Abstract
China has a rich resource of native sheep (Ovis aries) breeds associated with historical movements of several nomadic societies. However, the history of sheep and the associated nomadic societies in ancient China remains poorly understood. Here, we studied the genomic diversity of Chinese sheep using genome-wide SNPs, mitochondrial and Y-chromosomal variations in > 1,000 modern samples. Population genomic analyses combined with archeological records and historical ethnic demographics data revealed genetic signatures of the origins, secondary expansions and admixtures, of Chinese sheep thereby revealing the peopling patterns of nomads and the expansion of early pastoralism in East Asia. Originating from the Mongolian Plateau ∼5,000‒5,700 years ago, Chinese sheep were inferred to spread in the upper and middle reaches of the Yellow River ∼3,000‒5,000 years ago following the expansions of the Di-Qiang people. Afterwards, sheep were then inferred to reach the Qinghai-Tibetan and Yunnan-Kweichow plateaus ∼2,000‒2,600 years ago by following the north-to-southwest routes of the Di-Qiang migration. We also unveiled two subsequent waves of migrations of fat-tailed sheep into northern China, which were largely commensurate with the migrations of ancestors of Hui Muslims eastward and Mongols southward during the 12th‒13th centuries. Furthermore, we revealed signs of argali introgression into domestic sheep, extensive historical mixtures among domestic populations and strong artificial selection for tail type and other traits, reflecting various breeding strategies by nomadic societies in ancient China.
Keywords: Ovis aries, admixture, nomadic society, peopling pattern, early pastoralism
Introduction
Episodes of historical human migrations were influenced by multiple forces, and have always been a focus of research (Hellenthal et al. 2014; Pagani et al. 2016). For example, migration of nomadic societies over the past few thousand years in Central and East Asia was dominated by conquests, the rise of successive empires, large-scale warfare and interregional trading (Palstra et al. 2015; Yunusbayev et al. 2015). Living in proximity to humans, particularly nomads (Phillips 2001), domestic sheep (Ovis aries) have accompanied human migrations worldwide after their dispersal out from the Middle East c. 8,000–9,000 years before the present (BP) when the development of agriculture initiated the expansion of farming societies (Larson et al. 2007). During this process, sheep also acted as a key factor in the social transformation between sedentary agrarian, seminomadic, and nomadic pastoralism societies (Kamp and Yoffee 1980; Chang and Koster 1986).
In China, there has been a history of sheep farming for > 5,000 years (Cai et al. 2011), and the country harbors as many as 42 native breeds. The early history of Chinese sheep has rarely been documented and its reconstruction has mainly relied on paleontological findings and pictorial representations (Dodson et al. 2014). Only recently, have the genetic origin, population expansion, and admixture of Chinese sheep been characterized using a small number of genetic loci (Chen et al. 2006; Zhang et al. 2014; Lv et al. 2015) and/or populations (Wei et al. 2015; Yang et al. 2016b). On the other hand, the early peopling processes of Chinese nomadic ethnic groups are not yet well known owing to apparently contradictory evidence from genetic, linguistic, and historical data (Shou et al. 2010; Meyer et al. 2017). Given the close association between sheep and their early nomadic keepers (Ryder 1983), genomes of native sheep populations should bear genetic signatures that delineate these patterns in both the sheep and the associated nomadic societies.
Here, we generated a genome-wide SNP data set of > 1,000 samples of domestic and wild sheep (argali [Ovis ammon polii, a wild sheep that roams the highlands of Central Asia, the Himalaya, Tibet, and Altay] and European mouflon [Ovis aries musimon]; fig. 1A and supplementary table S1 and fig. S1, Supplementary Material online). We combined this data set with publicly available genomic data (ovine 50K array genotypes of 1,648 samples from 48 worldwide populations, supplementary table S1, Supplementary Material online; 55 ancient and 1,076 modern mtDNA fragments, supplementary tables S2 and S3, Supplementary Material online; and Y-chromosomal variations in 578 samples, supplementary table S4, Supplementary Material online; see also in Cai et al. 2007; Cai et al. 2011; Kijas et al. 2012; Demirci et al. 2013; Zhang et al. 2014; Lv et al. 2015) (supplementary table S5 and fig. S2, Supplementary Material online) and previous archeological (supplementary table S6, Supplementary Material online) and ethnohistorical records (supplementary information, Supplementary Material online). Our aims were 3-fold: 1) to provide a detailed picture of genomic landscape and demographic history of Chinese native sheep; 2) to track the spread of early pastoralism and the peopling patterns of nomads in East Asia; and 3) to trace the genetic legacy of centuries of artificial selection and breeding left on the sheep genomes.
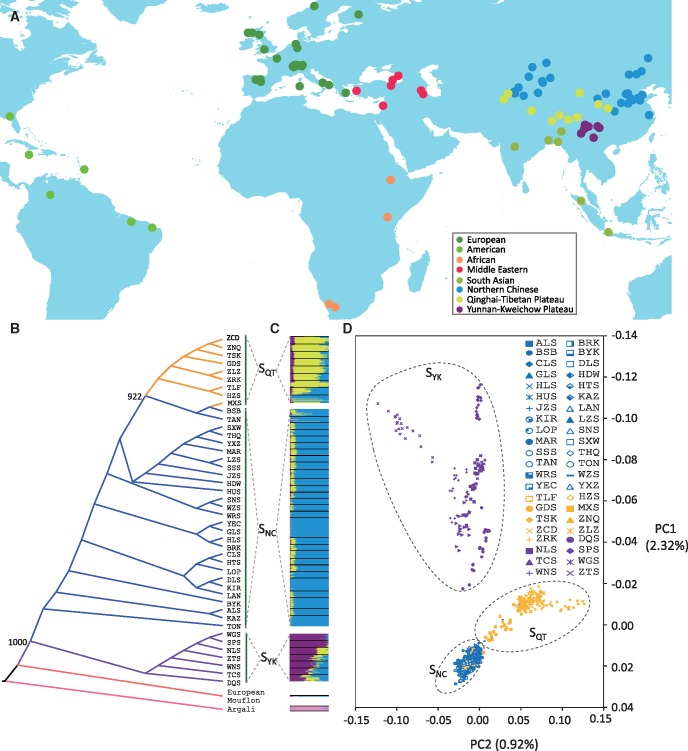
Fig. 1.
Locations and genetic structure of sheep (Ovis aries) populations. (A) Geographic distribution of the sheep populations studied. (B) Neighbor-joining (NJ) tree of Chinese sheep populations based on the Reynolds’ distances. (C) STRUCTURE analysis illustrates that the genetic clustering patterns of Chinese sheep populations are consistent with their geographic distributions at K = 4. (D) Visualization of principal components 1 and 2 from individual-based principal component analysis (PCA) of Chinese sheep. Full names of the sheep populations are detailed in supplementary table S1, Supplementary Material online. See supplementary fig. S1, Supplementary Material online for additional information.
