and this, together with a number of other lines of evidence, was interpreted as the fingerprint of a lateral gene transfer from Setaria or its close relatives (Christin et al. 2012; Dunning et al. 2017). Other incongruences were observed in genes encoding PEPC, PPDK, NAD(P)-malate dehydrogenase [NAD(P)-MDH], Sodium bile acid symporter family (SBAS), TPT, and NDH (supplementary fig S1, Supplementary Material online), and could stem from a combination of reticulate evolution during grass diversification and phylogenetic bias due to adaptive evolution. Gene duplicates specific to subgroups of grasses are evident for several genes, and can in some cases be associated to recent polyploidy (e.g. in Zea mays genes pck-1P1, ppc-1P4, ppdk-1P2, nadmdh-4P7; supplementary fig. S1, Supplementary Material online). Our analytical pipeline cannot estimate the expression level individually for each of these duplicates with very similar sequences, but these duplications specific to subgroups of grasses are relatively recent and occurred after the divergence of C3 and C4 clades (supplementary fig. S1, Supplementary Material online). The inferred evolutionary changes in expression patterns and co-option events are consequently not affected.
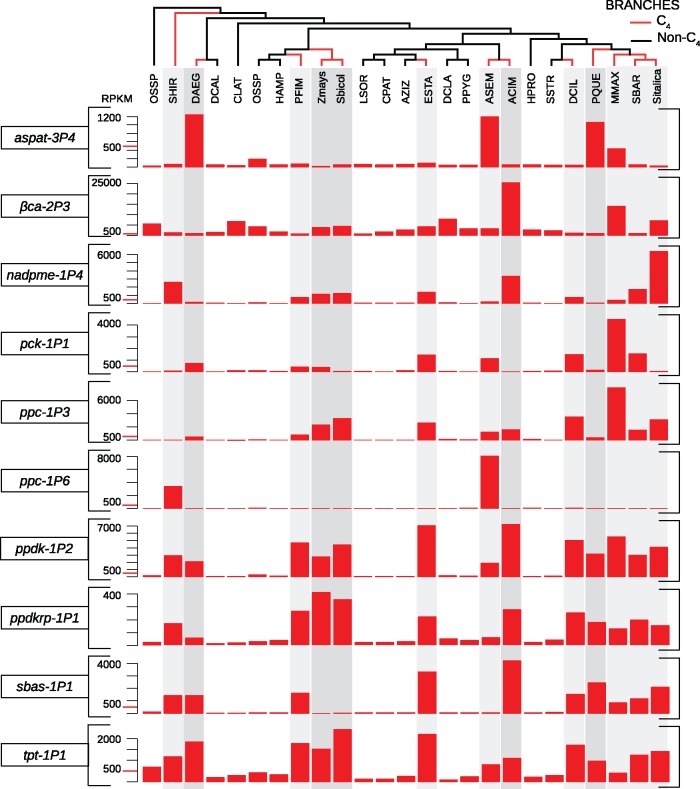
The most highly transcribed genes encoding C4-related proteins are those for β-carbonic anhydrase (βCA; fig. 2 and supplementary table S2, Supplementary Material online), an enzyme that acts in the cytosol of mesophyll cells in C4 plants. These genes are however equally abundant in non-C4 species (fig. 2), where the enzyme plays a key role in the chloroplasts of mesophyll cells (Tetu et al. 2007). Of the 31 other gene families encoding enzymes that can be related to the C4 pathway, 14 included gene lineages with transcript abundances above 500 rpkm in at least one C4 species (fig. 3; supplementary table S2, Supplementary Material online). The transcript abundance of ppa-4P4 reached 500 rpkm in some C4 species, but similar abundance was observed in a number of non-C4 taxa (supplementary table S2, Supplementary Material online), and the gene was consequently not counted as C4 specific. For the rest of the gene lineages, such high values were not found in non-C4 species (supplementary table S2, Supplementary Material online). Genes co-opted for C4 photosynthesis were identified in each C4 species for most core C4 enzymes, but putative C4 transporters and regulators were not always abundant in C4 leaves (supplementary table S2, Supplementary Material online). Genes for enzymes of the photorespiration pathway were downregulated in C4 species, as expected (supplementary table S2, Supplementary Material online).
Fig. 2.
Transcript abundances of the main C4 genes in C4 and non-C4 species. Barplots indicate rpkm values (reads per kilobase per million of reads). Phylogenetic relationships among species are indicated at the top, and C4 lineages are numbered as in figure 1, with branches colored in red. Species names are abbreviated as in supplementary tables S1 and S2, Supplementary Material online.
Fig. 3.
Gene expression profiles of C4-related genes in the studied taxa. Colors indicate leaf transcript abundance and leaf/ratio abundance ratio for C4-related genes in C4 and non-C4 species. Genes that have been co-opted at least once are at the top.
Factors Affecting Gene Co-Option
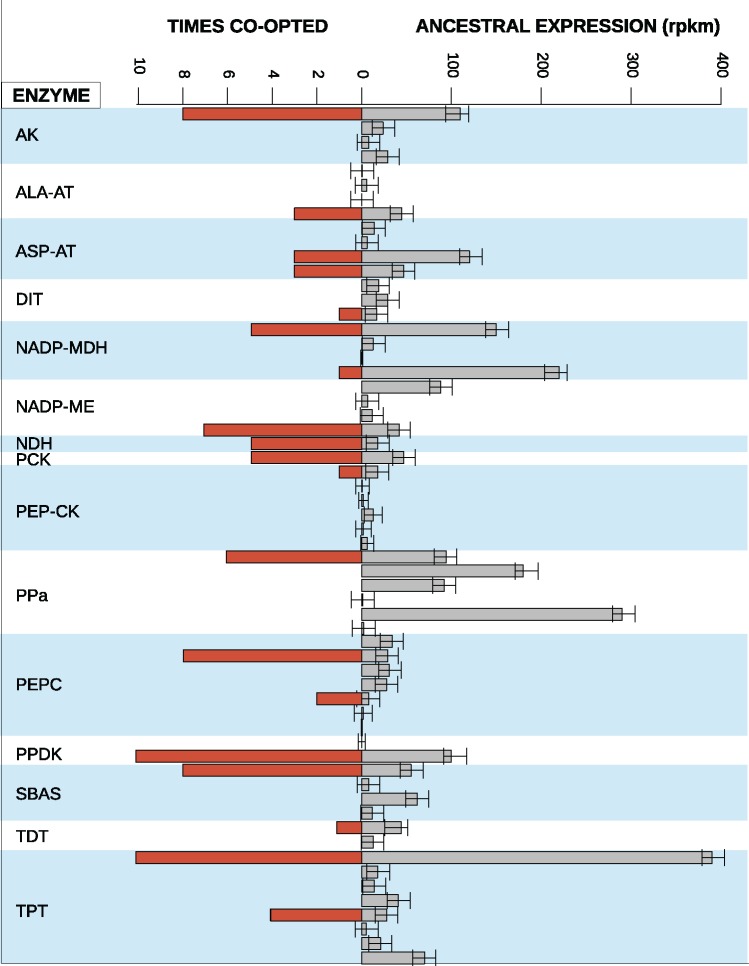
Out of 58 gene lineages encoding the 14 enzymes used by the C4 species sampled here, only 18 have been co-opted at least once, and up to ten times independently for ppdk–1P2 and tpt–1P1 and eight for ppc−1P3 (table 1). Given the size of the different gene families and the number of co-option events, fewer genes have been co-opted at least once than expected by chance (P-value < 0.00001). This confirms the existence of a co-option bias across the ten C4 origins considered here, a result previously reported for Caryophylalles and grasses (Christin et al. 2013, 2015).
