Abstract
Background:
Species with ‘young’ or nascent sex chromosomes provide unique opportunities to understand early evolutionary mechanisms (e.g. accumulation of repetitive sequences, cessation of recombination and gene loss) that drive the evolution of sex chromosomes. Among vertebrates, fishes exhibit highly diverse and a wide spectrum of sex-determining mechanisms and sex chromosomes, ranging from cryptic to highly differentiated ones, as well as, from simple to multiple sex chromosome systems. Such variability in sex chromosome morphology and composition not only exists within closely related taxa, but often within races/populations of the same species. Inside this context, the wolf fish Hoplias malabaricus offers opportunity to investigate the evolution of morphologically variable sex chromosomes within a species complex, as homomorphic to highly differentiated sex chromosome systems occur among its different karyomorphs.
Materials & Methods:
To discover various evolutionary stages of sex chromosomes and to compare their sequence composition among the wolf fish´s karyomorphs, we applied multipronged molecular cytogenetic approaches, including C-banding, repetitive DNAs mapping, Comparative Genomic Hybridization (CGH) and Whole Chromosomal Painting (WCP). Our study was able to characterize a cryptically differentiated XX/XY sex chromosome system in the karyomorph F of this species.
Conclusion:
The Y chromosome was clearly identified by an interstitial heterochromatic block on the short arms, primarily composed of microsatellite motifs and retrotransposons. Additionally, CGH also identified a male specific chromosome region in the same chromosomal location, implying that the accumulation of these repeats may have initiated the Y chromosome differentiation, as well as played a critical role towards the evolution and differentiation of sex chromosomes in various karyomorphs of this species.
Keywords: Fish, Early XY differentiation, Comparative genomic hybridization, Whole chromosome painting, Repetitive DNAs, Sex chromosomes
1. Introduction
Sex chromosomes evolve from an autosomal pair, when one chromosome acquired a sex determining locus [1, 2]. This process involves the accumulation of constitutive heterochromatin and/or the occurrence of structural changes (e.g. chromosomal inversions or translocations) leading to the suppression of recombination between the homologous chromosomes [3]. This evolutionary process can be better explored among lower vertebrates where the sex chromosomes exist either in incipient or in advanced forms.
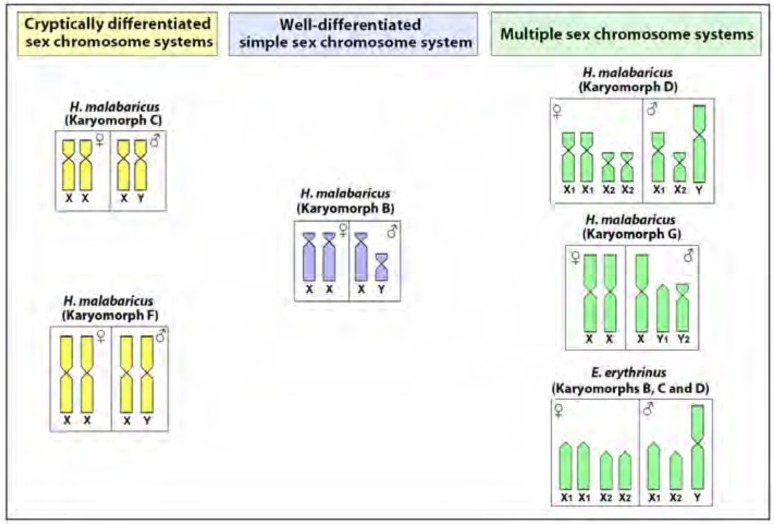
Notably, fishes exhibit the widest spectrum of sex-determining modes among vertebrates, ranging from environmental sex determination to a variety of chromosomal sex determining systems [4-8]. Neotropical fish species display diverse patterns of sex chromosome differentiation, ranging from simple to highly reorganized multiple sex chromosomes [9, 10], representing both XX/XY and ZZ/ZW sex systems. The Erythrinidae family (Teleostei, Characiformes) represents an ideal model group to understand the evolution of sex chromosomes, since different evolutionary stages, from cryptic to highly differentiated ones, can occur among closely related species (Fig. 1). Heteromorphic sex chromosomes in these species are hypothesized to be achieved by accumulation of heterochromatin and repetitive DNA sequences [10].
Fig. (1).
Overview of the different sex chromosome systems present in the Erythrinidae family. Data from [11, 13, 14] and present paper.
This family is composed by three genera, Hoplias, Hoplerythrinus and Erythrinus, being widespread throughout South America and presenting huge karyotype diversity among different populations of the same species, comprising distinct karyomorphs [11, 12]. Previous studies demonstrated that some karyomorphs of E. erythrinus and H. malabaricus have heteromorphic sex chromosomes, while others have undifferentiated ones (Fig. 1).
In particular, the H. malabaricus species complex not only has simple sex chromosomes with various evolutionary stages of differentiation, but also has multiple sex chromosomes. This species complex is composed of seven karyomorphs (A - G) with 2n ranging from 39 to 42 chromosomes, with four sex chromosome types already identified (Fig. 1). The karyomorphs A and E do not show noticeably differentiated sex chromosomes [11]. In contrast, a well-differentiated XY sex system is present in karyomorph B [13] and a cryptically differentiated one is found in karyomorph C [14], while XY multiple systems (X1X2Y and XY1Y2) occur in karyomorphs D and G, respectively [11]. In karyomorph F a discrete heterochromatic block was previously highlighted in one of the homologues of the pair 1, characterized as the largest chromosome in H. malabaricus, signaling a probable sex pair [15]. Indeed, the accumulation of heterochromatin and repetitive DNA sequences played an important role in the differentiation of the X chromosome in the karyomorphs B and C [14, 16].
In this study, we applied differential cytogenetic methods, such as C-banding, repetitive DNAs mapping, CGH and WCP to deeply investigate the occurrence of nascent sex chromosomes in the karyomorph F. These procedures have been applied successfully to unveil the mechanisms involved in the evolution of sex chromosomes even in early stages of differentiation [17-20]. Combinations of these techniques allowed us to identify a cryptic XX/XY sex chromosome system in karyomorph F involving the largest chromosomal pair. The X and Y chromosomes differ only slightly by the interstitial heterochromatic block present in the short arms of the nascent Y chromosome. We highlighted the possible roles of repetitive DNA accumulation in triggering the evolutionary process of the sex chromosomes differentiation.
2. Materials and Methods
2.1. Specimens, Chromosome Preparations, DNA Samples and C-banding
Twenty females and 21 males of H. malabaricus belonging to karyomorph F were collected from the São Francisco River (Três Marias, Minas Gerais State, Brazil) and from the Crixás-Açú River (Crixás, Goiás State, Brazil) under appropriate authorization of the Brazilian environmental agency ICMBIO/SISBIO (License number 48628-2). The specimens were deposited in the fish collection of the Cytogenetic Laboratory, Departamento de Genética e Evolução, Universidade Federal de São Carlos. Mitotic chromosomes were obtained by following protocols described in [21]. The experiments followed ethical and anesthesia conducts, in accordance with the Ethics Committee on Animal Experimentation of the Universidade Federal de São Carlos (Process number CEUA 1853260315). The C-positive heterochromatin was detected using barium hydroxide protocol according to [22]. Genomic DNA from male and female specimens was extracted according to standard phenol-chloroform procedures [23].
2.2. Probes for Chromosome Hybridization
A total of 17 repetitive DNA sequences, including: three multigene families (U2 snDNA, 5S and 18S rDNAs), one satellite DNA (5SHindIII-DNA), one transposable element (Rex 1), a telomeric (TTAGGG)n and 11 microsatellite repeat motifs, were used as probes.
Oligonucleotide probes containing microsatellite sequences (A)30, (C)30, (CA)15, (GA)15, (GC)15, (TA)15, (CAT)10, (CAC)10, (CGG)10, (GAA)10 and (GAG)10, were directly labelled with Cy5 during synthesis by Sigma (St. Louis, MO, USA), according to [24]. Telomeric (TTAGGG)n sequences were generated by PCR using (TTAGGG)5 and (CCCTAA)5 as primers, without DNA as template, according to [25]. These sequences were directly labelled with Spectrum Green-dUTP by nick translation, according to the manufacturer’s recommendations (Roche, Mannheim, Germany).
