Abstract
Background:
Gulf War Illness (GWI) impacts 25-30% of gulf war veterans. Due to its heterogeneity in both etiology and symptoms, it has been challenging to establish the commonly accepted case definition for GWI. Equally challenging are the understanding of the general mechanism of GWI and the development of biomarkers useful for its clinical diagnosis and treatment.
Objective:
We have observed that chromosome condensation defects can be detected in GWI patients. To document this phenomenon in GWI, we aim to describe and compare different types of chromosomal condensation defects in GWI patients, if possible. Since chromosomal condensation represents an important step of ensuring genome integrity, condensation defects could be used as a potential biomarker of GWI.
Methods:
Lymphocytes from GWI patients have been used for short term cell culture followed by chromosome slide preparation. Both Giemsa staining and multiple color spectral karyotyping (SKY) were applied to study chromosome aberrations, focusing on different types of condensation defects.
Results:
At least three subtypes of Defective Mitotic Figures (DMFs) were observed. Some individuals displayed elevated frequencies of DMFs. Another type of condensation defect identified as sticky chromosomes were also observed.
Conclusion:
Various types of condensation defects have been observed in GWI patients. It is rather surprising that some GWI patients exhibited a high level of chromosomal condensation defects. Previously, the elevated frequency of DMFs was only observed in cancer patients. Since chromosome condensation can be linked to other types of chromosome aberrations, as well as cellular stress conditions, the detailed mechanism and clinical impact should be further studied, especially with increased sample size.
Keywords: Chromosome aberrations, Defective Mitotic Figures (DMFs), Fuzzy inheritance, Genome instability, Genome heterogeneity, Gulf war illness (GWI), Stress, Sticky chromosomes
1. Introduction
It has been challenging to understand the general mechanism of Gulf War Illness (GWI) due to its highly heterogeneous characteristics from its etiology to symptoms [1, 2]. In recent years, many different types of molecular/cellular and phenotype defects have been identified including mitochondria defects, genetic network alterations, dysregulation of immunity, and neuro-images [3-5]. Meanwhile, many studies have focused on identifying Gulf War specific chemical or biological agents’ exposure over 25 years ago, including the use of animal models to mimic the original exposure [6]. A general viewpoint is that these unique exposures represent major causative factors for GWI, despite that it was challenging to directly identify them. The next question is how these initial trigger factors generate the diverse GWI symptoms which are still dominant in many patients after a quarter of a century. A subsequent and more challenging question is how to apply these molecular mechanisms to diagnose and treat GWI patients.
While the search for different gene-based biomarkers actively continues, the cytogenetic characterization of GWI is limited, possibly due to the dominance of various high resolution molecular methodologies over the past 25 years [7, 8]. For example, the frequency of chromosomal translocations including dicentric chromosomes has been linked to the Depleted Uranium (DU) exposure [9], however reports about this link have been inconsistent [10]. By using more advanced methods, our group has observed elevated frequencies of chromosomal aberrations from a few GWI patients including those not exposed to DU [11]. The elevated frequency of chromosomal aberrations suggests that GWI is linked to the overall genome instability, an observation which we have confirmed by analyzing more samples [11; Heng et al., submitted]. This observation explains the diverse molecular defects as well as the variable symptoms, as genome instability can unify a large number of molecular pathways, which has been demonstrated in cancer research [12, 13]. Interestingly, different types of chromosomal aberrations including translocations, aneuploidy, chromosome fragmentation (C-Frag), Defective Mitotic Figures (DMFs), and clusters of small nuclei have been observed in different GWI individuals [7, 14]. Many of these chromosomal/nuclear abnormalities have been ignored in cytogenetic studies as they are classified as Non Clonal Chromosome Aberrations, or NCCAs, rather than Clonal Chromosome Aberrations, or CCAs. Traditionally, many NCCAs were considered artifacts that were generated from slide preparation [14-16]. Clonal translocations are often reported, especially when they are likely involved with some fusion genes. Stochastic translocations that are present at low frequencies were often considered as genetic noise. Recently, by linking system inheritance or the genetic blueprint to karyotype, chromosomes no longer are just the vehicle of genes, but code for a new layer of genomic information, the interactive relationship among genes and other DNA sequences [7, 17, 18]. Furthermore, the genetic information that is defined by the genomic coding system is not precise but fuzzy, which explains well the high level of NCCAs detected, as well as the large numbers of the de novo gene mutation and epigenetic variations [19, 20]. Knowing the important role NCCAs play in monitoring genome instability, more attention should be paid to systematically characterize these under-reported aberration types, and to study their contribution to diseases.