Table 1.
Number of Times a Gene Lineage Was Co-Opted, For Genes Co-Opted at Least Once.
| Gene Lineage | Times Co-Opted | Main Catalytic Reaction |
|---|---|---|
| ak-1P1 | 8 | AMP→ADP |
| alaat-1P5 | 3 | Ala↔Pyruvate |
| aspat-2P3 | 3 | Asp↔OAA |
| aspat-3P4 | 3 | Asp↔OAA |
| dit-2P3 | 1 | Dicarboxylate transporter |
| nadpmdh-1P1 | 5 | Malate↔OAA |
| nadpmdh-3P4 | 1 | Malate↔OAA |
| nadpme-1P4 | 7 | Malate→pyruvate |
| nhd-1P1 | 5 | Sodium proton antiport |
| pck-1P1 | 5 | OAA→PEP |
| pepck-1P1 | 1 | ATP ADP/P antiport |
| ppa-1P2.1 | 6 | Pyrophosphate→phosphate |
| ppc-1P3 | 8 | PEP→OAA |
| ppc-1P6 | 2 | PEP→OAA |
| ppdk-1P2 | 10 | Pyruvate→PEP |
| ppt-1P5 | 4 | PEP phosphate antiport |
| sbas-1P1 | 8 | Pyruvate sodium symport |
| tpt-1P1 | 10 | 3-PGA TP antiport |
The ancestral state reconstructions inferred the abundance in leaves and leaf/root specificity in the last common ancestor of the sampled grasses for each C4-related gene (fig. 4). This approach comes with uncertainty, especially for deeper nodes in a tree, but the confidence intervals associated with the inferred values are small compared with the difference among members of the same gene family (fig. 4). The inferred values are moreover tightly correlated with averages of the values among C3 grasses (R2 = 0.98 for the leaf abundance and R2 = 0.91 for the leaf/root ratio), and were consequently used for modeling of gene co-option. Linear models showed that the ancestral transcript abundance in the leaf significantly affected the co-option frequency (F = 13.11, df = 56, P = 0.0006336; R2 = 0.19), and this stayed significant when the gene family was used as a co-factor (table 2). The effect of the ancestral leaf/root transcript abundance ratio on the co-option frequency was not significant when considered on its own (F = 0.40, df = 56, P = 0.54), or in combination with the ancestral leaf abundance and the gene family cofactor (table 2). Therefore, our modeling analyses indicate that genes were co-opted for C4 photosynthesis based on their transcription level in leaves (fig. 4), independently of the specificity of this expression in leaves compared with roots. The same conclusions were reached when using a threshold of 300, 1,000, and 1,500 rpkm for the identification of co-opted genes (see table 2).
Fig. 4.
Ancestral leaf transcript abundance and number of co-option events. Barplots on the left indicate the number of times each gene was co-opted, and those on the right indicate the inferred abundance in the non-C4 last common ancestor of grasses (see fig. 1), with the associated confidence intervals. Genes are sorted by enyzme, indicated on the left.
Table 2.
Results of Analyses of Variance on Linear Models of Number of Co-Option Events Based on Ancestral Leaf Abundance (ala), Leaf/Root Ratio, and Gene Family Identity (family), with Co-Opted Genes Identified with Different rpkm Thresholds.
| rpkm Threshold | 300 | 300 | 300 | 500 | 500 | 500 | 1,000 | 1,000 | 1,000 | 1,500 | 1,500 | 1,500 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Factors | ala | Leaf/Root | Family | ala | Leaf/Root | Family | ala | Leaf/Root | Family | ala | Leaf/Root | Family |
| P-value | 0.00 | 0.52 | 0.38 | 0.00 | 0.57 | 0.56 | 0.00 | 0.88 | 0.21 | 0.01 | 0.77 | 0.10 |
| df | 1, 42 | 1, 42 | 13, 42 | 1, 42 | 1, 42 | 13, 42 | 1, 42 | 1, 42 | 13, 42 | 1, 42 | 1, 42 | 13, 42 |
| F-statistics | 17.07 | 0.78 | 0.95 | 12.65 | 0.32 | 0.90 | 14.46 | 0.21 | 1.37 | 8.29 | 0.0.09 | 1.71 |
Note.—df, degrees of freedom. For each variable, the degrees of freedom for the residuals are given after the comma.
Transcriptome data sets for clades containing C3 and C4 species other than grasses are focused on small taxonomic groups, so that ancient evolutionary events cannot be inferred yet outside from grasses. A test using published transcriptomes for one C3 and C4 species within the eudicot family Cleomaceae failed to detect any effect of expression levels on the identity of genes co-opted for C4 (supplementary tables S3 and S4, Supplementary Material online), but the availability of a single C4 origin and only one C3 relative likely decreased statistical power. Although the same statistical limitations applied to the Flaveria data set, our preliminary investigation suggested that the effect of leaf abundance on the co-option probability might apply to multiple C4 origins across the angiosperms. Indeed, there was a significant effect of the leaf abundance in the close relatives on the co-option probability for Flaveria (supplementary table S4, Supplementary Material online).
Marked Differences in Transcript Abundance and CDSs
Although the ancestral transcript abundance significantly affects the probability of a gene being co-opted, the evolution of C4 photosynthesis is accompanied by major increases in transcript abundance. The transcripts of genes encoding C4 enzymes increase by a fold change of up to 480 for ppc–1P6 in A. semialata compared with related non-C4 taxa (fig. 2). In addition, their leaf specificity increases, to reach leaf/root ratios of up to 6,204 after their co-option into C4 photosynthesis, compared with a maximum of 257 in non-C4 taxa (fig. 3).