All the other repeats were isolated from the H. malabaricus genome. The 5S rDNA probe corresponded to copies of 120 base pair (bp) of the 5S rRNA encoding gene and 200 bp of the non-transcribed spacer (NTS) [26]. The 5SHindIII-DNA probe, which is a copy of repetitive satellite sequences with 360 bp, is composed of a 95-bp segment with similarity to the 5S rRNA gene and a 265-bp segment similar to the NTS [26]. The 18S rRNA probe containing copies of a 1,400 bp-segment of the 18S rRNA gene was obtained via PCR from nuclear DNA [27]. The retrotransposable element Rex 1 was obtained using primers described in [28]. The U2 snDNA sequence was produced by PCR, according to [29], using primers derived from the U2 coding sequences of several model organisms available in Genbank [30]. All these probes were directly labeled with Spectrum Orange-dUTP by nick translation (Roche, Mannheim, Germany), with the exception of 5S rDNA, that was labelled with Spectrum Green-dUTP also by nick translation (Roche, Mannheim, Germany).
2.3. Fluorescence In Situ Hybridization (FISH) for Repetitive DNA Mapping
Fluorescence In Situ Hybridization (FISH) was performed under high stringency conditions on metaphase chromosome spreads as described in [31]. The chromosome slides were incubated with RNAse (40 μg/ml) for 1.5 h at 37°C. After denaturation of the chromosomal DNA in 70% formamide/2x SSC at 70°C, spreads were dehydrated in ethanol series (70, 85 and 100%), 2 min each. 20 µl of the hybridization mixture (2.5 ng/μl probes, 2 μg/μl salmon sperm DNA, 50% deionized formamide, 10% dextran sulphate) were dropped on the slides, and the hybridization was performed for 14 h at 37°C in a moist chamber containing 2x SSC. The post-hybridization wash was carried out with 1x SSC for 5 min at 65oC. A final wash was performed at room temperature in 4x SSC for 5 min. Finally, the chromosomes were counterstained with DAPI (1.2 µg/ml) and mounted in antifade solution (Vector, Burlingame, CA, USA).
2.4. Chromosomal Microdissection, Probes Preparation and Labelling
Two distinct chromosomes were microdissected and used for the preparation of probes for chromosome painting. For this, 16 copies of the X chromosome of H. malabaricus (karyomorph B - XY system) and 18 copies of the largest chromosome pair (No 1) of H. malabaricus (karyomorph F) were microdissected, using the methodology described in [32]. We refer to these probes as HMA-X and HMA-1, respectively. The HMA-X probe was labelled via PCR with Spectrum Green-dUTP (Vysis, Downers Grove, USA) and the HMA-1 probe with Spectrum Orange-dUTP (Vysis, Downers Grove, USA) in a 30 cycle label-PCR with DOP primer, using 1 μl of the primary DOP-PCR products as template DNA, according to [32].
2.5. FISH for Whole Chromosome Painting (WCP)
Chromosomal preparations of males and females of the H. malabaricus (karyomorph F) were used for the FISH experiment, combining HMA-X and HMA-1 probes, according to [32]. To block the hybridization of high-copy repeat sequences, 50 µg of C0t1-DNA directly isolated from H. malabaricus male genome was used prepared according to [33]. Hybridization was performed for 16-18 h at 37°C in a moist chamber. The post-hybridization wash was carried out with 1x SSC for 5 min at 65ºC, and in 4x SSC/Tween using a shaker at RT and then rinsed quickly in 1x PBS. Subsequently, the slides were dehydrated in an ethanol series (70, 85 and 100%), 2 min each. Finally, the chromosomes were counterstained with DAPI (1.2 µg/ml) and mounted in antifade solution (Vector, Burlingame, CA, USA).
2.6. Preparation of Probes for Comparative Genome Hybridization (CGH)
For each probe, 1 μg of gDNA was used in the labeling procedure. The female gDNA was labeled with Digoxigenin-11-dUTP using DIG-nick-translation Mix (Roche, Mannheim, Germany), and the male gDNA was labeled with biotin-16-dUTP using BIO-nick-translation Mix (Roche). The hybridization solution for each slide (25 µl) was composed of 1 µg of male-specific labelled gDNA, 1 µg of female-specific labelled gDNA and 50 µg of C0t1-DNA directly isolated from H. malabaricus female genome, prepared according to [33].
2.7. FISH for CGH
The CGH experiments were performed according to [34]. The hybridization signal was detected with anti-digoxigenin-Rhodamin (Roche) diluted in 0.5% bovine serum albumin (BSA) in PBS; and avidin-FITC (Sigma) diluted in PBS containing 10% normal goat serum (NGS). The final washes were performed at 42°C in 4× SSC and 0.01% Tween, 7 min each for 3 times. Finally, the chromosomes were counterstained with DAPI (1.2 µg/ml) and mounted in antifade solution (Vector, Burlingame, CA, USA).
2.8. Microscopic Analyses
At least 30 metaphase spreads per individual were analyzed to confirm the 2n, karyotype structure and FISH results. Images were captured using an Olympus BX50 microscope (Olympus Corporation, Ishikawa, Japan) with CoolSNAP and the images processed using Image Pro Plus 4.1 software (Media Cybernetics, Silver Spring, MD, USA). Chromosomes were classified as metacentric (m) and submetacentric (sm), according to their arm ratios [35].
3. Results
3.1. Karyotypes and C-banding
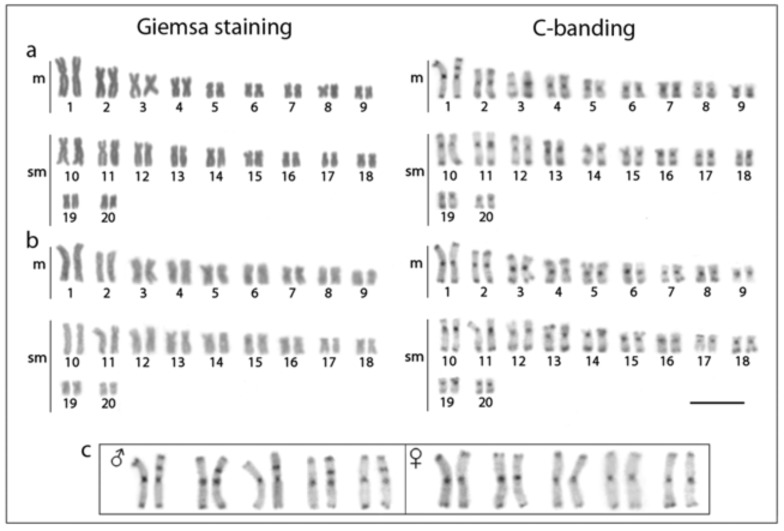
All specimens have 2n=40 chromosomes (10 m+10 sm) in both sexes. The large-sized metacentric pair (pair no. 1) constitutes a distinctive feature of the karyotype, representing the largest chromosome known for all H. malabaricus karyomorphs. C-positive heterochromatic bands are located in the centromeric regions, in addition to faint telomeric marks in some chromosomal pairs. However, C-banded male karyotypes always show an interstitial heterochromatic block in the short arms of one homologue of the first chromosome pair, whereas females lack this block, identifying the Y chromosome and the presence of a nascent XX/XY sex chromosome system in karyomorph F (Fig. 2), corroborating the findings of [15]. However, despite the presence of this exclusive heterochromatic block on the Y chromosome, both X and Y chromosomes are identical in size. For convenience, these chromosomes were named as XY chromosomes for the description of the following results.
Fig. (2).
Male (a) and female (b) karyotypes of Hoplias malabaricus (karyomorph F) under conventional Giemsa staining (left) and C-banding (right). In (c), distinct chromosome pairs no. 1 showing a conspicuous C-positive block in one of the homologues present only in males but not in females, identifying a cryptically Y chromosome and thus an XY sex chromosome system. Scale bar represents 5 µm.
3.2. Chromosomal Mapping of Repetitive DNAs
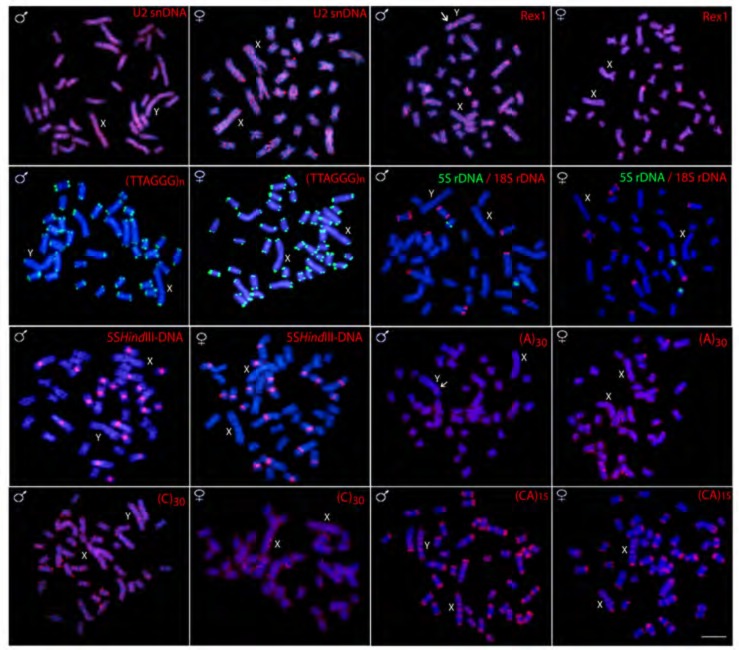
The 5S rDNA probe hybridized near the interstitial region of the long arms of only one chromosome pair, while 18S rDNA sites are found in four other pairs, including single and bi-telomeric sites. 5SHindIII satellite DNA is located in the centromeric regions of 10 chromosome pairs in both male and females (Fig. 3). No hybridization signals of theses repetitive DNAs were observed on the X and Y chromosomes (Fig. 3). These findings are in agreement with previous studies [36].