Chromosome condensation defects have been previously observed from various animal and human chromosome preparations. Following drug treatment in phase G2 of the cell cycle, partially condensed chromosomes can be obtained from both amphibian and human lymphocyte culture [21, 22]. In addition to the induced unit fibers, the partially condensed mitotic figures connect unit fibers and packed chromosomes, offering the intermediate structure to study the high order chromosomal packaging. It was assumed that the inhibition of topoisomerase II (Topo II) can interfere with the normal process of chromosome condensation, as Topo II is a major structure protein for chromosomes, generating the transitional structure. Initially, the “uncompleted-packing- mitotic figures” or UPM was used to describe these images with mixed condensed and uncondensed chromosomes. It was suggested that the condensation defect can explain a portion of total chromosomal aberrations generated from all phases of the cell cycle [23]. It is interesting to point out that the elongated chromosome within UPM has been successfully used to establish high resolution fiber FISH methodology [24]. The new term, Defective Mitotic Figures or DMFs, has been subsequently used when they were frequently detected from cancer samples without drug induction [8, 25]. This led to the realization that the increased frequency of DMFs reflected increased genome instability. DMFs can be linked to mitotic cell death as well as aneuploidy [26, 27]. Interestingly, sticky chromosomes, another type of chromosome aberration, might be related to the condensation process as well.
Here we report that the high frequency of DMFs can be detected from GWI patients with routine cytogenetic preparation from short term lymphocyte culture without the use of any drug treatment. Furthermore, high frequencies of other types of the chromosome condensation defects, including sticky chromosomes, can also be observed. Interestingly, even though different types of condensation defects can be observed from the same patients, some individuals will dominantly display one subtype. Following a description of different types of DMFs and sticky chromosomes, the potential mechanism of the condensation defects is discussed.
2. Materials and Methods
2.1. Sample Collection
Ten GWI patients and eight normal control individuals were included in this study. Patient samples were collected both from local VA clinics and through the patient network. The normal control samples were collected from VA clinics and the Detroit Medical Center. The patients were identified using the Fukuda case definition [28]. Patient consent was approved by Wayne State University, Veterans Affairs (VA), and the Department of Defense (DOD).
Fresh lymphocytes (5 ml each per subject) were collected for each individual. Short term blood culture was performed for each sample. Briefly, lymphocytes were isolated from 3-5 ml of fresh peripheral blood by low-speed centrifugation. 0.2 ml lymphocytes were collected using a transfer pipette, gently aspirated, and transferred into 5 mL of cell culture medium in a 15 mL cell culture tube. Lymphocytes were cultured for 52 hours at 37°C.
2.2. Cytogenetic Analyses
Following 52 hours cell culture, lymphocyte cells were harvested and treated with hypotonic solution (0.4% KCL, 10 minutes at 37°C), followed by three runs of fixation with methanol/acetic acid fixation (3:1). Chromosomal slides were made and subsequently air-dried [29].
Following Giemsa or DAPI staining, cytogenetic analyses were performed for all cases to study DMFs and sticky chromosomes. In addition, other types of abnormalities such as chromosome fragmentations (C-Frag) and chromatin bridges were studied. Digital imaging of chromosomal/nuclear abnormalities was captured using image system.
SKY (spectral karyotype) was performed according to published protocol [15, 30]. Briefly, the dehydrated chromosome slides were denatured in 70% formamide and 2xSSC and hybridized with denatured human painting probes (SKYPaint) for over 48 h at 37°C. Signals were detected using a detection kit following slide washing. Mitotic figures with sticky chromosomes were randomly captured using the image system.
The overall frequencies of DMFs and sticky chromosomes were measured by scoring the number of DMFs and sticky chromosomes among normal mitotic figures for all patients (based on Giemsa and DAPI staining). For each sample, 100-200 mitotic figures were analyzed and the frequencies of DMFs and sticky chromosomes were calculated for each sample.
3. Results
3.1. Different Types of DMFs were Observed
Different types of DMFs can be detected including: 1) the typical polarized shape where the condensed chromosomes grouped at one end and the un-condensed chromatin in the opposite direction; 2) the more random type where the de-condensed chromosomes were randomly distributed; and 3) the isolated de-condensed chromosomes within otherwise normal mitotic figures (often 1-3 defused chromosomes which differs from chromosome fragmentation). In addition, some isolated de-condensed chromosomes were also observed. A few examples of DMFs are listed in Fig. (1).