Results and Discussion
Within-Population Genomic Variability
We assessed the genomic variability of Chinese sheep by estimating the within-population diversity based on the whole-genome BeadChip SNPs (supplementary table S7, Supplementary Material online). Our results demonstrated that the level of genomic variability in Chinese sheep is comparable to that observed in other populations around the world (Kijas et al. 2012), but greater than that of the two wild species (supplementary tables S7 and S8, figs. S3‒S6, and supplementary information, Supplementary Material online). We observed congruent geographic patterns of genomic diversities assessed by various indices: northern Chinese populations (including those from North China, East China, Xinjiang, and Inner Mongolia) had the highest level of genomic variability, followed by the Qinghai-Tibetan populations and Yunnan-Kweichow populations (supplementary figs. S3‒S7 and supplementary information, Supplementary Material online). Similar geographic gradients among the three groups of populations were also found in the distribution of mtDNA (supplementary fig. S8, Supplementary Material online; Lv et al. 2015), Y-chromosomal (supplementary fig. S9, Supplementary Material online; Zhang et al. 2014), and whole-genome sequence variations in Chinese sheep (Yang et al. 2016b). The relatively high levels of genomic variability observed in northern Chinese sheep populations could be partially explained by the fact that these populations diverged earlier from the ancestral populations distributed in the Mongolian Plateau, a transportation hub of early sheep expansions in eastern Eurasia including China (Lv et al. 2015), whereas the low levels of genomic variability detected in the Yunnan-Kweichow populations could be the result of serial founder effect and strong genetic drift attributed to long-term genetic isolations among populations in the plateau. The genetic drift was also reflected by the increased drift parameters in the TreeMix analysis (fig. 2A and supplementary figs. S10 and S11, Supplementary Material online). We noted that the estimates of variability based on BeadChip SNPs might suffer from ascertainment bias in SNP discovery (Kijas et al. 2012). Nevertheless, the removal of SNPs in high levels of LD has been shown to counter the effect of ascertainment bias and generate meaningful interpopulation comparisons (Kijas et al. 2012). Thus, the ascertainment bias should have been greatly decreased by LD pruning in our quality control procedure.
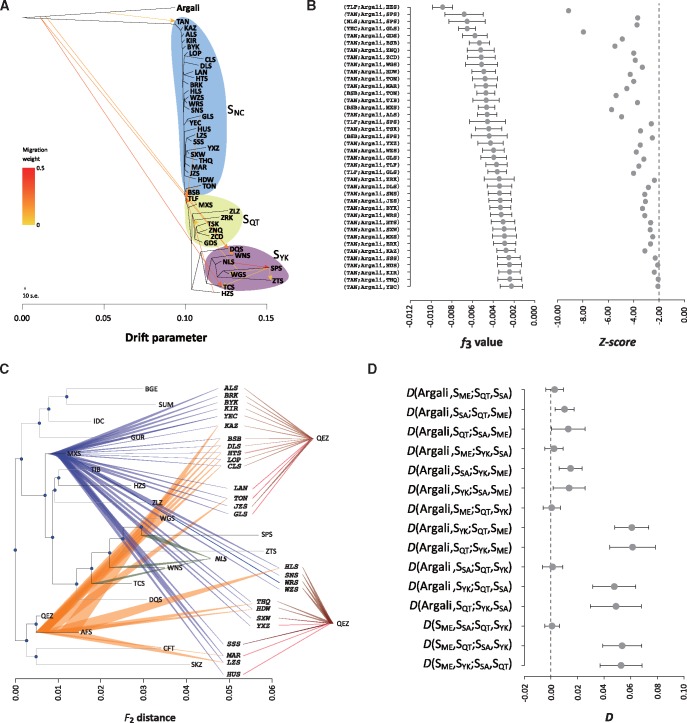
Fig. 2.
Migration and admixture of Chinese and Asian sheep. (A) Phylogenetic network from TreeMix v.1.12 showing the inferred relationships among Chinese native sheep populations. Up to 98.14% of the variance was explained by a model with 13 migration edges. The length of the branch is proportional to the drift of each population. ARG (argali) was used as the outgroup to root the tree. The scale bar shows ten units of standard error (s.e.) of the entries in the sample covariance matrix (supplementary fig. S10B, Supplementary Material online). The migration weight represents the fraction of ancestry derived from the migration edge. (B) Three-way admixture between ARG and Chinese domestic sheep populations. Only the population triples (C; ARG, B) with negative estimates of f3 value and Z-score < −2 (supplementary table S15, Supplementary Material online) are shown. (C) Inferred sources of ancestry for admixed Chinese sheep populations with the two-way model in the MixMapper v.2.0 program. Admixed populations, which are shown in bold type and located between the colored lines, were tested (supplementary table S13, Supplementary Material online) and added to the scaffold tree. (D) D statistics with the form D (W, X; Y, Z) for Asian sheep populations and ARG (argali). Full names of the sheep populations are detailed in supplementary table S1, Supplementary Material online.
Population Genetic Differentiation
To obtain a refined picture of interpopulation genetic relationships, we examined genetic differentiation among populations using the principal component (PC), STRUCTURE, and neighbor-joining (NJ) tree methods. The analyses revealed clear geographic patterns among domestic sheep populations at various scales (i.e., global, continental, and national) (fig. 1B‒D and supplementary table S9, figs. S12‒S18 and supplementary information, Supplementary Material online). In Chinese sheep, we identified three genetic population groups, which were consistent with their geographic distributions of origins, based on the first two PCs (PC1 = 2.32%, PC2 = 0.92%) and the STRUCTURE optimal fit model at K = 4: northern Chinese (including the populations from North China, East China, Xinjiang, and Inner Mongolia), Qinghai-Tibetan and Yunnan-Kweichow (fig. 1B‒D and supplementary figs. S17 and S18, Supplementary Material online). The geographic patterns of genomic diversity and population structure, which were further supported by the TreeMix (fig. 2A and supplementary fig. S10, Supplementary Material online) and D-statistics analyses (fig. 2D and supplementary table S10, Supplementary Material online), could be explained mainly by geographic isolation (e.g., the Qinghai-Tibetan Plateau and the Yunnan-Kweichow Plateau) and high levels of historical mixing among different domestic breeds within or between particular regions (fig. 2A and supplementary fig. S11 and supplementary information, Supplementary Material online; Kijas et al. 2012).