Fig. (3).
Male and female metaphase plates of Hoplias malabaricus (karyomorph F) showing the overall distribution of various repetitive sequences on the chromosomes. Arrows indicate Y specific signals. Scale bar represents 5 µm.
FISH with telomeric probe (TTAGGG)n hybridized on the telomeres of all chromosomes, however, no Interstitial Telomeric Sites (ITS) are detected. The U2 snRNA shows dispersed hybridization signals distributed widely across the whole chromosomal complement, including euchromatic and heterochromatic regions. This same pattern is also observed for the retrotransposon Rex 1, except some localized dense accumulation on the terminal region of five pairs of chromosomes (Fig. 3).
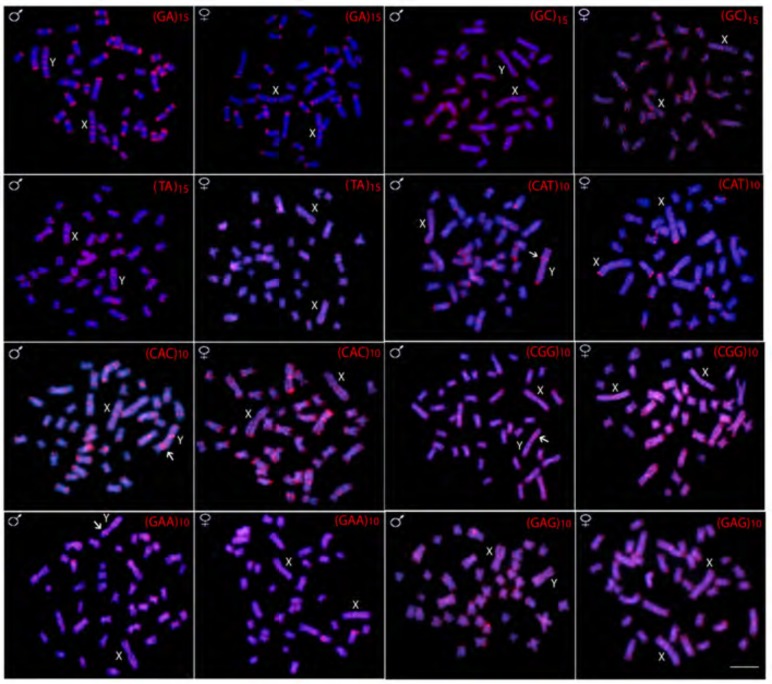
Variable patterns of hybridization signals were observed for microsatellite motifs-some discrete while others distributed uniformly along all chromosomes. For example, the (GA)n and (CA)n microsatellites provide a rich banding pattern in the subtelomeric region along almost all chromosomes, while (A)n, (C)n, (GC)n, (TA)n, (CGG)n, (GAA)n and (GAG)n provide strong dispersed signals across the entire length of all chromosomes, highlighting their widespread presence in the genome (Figs. 3 and 4). Microsatellites (CAT)n and (CAC)n present an intermediate pattern, having spread signals across the entire length of all chromosomes, together with a rich banding pattern in the subtelomeric region of some of them (Fig. 4).
Fig. (4).
Male and female metaphase plates of Hoplias malabaricus (karyomorph F) showing the overall distribution of various microsatellites repeats on the chromosomes. Arrows indicate Y specific signals. Scale bar represents 5 µm.
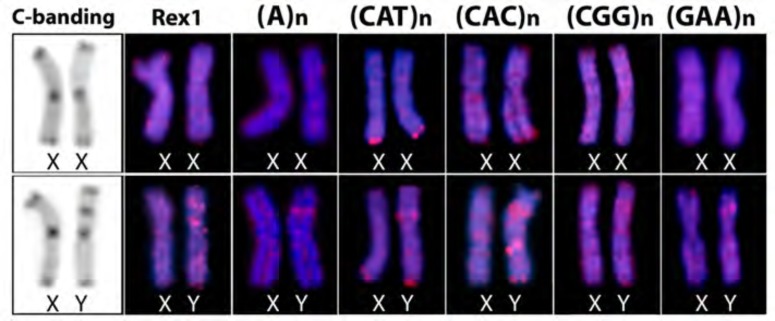
However, several microsatellite motifs [(A)n, (CAT)n, (CAC)n, (CGG)n and (GAA)n] and the Rex1 sequence showed Y specific amplification, being accumulated in the exclusive heterochromatic block, unlike the pattern found on the X chromosome (Fig. 5).
Fig. (5).
Comparison of the C-banding and the distribution of distinct repetitive DNAs on the X and Y chromosomes of Hoplias malabaricus (karyomorph F), highlighting the accumulation of some of them in the particular heterochromatic block of the Y chromosome.
3.3. Comparative Genomic Hybridization (CGH)
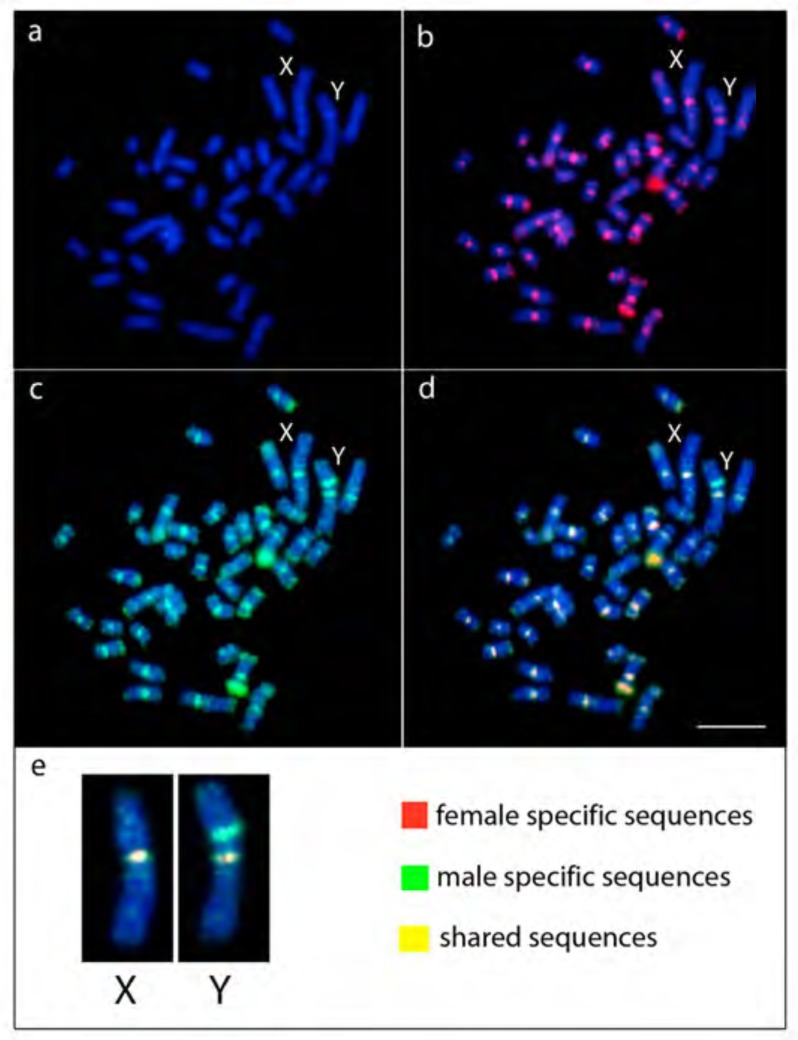
Remarkably, CGH experiments were able to show male specific sequences on the short arms of the Y chromosome (Fig. 6). All the remaining male chromosomes were uniformly stained with both gDNA probes, highlighting the centromeric region of all chromosomes and the telomeric region of several pairs. Additionally, with the male-specific gDNA probe, the Y chromosome stood out with an interstitial signal on its short arms. Sequences from male and females are shared in all chromosomes, with clear accumulation of male sequences on the Yp (Fig. 6e).
Fig. (6).