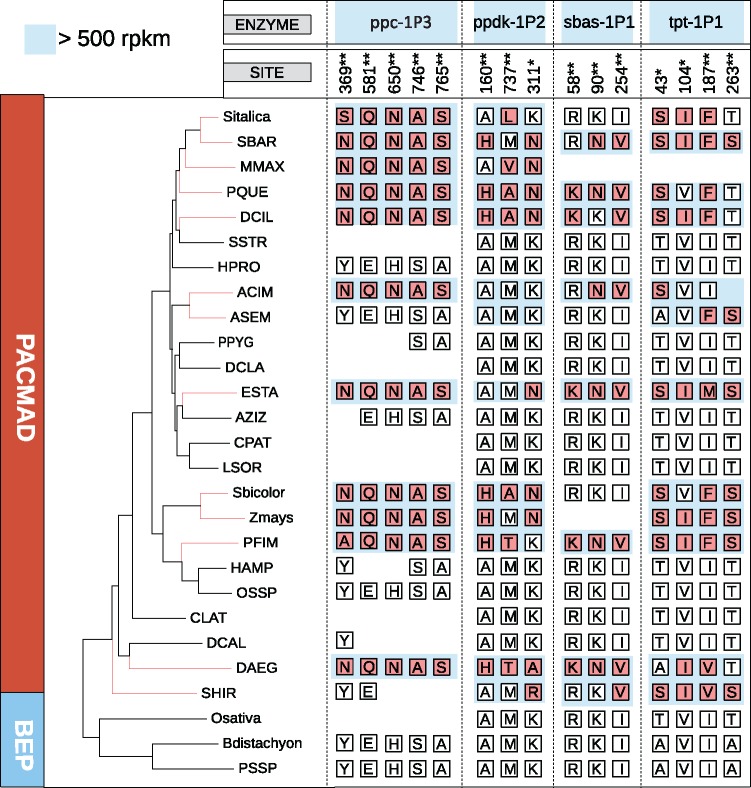
Besides these changes in transcript abundance, tests for positive selection revealed adaptive evolution in the CDSs of a number of genes during or slightly after their co-option into C4 photosynthesis. After correction for multiple testing, the test for a shift of selective pressures along C4 branches (A1 vs. M1a comparison) was significant for nine genes out of 19 (supplementary table S5, Supplementary Material online). The test specifically testing for a shift to positive selection as opposed to a relaxation of selection (A1 vs. A comparison) was also significant for four of these nine genes; ppc-1P3, ppdk-1P2, sbas-1P1, and tpt-1P1 (supplementary table S5, Supplementary Material online). The sites identified by the Bayes Empirical Bayes analysis as being under positive selection along C4 branches showed widespread cases of parallel amino acid replacements (fig. 5).
Fig. 5.
Patterns of convergent adaptive amino acid replacements. The phylogeny of the sampled species is indicated on the left, with species names abbreviated as in supplementary table S1, Supplementary Material online. Branches leading to C4 species in red. Amino acids at some of the sites under positive selection (*P < 0.05; **P < 0.01) are indicated on the right. Residues of co-opted genes are highlighted with a blue background.
Discussion
Expression Patterns Determined Which Genes Were Co-Opted for C4
In this study, we analyzed root and leaf transcriptomes from grass species representing ten independent origins of C4 photosynthesis as well as the close non-C4 relatives to each of them (fig. 1). As previously suggested based on smaller species samples (Christin et al. 2013, 2015), the co-option of genes for the C4 pathway has been a nonrandom process. Indeed, despite multiple gene lineages existing for most C4-related enzymes, a few of them were co-opted more frequently than expected by chance, while most were never used in the ten C4 lineages evaluated here (table 1 and figs. 3 and 4). A number of factors could explain the preferential co-option of some genes for a novel function, including their availability via genomic redundancy, the suitability of their kinetic properties, the fit of their expression patterns, and their evolvability (Aharoni et al. 2005; Landry et al. 2007; Christin et al. 2010, 2015; Stiffler et al. 2015; Huang, O’Donnell, et al. 2016). Our approach was specifically designed to test for the effects on co-option probability of two dimensions of the expression patterns inferred for non-C4 ancestors; the transcript abundance in leaves and the leaf versus root specificity. Thanks to the evolutionary-informed sampling (fig. 1), we were able to unambiguously show that the likelihood of gene co-option into C4 photosynthesis was determined in a large part by their transcript abundance in leaves prior to C4 evolution (fig. 4), with no apparent effect of the leaf to root specificity (table 2).
The C4 biochemical pathway, like any complex pathway, is assumed to result from many rounds of fixation of adaptive mutations (Sage et al. 2012; Heckmann et al. 2013; Dunning et al. 2017). However, natural selection cannot gradually improve a pathway before it exists, even in a rudimentary stage (Huang, O’Donnell, et al. 2016). It is likely that a primitive, weak C4 cycle initially emerged in some species via a slight upregulation of few genes, as observed in intermediate plants accumulating only part of their CO2 via the C4 cycle (Mallmann et al. 2014; Dunning et al. 2017). We show here that some genes were already moderately abundant in leaves of non-C4 plants (fig. 4), a pattern that likely evolved for a number of reasons not related to C4 photosynthesis, but eased its later evolution. This facilitator effect would have been even stronger if C4-related genes were upregulated in the low-CO2 conditions that prevailed until the Industrial Revolution, as has been suggested for the distantly related Arabidopsis (Li et al. 2014). The encoded enzymes, present in the leaves of the non-C4 ancestors, constituted the building blocks needed to generate a weak, yet functional, C4 pathway following key mutations. These could have included further upregulation of key C4 enzymes or alterations of the leaf structural arrangements, pushing the system beyond a tipping point where the C4 pathway could emerge. Models predict that, once a C4 pathway is in place, any increase in the rate of the C4 pathway will increase productivity in warm conditions (Heckmann et al. 2013; Mallmann et al. 2014). Any rudimentary C4 pathway based on ancestrally abundant enzymes would therefore have created the selective impetus for upregulation of enzymes, generating the striking patterns observed in derived C4 plants (figs. 2 and 3).
Besides elevated abundance of numerous enzymes, the C4 trait is characterized by a precise compartmentalization of the biochemical reactions in different parts of the leaves (Hatch and Osmond 1976; Hatch 1987; John et al. 2014). Interestingly, transcript abundance in nonphotosynthetic tissues, such as roots, did however not prevent the co-option of a gene lineage for C4 photosynthesis (table 2 and fig. 3), and previous pairwise comparisons have established that orthologs to C4 genes have a diversity of expression patterns in non-C4 species (Külahoglu et al. 2014). We conclude that being abundant in leaves was a sufficient condition for the C4 function, independently of the presence in other tissues. Cellular and subcellular localization, which was not captured by our whole-leaf transcriptomes, probably still contributed to determining which genes were co-opted for C4. For instance, only one of the four gene lineages for NADP-ME present in grasses encodes a chloroplast-specific isoform, and this gene lineage has been recurrently co-opted for C4 despite an ancestral abundance of a second gene (fig. 4; Christin et al. 2009). Similarly, the product of ppc-1P2, the most highly expressed gene for PEPC in non-C4 plants (fig. 4), is chloroplast-specific (Masumoto et al. 2010), which very likely prevented a function in C4 photosynthesis, since this enzyme is cytosolic in the C4 pathway. Independently of these specific cases, the mere moderate abundance in leaves explains a large fraction of the co-option probability.