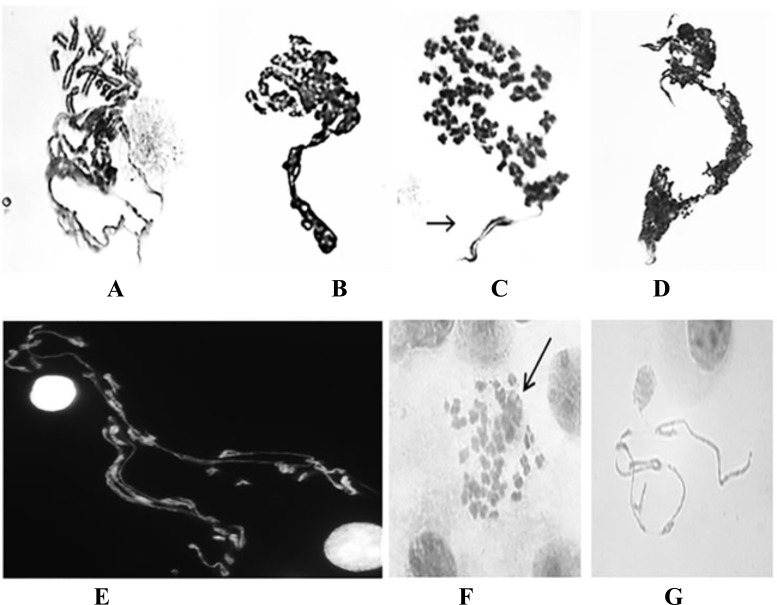
Fig. (1).
Examples of DMFs detected from GWI patients. A-C) Type 1 DMFs with the typical polarizing shape where the condensed chromosomes group at one end and the un-condensed chromatin in the opposite direction (Giemsa staining). In C) an arrow indicates a less condensed chromosome. D-E) Type 2 DMFs with more random distribution of de-condensed chromosomes. In D), there is a mixture of DMFs and sticky chromosomes (Giemsa staining). In E), there is a mixture of condensed and de-condensed chromosomes (DAPI staining). F) Type 3 DMF, a few chromosomes display diffused morphology among most seemingly normal chromosomes (Giemsa staining). Note that the morphology of defused chromosomes differs from chromosome fragmentation. G) A few isolated de-condensed chromosomes (Giemsa staining).
Of 10 randomly selected GWI patients, 2 displayed highly elevated frequencies of DMFs (over 10%), and 7 displayed 3-4%. In contrast, the control group usually had frequencies of 0-2% (Table 1).
Table 1.
Different types of DMFs were observed.
| Samples | DMF | Sticky Chromosome |
|---|---|---|
| #1 | 3% | 10% |
| #2 | 11% | 11% |
| #3 | 2% | 26% |
| #4 | 3% | 0% |
| #5 | 10% | 11% |
| #6 | 3% | 21% |
| #7 | 3% | 3% |
| #8 | 4% | 3% |
| #9 | 3% | 6% |
| #10 | 4% | 11% |
| Control | 0-2% | 1-2% |
Morphologically speaking, the de-condensed chromatin fibers from this study were less distinctive than those induced DMFs observed in frogs [21, 22], where the sister unit fibers were clearly visible. This difference may be due to the fact that the degree of chromosome condensation in frog chromosomes is about 3 times higher than human chromosomes, and it is easier to detect unit fibers. In addition, the DMFs observed from GWI samples that were not drug induced, while the DMFs observed from frog were drug induced.
3.2. Different Types of Sticky Chromosomes were Observed
Various sticky chromosomes were detected from GWI patients. Similar to our previous studies, sticky chromosomes either existed in a limited region of a given mitotic figure, or affect all or most chromosomes (Figs. 2-3). The identification of sticky chromosomes is straightforward. The chromosomes were already formed, but some chromatin fibers were visible among different chromosomes. During slide preparation, chromosomes will spread out evenly with minimal overlap between chromosomes. Sticky chromosomes are present if some mitotic figures display a phenotype where chromosomes are stuck together (either partially or wholly). They should be classified as exhibiting some defect as opposed to its designation as an artifact from slide preparation. Among 10 GWI patients, six patients displayed a higher frequency of mitotic figures with sticky chromosomes (10-26%) compared with control samples (1-2%) (Table 1). Two patients display elevated DMFs and sticky chromosomes. Previously, sticky chromosomes have been observed following drug treatment including ethidium bromide (EB) and mitomycin C (MMC). Sticky chromosomes in GWI samples, however, were detected without any drug treatment. To confirm that some chromosome bundles were caused by sticky chromosomes rather than uncondensed chromatin fibers, SKY was used to visualize the condensed chromosomes. If the defined signals can be observed within these bundles, it suggests the presence of condensed chromosomes (Fig. 3A-B). It should be pointed out that various stressful conditions (physiological or pathological) are linked to the observations of sticky chromosomes. The elevated frequencies of sticky chromosomes in GWI patients might reflect their cellular stress conditions.
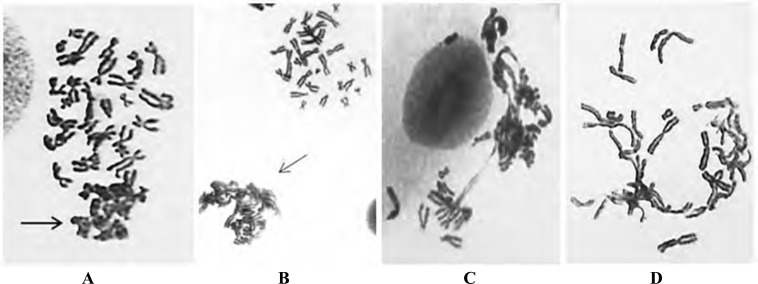
Fig. (2).