Comparative genomic hybridization (CGH) on male metaphase of Hoplias malabaricus (karyomorph F), with emphasis on the X and Y chromosomes. DAPI staining (a). Hybridization with female gDNA probe (red) (b). Hybridization with male gDNA probe (green) (c). Merged (d). Enlarged merged image of X and Y chromosomes highlighting the distribution of sex chromosome specific sequences (e). Scale bar represents 5 µm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this paper.)
3.4. Whole Chromosome Painting (WCP) with HMA-X and HMA-1 Probes
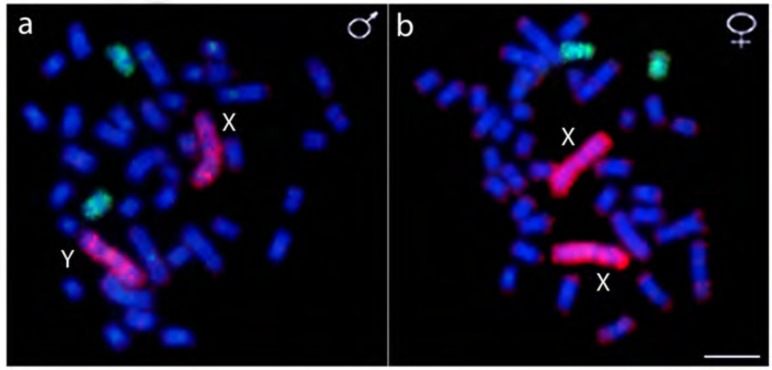
The HMA-1 probe hybridized completely to the largest chromosomal pair in both females (XX) and males (XY), while the HMA-X probe hybridized to 2 other sm chromosomes in both sexes (Fig. 7).
Fig. (7).
Whole chromosome painting in male (a) and female (b) metaphase plates of Hoplias malabaricus (karyomorph F) combining the HMA-X probe (X chromosome of karyomorph B - green) and the HMA-1 probe (chromosome 1 of karyomorph F - red) highlighting the independent origin of the sex chromosomes of these karyomorphs. Scale bar represents 5 µm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this paper.)
4. Discussion
Our overall data show that the largest metacentric pair (chromosome 1) found in the karyomorph F of H. malabaricus really represents nascent XY sex chromosomes, where the Y is still at an early stage of differentiation. Indeed, our CGH experiments clearly demonstrated that the Y specific interstitial heterochromatic block in the short arms and thus identified male-specific chromosomal region (Fig. 6d, e). The particular accumulation of repeated DNAs in such region reflects the initial steps shaping the evolutionary differentiation of the Y chromosome and probably the starting point to impair the recombination between the undifferentiated sex pair. Similarly, in the plant Silene latifolia (Caryophyllales, Caryophyllaceae), it was found that the accumulation of a specific satellite DNA sequence (TRAYC) in the young Y chromosome also had a key role in its early stage of differentiation [37]. However, in hermaphrodite and dioecious species of the genus Rumex (Caryophyllales, Polygonaceae) the distribution of satellite DNAs suggests that the amplification of tandem repeats in the Y chromosome was not necessary to suppress recombination but, in turn, accelerated the sex specific differentiation at earlier stages of its evolution [38]. Microsatellite repeat motifs have also a central role in the evolution of sex chromosomes in several Sauropsida [39-41] and fish [42-44] species. Nevertheless, in Christinus marmoratus, a species of Australian gecko with a heterochromatic W chromosome, no sex specific amplification of microsatellites repeats was observed, suggesting accumulation of unknown novel repeats [40]. Thus, it is highlighted that repetitive DNAs may play significant roles in the evolutionary process of sex chromosomes differentiation among different species.
Investigation of ‘young’ sex chromosomes helps us to understand how sex chromosomes first become non-recombining and the evolutionary consequences of the loss of recombination. Contrary to higher vertebrates, cryptically differentiated sex chromosomes can be found among amphibians [45-48], reptiles [49-52] and particularly in distinct fish species [14, 53-60]. In fact, among fishes, approximately 90% of the species studied so far present undifferentiated or cryptic sex chromosomes [4, 9]. In the well-studied medaka fish, Oryzias latipes, the X and Y chromosomes are morphologically indistinguishable [53], where the Y chromosome is only 5 to 10 mya and shows only slight molecular differences in relation to the X [57]. Among salmonids, an XY sex chromosome system is also found [60], in which the Y chromosome contains a small sex-specific region and with minor morphological differentiation from the X chromosome in some species [61]. In the nascent XY system of Xiphophorus maculatus, the expansion of a specific repeat (XIR) was highlighted as the first molecular event linked with the divergence of the Y chromosome and the isolation of the sex-determining locus [62]. In addition, nascent sex chromosomes can also evolve and operate via epigenetic regulation, as discovered in half-smooth tongue sole [63].
Heterochromatin accumulation has been reported as a major event in the sex chromosome differentiation. In the glass knifefish, Eigenmannia virescens, the acrocentric X chromosome diverges from the Y due to a particular heterochromatin accumulation on its distal arms [64]. In other Poeciliidae species, a heteromorphism in the heterochromatin between two homologous chromosomes allowed the identification of the sex chromosomes of ZZ/ZW type in Poecilia schenops var. melanistica [65] and of the XY system in P. reticulata [66]. Additionally, in Dicentrarchus labrax, the amount and patterns of the heterochromatin distribution in one of the two smallest chromosomes of the male karyotype have indicated in early stage of the sex chromosome differentiation [67]. A particular scenario was observed for the XY systems of karyomorph B of H. malabaricus [16] and of E. virescens [68], representing odd examples of accumulation of repetitive DNAs on the non-sex specific chromosome. In both cases, the accumulation of repetitive DNAs had a central role in triggering the process of morphological differentiation between the sex pair, making possible its identification even in an early stage of differentiation, as in E. virescens, or in a highly differentiated system, as in H. malabaricus. Remarkably, the distribution of repetitive DNAs in the sex chromosomes indicates that they follow distinct evolutionary pathways among the H. malabaricus karyomorphs (Fig. 8) [69]. In fact, this accumulation does not follow a general rule and appears to be independent processes even among closely related species. Particularly in karyomorph F, an accumulation of DNA repeats (especially microsatellite motifs and transposable elements) was observed in the Y-specific region, revealing the early stage of differentiation of this chromosome. Indeed, these sequences are considered early colonizers of sex chromosomes and their likely key role in the sex-specific chromosome differentiation have been suggested for several species [24, 40-42].
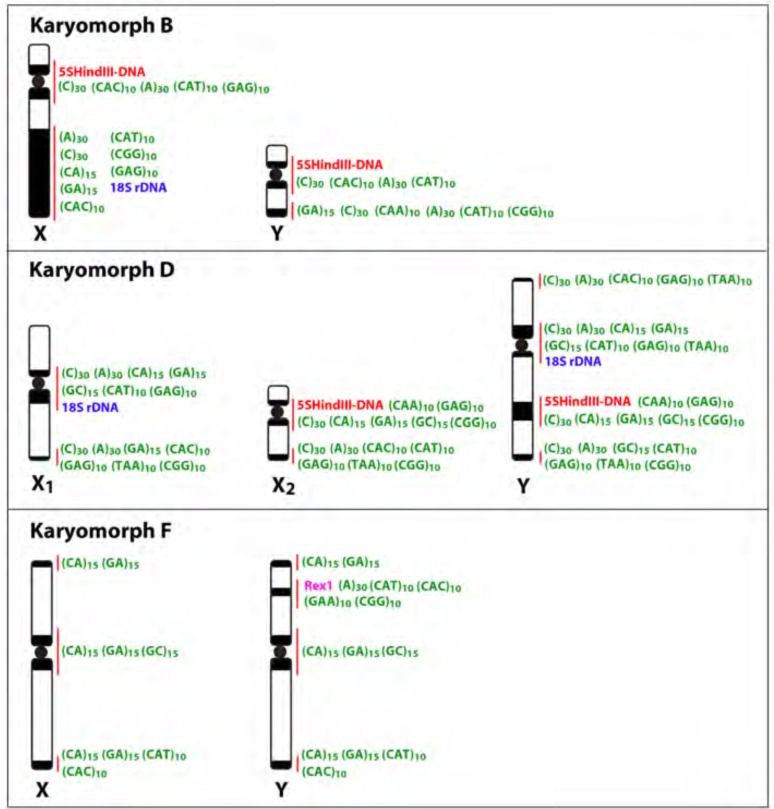
Fig. (8).
Idiograms of sex chromosomes of karyomorphs B, D and F highlighting the marked accumulation of distinct types of repetitive DNA sequences along these chromosomes. Data from [14, 16] and present paper.