Analyses of Uniparental Genetic Markers
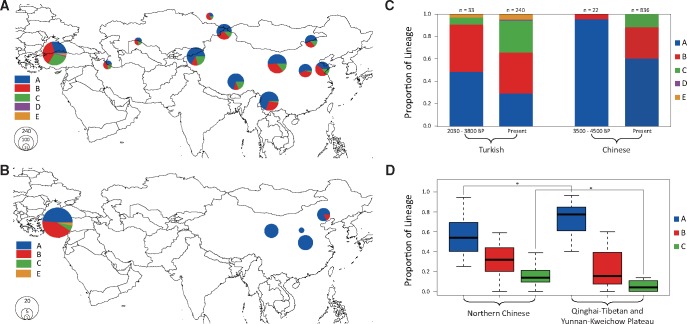
To investigate the historical mixing among populations from different regions, we analyzed hypervariable mtDNA fragments from 55 publicly available ancient remains (22 ancient Chinese sheep remains at c. 3,500‒4,500 BP and 33 ancient Turkish sheep remains at c. 2,030‒3,800 BP; Cai et al. 2007; Cai et al. 2011; Demirci et al. 2013) and 85 newly generated and 991 publicly available modern samples (Demirci et al. 2013; Lv et al. 2015). We demonstrated that the proportion of lineage B in Chinese sheep significantly increased from 5% in ancient remains (3,500‒4,500 BP) to 27% in modern samples (χ2 test, df = 1, P = 0.034), and lineage C, which was absent in the ancient remains, increased to 13% in modern samples (fig. 3B and C and supplementary tables S2 and S3, Supplementary Material online). Moreover, the present northern Chinese populations exhibited significantly higher proportions of lineages B and C but a lower proportion of lineage A than those from the Qinghai-Tibetan and Yunnan-Kweichow plateaus (fig. 3A and D and supplementary table S3 and fig. S8, Supplementary Material online). The observed temporal and spatial patterns indicated that maternal genetic introgressions of the sheep with high proportions of lineages C (e.g., fat-tailed sheep) and B in early Chinese sheep populations have mainly occurred in northern China (Tapio et al. 2006; Lv et al. 2015). In addition, we observed a relatively high level of Y-chromosomal haplotype diversity in populations from Inner Mongolia compared with other regions (supplementary table S4 and fig. S9, Supplementary Material online). This observation provided an indicial piece of evidence for the previous finding that the Mongolian Plateau was a transportation hub of early sheep expansions in eastern Eurasia including China (Lv et al. 2015).
Fig. 3.
MtDNA lineage frequencies in ancient and modern samples. (A, B) The frequency distribution of the mtDNA lineages across Eurasia based on partial control region sequences in (A) modern samples and (B) ancient remains. (C, D) The proportions of mtDNA lineages in ancient and modern samples from (C) Turkey and China and (D) various regions of China (*P < 0.05). The time ranges of the time bins were determined by the Accelerator Mass Spectrometry (AMS) radiocarbon dating method (Demirci et al. 2013) for the ancient Turkish samples and by the culture which the archeological site belonged to (Cai et al. 2011) for the ancient Chinese samples.
Genetic Admixture among Domestic Sheep Populations
We used TreeMix, Mixmapper, and f3-statistics analyses to test for the occurrence of gene flow among domestic sheep populations. In the test, we found extensive admixture in northern Chinese populations (fig. 2C and supplementary tables S11‒S14, Supplementary Material online). Among the ten northern Chinese populations (ALS, BSB, BYK, JZS, KAZ, KIR, LOP, TLF, WRS, and YEC) with negative f3-statistic, eight (ALS, BSB, BYK, KAZ, KIR, TLF, WRS, and YEC) showed significant evidence of admixture (Z < −2; supplementary table S11, Supplementary Material online; Reich et al. 2009). Also, 28 northern Chinese populations were best fitted into the two-way admixture model in the MixMapper analysis, which most likely descended from the admixture between the fat-tailed sheep (as represented by AFS and QEZ) and early Chinese sheep populations as represented by the MXS (fig. 2C and supplementary table S13, Supplementary Material online). Moreover, one sheep population geographically from Xinjiang, the TLF, showed significant evidence of admixture from multiple source populations (i.e., SME, SQT, SSA, and SNC; supplementary table S11, Supplementary Material online) in the f3-statistics test and was best fitted into the three-way admixed model in the MixMapper analysis (supplementary table S14, Supplementary Material online). In addition, the TreeMix analysis illustrated extensive migrations among Asian populations (supplementary fig. S11, Supplementary Material online), in particular between the Middle Eastern and northern Chinese populations. These findings, together with previously inferred maternal introgressions of mtDNA lineages (mainly lineages B and C) in early Chinese sheep in northern China (fig. 3), suggest that modern northern Chinese populations were extensively influenced by genetic admixture from the fat-tailed sheep.
Genetic Introgressions of Argali in Domestic Sheep
Hybridization between wild and local domestic sheep populations has been reported (Larson and Burger 2013), but little is known about the geographic scales and genomic levels of such introgression. Our TreeMix and f3-statistic results revealed clear signs of argali genetic introgression in several domestic populations (e.g., TAN, BSB, TLF, YEC, TCS, WNS, and DQS) from northern China and the Yunnan-Kweichow Plateau, which argali has been known to inhabit (fig. 2A and B and supplementary table S15, fig. S11 and supplementary information, Supplementary Material online). From the TreeMix analysis of Asian sheep, we also observed argali introgression in South Asian populations (supplementary fig. S11, Supplementary Material online), which could probably occur during the migrations of domestic sheep from the Tibetan Plateau and the Himalaya Range regions to South Asia (Lv et al. 2015). Also, we detected admixture from argali into domestic sheep (e.g., TAN, TCS, WNS, and DQS) via STRUCTURE analysis. The STRUCTURE analysis identified that an average of 3.26%, 2.20%, 0.81%, and 1.38% of the TAN, TCS, WNS, and DQS genomes could be attributed to the argali introgression, respectively (fig. 1C and supplementary fig. S17, Supplementary Material online). Our results clearly supported extensive argali genomic introgressions into domestic sheep, which could be due to the interspecific hybridization with wild relatives adopted in early sheep breeding practice. Similar introgressions from wild to domestic populations have also been observed in other domestic animals such as pig (Frantz et al. 2015) and cattle (Węcek et al. 2017). In addition, a genetic differentiation test between argali and domestic sheep detected five putatively adaptive genes (e.g., BMPR2, FGF9, FGF7, KGFLP2, and NFIB; supplementary table S16 and fig. S19, Supplementary Material online), whose functions are associated with the developments of lung, respiratory tube, and respiratory system.