Images of sticky chromosomes. A) a portion of the mitotic figure displays sticky chromosomes where multiple sticky chromosomes form a cluster (as indicated by the arrows) (Gimesa staning); B) a comparison between non-sticky chromosomes (top right) and sticky chromosomes (indicated by an arrow), which is different from 2A as the sticky chromosome cluster likely belongs to a different mitotic figure; C-D) sticky chromosomes are detected across the entire mitotic figure. In picture D, many sticky chromosomes have fused together.
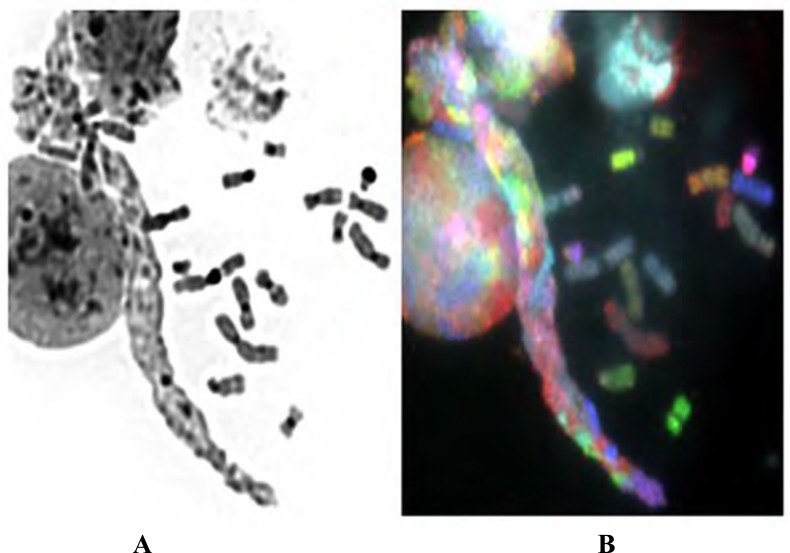
Fig. (3).
Reversed DAPI (A) and SKY image (B) of sticky chromosomes, normal chromosomes, and interphase nuclei. Many sticky chromosomes form a chromosome bundle. The reverse-DAPI image provides less detailed information, while the SKY image shows the chromosomes sticking together (this is not a bundle of de-condensed chromatin fibers), as the defined color domain is visible, which is different from the interphase signals, revealing the condensed chromosomal status.
3.3. Various Types of Chromosomal/Nuclear Abnormalities were Observed
In addition to DMFs and sticky chromosomes, chromosome translocations, chromosome breakages, C-Frag, chromosome bridges, free chromatin, and clusters of small nuclei were also observed in different frequencies for each individual sampled. A systematic study was performed that included all types of abnormalities in measuring the overall genome instability (Ye et al., manuscript in preparation). Even though the focus of this specific report is on the morphological description of chromosomal condensation defect subtypes observed in GWI patients (DMFs and sticky chromosomes), readers need to be aware that there are many other chromosomal aberration subtypes in GWI patients. An in depth analysis is needed to compare these different subtypes, as well as types of chromosomal/nuclear abnormalities, and link them to GWI. For example, chromosome condensation subtypes may be used for classifying GWI patients into different subgroups. In that case, more samples will be needed.
4. Discussion
The identification of new chromosomal aberration subtypes such as DMFs and sticky chromosomes occurring in high frequencies in GWI patients holds significant importance. It has been challenging to identify biomarkers for GWI diagnosis, and any new development in this front is welcome. Our studies not only link the specific types of chromosomal aberrations to some GWI patients, but also link increased genome instability to a majority of GWI patients. Both DMFs and sticky chromosomes are classified as non clonal chromosome aberrations or NCCAs [16], and the rate of NCCAs has been used to measure genome instability [13]. This is particularly true for those patients who failed to display a high frequency of chromosomal translocations but did exhibit an increased frequency of DMFs and/or sticky chromosomes. Increased frequencies of DMFs and sticky chromosome have also been observed in cancer patients. The observation that they also can be detected in GWI patients was initially very surprising, as cancer and GWI share little similarities. Our conceptual realization that genome instability-mediated cellular evolution is the basis for many common and complex diseases/illness can explain rather well why a high level of DMFs and sticky chromosomes were detected in GWI samples. A broader implication of this finding is to study other diseases/illness conditions to investigate if chromosome condensation defects are also involved. Based on the importance of chromosome condensation in cell cycle regulation and the integrity of the genome, we anticipate that will be the case.