Suppression of recombination between the sex chromosomes stretches out to neighboring regions permitting a large number of Y-linked genes to degenerate and create a male-specific region on the nascent Y chromosome [70]. Here, this process appears to have started mainly through the accumulation and amplification of microsatellite motifs, probably due to the ability of these sequences for rapid expansion over other classes of repetitive DNAs in the sex-specific chromosome, as demonstrated in distinct animal and plant species [44, 71, 72]. Among fishes, male (or female) specific regions are normally enriched by different types of Transposable Elements (TEs) (reviewed in [73]). TEs are also considered major drivers of sex chromosome differentiation through the accumulation of functional insertions, promoting genome rearrangements and also playing an active role in the formation of new sex determination (SD) genes [73, 74].
Why are sex chromosomes so often homomorphic in lower vertebrates? One possible explanation is the high rate of turnover events, where new master sex-determining genes appear and replace previously established sex chromosomes [8, 75]. Indeed, contrary to birds and mammals, sex determining mechanisms in reptiles, amphibians and fishes show a high turnover and new sex chromosomes appear again and again [76-79]. There is evidence that turnovers recently occurred in several fish groups, such as Oryzias [80], Eigenmannia [19], Salvelinus [81] and sticklebacks [82]. In amphibians, different populations from the same species have been shown to display different sex chromosomes systems [83] or even several heterogametic transitions (i.e., transitions from male to female heterogamety or vice versa) [84, 85]. In Australian dragon lizards, non homologous sex chromosomes have evolved within a very short evolutionary time frame [52], where numerous transitions between sex chromosomes have been documented likely via independent evolution of nascent sex chromosomes [79].
A variety of sex chromosomes have also evolved among the H. malabaricus karyomorphs, where X and Y emerged independently and followed distinct patterns of differentiation [20]. The lack of hybridization signals with HMA-X on the nascent XY chromosomes of karyomorph F highlighted that the XY chromosomes of karyomorphs B and F have evolved independently, though recruitment of non-homologous autosomal pairs (Fig. 7). Why in H. malabaricus the path of the genetic sex determination has occurred in so many different ways and at so many different times? This species is well adapted to life in small populations with low vagility [86], which increase the probability of fixation of chromosomal rearrangements. In fact, it is noteworthy that karyomorphs possessing well-differentiated sex chromosomes (B, D and G) show a more restricted geographical distribution, indicating their derived origin [11, 12] and the highlighting the potential of small populations in accelerating the genomic diversity and in triggering rapid degeneration of the sex specific chromosome.
CONCLUSION
Our study clearly demonstrated that a nascent XX/XY sex chromosome system can be actually assigned to karyomorph F of H. malabaricus, as clearly evidenced by the particular distribution of distinct tandem DNA repeats in a male specific region on the young Y chromosome. It is noteworthy that our study added another example of independent origin of sex systems within this species complex, which is already characterized by several karyomorphs, ranging from single, multiple, cryptic and highly differentiated sex chromosomes. In fact, H. malabaricus represents an assemblage of species with unresolved taxonomy, in which such frequent sex chromosome turnover might have played a central role in speciation, as well documented for other fish species [87-91]. Therefore, this species complex provides a unique opportunity to gain insights into the evolutionary forces that drive the evolution of nascent sex chromosomes, the relationships among distinct sex chromosomes systems, speciation and the role of accumulation and amplification of distinct types of repetitive sequences in the origin and differentiation of sex chromosomes.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The experiments followed ethical and anesthesia conducts, in accordance with the Ethics Committee on Animal Experimentation of the Universidade Federal de São Carlos (Process number CEUA 1853260315).
HUMAN AND ANIMAL RIGHTS
No humans were involved in this study. All the reported experiments involving animals were in accordance with the standards set for the Brazilian agency Conselho Nacional de Experimentação Animal (CONCEA) (http://www.sbcal.org. br/conteudo/view?ID_CONTEUDO=41) as approved by the Ethics Committee on Animal Experimentation of the Universidade Federal de São Carlos.
CONSENT FOR PUBLICATION
Not applicable.
Acknowledgements
This study was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico - CNPq (Proc. no 304992/2015-1 and 306896/2014-1) and Fundação de Amparo à Pesquisa do Estado de São Paulo- FAPESP (Proc. No 2014/22532-7). Natália Lourenço de Freitas is an undergraduate student in Biotechnology at Universidade Federal de São Carlos and sponsored by PIBIC/CNPq/UFSCar. TE is partially supported by an Australian Research Council (ARC) Future Fellowship (FT110100733).
LIST OF ABBREVIATIONS
- 2n
Diploid number
- BSA
Bovine Serum Albumin
- CGH
Comparative Genomic Hybridization
- DAPI
4',6-diamidino-2-phenylindole
- dUTP
2'-Deoxyuridine-5'-Triphosphate
- FISH
Fluorescence In Situ Hybridization
- gDNA
Total genomic DNA
- ITS
Interstitial Telomeric Sites
- m
Metacentric chromosome
- mya
Millions of years
- NGS
Normal Goat Serum
- NOR
Nucleolar Organizing Regions
- NTS
Non-transcribed Spacer
- PCR
Polymerase Chain Reaction
- PBS
Phosphate-buffered saline
- rDNA
Ribosomal DNA
- rRNA
Ribosomal RNA
- SD
Sex Determination
- sm
Submetacentric chromosome
- TEs
Transposable Elements
- WCP
Whole Chromosomal Painting
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
References
- 1.Muller H.J. A gene for the fourth chromosome of Drosophila. J. Exp. Zool. 1914;17(3):325–336. [Google Scholar]
- 2.Ohno S. Sex chromosome and sex-linked genes. Berlin, Heidelberg: Springer-Verlag; 1967. pp. 1–9. [Google Scholar]
- 3.Charlesworth D., Charlesworth B., Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity. 2005;95(2):118–128. doi: 10.1038/sj.hdy.6800697. [DOI] [PubMed] [Google Scholar]
- 4.Devlin R.H., Nagahama Y. Sex determination and sex differentiation in fish: An overview of genetic, physiological, and environmental influences. Aquaculture. Available from: https://www.sciencedirect.com/science/article/pii/S0044848. 2002;208(2):191–136. [Google Scholar]
- 5.Penman D.J., Piferrer F. Fish Gonadogenesis. Part I: Genetic and environmental mechanisms of sex determination. Rev. Fish. Sci. 2008;16:16–34. http://www.tandfonline.com/doi/ abs/10.1080/10641260802324610?src=recsys&journalCode=brfs20 [Google Scholar]
- 6.Kobayashi Y., Nagahama Y., Nakamura M. Diversity and plasticity of sex determination and differentiation in fishes. Sex Dev. 2013;7(1-3):115–125. doi: 10.1159/000342009. [DOI] [PubMed] [Google Scholar]
- 7.Schartl M., Schmid M., Nanda I. Dynamics of vertebrate sex chromosome evolution: From equal size to giants and dwarfs. Chromosoma. 2016;125(3):553–571. doi: 10.1007/s00412-015-0569-y. [DOI] [PubMed] [Google Scholar]
- 8.Volff J.N., Nanda I., Schmid M., Schartl M. Governing sex determination in fish: Regulatory putsches and ephemeral dictators. Sex Dev. 2007;1(2):85–99. doi: 10.1159/000100030. [DOI] [PubMed] [Google Scholar]
- 9.Oliveira R.R., Feldberg E., Anjos M.B., Zuanon J. Karyotype characterization and ZZ/ZW sex chromosome heteromorphism in two species of the catfish genus Ancistrus Kner, 1854 (Siluriformes: Loricariidae) from the Amazon basin. Neotrop. Ichthyol. Available from: www.scielo.br/scielo.php?script. 2007;(5):301–306.