Population History Reconstruction
We used the approximate Bayesian computation (ABC) approach and Ovine SNP50K BeadChip data to reconstruct the population history of Chinese sheep. We tested four alternative demographic models (supplementary table S17 and fig. S20, Supplementary Material online) based on 12 uncorrelated summary statistics (supplementary table S18, fig. S21, and supplementary information, Supplementary Material online). The best fit to the data was obtained in Model-2 (supplementary table S19, Supplementary Material online), in which the split of northern Chinese sheep from their ancestral populations predate the divergence between the Qinghai-Tibetan sheep and the Yunnan-Kweichow sheep (fig. 4A and supplementary fig. S20, Supplementary Material online). This model also included a later admixture (6.31%, 50% highest posterior density [HPD]: 1.45‒12.21%) with Middle Eastern sheep (fig. 4A and supplementary table S20 and fig. S20, Supplementary Material online), which is consistent with the admixed signatures observed in the MixMapper and f3-statistics tests (fig. 2C and supplementary tables S11 and S13, Supplementary Material online). Under the best-supported model, the simulations produced realistic posterior distributions for all the parameters (supplementary figs. S22 and S23, Supplementary Material online) and yielded the estimates of the divergence time of northern Chinese sheep from their ancestral populations to be c. 4,000 BP (T1, 50% HPD: 3,513‒4,542; supplementary table S20, Supplementary Material online) and that of the divergence between the Qinghai-Tibetan sheep and the Yunnan-Kweichow sheep to be c. 2,500 BP (T2, 50% HPD: 1,881‒3,564; supplementary table S20, Supplementary Material online). Furthermore, the time of the admixture events between the Middle Eastern sheep and northern Chinese sheep was dated to be c. 1,600 BP (Tm, 50% HPD: 930‒2,652; supplementary table S20, Supplementary Material online). The optimal scenario inferred was consistent with the divergence pattern and timing of Chinese sheep reported in a recent study based on whole-genome sequences (Yang et al. 2016b).
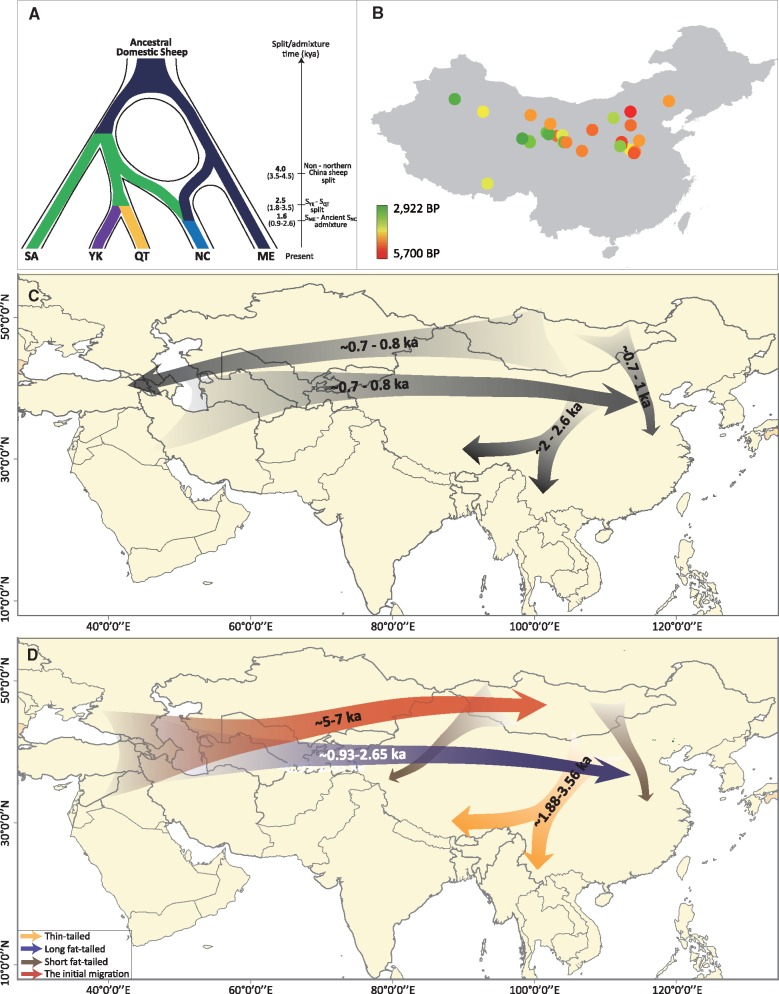
Fig. 4.
Integrated diagram of inferred demographic history of the nomadic ethnic groups and domestic sheep in East Asia, particularly in China. (A) The inferred divergence mode of Asian sheep populations. The abbreviations represent sheep populations from the Middle East (ME), South Asia (SA), northern China (NC), the Qinghai-Tibetan Plateau (QT) and the Yunnan-Kweichow Plateau (YK). (B) Approximate dates for ancient Chinese sheep remains obtained from archeological records (supplementary table S6, Supplementary Material online). (C) The schematic summarizes the recorded migration routes of the nomadic ethnic groups in China from historical knowledge (supplementary information, Supplementary Material online). (D) The schematic summarizes the inferred migration routes of Chinese sheep from genetic data.
We tested the colonization processes by calculating the correlation between the multidimensional scaling (MDS) dimension λ1 values (fig. 5A and supplementary table S21, Supplementary Material online) and geographic distances from the putative regions of origins (Hanotte et al. 2002). A synthetic map constructed with the use of interpolated λ1 values allowed us to show gradients of admixture that peaked in the northwesternmost or northernmost regions of the Xinjiang and Inner Mongolian Provinces. We observed a strong and positive correlation (long fat-tailed/fat-rumped: Spearman’s r = 0.706, P < 0.01; short fat-tailed, Spearman’s r = 0.709, P < 0.01) between the λ1 values of the populations and the distances from site of the peak (supplementary fig. S24B, Supplementary Material online). The correlation indicated the presence of two different patterns of fat-tailed sheep admixture: one was from Xinjiang Province in northwestern China eastward, and the other from the Mongolian Plateau southward.
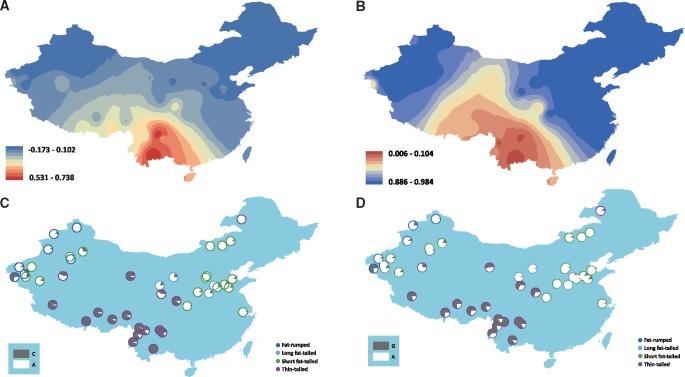
Fig. 5.