Despite Genetic Enablers, C4 Evolution Required Massive Changes
Our study is the first to scan the transcriptomes of a number of non-C4 grasses closely related to C4 species, and showed that genes co-opted for C4 tended to already be abundant in non-C4 ancestors (figs. 3 and4). Although transcriptomes in other groups are not available for multiple C4 origins and their C3 relatives, our reanalysis of eudicot data sets suggested that the preferential co-option of the most abundant genes might underly C4 origins in groups other than grasses (supplementary table S4, Supplementary Material online). This suggests that the abundance of some enzymes able to fulfil a C4 function facilitated the emergence of a C4 pathway. However, massive changes in gene expression are still observed between non-C4 and C4 relatives (e.g. Bräutigam et al. 2011, 2014; Külahoglu et al. 2014). Indeed, genes encoding C4 enzymes are orders of magnitude more abundant in C4 leaves, and leaf specificity strongly increased after the co-option of genes for C4 (figs. 2 and3). In addition, evidence for widespread adaptive evolution of CDSs for the C4 context, obtained here and in other studies (fig. 5; Besnard et al. 2009; Christin et al. 2009; Wang et al. 2009; Huang, Studer, et al. 2016), suggests important modifications of the kinetic properties, shown for some enzymes (Bläsing et al. 2000; Tausta et al. 2002). Instead of being involved in the initial emergence of a C4 cycle, we propose that these massive changes were involved in the transition from a weak to a strong C4 pathway able to match the high rates of the Calvin cycle, as suggested for specific study systems (Svensson et al. 2003; Mallmann et al. 2014; Dunning et al. 2017).
Since the major requirement for a C4 function was sufficient abundance in leaves, the co-opted genes were not necessarily the best suited for the C4 function, in terms of the tissue specificity or kinetic properties of the encoded enzyme. The ancestral abundance might therefore have constrained the initial emergence of a weak C4 cycle based on specific sets of genes, forcing natural selection to later adapt their properties to those required for a high-flux strong C4 cycle. The recurrent co-option of the same co-orthologs would have increased the likelihood of adaptation via similar changes, explaining the observed parallel amino acid replacements among C4 origins in grasses (fig. 5; Christin et al. 2007). It has been shown that C4 lineages belonging to distant groups of angiosperms in some cases co-opted distinct genes (Christin et al. 2015; supplementary table S4, Supplementary Material online). Because of the large evolutionary distances separating these groups, which are further increased when different co-orthologs are co-opted (supplementary table S4, Supplementary Material online), the encoded enzymes likely varied in their kinetic properties in addition to their leaf and cell specificities. The amount of optimizing adaptive changes might have varied among major C4 groups as a consequence, explaining that the frequency and identity of selection-driven amino acid replacements shows high convergence among closely related C4 lineages (fig. 5), but varies between C4 origins in grasses and those in the distantly related sedges and eudicots (Besnard et al. 2009).
Conclusions
In this study, we sequenced the transcriptomes of species from the main C4 grass lineages as well as their close non-C4 relatives, and used models to show that the identity of genes co-opted for C4 photosynthesis was largely explained by transcript abundance before C4 evolution. The co-option, likely dictated by the mere presence of each protein in leaves, was followed by massive upregulation and widespread adaptation of CDSs. Both of these processes likely accelerated and optimized a C4 pathway that initially emerged from the combined action of enzymes already present in leaves. It is currently unknown why some gene lineages came to be more expressed than others in non-C4 plants but, despite variation among species, the increased abundance of these genes seems to date back to at least the last common ancestor of grasses. Comparison among distant groups of angiosperms indicates that the preferential co-option of the most abundant gene lineages might be a recurrent pattern, but the sampling is not yet dense enough across angiosperms to precisely determine when increased transcript abundance first happened, among the ancestors of grasses and other groups that recurrently evolved C4 photosynthesis. When this information is available, we might be able to test whether gene abundance combined with anatomical variation determined which plant lineages were more likely to evolve C4 photosynthesis, once environmental changes created the selective pressure for this physiological novelty.
Materials and Methods
Species Sampling
Grass species were selected for analyses based on their photosynthetic type to include multiple C4 origins and their non-C4 relatives, based on previous phylogenetic analyses (GPWG II 2012). We sequenced eight C4 species and eleven non-C4 species, which separate them in the phylogenetic tree of grasses (GPWG II 2012, fig. 1). Most of these belong to the PACMAD clade (subfamilies Panicoideae, Arundinoideae, Chloridoideae, Micrairoideae, Aristidoideae, and Danthonioideae), which contains all C4 origins in grasses, and one non-C4 Pooideae species was added as an outgroup for comparisons.
The selected species were grown from seeds, using the material from Atkinson et al. (2016) and Lundgren et al. (2015). Plants were maintained in controlled environment growth chambers (Conviron BDR16; Manitoba, Canada), with 60% relative humidity, 500 µmol m−2 s−1 photosynthetic photon flux density, and 25/20 °C day/night temperatures, with a 14-h photoperiod. John Innes No. 2 potting compost (John Innes Manufacturers Association, Reading, England) was used. Plants were watered three times a week to keep the soil damp, and were fertilized every 2 weeks with Scotts Evergreen Lawn Food (The Scotts Company, Surrey, England). After a minimum of 30 days in these controlled conditions, two young roots and the most photosynthetically active distal half of fully expanded leaves were sampled from two individuals of each species (biological replicates) during the middle of the photoperiod, and immediately frozen in liquid nitrogen. All samples were stored at −80 °C until RNA extraction.
RNA Extraction, Sequencing, and Transcriptome Assembly
Samples were homogenized in liquid nitrogen using a pestle and a mortar, and RNA was extracted using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions. The isolated RNA was DNA digested on-column using the RNase-Free Dnase Set (Qiagen, Hilden, Germany) and eluted in RNAse-free water with 20 U/µl of SUPERase-IN RNase Inhibitor (Life Technologies, Carlsbad, CA). Extractions that yielded an RNA integrity number (RIN) >6.5 and at least 0.5 µg of total RNA, as determined with the RNA 6000 Nano kit with an Agilent Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA), were used for upstream procedures. Individual RNA libraries were prepared using TruSeq RNA Library Preparation Kit v2 (Illumina, San Diego, CA), following the manufacturer’s protocol with a target median insert length of 155 bp. A total of 24-indexed libraries were pooled per lane of flow cell and sequenced on an Illumina HiSeq 2500 platform with 100 cycles in rapid mode generating 100 bp paired-end reads, at the Sheffield Diagnostic Genetics Service.
Reads were filtered and assembled using the Agalma pipeline version 0.5.0, with default parameters (Dunn et al. 2013). This pipeline removes low quality reads (Q < 33), and those that are adaptor-contaminated or correspond to ribosomal RNA. The filtered reads are then used for de novo assembly using Trinity (version trinityrnaseq_r20140413p1; Grabherr et al. 2011). One assembly was generated per species, using all the libraries available. Leaf assembly and reads in duplicates from the C4Alloteropsis cimicina were retrieved from Dunning et al. (2017), and reads for the C4Megathyrsus maximus and the non-C4Dichanthelium clandestinum, in triplicates and without replicate, respectively, were retrieved from Bräutigam et al. (2014). RNA-seq reads for C4 grasses with a completely sequenced genome were also retrieved from the literature (Setaria italica without replicate from Zhang et al. [2012], Z. mays without replicate from Liu et al. [2015], and Sorghum bicolor in duplicates from Fracasso et al. [2016]). The final RNA expression data set included 12 non-C4 species and 13 C4 species of grasses.