- 10.Cioffi M.B., Moreira-Filho O., Almeida-Toledo L.F., Bertollo L.A.C. The contrasting role of heterochromatin in the differentiation of sex chromosomes. An overview from Neotropical fish. J. Fish Biol. 2012;80(6):2125–2139. doi: 10.1111/j.1095-8649.2012.03272.x. [DOI] [PubMed] [Google Scholar]
- 11.Bertollo L.A.C., Born G.G., Dergam J.A., Fenocchio A.S., Moreira-Filho O. A biodiversity approach in the neotropical fish Hoplias malabaricus. Karyotypic survey, geographic distribution of cytotypes and cytotaxonomic considerations. Chromosome Res. 2000;8(7):603–613. doi: 10.1023/a:1009233907558. [DOI] [PubMed] [Google Scholar]
- 12.Cioffi M.B., Molina W.F., Artoni R.F., Bertollo L.A.C. Chromosomes as tools for discovering biodiversity. The case of Erythrinidae fish family. In: Tirunilai P., editor. Recent Trends in Cytogenetic Studies - Methodologies and Applications. Vol. 1. Rijeka: InTech Open Access Publisher; 2012. pp. 125–146. [Google Scholar]
- 13.Born G.G., Bertollo L.A.C. An XX/XY sex chromosome system in a fish species, Hoplias malabaricus, with a polymorphic NOR-bearing X chromosome. Chromosome Res. 2000;8(2):111–118. doi: 10.1023/a:1009238402051. [DOI] [PubMed] [Google Scholar]
- 14.Cioffi M.B., Bertollo L.A.C. Initial steps in XY chromosome differentiation in Hoplias malabaricus and the origin of an X1X2Y sex chromosome system in this fish group. Heredity. Available from: https://www.nature.com/articles/ 2010 doi: 10.1038/hdy.2010.18. [DOI] [PubMed]
- 15.Santos U., Volcker C.M., Belei F.A., Cioffi M.B., Bertollo L.A.C., Paiva R., Dergam J.A. Molecular and karyotypic phylogeography in the Neotropical Erythrinidae fish, Hoplias malabaricus (Bloch, 1794). J. Fish Biol. 2009;75(9):2326–2343. doi: 10.1111/j.1095-8649.2009.02489.x. [DOI] [PubMed] [Google Scholar]
- 16.Cioffi M.B., Martins C., Vicari M.R., Rebordinos L., Bertollo L.A.C. Differentiation of the XY sex chromosomes in the fish Hoplias malabaricus (Characiformes, Erythrinidae). Unusual accumulation of repetitive sequences on the X chromosome. Sex Dev. 2010;4(3):176–185. doi: 10.1159/000309726. [DOI] [PubMed] [Google Scholar]
- 17.Traut W., Sahara K., Otto T.D., Marec F. Molecular differentiation of sex chromosomes probed by comparative genomic hybridization. Chromosoma. 1999;108(3):173–180. doi: 10.1007/s004120050366. [DOI] [PubMed] [Google Scholar]
- 18.Vítková M., Fuková I., Kubíčková S., Marec F. Molecular divergence of the W chromosomes in pyralid moths (Lepidoptera). Chromosome Res. 2007;15(7):917–930. doi: 10.1007/s10577-007-1173-7. [DOI] [PubMed] [Google Scholar]
- 19.Henning F., Trifonov V., Ferguson-Smith M.A., de Almeida-Toledo L. Non-homologous sex chromosomes in two species of the genus Eigenmannia (Teleostei: Gymnotiformes). Cytogenet. Genome Res. 2008;121(1):55–58. doi: 10.1159/000124382. [DOI] [PubMed] [Google Scholar]
- 20.Cioffi M.B., Liehr T., Trifonov V., Molina W.F., Bertollo L.A.C. Independent sex chromosome evolution in lower vertebrates: A molecular cytogenetic overview in the Erythrinidae Fish Family. Cytogenet. Genome Res. 2013;141(2-3):186–194. doi: 10.1159/000354039. [DOI] [PubMed] [Google Scholar]
- 21.Bertollo L.A.C., Moreira-Filho O., Cioffi M.B. Direct chromosome preparations from freshwater Teleost fishes. In: Ozouf-Costaz C., Pisano E., Foresti F., Foresti de Almeida Toledo L., editors. Fish techniques, Ray-Fin Fishes and Chondrichthyans. Vol. 1. Boca Raton: CRC Press; 2015. pp. 21–26. [Google Scholar]
- 22.Sumner A.T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 1972;75(1):304–306. doi: 10.1016/0014-4827(72)90558-7. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J., Russell D.W. Molecular cloning - A Laboratory Manual. 3rd ed. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 24.Kubat Z., Hobza R., Vyskot B., Kejnovsky E. Microsatellite accumulation in the Y chromosome of Silene Latifolia. Genome. 2008;51(5):350–356. doi: 10.1139/G08-024. [DOI] [PubMed] [Google Scholar]
- 25.Ijdo J.W., Wells R.A., Baldini A., Reeders S.T. Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucleic Acids Res. 1991;19(17):47–80. doi: 10.1093/nar/19.17.4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martins C., Ferreira I.A., Oliveira C., Foresti F., Galetti P.M., Jr A tandemly repetitive centromeric DNA sequence of the fish Hoplias malabaricus (Characiformes: Erythrinidae) is derived from 5S rDNA. Genetica. 2006;127(1-3):133–141. doi: 10.1007/s10709-005-2674-y. [DOI] [PubMed] [Google Scholar]
- 27.Cioffi M.B., Martins C., Centofante L., Jacobina U., Bertollo L.A.C. Chromosomal variability among allopatric populations of Erythrinidae fish Hoplias malabaricus: Mapping of three classes of repetitive DNAs. Cytogenet. Genome Res. 2009;125(2):132–141. doi: 10.1159/000227838. [DOI] [PubMed] [Google Scholar]
- 28.Volff J.N., Korting C., Sweeney K., Schartl M. The non-LTR retrotransposon Rex3 from the fish Xiphophorus is widespread among Teleosts. Mol. Biol. Evol. 1999;16(11):1427–1438. doi: 10.1093/oxfordjournals.molbev.a026055. [DOI] [PubMed] [Google Scholar]
- 29.Úbeda-Manzanaro M., Merlo M.A., Palazón J.L., Cross I., Sarasquete C., Rebordinos L. Chromosomal mapping of the major and minor ribosomal genes, (GATA)n and U2 snRNA gene by double-colour FISH in species of the Batrachoididae family. Genetica. 2010;138(7):787–794. doi: 10.1007/s10709-010-9460-1. [DOI] [PubMed] [Google Scholar]
- 30.Cross I., Merlo A., Manchado M., Infante C., Canãvate J.P., Rebordinos L. Cytogenetic characterization of the Solea senegalensis (Teleostei: Pleurenectiformes. Soleidae): Ag-NOR, (GATA)n, (TTAGGG)n and ribosomal genes by one-color and two-color FISH. Genetica. 2006;128(1-3):253–259. doi: 10.1007/s10709-005-5928-9. [DOI] [PubMed] [Google Scholar]
- 31.Pinkel D., Gray J., Trask B., Van den Engh G., Fuscoe J., Van Dekken H. Cytogenetic analysis by in situ hybridization with fluorescently labeled nucleic acid probes. Cold Spring Harb. Symp. Quant. Biol. 1986;51(Pt 1):151–157. doi: 10.1101/sqb.1986.051.01.018. [DOI] [PubMed] [Google Scholar]
- 32.Yang F., Graphodatsky A.S. Animal probes and ZOO-fish. In: Liehr T., editor. Fluorescence in situ hybridization (FISH). Berlin: Springer; 2009. pp. 323–346. [Google Scholar]
- 33.Zwick M.S., Hanson R.E., Islam-Faridi M.N., Stelly D.M., Wing R.A., Price H.J., McKnight T.D. A rapid procedure for the isolation of C0t-1 DNA from plants. Genome. 1997;40(1):138–142. doi: 10.1139/g97-020. [DOI] [PubMed] [Google Scholar]
- 34.Symonová R., Sember A., Majtánová Z., Ráb P. Characterization of fish genomes by GISH and CGH. In: Ozouf-Costaz C., Pisano E., Foresti F., Foresti de Almeida Toledo L., editors. Fish techniques, Ray-Fin Fishes and Chondrichthyans. Vol. 1. Boca Raton: CRC Press; 2015. pp. 118–131. [Google Scholar]
- 35.Levan A., Fredga K., Sandberg A.A. Nomenclature for centromeric position on chromosomes. Hereditas. 1964;52(2):201–220. [Google Scholar]
- 36.Blanco D.R., Lui R.L., Bertollo L.A.C., Diniz D., Moreira-Filho O. Characterization of invasive fish species in a river transposition region: Evolutionary chromosome studies in the genus Hoplias (Characiformes, Erythrinidae). Rev. Fish Biol. Fisher. Available from: http://www.academia.edu/7373263 /Characterization_of_invasive_fish_species_in_a_river_. 2010
- 37.Hobza R., Lengerova M., Svoboda J., Kubekova H., Kejnovsky E., Vyskot B. An accumulation of tandem DNA repeats on the Y chromosome in Silene latifolia during early stages of sex chromosome evolution. Chromosoma. 2006;115(5):376–382. doi: 10.1007/s00412-006-0065-5. [DOI] [PubMed] [Google Scholar]