Geographic distribution of genetic variability in Chinese native sheep. (A) Synthetic map illustrating geographic variation of multidimensional scaling (MDS) dimension 1 (λ1) values (supplementary table S21, Supplementary Material online). (B) Synthetic map illustrating geographic variation of the genetic ancestries of fat-tailed sheep obtained from the STRUCTURE analysis (K = 4; see supplementary table S25 and fig. S25, Supplementary Material online) based on the top 1% selective SNPs (supplementary table S22, Supplementary Material online). (C) Frequency distribution of the SNP alleles of the most significant loci rs422598859 on chromosome 13 in the selection test for tail type in Chinese native sheep populations. (D) Frequency distribution of the SNP alleles of the most significant loci rs400618975 on chromosome 15 in the selection test for tail type in Chinese native sheep populations. The colored cycles represent different tail types.
The explicit geographic patterns of tail type were further evidenced by the distribution of genetic ancestries from fat-tailed sheep as inferred based on the STRUCTURE and interpolation analyses (fig. 5B and supplementary figs. S25 and S26, Supplementary Material online) of the selective SNPs associated with tail type (supplementary table S22, Supplementary Material online) as well as allele frequencies of two selective SNPs under strong selection (rs422598859 and rs400618975; fig. 5C and D and supplementary table S23, Supplementary Material online). We performed the STRUCTURE analysis using the selective SNPs (supplementary table S22, Supplementary Material online). At K = 4, we detected significant genetic differentiation (FST = 0.00951‒0.289, P < 0.001; supplementary table S24, Supplementary Material online) among the four groups of populations of various geographic origins: long fat-tailed and fat-rumped sheep in Northwest China, short fat-tailed sheep in North China, and thin-tailed sheep in the Qinghai-Tibetan Plateau and Yunnan-Kweichow Plateau (supplementary table S25 and fig. S25, Supplementary Material online). In northern Chinese populations, the genetic ancestry of long fat-tailed and fat-rumped sheep was reduced following a northwest-to-east route, whereas ancestry of short fat-tailed sheep exhibited a gradual north-to-south decrease over North China (supplementary figs. S25 and S26, Supplementary Material online). A synthetic map (fig. 5B and supplementary fig. S26, Supplementary Material online) showed a pattern similar to that based on genome-wide SNPs (fig. 5A and supplementary fig. S24, Supplementary Material online).
In addition, we observed increased λ1 values with the geographic distance from the Shihushan archeology site in the Mongolian Plateau in the thin-tailed populations, following two different directions for the Qinghai-Tibetan populations (Spearman’s r = 0.214, P = 0.645) and the Yunnan-Kweichow populations (Spearman’s r = 0.455, P = 0.187; supplementary fig. S24C, Supplementary Material online). The two regression lines supported a pattern of dispersal in different directions from a single geographic region of origin (Hanotte et al. 2002).
Archeological Evidences
To assess the consilience between our results and the archeological records, we compiled evidence based on ancient sheep remains across China (fig. 4B and supplementary table S6, Supplementary Material online). We found that the earliest sheep remains were located in Inner Mongolia. These have been dated at c. 5,000‒5,700 BP (Cai et al. 2011; Dodson et al. 2014), and are associated with the introduction of Neolithic domesticates (e.g., sheep, goats, and barley) into China c. 5,500 BP, originating from West Asia (Aldenderfer 2011; Jeong et al. 2016). This coincides with the historical records of the first sheep farming and breeding community, the “Di-Qiang” tribes (DQs), who originally resided in the regions closely neighboring Inner Mongolia (i.e., the upper valleys of the Yellow and Weihe Rivers in the Loess Plateau) over 5,000 BP (Wagner et al. 2011; Dong et al. 2013). The wide geographic area was covered by a common culture, known as the Yangshao culture, from 5,000‒7,000 BP (Barton et al. 2009).
Ethnohistorical Demographics
We compiled evidence regarding nomadic demographics in ancient China to evaluate the concordance between our results and ethnohistorical records (supplementary information, Supplementary Material online). The date for the split of northern Chinese sheep at c. 4,000 BP (T1, 50% HPD: 3,513‒4,542; supplementary table S20, Supplementary Material online) yielded by the ABC modeling framework was in rough agreement with the historical evidence for the migration of the DQs outward in the upper and middle reaches of the Yellow River in northern China during the middle of the Shang Dynasty (c. 3,400 BP; Fan 1131; Si 1131). The genetic-based estimates of divergence time of the Qinghai-Tibetan sheep and the Yunnan-Kweichow sheep at c. 2,500 BP (T2, 50% HPD: 1,881‒3,564; supplementary table S20, Supplementary Material online) also coincided largely with the massive population movements of the DQs toward Southwest China (c. 2,000‒2,600 BP) and the subsequent formation of Tibetans and other ethnic groups in Southwest China from historical knowledge (Fan 1131; Si 1131; Ou 1936). Thus, among the proposed “Di-Qiang”, “Han Chinese”, “Aboriginal”, and “South Asia” origin hypotheses for Tibetans and other southwestern ethnic groups (He 2000), our sheep genetic data support a “Di-Qiang” origin from northern China, most likely via the “Tibetan-Yi” corridor and the Yalong and Dadu Rivers route (fig. 4C and D; He 2000). The northern origin for Tibetans and other southwestern ethnic groups was also supported by a recent human genomics study, which found gene flow into the Himalaya region from the north but not from the south (Jeong et al. 2016).
The estimated time for the extensive admixture from the long fat-tailed sheep into northern Chinese sheep by the ABC modeling (Tm = c. 1,600 BP, 50% HPD: 930‒2,652 BP; supplementary table S20, Supplementary Material online) and the revealed zone of introgression in northern China (supplementary fig. S25A, Supplementary Material online) indicated that the admixture occurred generally commensurate with the known massive historical westward invasions of the Mongols (c. 1,219‒1,260 A.D.; supplementary information, Supplementary Material online). During their westward invasions, a substantial number of Islamic peoples from the Arabian Peninsula, Persia, and Central Asia were forced to move eastwards into northern China (Grousset 1941; Song and Wang 1983) through the northern route of the Silk Road (e.g., from Central Asia to Xi’an via Xinjiang Province, China; Christian 2000), possibly resulting in west-to-east migrations and introgression of long fat-tailed sheep into northern Chinese populations. Similarly, the inferred migrations of long fat-tailed sheep may mirror the history of the Hui Muslims in China, who were formed by a fusion of western Islamic immigrants, Han Chinese, and Mongolians (Li 2008).
In addition, the southward expansion of short fat-tailed sheep inferred from the MDS analysis (fig. 4D and supplementary fig. S25A, Supplementary Material online) was likely attributed to the historical invasions of the nomadic Mongols and the seminomadic Jurchen from Northwest China (fig. 4C) during the 12th and 13th centuries according to the historical demographics, conquering northern regions (including what is now known as North China) of the Song Dynasty (supplementary information, Supplementary Material online; Grousset 1941; Song and Wang 1983). Moreover, the westward expansion of short fat-tailed sheep (fig. 4D) generally coincided with the massive westward invasions of the Mongols from the Mongolian Plateau (fig. 4C) during the 12th and 13th centuries (supplementary information, Supplementary Material online). Because they mainly reared short fat-tailed sheep, the invasions may have led to a north-to-south and a westward expansion of short fat-tailed sheep (fig. 4C and D).