Inference of a Species Tree Based on Core Orthologs
CDSs were predicted from the assembled contigs and those retrieved from the literature using the standalone version of OrfPredictor (Min et al. 2005). Protein sequences of eight publicly available genomes (Arabidopsis thaliana, Brachypodium distachyon, Glycine max, Oryza sativa, Populus trichocarpa, S. italica, So. bicolor, and Z. mays) were used as references to improve the identification of open reading frames by providing the program with a precomputed BLASTX output file, using parameters suggested by the authors (Min et al. 2005). CDS from contigs with “no hit” in the BLASTX output were predicted ab initio. The predicted CDS were used for subsequent analyses.
CDS homologous to an a priori defined set of plant genes were retrieved using a Hidden Markov Model based search tool (HaMSTR v.13.2.3; Ebersberger et al. 2009). The set of genes includes 581 single copy core-orthologs from plants and is derived from the Inparanoid ortholog database (Sonnhammer and Östlund 2014), using five high quality genomes (A. thaliana, Vitis vinifera, O. sativa, So. bicolor, and Ostreococcus lucimarinus). Sequences were aligned as described in Dunning et al. (2017); alignments shorter than 100 bp after trimming were discarded, and alignments including sequences from at least ten species were concatenated. The resulting alignment was used to infer a maximum likelihood tree with Phyml (Guindon and Gascuel 2003), using a GTR + G + I nucleotide substitution model, which was identified as the best model using the Smart Model Selection (Lefort et al. 2017). Support was evaluated by 100 bootstrap pseudoreplicates.
Identification of Homologs and Grass Co-Orthologs Encoding C4-Related Enzymes
For each gene family that encodes enzymes related to the C4 pathway (identified based on the literature; Mallmann et al. 2014; Li et al. 2015), homologous CDS were retrieved from three publicly available genomes (S. italica, So. bicolor, and A. thaliana), based on the annotation and previously inferred homology (Vilella et al. 2009). The same approach was used to analyze genes of the photorespiration pathway, which are expected to be downregulated during C4 evolution (Mallmann et al 2014). CDS from the sequenced transcriptomes or retrieved from the literature that were homologous to any sequence in each gene family were identified via BLAST searches. Positive matches with a minimal e-value of 0.01 and minimal mapping length of 500 bp were retrieved and added to the data sets. Only the first transcript model was considered for complete genomes, and the longest CDS from each set of Trinity gene isoforms was used.
A new alignment was produced for each gene family ensuring high-quality alignments while maintaining as many sites as possible. This approach requires manual curation, and was consequently not used for the 581 sets of core orthologs described earlier. A preliminary alignment was obtained for each gene family using MUSCLE (Edgar 2004). The alignment was manually inspected in MEGA version 6 (Tamura et al. 2013), and potential chimeras and sequences of ambiguous homology (false positives) identified through visual inspection and comparison with other sequences were removed. The remaining sequences were re-aligned as codons using ClustalW (Thompson et al. 1994), and the alignments were manually refined. For each gene family, the alignment was used to compute a maximum likelihood phylogenetic tree, using PhyML (Guindon and Gascuel 2003), and the GTR + G + I substitution model as best-fit model identified previously for most of the gene families in this study (Christin et al. 2015). Support values were evaluated with 100 bootstrap pseudoreplicates.
Groups of grass co-orthologs, which include all the genes that descend from a single gene in the last common ancestor of grasses through speciation and gene or genome duplications (including the ancient polyploidy in the common ancestor of grasses; Tang et al. 2010), were identified based on the phylogenetic trees inferred for each gene family. Duplicates specific to some groups of grasses, which might have emerged via gene or genome duplication (whether via auto- or allopolyploidy) after the diversification of grasses, would be grouped in the same co-orthologs, so that our orthology assessment and subsequent expression analyses are not influenced by polyploidization events. Cleaned reads were mapped back to sequences belonging to any of the gene families as single reads, using the local alignment option in Bowtie2 (Langmead and Salzberg 2012). Our approach allows reads to map back to sequences from the same species, but also allows sequences from other closely related species to serve as the reference. The number of reads mapped to each group of co-orthologs was reported as reads per kilobase of aligned exons per million of cleaned reads (rpkm). These proxies for transcript abundances were obtained for each replicate.
Identification of Co-Opted Genes and Factors Increasing Co-Option Rates
Enzymes of the C4 pathway are abundant in the leaves of C4 species because high catalytic rates are needed to match the fluxes of the Calvin cycle (Furbank et al. 1997, Mallmann et al. 2014). Transcripts encoding enzymes known to act in the C4 pathway were consequently identified as those that reached an abundance of at least 500 rpkm in leaves of a given C4 species. Because this threshold is arbitrary, subsequent analyses were repeated with other thresholds (300, 1,000, and 1,500 rpkm), which did not affect our conclusions (see “Results” section). Previous investigations comparing a limited number of species have shown that, within a given taxonomic group, independent C4 origins tend to co-opt the same gene lineages (Christin et al. 2013, 2015; Emms et al. 2016). To test this expectation across our larger species sample, the number of genes co-opted at least once in our data set was compared with the number expected by chance given the size of the different gene lineages and the number of co-option events, following the resampling approach of Christin et al. (2015).
Once a bias in gene co-option was confirmed (see Results), we tested for factors potentially affecting the probability of a given group of co-orthologs being co-opted for C4. We used the values inferred for the last common ancestor of grasses as proxies for the condition before C4 evolved, with two different dimensions of the expression patterns. First, we inferred the leaf transcript abundance. Second, we inferred the leaf/root ratio of abundances as a proxy for leaf specificity. For each group of co-orthologs, the values of these variables in the common ancestor of grasses were estimated using the phylogeny obtained with HaMSTR and the “ace” function in the R package “ape” version 3.5 (Paradis et al. 2004). The maximum likelihood method was selected, with a Brownian motion model. In this approach, the value of the continuous variable that maximizes the likelihood is calculated for each node, with the associated confidence intervals. Only non-C4 species were included in the ancestral state analyses to avoid biases caused by high levels in C4 taxa. Considering only the gene families co-opted at least once, linear models, as implemented in the “lm” function in R version 3.3.2 (R Development Core Team 2016), were used to test independently for an effect of ancestral leaf transcription abundance and of ancestral leaf/root ratio on the number of times each group of co-orthologs has been co-opted. An analysis of variance on multiple linear models was then used to determine whether the effect of ancestral leaf abundance and/or leaf/root ratio remain when the gene family is included as a co-factor.