- 38.Mariotti B., Manzano S., Kejnovský E., Vyskot B., Jamilena M. Accumulation of Y-specific satellite DNAs during the evolution of Rumex acetosa sex chromosomes. Mol. Genet. Genomics. 2009;281(3):249–259. doi: 10.1007/s00438-008-0405-7. [DOI] [PubMed] [Google Scholar]
- 39.Pokorná M., Giovannotti M., Kratochvíl L., Kasai F., Trifonov V.A., O’Brien P.C.M., Caputo V., Olmo E., Ferguson-Smith M.A., Rens W. Strong conservation of the bird Z chromosome in reptilian genomes is revealed by comparative painting despite 275 million years divergence. Chromosoma. 2011;120(5):455–468. doi: 10.1007/s00412-011-0322-0. [DOI] [PubMed] [Google Scholar]
- 40.Matsubara K., Sarre S.D., Georges A., Matsuda Y., Graves J.A.M., Ezaz T. Highly differentiated ZW sex chromosomes in the Australian Varanus species evolved through rapid amplification of repetitive sequences. PLoS One. 2014;9(4):1–9. doi: 10.1371/journal.pone.0095226. journals.plos.org/plosone/article?id=10.1371/journal.pone.0095226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsubara K., O’Meally D., Azad B., Georges A., Sarre S.D., Graves J.A.M., Matsuda Y., Ezaz T. Amplification of microsatellite repeat motifs is associated with the evolutionary differentiation and heterochromatinization of sex chromosomes in Sauropsida. Chromosoma. 2016;125(1):111–123. doi: 10.1007/s00412-015-0531-z. [DOI] [PubMed] [Google Scholar]
- 42.Cioffi M.B., Kejnovsky E., Marquioni V., Poltronieri J., Molina W.F., Diniz D., Bertollo L.A.C. The key role of repeated DNAs in sex chromosome evolution in two fish species with ZW sex chromosome system. Mol. Cytogenet. 2012;5(28):1–7. doi: 10.1186/1755-8166-5-28. https://molecularcytogenetics.biomedcentral.com/articles/ 10.1186/1755-8166-5-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Terencio M.L., Schneider C.H., Gross M.C., Nogaroto V., de Almeida M.C., Artoni R.F., Vicari M.R., Feldeberg E. Repetitive sequences associated with differentiation of W chromosome in Semaprochilodus taeniurus. Genetica. 2012;140(10-12):505–512. doi: 10.1007/s10709-013-9699-4. [DOI] [PubMed] [Google Scholar]
- 44.Yano C.F., Bertollo L.A.C., Liehr T., Troy W.P., Cioffi M.B. W Chromosome dynamics in Triportheus species (Characiformes, Triportheidae): An ongoing process narrated by repetitive sequences. J. Hered. 2016;107(4):342–348. doi: 10.1093/jhered/esw021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schempp W., Schmid M. Chromosome banding in amphibia. Chromosoma. 1981;83(5):697–710. doi: 10.1007/BF00328528. [DOI] [PubMed] [Google Scholar]
- 46.Iturra P., Veloso A. Further evidence for early sex chromosome differentiation of Anuran species. Genetica. 1986;78(1):25–31. doi: 10.1007/BF00058671. [DOI] [PubMed] [Google Scholar]
- 47.Berset-Brändli L., Jaquiéry J., Broquet T., Ulrich Y., Perrin N. Extreme heterochiasmy and nascent sex chromosomes in European tree frogs. Proc. R. Soc. Lond. B Biol. Sci. 2008;275(1642):1577–1585. doi: 10.1098/rspb.2008.0298. http://rspb.royalsocietypublishing.org/content/275/1642/1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abramyan J., Ezaz T., Marshall Graves J.A., Koopman P. Z and W sex chromosomes in the cane toad (Bufo marinus). Chromosome Res. 2009;17(8):1015–1024. doi: 10.1007/s10577-009-9095-1. [DOI] [PubMed] [Google Scholar]
- 49.Schmid M., Steinlein C., Haaf T., Mijares-Urrutia A. Nascent ZW sex chromosomes in Thecadactylus rapicauda (Reptilia, Squamata, Phyllodactylidae). Cytogenet. Genome Res. Available from: https://www.karger.com/Article/Pdf/ 2014;(143):259–267. doi: 10.1159/000366212. [DOI] [PubMed]
- 50.Ezaz T., Quinn A.E., Miura I., Sarre S.D., Georges A., Marshall Graves J.A. The dragon lizard Pogona vitticeps has ZZ/ZW micro-sex chromosomes. Chromosome Res. 2005;13(8):763–776. doi: 10.1007/s10577-005-1010-9. [DOI] [PubMed] [Google Scholar]
- 51.Martinez P., Ezaz T., Valenzuela N., Georges A., Marshall Graves J.A. An XX/XY heteromorphic sex chromosome system in the Australian chelid turtle Emydura macquarii: A new piece in the puzzle of sex chromosome evolution in turtles. Chromosome Res. 2008;16(6):815–825. doi: 10.1007/s10577-008-1228-4. [DOI] [PubMed] [Google Scholar]
- 52.Ezaz T., Quinn A.E., Sarre S.D., O’Meally D., Georges A., Graves J.A. Molecular marker suggests rapid changes of sex-determining mechanisms in Australian dragon lizards. Chromosome Res. 2009;17(1):91–98. doi: 10.1007/s10577-008-9019-5. [DOI] [PubMed] [Google Scholar]
- 53.Matsuda M., Matsuda C., Hamaguchi S., Sakaizumi M. Identification of the sex chromosomes of the medaka, Oryzias latipes, by fluorescence in situ hybridization. Cytogenet. Genome Res. 1998;82(3-4):257–262. doi: 10.1159/000015113. [DOI] [PubMed] [Google Scholar]
- 54.Traut W., Winking H. Meiotic chromosomes and stages of sex chromosome evolution in fish: zebrafish, platyfish and guppy. Chromosome Res. 2001;9(8):659–672. doi: 10.1023/a:1012956324417. [DOI] [PubMed] [Google Scholar]
- 55.Griffin D.K., Harvey S.C., Campos-Ramos R., Ayling L-J., Bromage N.R., Masabanda J.S., Penman D.J. Early origins of the X and Y chromosomes: Lessons from tilapia. Cytogenet. Genome Res. 2002;99:157–163. doi: 10.1159/000071588. https://www.karger.com/ Article/PDF/71588 [DOI] [PubMed] [Google Scholar]
- 56.Harvey S.C., Masabanda J., Carrasco L.A.P., Bromage N.R., Penman D.J., Griffin D.K. Molecular-cytogenetic analysis reveals sequence differences between the sex chromosomes of Oreochromis niloticus: Evidence for an early stage of sex-chromosome differentiation. Cytogenet. Genome Res. 2002;97(1-2):76–80. doi: 10.1159/000064036. [DOI] [PubMed] [Google Scholar]
- 57.Kondo M., Nanda I., Hornung U., Schmid M., Schartl M. Evolutionary origin of the medaka Y chromosome. Curr. Biol. 2004;14(18):1664–1669. doi: 10.1016/j.cub.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 58.Vicari M.R., Artoni R.F., Moreira-Filho O., Bertollo L.A.C. Diversification of a ZZ/ZW sex chromosome system in Characidium fish (Crenuchidae, Characiformes). Genetica. 2008;134:311–317. doi: 10.1007/s10709-007-9238-2. europepmc.org/abstract/med/18175199 [DOI] [PubMed] [Google Scholar]
- 59.Schemberger M.O., Bellafronte E., Nogaroto V., Almeida M.C., Schühli G.S., Artoni R.F., Moreira-Filho O., Vicari M.R. Differentiation of repetitive DNA sites and sex chromosome systems reveal closely related group in Parodontidae (Actinopterygii: Characiformes). Genetica. 2011;139(11-12):1499–1508. doi: 10.1007/s10709-012-9649-6. [DOI] [PubMed] [Google Scholar]
- 60.Phillips R.B. Evolution of the sex chromosomes in salmonid fishes. Cytogenet. Genome Res. 2013;141(2-3):177–185. doi: 10.1159/000355149. [DOI] [PubMed] [Google Scholar]
- 61.Davidson W.S., Huang T-K., Fujiki K., Von Schalburg K.R., Koop B.F. The sex determining loci and sex chromosomes in the family Salmonidae. Sex Dev. 2009;3(2-3):78–87. doi: 10.1159/000223073. [DOI] [PubMed] [Google Scholar]
- 62.Nanda I., Volff J.N., Weis S., Korting C., Froschauer A., Schmid M., Schartl M. Amplification of a long terminal repeat-like element on the Y chromosome of the platyfish, Xiphophorus maculatus. Chromosoma. 2000;109(3):173–180. doi: 10.1007/s004120050425. [DOI] [PubMed] [Google Scholar]