As different individuals can display different subtypes of chromosome condensation defects dominantly, it suggests the possibility that different molecular mechanisms might be involved for different GWI patients. There are a few lines of thought on the mechanisms of DMFs and sticky chromosomes, even though they all result in impaired condensation. For example, while the main mechanism for DMFs may involve the G2 checkpoint and a chromosome condensation defect, as it is easy to induce DMFs in ATM -/- cells with topoisomerase II inhibitors [8, 16], it can also be generated by delaying replication time and then delaying chromosome condensation [31]. DMFs can also be induced by heterochromatin under-condensation following the treatment of 5-Aza-dC [32]. As there is a chromosome cycle involving replication, condensation, segregation and de-condensation, mistakes or variations among these many steps can impact condensation [23]. Therefore, there are direct and indirect mechanisms that can lead to condensation defects. Similarly, there are many factors that can contribute to the generation of sticky chromosomes, from drug induced sticky chromosome (such as treatment of ethidium bromide or MMC), to DNA methylation status [33], and chromosome slide preparation. In fact, the term “stickiness” was first used by Beadle in 1932 to describe the sticky aspect of chromosomes in mutated maize cells [34]. Since then, genetically controlled stickiness was also reported for different plant species as well as in animal cells [35]. Various factors have been linked to this phenomenon, including radiation exposure, temperature changes, different chemical treatments, hybridization, and chromosomal dynamics [36], (Heng et al., unpublished observations). However, the common cause and biochemical basis of this phenomenon are unknown. Recently, to solve the puzzle of why there are high levels of genetic “noise” or heterogeneity at all genomic levels, a new concept of “fuzzy inheritance” was introduced [13, 14, 18]. Different from traditional inheritance concepts, fuzzy inheritance defines a range of genetic information or potential, rather than a precise status to be genetically passed on. Equally importantly, the boundaries of fuzzy inheritance can be altered under stressful conditions. Based on the observation that sticky chromosomes are often associated with stress, the elevated frequencies of sticky chromosomes observed in GWI patients likely reflect a cellular stress status.
We also noticed that, for some normal individual, prolonged hypotonic treatment can increase the frequency of sticky chromosomes as well. Interestingly, some cancer cell lines extensively display the phenomenon of sticky chromosomes (Anja Weise and Thomas Liehr, personal communication). These cells seem to have high plasticity with respect to the gain or loss of genetic material during cell division, which might just represent a mechanism of fuzzy inheritance at the chromosome level (manuscript in preparation). Recently, a specific chromosome surface protein has been identified, which may reduce the stickiness of chromatin to prevent chromosomes from sticking together [37]. Specifically, Ki-67 protein likely forms a steric and electrostatic charge barrier to avoid chromatin on different chromosomes from tangling with each other. Since interfering with chromatin condensation also can generate a similar phenotype for some cells, it is clear that many molecular mechanisms can lead to the same phenotype of sticky chromosomes. It is interesting to point out that, during genome chaos (which can be triggered by cancer drug treatment) there are increased frequencies of the sticky nuclei as well [8, 30], (Ye et al., unpublished observations). A similar phenomenon can also be observed in GWI patients at low frequencies.
There are other types of chromosomal/nuclear abnormalities detected from GWI patients as well. A systematic comparison is needed for future research to study their contribution to final structural and numerical changes of the chromosomes, and the relationship among different types of abnormalities. For example: whether there is any direct link between DMFs and chromosomal translocations; or whether there is a link between sticky chromosomes and the C-Frag. Knowing that C-Frag represents an effective way for mitotic cell death [26, 27], and some sticky chromosomes can lead to cell death when they block the cell cycle, one might anticipate the coupled frequency of both C-Frag and sticky chromosomes. Similarly, C-Frag, DMFs and sticky chromosomes can be linked to aneuploidy. Equally important is how these aberrations can be used for grouping patients, and linking them to a diagnostic and treatment value. It should be pointed out that, both DMFs and sticky chromosomes can even be observed in normal individuals. However, their frequencies are usually low. These few cases we reported here display much higher frequencies. One challenge for the diagnosis of GWI and some other common and complex illnesses is that, often, there is overlap of a given biomarker between patients with moderate symptoms and normal control (it is relatively easier to diagnose patients with severe symptoms who often display a high level of chromosomal instability). Recently, we have proposed that a certain degree of genetic variation including at the chromosome level is essential for cellular adaptation [19], it is thus not surprising to observe DMFs and sticky chromosome in normal individual, as long as the frequencies are not high. A related clinical question for GWI patients is whether GWI patients with high frequencies of DMFs is associated with cancer incidence. As the high frequency of DMFs can be frequently observed in the cancer genome, developing a quantitative model that uses the profile of DMFs and sticky chromosomes for clinical use is essential.