Transcriptome data sets available for groups of closely related C3 and C4 species outside of grasses were used to assess whether the observed patterns are valid across flowering plants. Data for one C3 and one C4 Cleomaceae were retrieved from Bräutigam et al. (2011), and the phylogenetic annotation of C4-related genes in these data sets was deduced from the identity of orthologs from the closely related Arabidopsis and the phylogenetic trees from Christin et al. (2015). For Flaveria, RNAseq data were retrieved for two C3 species from Mallmann et al. (2014) and for one C4 species from Lyu et al. (2015). The reads were annotated in the original study based on their similarity to Arabidopsis sequences, but the evolutionary distance between Flaveria and Arabidopsis can potentially mislead orthology assessments. We consequently performed de novo assemblies using the published reads, and obtained the transcript abundance for C4-related genes using the previously published phylogenetic annotation pipeline (Christin et al. 2015). Groups of co-orthologs co-opted for C4 by Flaveria or Cleomaceae were identified based on the literature (reviewed in Christin et al. 2015) or based on leaf abundance reaching 500 rpkm in C4 species for the genes not included in previous reviews. The effect of the abundance in the C3 relatives on the co-option probability was modeled as for grasses, independently for Cleomaceae and Flaveria. Because two C3 species are available for Flaveria, their average abundance was used. Root abundance was not available for the same species, so that the effect of leaf specificity in these groups of eudicots could not be tested.
Positive Selection Tests
Codon models were used to test for positive selection following the co-option of genes for C4 photosynthesis. For each group of co-orthologs that has been co-opted at least once for C4, the inferred alignment was truncated as needed to remove poorly aligning ends and a new phylogenetic tree was inferred with phyML, considering only third positions of codons to remove potential biases due to adaptive evolution. The inferred topology was used to optimize three different codon models, using codeml as implemented in PAML (Yang 2007). These models rely on the ratio of nonsynonymous mutation rate per synonymous mutation rate (ω; Yang and Nielsen 2002, 2008; Yang and Swanson 2002). In the null model M1a, codons evolve under either purifying or relaxed selection in all branches (ω smaller than and equal to one, respectively). In the branch-site models, some codons still evolve under neutral or purifying selection in all branches, but others shift from purifying or relaxed selection in background branches to relaxed (in model A) or positive (in model A1) selection in foreground branches. These foreground branches are defined a priori. In our case, all branches descending from each C4 co-opted gene (identified above for the species sequenced here and from the literature for the rest of species) were set as the foreground branches. Because genes for βCA were present at similar abundance in non-C4 and C4 species (see “Results” section), but these are known to be part of the C4 pathway (Budde et al.1985; Hatch and Burnell 1990), all branches leading to C4 species in these gene families were selected as foreground branches. The fit improvement of the model assuming changes in selection pressures was evaluated using likelihood ratio tests. The model A1 was first compared with the model M1a, to test for selective shifts following the co-option event, and then to the model A to specifically test whether the shift corresponded to positive selection. P-values were corrected for multiple testing.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the Royal Society (RG130448, URF120119) and the Natural Environment Research Council (NE/M00208X/1). All sequences generated in this work have been submitted to NCBI Sequence Read Archive and Transcriptome Shotgun Assembly repository (BioProject PRJNA395007).
References
- Aharoni A, Gaidukov L, Khersonsky O, Gould SM, Roodveldt C, Tawfik DS.. 2005. The ‘evolvability’ of promiscuous protein functions. Nat Genet. 37:73–76. [DOI] [PubMed] [Google Scholar]
- Atkinson RRL, Mockford EJ, Bennett C, Christin PA, Spriggs EL, Freckleton RP, Thompson K, Rees M, Osborne CP.. 2016. C4 photosynthesis boosts growth by altering physiology, allocation and size. Nat Plants. 2(5):16038.. [DOI] [PubMed] [Google Scholar]
- Aubry S, Brown NJ, Hibberd JM.. 2011. The role of proteins in C3 plants prior to their recruitment into the C4 pathway. J Exp Bot. 62(9):3049–3059. [DOI] [PubMed] [Google Scholar]
- Aubry S, Kelly S, Kümpers BMC, Smith-Unna RD, Hibberd JM, Bomblies K.. 2014. Deep evolutionary comparison of gene expression identifies parallel recruitment of trans-factors in two independent origins of C4 photosynthesis. PLoS Genet. 10(6):e1004365.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard G, Muasya AM, Russier F, Roalson EH, Salamin N, Christin PA.. 2009. Phylogenomics of C4 photosynthesis in sedges (Cyperaceae): multiple appearances and genetic convergence. Mol Biol Evol. 26(8):1909–1919. [DOI] [PubMed] [Google Scholar]
- Bläsing OE, Westhoff P, Svensson P.. 2000. Evolution of C4 phosphoenolpyruvate carboxylase in Flaveria-a conserved serine residue in the carboxyterminal part of the enzyme is a major determinant for C4-specific characteristics. J Biol Chem. 275:27917–27923. [DOI] [PubMed] [Google Scholar]
- Blount ZD, Barrick JE, Davidson CJ, Lenski RE.. 2012. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature 489(7417):513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A, Kajala K, Wullenweber J, Sommer M, Gagneul D, Weber KL, Carr KM, Gowik U, Mass J, Lercher MJ, et al. 2011. An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiol. 155(1):142–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A, Schliesky S, Külahoglu C, Osborne CP, Weber APM.. 2014. Towards an integrative model of C4 photosynthetic subtypes: insights from comparative transcriptome analysis of NAD-ME, NADP-ME, and PEP-CK C4 species. J Exp Bot. 65(13):3579–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NJ, Newell CA, Stanley S, Chen JE, Perrin AJ, Kajala K, Hibberd JM.. 2011. Independent and parallel recruitment of preexisting mechanisms underlying C4 photosynthesis. Science 331(6023):1436–1439. [DOI] [PubMed] [Google Scholar]
- Budde RJA, Holbrook GP, Chollet R.. 1985. Studies on the dark/light regulation of maize leaf pyruvate, orthophosphate dikinase by reversible phosphorylation. Arch Biochem Biophys. 242(1):283–290. [DOI] [PubMed] [Google Scholar]
- Byrt CS, Grof CPL, Furbank RT.. 2011. C4 plants as biofuel feedstocks: optimising biomass production and feedstock quality from a lignocellulosic perspective. J Integr Plant Biol. 53(2):120–135. [DOI] [PubMed] [Google Scholar]
- Carroll SB. 2008. Evo-Devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134(1):25–36. [DOI] [PubMed] [Google Scholar]