- 63.Chen S., Zhang G., Shao C., Huang Q., Liu G., Zhang P., Song W., An N., Chalopin D., Volff J.N., Hong Y., Li Q., Sha Z., Zhou H., Xie M., Yu Q., Liu Y., Xiang H., Wang N., Wu K., Yang C., Zhou Q., Liao X., Yang L., Hu Q., Zhang J., Meng L., Jin L., Tian Y., Lian J., Yang J., Miao G., Liu S., Liang Z., Yan F., Li Y., Sun B., Zhang H., Zhang J., Zhu Y., Du M., Zhao Y., Schartl M., Tang Q., Wang J. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 2014;46(3):253–260. doi: 10.1038/ng.2890. [DOI] [PubMed] [Google Scholar]
- 64.Almeida Toledo L.F., Foresti F. Morphologically differentiated sex chromosomes in Neotropical freshwater fish. Genetica. 2001;111(1-3):91–100. doi: 10.1023/a:1013768104422. [DOI] [PubMed] [Google Scholar]
- 65.Haaf T., Schmid M. An early stage of ZW/ZZ sex chromosome differentiation in Poecilia schenops var. melanistica (Poeciliidae, Cyprinodontiformes). Chromosoma. 1984;89:37–41. [Google Scholar]
- 66.Nanda I., Schartl M., Epplen J.T., Feichtinger W., Schmid M. Primitive sex chromosomes in Poeciliid fishes harbor simple repetitive DNA sequences. J. Exp. Zool. 1993;265(3):301–308. doi: 10.1002/jez.1402650311. [DOI] [PubMed] [Google Scholar]
- 67.Cano J., Pretel A., Melendez S., Garcia F., Caputo V., Fenochio A.F., Bertollo L.A.C. Determination of early stages of sex chromosome differentiation in the sea bass Dicentrarchus labrax L. (Pisces: Perciformes). Cytobios. 1996;87:45–59. agris.fao.org/agris-search/search.do?recordID=GB9717369 [Google Scholar]
- 68.Almeida-Toledo L.F., Foresti F., Pequignot E.V., Daniel-Silva M.F. XX: XY sex chromosome system with X heterochromatinization: An early stage of sex chromosome differentiation in the Neotropic electric eel Eigenmannia virescens. Cytogenet. Cell Genet. 2001;95(1-2):73–78. doi: 10.1159/000057020. [DOI] [PubMed] [Google Scholar]
- 69.Pokorná M., Giovannotti M., Kratochvíl L., Kasai F., Trifonov V.A., O’Brien P.C.M., Caputo V., Olmo E., Ferguson-Smith M.A., Rens W. Strong conservation of the bird Z chromosome in reptilian genomes is revealed by comparative painting despite 275 million years divergence. Chromosoma. 2011;120(5):455–468. doi: 10.1007/s00412-011-0322-0. [DOI] [PubMed] [Google Scholar]
- 70.Ming R., Bendahmane A., Renner S.S. Sex chromosomes in land plants. Annu. Rev. Plant Biol. 2011;62:485–514. doi: 10.1146/annurev-arplant-042110-103914. www.annualreviews.org/doi/pdf/10.1146/annurev-arplant-042110-103914 [DOI] [PubMed] [Google Scholar]
- 71.Cermak T., Kubat Z., Hobza R., Koblizkova A., Widmer A., Macas J., Vyskot B., Kejnovsky E. Survey of repetitive sequences in Silene latifolia with respect to their distribution on sex chromosomes. Chromosome Res. 2008;16(7):961–976. doi: 10.1007/s10577-008-1254-2. [DOI] [PubMed] [Google Scholar]
- 72.Kejnovský E., Michalovova M., Steflova P., Kejnovska I., Manzano S., Hobza R., Kubat Z., Kovarik J., Jamilena M., Vyskot B. Expansion of microsatellites on evolutionary young Y chromosome. PLoS One. 2013;8(1):e45519. doi: 10.1371/journal.pone.0045519. journals.plos.org/plosone/article?id=10.1371/journal.pone.0045519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chalopin D., Naville M., Plard F., Galiana D., Volff J.N. Comparative analysis of transposable elements highlights mobilome diversity and evolution in vertebrates. Genome Biol. Evol. 2015;7(2):567–580. doi: 10.1093/gbe/evv005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Volff J.N. Turning junk into gold: Domestication of transposable elements and the creation of new genes in eukaryotes. BioEssays. 2006;28(9):913–922. doi: 10.1002/bies.20452. [DOI] [PubMed] [Google Scholar]
- 75.Schartl M. Sex chromosome evolution in non-mammalian vertebrates. Curr. Opin. Genet. Dev. 2004;14(6):634–641. doi: 10.1016/j.gde.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 76.Ellegren H. Hens, cocks and avian sex determination. A quest for genes on Z or W? EMBO Rep. 2001;2(3):192–196. doi: 10.1093/embo-reports/kve050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ezaz T., Stiglec R., Veyrunes F., Graves J.A.M. Relationships between vertebrate ZW and XY sex chromosome systems. Curr. Biol. 2006;16(17):736–743. doi: 10.1016/j.cub.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 78.Mank J.E., Hall D.W., Kirkpatrick M., Avise J.C. Sex chromosomes and male ornaments: A comparative evaluation in ray-finned fishes. Proc. Biol. Sci. 2006;273(1583):233–236. doi: 10.1098/rspb.2005.3334. http://rspb.royalsocietypublishing.org/content/273/1583/233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gamble T., Coryell J., Ezaz T., Lynch J., Scantlebury D.P., Zar-kower D. Restriction site-associated DNA sequencing (RAD-seq) reveals an extraordinary number of transitions among gecko sex-determining systems. Mol. Biol. Evol. 2015;32(5):1296–1309. doi: 10.1093/molbev/msv023. [DOI] [PubMed] [Google Scholar]
- 80.Takehana Y., Naruse K., Hamaguchi S., Sakaizumi M. Evolution of ZZ/ZW and XX/XY sex-determination systems in the closely related medaka species, Oryzias hubbsi and O. dancena. Chromosoma. 2007;116(5):463–470. doi: 10.1007/s00412-007-0110-z. [DOI] [PubMed] [Google Scholar]
- 81.Phillips R.B., Konkol N.R., Reed K.M., Stein J.D. Chromosome painting supports lack of homology among sex chromosomes in Oncorhynchus, Salmo and Salvelinus (Salmonidae). Genetica. 2001;111(1-3):119–123. doi: 10.1023/a:1013743431738. [DOI] [PubMed] [Google Scholar]
- 82.Ross J.A., Urton J.R., Boland J., Shapiro M.D., Peichel C.L. Turnover of sex chromosomes in the stickleback fishes (Gasterosteidae). PLoS Genet. Available from: journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen. 2009 doi: 10.1371/journal.pgen.1000391. [DOI] [PMC free article] [PubMed]
- 83.Miura I. An evolutionary witness: The frog Rana rugosa underwent change of heterogametic sex from XY male to ZW female. Sex Dev. 2007;1(6):323–331. doi: 10.1159/000111764. [DOI] [PubMed] [Google Scholar]
- 84.Perrin N. Sex reversal: A fountain of youth for sex chromosomes? Evolution. 2009;63(12):3043–3049. doi: 10.1111/j.1558-5646.2009.00837.x. [DOI] [PubMed] [Google Scholar]
- 85.Hillis D.M., Green D.M. Evolutionary changes of heterogametic sex in the phylogenetic history of amphibians. J. Evol. Biol. 1990;3(1-2):49–64. [Google Scholar]
- 86.Faria R., Navarro A. Chromosomal speciation revisited: Rearranging theory with pieces of evidence. Trends Ecol. Evol. 2010;25(11):660–669. doi: 10.1016/j.tree.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 87.Kitano J., Peichel C.L. Turnover of sex chromosomes and speciation in fishes. Environ. Biol. Fishes. 2012;94(3):549–558. doi: 10.1007/s10641-011-9853-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yamada M., Higuchi M., Goto A. Extensive introgression of mitochondrial DNA found between two genetically divergent forms of threespine stickleback, Gasterosteus aculeatus, around Japan. Environ. Biol. Fishes. 2001;61(3):269–284. [Google Scholar]
- 89.Yamada M., Higuchi M., Goto A. Long-term occurrence of hybrids between Japan Sea and Pacific Ocean forms of threespine stickleback, Gasterosteus aculeatus, in Hokkaido Island, Japan. Environ. Biol. Fishes. 2007;80(4):435–443. [Google Scholar]
- 90.Kitano J., Mori S., Peichel C.L. Sexual dimorphism in the external morphology of the threespine stickleback (Gasterosteus aculeatus). Copeia. 2007;2007(2):336–349. https://www.jstor.org/stable/25140637 [Google Scholar]
- 91.Bertollo L.A.C., Oliveira C., Molina W.F., Margarido V.P., Fontes M.S., Pastori M.S., Falcão J.N., Fenocchio A.S. Chromosome evolution in the erythrinid fish, Erythrinus erythrinus (Teleostei: Characiformes). Heredity. 2004;93(2):228–233. doi: 10.1038/sj.hdy.6800511. [DOI] [PubMed] [Google Scholar]