Systematically characterize different chrosmomal aberrations and their combinational contribution to diseases also represents a new direction for cytogenetics and cytogenomics [16, 38]. Prior to the genome sequencing era, the highest hope for treating genetic disorders was the characterization of individual genes that cause a given disease. Cytogenetic studies provided the needed help to narrow down the physical regions on the chromosome to clone these genes. The gene centric mechanistic understanding of the disease requires the link of a specific gene mutation to the disease phenotype, a so-called molecular mechanism. A good example is the identification of Philadelphia chromosome which led to the identification of the BCR-ABL fusion gene found in Chronic Myelogenous Leukemia (CML) [39, 40]. Unfortunately, it has been difficult to clone commonly shared key driver gene mutations in a majority of cancer cases, as most driver genes exist in low frequencies within the patient population. Furthermore, many well-known driver mutations often are accompanied by hundreds if not thousands of other gene mutations in a stochastic fashion, and the clinical prediction based on individual genes is rather less certain. Together, the massive amount of sequencing data cataloging the thousands of gene mutations has ultimately challenged the gene mutation theory for cancer and many other diseases, and calls for a new framework to explain the confusion in genomics [7, 8, 12, 13, 20]. One of the new ideas is the re-evaluation of the significance of the chromosomal sets or karyotype in genomics. According to genome theory, the karyotype represents a new chromosome based genetic coding. The heterogeneity at the chromosome level is essential for understanding somatic cell evolution, which is the evolutionary and general cellular basis for many common and complex diseases/illnesses. How different types of chromosomal/nuclear variations can impact cellular adaptation and evolution (rather than individual genes) will initiate a new and exciting phase of genetic/genomic research [19]. The documentation of chromosome heterogeneity in various disease/illness conditions including GWI thus represents a timely effort.
CONCLUSION
Various types of chromosomal condensation defects including DMFs and sticky chromosomes were observed from the cultured lymphocytes cells of GWI patients with elevated frequencies. Since chromosomal condensation defects represent important subtypes of chromosomal aberrations, and the elevated number of sticky chromosomes may reflect increased cellular stress, further studies are needed to illustrate the relationship between these chromosomal condensation defects and other subtypes of chromosomal aberrations. In particular, additional research is needed to test if chromosomal aberrations can be used as potential GWI diagnosis biomarkers.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The use of human samples was approved by the Wayne State University, Veterans Affairs (VA) and the Department of Defense (DOD).
HUMAN AND ANIMAL RIGHTS
No animals were used for studies that are base of this research. All human research procedures followed were in accordance with Institutional approvals, and the laws which apply to them were followed.
CONSENT FOR PUBLICATION
Patient consent was approved by Wayne State University, Veterans Affairs (VA), and the Department of Defense (DOD).
ACKNOWLEDGEMENTS
This work was supported by a grant from the DOD (GW093028). We thank the Discovery Channel and Dr. Asaf Durakovic for introducing GWI studies to us. We also appreciate the support of many GWI patients’ in our effort to characterize the chromosome defects of GWI.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Heng H.H. Challenges and new strategies for gulf war illness research. Environ. Dis. 2016;1(4):118–125. [Google Scholar]
- 2.RAC GWVI . Gulf War Illness and the Health of Gulf War Veterans: Research Update and Recommendations, 2009-2013. V. S. Affairs. Boston: Government Printing Office; 2014. [Google Scholar]