- Christin PA, Arakaki M, Osborne CP, Edwards EJ.. 2015. Genetic enablers underlying the clustered evolutionary origins of C4 photosynthesis in angiosperms. Mol Biol Evol. 32(4):846–858. [DOI] [PubMed] [Google Scholar]
- Christin PA, Boxall SF, Gregory R, Edwards EJ, Hartwell J, Osborne CP.. 2013. Parallel recruitment of multiple genes into C4 photosynthesis. Genome Biol Evol. 5(11):2174–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin PA, Edwards EJ, Besnard G, Boxall SF, Gregory R, Kellogg EA, Hartwell J, Osborne CP.. 2012. Adaptive evolution of C4 photosynthesis through recurrent lateral gene transfer. Curr Biol. 22(5):445–499. [DOI] [PubMed] [Google Scholar]
- Christin PA, Salamin N, Savolainen V, Duvall MR, Besnard G.. 2007. C4 photosynthesis evolved in grasses via parallel adaptive genetic changes. Curr Biol. 17(14):1241–1247. [DOI] [PubMed] [Google Scholar]
- Christin PA, Weinreich DM, Besnard G.. 2010. Causes and evolutionary significance of genetic convergence. Trends Genet. 26(9):400–405. [DOI] [PubMed] [Google Scholar]
- Christin PA, Samaritani E, Petitpierre B, Salamin N, Besnard G.. 2009. Evolutionary insights on C4 photosynthetic subtypes in grasses from genomics and phylogenetics. Genome Biol Evol. 1:221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboule D, Wilkins AS.. 1998. The evolution of “bricolage.” Trends Genet. 14(2):54–59. [DOI] [PubMed] [Google Scholar]
- Dunn CW, Howison M, Zapata F.. 2013. Agalma: an automated phylogenomics workflow. BMC Bioinformatics 14:330.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning LT, Lundgren MR, Moreno-Villena JJ, Namaganda M, Edwards EJ, Nosil P, Osborne CP, Christin PA.. 2017. Introgression and repeated co-option facilitated the recurrent emergence of C4 photosynthesis among close relatives. Evolution 71(6):1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D.. 2000. Determinants of substitution rates in mammalian genes: expression pattern affects selection intensity but not mutation rate. Mol Biol Evol. 17(1):68–70. [DOI] [PubMed] [Google Scholar]
- Ebersberger I, Strauss S, von Haeseler A.. 2009. HaMStR: profile hidden markov model based search for orthologs in ESTs. BMC Evol Biol. 9:157.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehleringer JR, Cerling TE, Helliker BR.. 1997. C4 photosynthesis, atmospheric CO2, and climate. Oecologia 112(3):285–299. [DOI] [PubMed] [Google Scholar]
- Emms DM, Covshoff S, Hibberd JM, Kelly S.. 2016. Independent and parallel evolution of new genes by gene duplication in two origins of C4 photosynthesis provides new insight into the mechanism of phloem loading in C4 species. Mol Biol Evol. 33(7):1796–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fracasso A, Trindade LM, Amaducci S.. 2016. Drought stress tolerance strategies revealed by RNA-Seq in two sorghum genotypes with contrasting WUE. BMC Plant Biol. 16(1):115.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furbank RT, Chitty JA, Jenkins CLD, Taylor WC, Trevanion SJ, von Caemmerer S, Ashton AR.. 1997. Genetic manipulation of key photosynthetic enzymes in the C4 plant Flaveria bidentis. Aus J Plant Physiol. 24(4):477–485. [Google Scholar]
- GPWGII – Grass Phylogeny Working Group II. 2012. New grass phylogeny resolves deep evolutionary relationships and discovers C4 origins. New Phytol. 193(2):304–312. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnol. 29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O.. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52(5):696–704. [DOI] [PubMed] [Google Scholar]
- Hatch MD. 1987. C4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Biochim Biophys Acta 895(2):81–106. [Google Scholar]
- Hatch MD, Osmond CB.. 1976. Compartmentation and transport in C4 photosynthesis In: Stocking CR, Heber U editors. Transport in Plants III. Berlin, Heidelberg: Springer Berlin Heidelberg; p. 144–184. [Google Scholar]
- Hatch MD, Burnell JN.. 1990. Carbonic anhydrase activity in leaves and its role in the first step of C4 photosynthesis. Plant Physiol. 93(2):825–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann D, Schulze S, Denton A, Gowik U, Westhoff P, Weber APM, Lercher MJ.. 2013. Predicting C4 photosynthesis evolution: modular, individually adaptive steps on a Mount Fuji fitness landscape. Cell 153(7):1579–1588. [DOI] [PubMed] [Google Scholar]
- Hibberd JM, Quick WP.. 2002. Characteristics of C4 photosynthesis in stems and petioles of C3 flowering plants. Nature 415(6870):451–454. [DOI] [PubMed] [Google Scholar]
- Hibberd JM, Covshoff S.. 2010. The regulation of gene expression required for C4 photosynthesis. Annu Rev Plant Biol. 61:181–207. [DOI] [PubMed] [Google Scholar]
- Huang P, Studer AJ, Schnable JC, Kellogg EA, Brutnell TP.. 2016. Cross species selection scans identify components of C4 photosynthesis in the grasses. J Exp Bot. 68:127–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, O’Donnell AJ, Barboline JJ, Barkman TJ.. 2016. Convergent evolution of caffeine in plants by co-option of exapted ancestral enzymes. Proc Natl Acad Sci USA. 113:10613–10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins CD, Wallbank RWR, Hanly JJ.. 2017. Waiting in the wings: what can we learn about gene co-option from the diversification of butterfly wing patterns? Philos Trans R Soc Lond B Biol Sci. 372(1713):20150485.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John CR, Smith-Unna RD, Woodfield H, Covshoff S, Hibberd JM.. 2014. Evolutionary convergence of cell-specific gene expression in independent lineages of C4 grasses. Plant Physiol. 165(1):62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajala K, Brown NJ, Williams BP, Borrill P, Taylor LE, Hibberd JM.. 2012. Multiple Arabidopsis genes primed for recruitment into C4 photosynthesis. Plant J. 69(1):47–56. [DOI] [PubMed] [Google Scholar]
- Külahoglu C, Denton AK, Sommer M, Maß J, Schliesky S, Wrobel TJ, Berckmans B, Gongora-Castillo E, Buell CR, Simon R, et al. 2014. Comparative transcriptome atlases reveal altered gene expression modules between two Cleomaceae C3 and C4 plant species. Plant Cell 26(8):3243–3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CR, Lemos B, Rifkin SA, Dickinson WJ, Hartl DL.. 2007. Genetic properties influencing the evolvability of gene expression. Science 317(5834):118–121. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort V, Longueville JE, Gascuel O.. 2017. SMS: Smart Model Selection in PhyML. Mol Biol Evol. 34(9):2422–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xu J, Ul Haq N, Zhang H, Zhu XG.. 2014. Was low CO2 a driving force for C4 evolution: Arabidopsis responses to long-term low CO2 stress. J Exp Bot. 65(13):3657–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Ma X, Zhao J, Xu J, Shi J, Zhu XG, Zhao Y, Zhang H.. 2015. Developmental genetic mechanisms of C4 syndrome based on transcriptome analysis of C3 cotyledons and C4 assimilating shoots in Haloxylon ammodendron. Plos One 10(2):e0117175.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhou M, Gao Z, Ren W, Yang F, He H, Zhao J.. 2015. RNA-Seq analysis reveals MAPKKK family members related to drought tolerance in maize. Plos One 10(11):e0143128.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu MA, Gowik U, Kelly S, Covshoff S, Mallmann J, Westhoff P, Hibberd JM, Stata M, Sage RF, Lu H, et al. 2015. RNA-Seq based phylogeny recapitulates previous phylogeny of the genus Flaveria (Asteraceae) with some modifications. BMC Evol Biol. 15(1):116.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren MR, Besnard G, Ripley BS, Lehmann CER, Chatelet DS, Kynast RG, Namaganda M, Vorontsova MS, Hall RC, Elia J, et al. 2015. Photosynthetic innovation broadens the niche within a single species. Ecol Lett. 18(10):1021–1029. [DOI] [PubMed] [Google Scholar]