Altogether, the inferred expansion, divergence and admixture of Chinese sheep based on genomic data are roughly congruent with the migrations of ancient nomadic ethnic groups (e.g., the Mongols, Jurchen, Hui Muslims, Tibetans, and other ethnic groups like the “Yi” in Southwest China) in East Asia, as recorded in authoritative historical literature in terms of time and routes (fig. 4C and D and supplementary information, Supplementary Material online). Although the migration of pastoralists with sheep is the most likely mechanism for interpretation of our findings, it is possible that sheep were transferred across regions without the accompanying migration of human populations, e.g., through mechanisms such as trade, trophy, tribute, and introduction of breeding stock (Ryder 1983; Yunusbayev et al. 2015). Therefore, these potential caveats should be noted regarding making inferences about human migration using nonhuman data. Also, it should be noted that our prior modeling analysis (supplementary table S20, Supplementary Material online) revealed earlier times for the divergence and admixture than the relevant historical events. The detected admixture signatures from wild sheep into domestic population most likely pushed the estimated time deeper into the past as was observed in similar cases for the older estimates of divergence among dogs (Frantz et al. 2016). Alternatively, this observation could be explained by potential, although very limited, gene flow between Chinese and South Asian populations (supplementary table S11, Supplementary Material online), which have an early Middle Eastern ancestral origin (Lv et al. 2015). Reconstruction using high-depth whole genomes will improve the resolution, particularly for recent time periods (Frantz et al. 2016).
Signal of Selection on Diverse Phenotypes
Chinese native sheep exhibits rich phenotypic diversity including morphological and production traits (supplementary table S26, Supplementary Material online), and no previous studies have involved selective sweep test associated with some of the phenotypes (e.g., number of vertebra and fur characteristics). In the FST-based selective test across groups of populations with different phenotypes, a total of 24 genomic regions encompassing 103 candidate genes were identified to be under directional selection (supplementary table S27 and fig. S27, Supplementary Material online). In particular, strong selective signals were mapped to the genes RXFP2, BMP2, PDGFD, and VRTN (supplementary table S27 and fig. S27, Supplementary Material online), which were associated with the main phenotypes of the Ovis species, such as type, size and presence or absence of horn (Johnston et al. 2011; Kijas et al. 2012), skeletal morphology and body size (Kijas et al. 2012), tail types (Tang et al. 2007; Xie et al. 2008; Moioli et al. 2015; Wei et al. 2015), and number of vertebra (Fan et al. 2013; Yang et al. 2016a).
In the selection tests between fat-tailed (including long fat-tailed, fat-rumped, and short fat-tailed) and thin-tailed populations, we detected strong selective signals that spanned the BMP2, PDGFD, and VRTN genes as well (supplementary table S26 and fig. S28, Supplementary Material online). Two (BMP2 and VRTN) of the genes are overlapping with the study by Moioli et al. (2015), and were found to be associated with the fat tail phenotype of sheep. Besides these genes, we detected a number of novel genes (e.g., NPC2; supplementary table S27 and fig. S28, Supplementary Material online) related to lipid metabolism and not previously identified as under selection in sheep. NPC2 plays an important role in cholesterol homoeostasis, and the recombinant of NPC2 protein increases triglyceride level in body fat (Adachi et al. 2014). We also detected that a set of important and/or novel genes associated with other major phenotypic traits (e.g., high fertility, coat color, and wool quality) were putatively selected. For example, we identified genes of IGF1, BMPR1B, and BRCA2 for high fertility, MITF and MCAM for coat colors, and KRT for wool quality (supplementary table S27, Supplementary Material online). Overall, the extensive selective signatures detected here could be explained by the effect of long-term artificial selection on diverse phenotypes and selective pressures under various local environmental conditions (e.g., different altitudes and precipitations).
Conclusions
We conducted a multidisciplinary investigation in which the movements of the early pastoralist communities in China has been clarified using genomic data regarding domestic animals and in the context of the spread of domestic animals. The investigation involved primarily genomic analyses combined an extensive review of the archeological and historical literature. We unveiled the history of migration, differentiation and admixture in Chinese sheep, which provides novel insights into the peopling patterns of associated nomadic ethnic groups in ancient East Asia. These findings, together with the signs of extensive argali genetic introgressions into domestic sheep, high levels of historical mixture and strong artificial selection for different traits, offered evidence of various sheep breeding practice and expansion of early pastoralism. Also, the newly generated genome-wide data are valuable resources for the future conservation and genetic improvement of domestic sheep.
Materials and Methods
Sample Collection and DNA Extraction
We sampled a total of 1,016 animals representing 44 Chinese native sheep populations (Ovis aries, 998 individuals) and 2 wild species (argali or Ovis ammon polii, 15 individuals from Kashgar, Xinjiang, China; and European mouflon or Ovis aries musimon, 3 individuals from the Island of Sappi, Finland) in this study. Marginal ear tissues were collected and preserved in tubes with 95‒100% ethanol and stored at -80 °C before DNA extraction. In all cases, knowledge from local herdsmen and pedigree information were used to ensure that sampled animals were purebred and unrelated. Summary information of the samples, including species/populations, names, sampling sites and sampling sizes, are detailed in fig. 1A and supplementary table S1 and fig. S1, Supplementary Material online. Genomic DNA was extracted from the ear tissues following a standard phenol-chloroform extraction procedure (Sambrook and Russell 2000).
SNP BeadChip Genotyping
The 1,016 samples were genotyped with the Illumina Ovine SNP50K (54,241 SNPs) or Illumina Ovine Infinium HD SNP (685,734 SNPs) BeadChips, both of which were developed by the International Sheep Genomics Consortium (ISGC; http://www.sheephapmap.org; last accessed April 9, 2014). Details regarding SNP discovery, the design of the ovine array and genotyping procedures for the Illumina Ovine SNP50K or Illumina Ovine Infinium HD SNP BeadChips can be found at the following site: http://www.sheephapmap.org/hapmap.php (last accessed April 9, 2014), and in Kijas et al. (2012) for the SNP50K BeadChip and in Anderson et al. (2014) for the HD SNP BeadChip. Specifically, 874 individuals from 39 domestic sheep populations and 18 wild sheep (15 argali and 3 European mouflon) were genotyped using the Ovine SNP50K BeadChip, which are referred to as “data set I-Chinese data set” and “data set II-wild data set”, respectively (supplementary tables S1 and S5, Supplementary Material online). The remaining 124 individuals from five domestic sheep populations (i.e., Hu, Wadi, Sishui Fur, Large-tailed Han, and Small-tailed Han sheep) were genotyped using the Ovine HD SNP BeadChip, which hereafter are referred to as “data set III-Chinese HD data set” (supplementary tables S1 and S5, Supplementary Material online).