- 3.Koslik H.J., Hamilton G., Golomb B.A. Mitochondrial dysfunction in Gulf War illness revealed by 31Phosphorus Magnetic Resonance Spectroscopy: A case-control study. PLoS One. 2014;9(3):e92887. doi: 10.1371/journal.pone.0092887. journals.plos.org/plosone/article?id= 10.1371/journal.pone.0092887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craddock T.J., Harvey J.M., Nathanson L., Barnes Z.M., Klimas N.G., Fletcher M.A., Broderick G. Using gene expression signatures to identify novel treatment strategies in gulf war illness. BMC Med. Genomics. 2015;8:36. doi: 10.1186/s12920-015-0111-3. europepmc.org/abstract/med/26156520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parihar V.K., Hattiangady B., Shuai B., Shetty A.K. Mood and memory deficits in a model of Gulf War illness are linked with reduced neurogenesis, partial neuron loss, and mild inflammation in the hippocampus. Neuropsychopharmacol. 2013;38(12):2348–2362. doi: 10.1038/npp.2013.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White R.F., Steele L., O’Callaghan J.P., Sullivan K., Binns J.H., Golomb B.A., Bloom F.E., Bunker J.A., Crawford F., Graves J.C., Hardie A., Klimas N., Knox M., Meggs W.J., Melling J., Philbert M.A., Grashow R. Recent research on Gulf War illness and other health problems in veterans of the 1991 Gulf War: Effects of toxicant exposures during deployment. Cortex. 2016;74:449–475. doi: 10.1016/j.cortex.2015.08.022. https://www.sciencedirect.com/science/ article/pii/S0010945215003329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heng H.H., Liu G., Stevens J.B., Bremer S.W., Ye K.J., Abdallah B.Y., Horne S.D., Ye C.J. Decoding the genome beyond sequencing: The new phase of genomic research. Genomics. 2011;98:242–252. doi: 10.1016/j.ygeno.2011.05.008. https://www.sciencedirect.com/science/article/ pii/S0888754311001327 [DOI] [PubMed] [Google Scholar]
- 8.Heng H.H., Liu G., Stevens J.B., Abdallah B.Y., Horne S.D., Ye K.J., Bremer S.W., Chowdhury S.K., Ye C.J. Karyotype heterogeneity and unclassified chromosomal abnormalities. Cytogenet. Genome Res. 2013;139(3):144–157. doi: 10.1159/000348682. [DOI] [PubMed] [Google Scholar]
- 9.Schröder H., Heimers A., Frentzel-Beyme R., Schott A., Hoffmann W. Chromosome aberration analysis in peripheral lymphocytes of Gulf War and Balkans War veterans. Radiat. Prot. Dosimetry. 2003;103(3):211–219. doi: 10.1093/oxfordjournals.rpd.a006135. [DOI] [PubMed] [Google Scholar]
- 10.Bakhmutsky M.V., Squibb K., McDiarmid M., Oliver M., Tucker J.D. Long-term exposure to depleted uranium in Gulf-War veterans does not induce chromosome aberrations in peripheral blood lymphocytes. Mutat. Res. 2013;757(2):132–139. doi: 10.1016/j.mrgentox.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 11.2007 https://www.youtube.com/watch?v=jhvdkd
- 12.Heng H.H., Bremer S.W., Stevens J.B., Horne S.D., Liu G., Abdallah B.Y., Ye K.J., Ye C.J. Chromosomal Instability (CIN): What it is and why it is crucial to cancer evolution. Cancer Met. Rev. 2013;32(3-4):325–340. doi: 10.1007/s10555-013-9427-7. [DOI] [PubMed] [Google Scholar]
- 13.Horne S.D., Ye C.J., Heng H.H. 2015 http://onlinelibrary. wiley.com/doi/10.1002/9780470015902.a0006069.pub2/full
- 14.Heng H.H., Horne S.D., Stevens J.B., Abdallah B.Y., Liu G., Chowdhury S.K., Bremer S.W., Zhang K., Ye C.J. Heterogeneity mediated system complexity: the ultimate challenge for studying common and complex diseases [Google Scholar]
- 15.Heng H.H., Stevens J.B., Liu G., Bremer S.W., Ye K.J., Reddy P.V., Wu G.S., Wang Y.A., Tainsky M.A., Ye C.J. Stochastic cancer progression driven by non-clonal chromosome aberrations. J. Cell. Physiol. 2006;208(2):461–472. doi: 10.1002/jcp.20685. [DOI] [PubMed] [Google Scholar]
- 16.Heng H.H., Regan S.M., Liu G., Ye C.J. Why it is crucial to analyze non clonal chromosome aberrations or NCCAs? 2016 doi: 10.1186/s13039-016-0223-2. tics.biomedcentral.com/articles/10.1186/s13039-016-0223-2 [DOI] [PMC free article] [PubMed]
- 17.Heng H.H. The genome-centric concept: Resynthesis of evolutionary theory. BioEssays. 2009;31(5):512–525. doi: 10.1002/bies.200800182. [DOI] [PubMed] [Google Scholar]
- 18.Heng H.H. Debating Cancer: The paradox in cancer research. Singapore: World Scientific Publishing Co.; 2015. [Google Scholar]
- 19.Horne S.D., Chowdhury S.K., Heng H.H. Stress, genomic adaptation, and the evolutionary trade-off. Front. Genet. 2014;5:92. doi: 10.3389/fgene.2014.00092. https://www.frontiersin.org/articles/10.3389/ fgene.2014.00092/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heng H.H., Regan S., Ye C.J. Genotype, environment, and evolutionary mechanism of diseases. Environ. Dis. 2016;1(1):14–23. [Google Scholar]
- 21.Heng H.Q., Chen W.Y. The study of the chromatin and the chromosome structure for Bufo gargarizans by the light microscope. J. Sichuan Univ. Nat. Sci. 1985;2:105–109. [Google Scholar]