- Mallmann J, Heckmann D, Bräutigam A, Lercher MJ, Weber APM, Westhoff P, Gowik U.. 2014. The role of photorespiration during the evolution of C4 photosynthesis in the genus Flaveria. Elife 3:e02478.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazzi B, Ané C, Simon MF, Delgado-Salinas A, Luckow M, Sanderson MJ.. 2012. Locating evolutionary precursors on a phylogenetic tree. Evolution 66(12):3918–3930. [DOI] [PubMed] [Google Scholar]
- Masumoto C, Miyazawa SI, Ohkawa H, Fukuda T, Taniguchi Y, Murayama S, Kusano M, Saito K, Fukayama H, Miyao M.. 2010. Phosphoenolpyruvate carboxylase intrinsically located in the chloroplast of rice plays a crucial role in ammonium assimilation. Proc Natl Acad Sci USA. 107(11): 5226–5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min XJ, Butler G, Storms R, Tsang A.. 2005. OrfPredictor: predicting protein-coding regions in EST-derived sequences. Nucleic Acids Res. 33(Web Server):W677–W680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monson RK. 2003. Gene duplication, neofunctionalization, and the evolution of C4 photosynthesis. Int J Plant Sci. 164(S3): S43–S54. [Google Scholar]
- Monteiro A, Podlaha O.. 2009. Wings, horns, and butterfly eyespots: How do complex traits evolve? Plos Biol. 7(2):0209–0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montiel EE, Badenhorst D, Tamplin J, Burke RL, Valenzuela N.. 2017. Discovery of the youngest sex chromosomes reveals first case of convergent co-option of ancestral autosomes in turtles. Chromosoma 126(1):105–113. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K.. 2004. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 20(2):289–290. [DOI] [PubMed] [Google Scholar]
- Pearcy RW, Ehleringer J.. 1984. Comparative ecophysiology of C3 and C4 plants. Plant Cell Environ. 7(1):1–13. [Google Scholar]
- R Development Core Team. 2016. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/.
- Sage RF, Christin PA, Edwards EJ.. 2011. The C4 plant lineages of planet Earth. J Exp Bot. 62(9):3155–3169. [DOI] [PubMed] [Google Scholar]
- Sage RF, Sage TL, Kocacinar F.. 2012. Photorespiration and the evolution of C4 photosynthesis. Annu Rev Plant Biol. 63:19–47. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, Östlund G.. 2014. In Paranoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 43(D1):D232–D239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiffler MA, Hekstra DR, Ranganathan R.. 2015. Evolvability as a function of purifying selection in TEM-1 β-lactamase. Cell 160(5):882–892. [DOI] [PubMed] [Google Scholar]
- Still CJ, Berry JA, Collatz GJ, DeFries RS.. 2003. Global distribution of C3 and C4 vegetation: carbon cycle implications. Glob Biogeochem Cycles. 17(1):6-1–6–14. [Google Scholar]
- Svensson P, Bläsing OE, Westhoff P.. 2003. Evolution of C4 phosphoenolpyruvate carboxylase. Arch Biochem Biophys. 414(2):180–188. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S.. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Bowers JE, Wang X, Paterson AH.. 2010. Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. Proc Natl Acad Sci U S A. 107(1):472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tausta SL, Miller Coyle H, Rothermel B, Stiefel V, Nelson T.. 2002. Maize C4 and non-C4 NADP-dependent malic enzymes are encoded by distinct genes derived from a plastid-localized ancestor. Plant Mol Biol. 50:635–652. [DOI] [PubMed] [Google Scholar]
- Tenaillon O, Rodríguez-Verdugo A, Gaut RL, McDonald P, Bennett AF, Long AD, Gaut BS.. 2012. The molecular diversity of adaptive convergence. Science 335(6067):457–461. [DOI] [PubMed] [Google Scholar]
- Tetu SG, Tanz SK, Vella N, Burnell JN, Ludwig M.. 2007. The Flaveria bidentis beta-carbonic anhydrase gene family encodes cytosolic and chloroplastic isoforms demonstrating distinct organ-specific expression patterns. Plant Physiol. 144(3):1316–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ.. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22(22):4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True JR, Carroll SB.. 2002. Gene Co-Option in Physiological and Morphological Evolution. Annu Rev Cell Dev Biol. 18:53–80. [DOI] [PubMed] [Google Scholar]
- Vicentini A, Barber JC, Aliscioni SS, Giussani LM, Kellogg EA.. 2008. The age of the grasses and clusters of origins of C4 photosynthesis. Global Change Biol. 14(12):2963–2977. [Google Scholar]
- Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E.. 2009. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 19(2):327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Gowik U, Tang H, Bowers JE, Westhoff P, Paterson AH.. 2009. Comparative genomic analysis of C4 photosynthetic pathway evolution in grasses. Genome Biol. 10(6):R68.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich DM, Delaney NF, Depristo MA, Hartl DL.. 2006. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312(5770):111–114. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic Analysis by Maximum Likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R.. 2002. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol. 19(6):908–917. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R.. 2008. Mutation-selection models of codon substitution and their use to estimate selective strengths on codon usage. Mol Biol Evol. 25(3):568–579. [DOI] [PubMed] [Google Scholar]
- Yang Z, Swanson WJ.. 2002. Codon-substitution models to detect adaptive evolution that account for heterogeneous selective pressures among site classes. Mol Biol Evol. 19(1):49–57. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Yura K, Ogura A, Furuya H.. 2014. Cephalopod eye evolution was modulated by the acquisition of Pax-6 splicing variants. Sci Rep. 4:4256.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Liu X, Quan Z, Cheng S, Xu X, Pan S, Xie M, Zeng P, Yue Z, Wang W, et al. 2012. Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat Biotechnol. 30(6):549–554. [DOI] [PubMed] [Google Scholar]
- Zinovieva RD, Piatigorsky J, Tomarev SI.. 1999. O-Crystallin, arginine kinase and ferritin from the octopus lens. Biochim Biophys Acta. 1431(2):512–517. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.