To investigate the genetic landscape of Chinese native sheep in the continental and global context, we retrieved publicly available Ovine SNP50K data from 48 populations (referred to as “data set IV-worldwide reference data set”; fig. 1A and supplementary tables S1 and S5, Supplementary Material online; Kijas et al. 2012), which consisted of 16 Asian (represented by South Asian and Middle Eastern populations; named “data set V-Asian reference data set”; supplementary tables S1 and S5, Supplementary Material online), 22 European, 4 African, and 6 American populations. Different integrations of the data sets were used for various analyses, as detailed in below and summarized in supplementary table S5 and fig. S2, Supplementary Material online.
Quality Control
The Ovine SNP BeadChip data sets (data sets I–IV) were merged to apply the quality control procedures (supplementary fig. S2, Supplementary Material online) using the PLINK v.1.07 software (Purcell et al. 2007). The quality control measures have also been used elsewhere (Meadows and Kijas 2009; Miller et al. 2011; Kijas et al. 2012; Lv et al. 2014). Briefly, the following criteria were applied, and SNPs or individuals that did not meet any of the criteria were removed: 1) Individuals with a SNPs call rate > 98%; 2) SNPs with a minor allele frequency (MAF) > 0.01; and 3) SNPs with physical locations on autosomes (i.e., not on the mitochondria, X or Y chromosomes) based on the sheep reference genome assembly Oar v.4.0 (http://www.ncbi.nlm.nih.gov/assembly/GCF_000298735.2; last accessed March 31, 2016). Further, we implemented linkage disequilibrium (LD) pruning using the function indep-pairwise (100 25 0.05). This function calculates pairwise LD estimate r2 in a 100-SNPs-window, shifts at a pace of 25 SNPs and excludes one of a pair of SNPs when r2 > 0.05. Nevertheless, the X chromosomal SNPs remained in the detection of SNPs under selection for the prolificacy trait. The number of SNPs and individuals retained was given in supplementary fig. S2, Supplementary Material online.
Genetic Variability
Within-population genetic diversity for Chinese sheep populations and two wild species was assessed using genome-wide SNPs data and various metrics, including the proportion of polymorphic SNPs (Pn), observed (Ho) and expected heterozygosity (He), minor allele frequency (MAF), inbreeding coefficient (F), allelic richness (Ar), and private allele richness (pAr). Estimates of Pn, Ho, He, MAF, and F were computed using the PLINK v1.07 software (Purcell et al. 2007), and values of Ar and pAr were calculated using the ADZE v1.0 software (Szpiech et al. 2008). To evaluate the relationship between genetic diversity and geographic distance from the sheep domestication center, the He values of SNPs data of Asian native sheep populations were plotted against geographic distance from the archeological site Oylum Höyük in Kilis Province in Southeastern Turkey (supplementary information, Supplementary Material online; Arslan et al. 2011; Demirci et al. 2013; Lv et al. 2015). The LD between pairs of autosomal SNPs was calculated with the r2 estimate (Hill and Robertson 1968) using PLINK v1.07 and parameters “–r2-ld-window 99999 –ld-window-r2 0” (supplementary information, Supplementary Material online). Runs of homozygosity (ROHs) for each population of domestic and wild sheep, including number of ROHs, size per ROHs, size of ROHs per individual, and genomic coverage of ROHs per individual, were estimated using the program PLINK v1.07 with the following parameters “–homozyg-density 1000, –homozyg-window-het 1, –homozyg-kb 500, –homozyg-window-snp 50”.
Maternal variability of Chinese sheep was assessed using ancient and modern mtDNA sequences. Partial control region fragments (749-bp) of mtDNA were sequenced in 85 animals from three modern populations of Tibetan sheep (supplementary table S3 and supplementary information, Supplementary Material online). Publicly available haplotype diversities and lineage frequencies of the mtDNA control region in 751 Chinese native sheep representing 40 modern populations (Lv et al. 2015) and 240 Turkish native sheep (Demirci et al. 2013) were also retrieved and included in the integrated analyses (supplementary table S3, Supplementary Material online). To further investigate the temporal variation in the genetic makeup of Chinese and Turkish native sheep, we compared the proportion of mtDNA lineages between modern samples and ancient remains. We used publicly available control region segments from 22 ancient Chinese sheep remains (c. 3,500‒4,500 years before the present [BP]) excavated at four archeological sites (Taosi in Shanxi Province, c. 3,900‒4,500 BP, 1 remain; Changning in Qinghai Province, c. 3,700‒4,200 BP, 7 remains; Dashanqian in Liaoning Province, c. 3,500‒4,000 BP, 6 remains; and Erlitou in Henan Province, c. 3,800‒4,100 BP, 8 remains; supplementary table S2, Supplementary Material online; Cai et al. 2007; Cai et al. 2011) and from 33 ancient Turkish sheep remains (c. 2,030‒3,800 BP; supplementary table S2, Supplementary Material online; Demirci et al. 2013). These represent all known ancient mtDNA data for Chinese and Turkish native sheep. All the modern and ancient mtDNA sequences have been assigned to one of the three major mtDNA lineages (i.e., lineages A, B, and C) (Cai et al. 2007, 2011; Demirci et al. 2013; Lv et al. 2015).
To investigate paternal variation, three male-specific genetic markers, one microsatellite SRYM18 and two SNPs oY1 and oY2 on the Y chromosome (Meadows et al. 2006), were genotyped in 40 rams from four populations of Tibetan sheep (supplementary table S4 and supplementary information, Supplementary Material online). Publicly available haplotype diversities of 538 rams from 40 Chinese native sheep populations were also retrieved (supplementary table S4, Supplementary Material online; Zhang et al. 2014) and included in the analyses. Median-joining networks of inferred haplotypes were constructed using the NETWORK 4.0c program (Bandelt et al. 1999).