- 22.Heng H.Q., Lin R., Zhao X.L., Chen W.Y. Structure of the chromosome and its formation. II. Studies on the sister unit fibers. Nucleus. 1988;30:2–9. [Google Scholar]
- 23.Heng H.Q., Chen W.Y., Wang Y.C. Effects of pingyanymycin on chromosomes: A possible structural basis for chromosome aberration. Mutat. Res. 1988;199(1):199–205. doi: 10.1016/0027-5107(88)90246-1. [DOI] [PubMed] [Google Scholar]
- 24.Heng H.H., Squire J., Tsui L.C. High-resolution mapping of mammalian genes by in situ hybridization to free chromatin. Proc. Natl. Acad. Sci. USA. 1992;89(20):9509–9513. doi: 10.1073/pnas.89.20.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heng H.H., Stevens J.B., Liu G., Bremer S.W., Ye C.J. Imaging genome abnormalities in cancer research. Cell Chromosome. 2004;3(1):1. doi: 10.1186/1475-9268-3-1. https://cellandchromosome.biomedcentral.com/ articles/10.1186/1475-9268-3-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stevens J.B., Liu G., Bremer S.W., Ye K.J., Xu W., Xu J., Sun Y., Wu G.S., Savasan S., Krawetz S.A., Ye C.J., Heng H.H. Mitotic cell death by chromosome fragmentation. Cancer Res. 2007;67(16):7686–7694. doi: 10.1158/0008-5472.CAN-07-0472. [DOI] [PubMed] [Google Scholar]
- 27.Stevens J.B., Abdallah B.Y., Regan S.M., Liu G., Bremer S.W., Ye C.J., Heng H.H. Comparison of mitotic cell death by chromosome fragmentation to premature chromosome condensation. 2010 doi: 10.1186/1755-8166-3-20. tics.biomedcentral.com/articles/10.1186/1755-8166-3-20 [DOI] [PMC free article] [PubMed]
- 28.Fukuda K., Nisenbaum R., Stewart G., Thompson W.W., Robin L., Washko R.M., Noah D.L., Barrett D.H., Randall B., Herwaldt B.L., Mawle A.C., Reeves W.C. Chronic multisymptom illness affecting Air Force veterans of the Gulf War. 1998. [DOI] [PubMed]
- 29.Heng H.H., Tsui L.C. Modes of DAPI banding and simultaneous in situ hybridization. Chromosoma. 1993;102(5):325–332. doi: 10.1007/BF00661275. [DOI] [PubMed] [Google Scholar]
- 30.Liu G., Stevens J.B., Horne S.D., Abdallah B.Y., Ye K.J., Bremer S.W., Ye C.J., Chen D.J., Heng H.H. Genome chaos: Survival strategy during crisis. Cell Cycle. 2014;13(4):528–537. doi: 10.4161/cc.27378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith L., Plug A., Thayer M. Delayed replication timing leads to delayed mitotic chromosome condensation and chromosomal instability of chromosome translocations. Proc. Natl. Acad. Sci. USA. 2001;98(23):13300–13305. doi: 10.1073/pnas.241355098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haaf T., Schmid M. 5-Azadeoxycytidine induced undercondensation in the giant X chromosomes of Microtus agrestis. Chromosoma. 1989;98:93–98. doi: 10.1007/BF00291043. link.springer.com/article/10.1007/BF00291043 [DOI] [PubMed] [Google Scholar]
- 33.Flagiello D., Bernardino-Sgherri J., Dutrillaux B. Complex relationships between 5-aza-dC induced DNA demethylation and chromosome compaction at mitosis. Chromosoma. 2002;111:37–44. doi: 10.1007/s00412-001-0180-2. https://link.springer.com/article/10.1007/s00412-001-0180-2 [DOI] [PubMed] [Google Scholar]
- 34.Beadle G.W. A gene for stiky chromosomes in Zea mays. Indukt. Abstamm. Vererbungle. 1932;63(1):195–217. [Google Scholar]
- 35.Al Achkar W., Sabatier L., Dutrillaux B. How are sticky chromosomes formed? Ann. Genet. 1989;32(1):10–15. [PubMed] [Google Scholar]
- 36.Klásterská I., Natarajan A.T. Stickiness in Rosa meiosis induced by hybridization. Caryologia. 1975;28(1):81–88. [Google Scholar]
- 37.Cuylen S., Blaukopf C., Politi A.Z., Müller-Reichert T., Neumann B., Poser I., Ellenberg J., Hyman A.A., Gerlich D.W. Ki-67 acts as a biological surfactant to disperse mitotic chromosomes. Nature. 2016;535(7611):308–312. doi: 10.1038/nature18610. https://www.nature.com/articles/nature18610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heng H.H., Regan S. A systems biology perspective on molecular cytogenetics. Curr. Bioinfomat. 2016;11(3):4–10. [Google Scholar]
- 39.Rowley J.D. A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243(5405):290–293. doi: 10.1038/243290a0. https://www.nature.com/articles/243290a0 [DOI] [PubMed] [Google Scholar]
- 40.Horne S.D., Stevens J.B., Abdallah B.Y., Liu G., Bremer S.W., Ye C.J., Heng H.H. Why imatinib remains an exception of cancer research. J. Cell. Physiol. 2013;228(4):665–670. doi: 10.1002/jcp.24233. [DOI] [PubMed] [Google Scholar]