Genetic Relationships and Population Structure
To investigate the genetic relationships and population structure among domestic sheep populations, PCA, NJ tree construction, and genetic structure analysis were performed at the national, continental, and global scales (supplementary information, Supplementary Material online). PCA was conducted using the SmartPCA program in the EIGENSOFT v.6.0beta package (Patterson et al. 2006). Pairwise Reynolds’ distances (Reynolds et al. 1983) between populations were calculated based on the SNP allele frequencies using PEAS v.1.0 (Xu et al. 2010). The distances were then used to construct NJ trees for Chinese, Asian, and worldwide sheep populations with the PHYLIP v.3.695 software package (Felsenstein 1989). The NJ trees were rooted with argali and visualized by FigTree v.1.4.2 (available at http://tree.bio.ed.ac.uk/software/figtree/), and the robustness of tree topologies was tested by 1,000 bootstraps. Similarly, the NJ tree for all the individuals was constructed using SplitsTree v.4.0 (Huson and Bryant 2006) based on pairwise Reynolds’ distances (Reynolds et al. 1983) calculated using Arlequin v.3.5.1.2 (Excoffier and Lischer 2010). To provide more detailed information of the individual-based NJ tree, individual relationships among the 1,001 individuals, which corresponded to the tree clusters and associated individuals, were summarized in a table. The population genetic structure was assessed using a Bayesian model-based clustering approach as implemented in the STRUCTURE v.2.3 program (Pritchard et al. 2000). The number of assumed genetic clusters K ranged from 2 to 10, and ten independent runs were operated and checked for convergence. All the runs were performed using a model of admixture and correlated allele frequencies (Falush et al. 2003; Excoffier et al. 2005), with a burn-in length of 15,000 followed by 35,000 iterations. Prior information about the population origin of the animals was not used. Outputs were postprocessed with Structure Harvester v0.6.93 (Earl and vonHoldt 2012) and Clumpp v1.1.2 (Jakobsson and Rosenberg 2007) and visualized using the Distruct v1.1 program (Rosenberg 2004).
Further, pairwise FST values (Weir and Cockerham 1984) between Chinese sheep populations were calculated using Arlequin v.3.5.1.2 (Excoffier and Lischer 2010). Multidimensional scaling (MDS) analysis was then performed based on the matrix of FST values and visualized using the first two components λ1 and λ2 with R v.3.3.0 (R Development Core Team 2015). Subsequently, the λ1 and λ2 values were interpolated into geographic maps with the ArcMap tool in ArcGIS software v.10.1 (Environmental Systems Research Institute (ESRI), Redlands, CA). The inverse distance weighted (IDW) option with a power of 2 was used for the surface interpolation (Hanotte et al. 2002; Lv et al. 2015). Moreover, the λ1 and λ2 produced by MDS were plotted against the geographic distance from a putative location of origin for Chinese sheep populations with different tail types (i.e., thin tail, short fat-tail, and long fat-tail/fat-rump; supplementary table S21 and fig. S24, Supplementary Material online): the earliest archeological site of Chinese sheep remains—the Shihushan archeology site in the Mongolian Plateau (fig. 4B and supplementary table S6, Supplementary Material online; Dodson et al. 2014) for the thin-tailed populations, the sampling site of Hulun Buir sheep (HLS) in the Inner Mongolia for the short fat-tailed populations, and the sampling site of Bashbay sheep (BSB) in Xinjiang Province for the long fat-tailed/fat-rumped populations (fig. 1A and supplementary fig. S1, Supplementary Material online).
Inference of Population Divergence and Admixture
To reconstruct the dispersal and admixture patterns of Asian sheep, in particular Chinese sheep, we conducted several statistical tests that determine population splitting as well as migration and admixture events using genome-wide allele frequency data (Patterson et al. 2012; Pickrell and Pritchard 2012; Skoglund et al. 2012; Lipson et al. 2014), such as TreeMix, f3-statistics, MixMapper, and D-statistics analyses (supplementary information, Supplementary Material online).
We inferred the introgression of argali into domestic sheep as well as among domestic sheep populations using the TreeMix v.1.12 program (Pickrell and Pritchard 2012). We implemented two sets of TreeMix analyses. The first analysis was conducted using the 60 Asian native populations (data sets I, II, III, and V; supplementary table S1, Supplementary Material online), and the second analysis was performed using the 44 Chinese populations (data sets I, II, and III). In both analyses, the ML trees were constructed using blocks of 1,000 SNPs and argali as the root. The number of tested migration events (M) varied from 1 to 40 and from 1 to 20 for the first and second analyses, respectively, and the model fit was quantified by the covariance for each migration event. Bootstrap replicates were generated using blocks of 1,000 SNPs to evaluate the robustness of the tree topology (Pickrell and Pritchard 2012). The plotting function in the R software package was used to visualize the graph results.
To test the admixture of Asian sheep, we computed the three-population statistic (f3-statistic) using the qp3Pop program in the AdmixTools package with the default parameters (Patterson et al. 2012). Using the national data set (the combination of data sets I, II, and III), we first computed all possible f3-statistics f3(C; argali, B) to investigate the genetic introgressions of wild argali sheep into Chinese domestic sheep, with populations B and C representing any two of the 44 Chinese populations (supplementary table S15, Supplementary Material online). Next, all possible f3-statistics f3(C; A, B) were calculated to test the potential admixture among populations from different geographic regions, with populations A, B, and C denoting Asian populations from five different regions (i.e., SME, SSA, SNC, SQT, and SYK; supplementary table S11, Supplementary Material online). The admixture among populations was also examined with populations A, B, and C representing populations from the same region (supplementary table S12, Supplementary Material online). Observed negative value of the f3 statistic and Z-score < −2 may indicate historical events of admixture (Reich et al. 2009).
Based on the results of the f3-statistic analysis, admixture events among Asian native sheep populations were further examined using the MixMapper v.2.0 program (Lipson et al. 2013, 2014). We first built the nonadmixed scaffold tree using the NJ method based on the pairwise f2-statistics (Lipson et al. 2013) of 18 relatively nonadmixed populations (i.e., BGE, SUM, IDC, GUR, WGS, SPS, ZTS, WNS, TCS, DQS, TIB, HZS, ZLZ, MXS, QEZ, AFS, CFT, and SKZ; fig. 2C), which represent all genetic components of Asian native sheep. Admixed northern Chinese populations were then added to the scaffold tree using the MixMapper program to infer admixture under two-way and three-way mixture-fitting scenarios, compare full optimization between the two-way and three-way scenarios, and determine the best fit among alternative admixture models (Lipson et al. 2014).
Additionally, we calculated D-statistics among the tested populations (Green et al. 2010; Skoglund et al. 2012) to clarify the dispersal and phylogenetic relationships of Asian sheep populations. In particular, to test whether the sheep migration from the Middle East (i.e., the domestication center) to South Asia preceded their dispersal to China, all possible D-statistics of forms D (Outgroup, SSA; SME, SYK/SQT) and D (Outgroup, SME; SSA, SYK/SQT) were computed, with argali as the outgroup. From each geographic region, we selected genetically representative populations which can represent the genetic components of the groups based on the results of STRUCTURE analysis (K = 5; supplementary fig. S16, Supplementary Material online): TIB and HZS from SQT; DQS, TCS, WNS, WGS, ZTS, and SPS from SYK; GUR, IDC, SUM, BGE, and GAR from SSA; and QEZ and AFS from SME (supplementary table S10, Supplementary Material online). To further examine the phylogenetic relationships between Chinese and South Asian sheep populations, all possible D (SME, SSA; SYK, SQT) for populations from South Asia, the Yunnan-Kweichow Plateau and the Qinghai-Tibetan Plateau were computed (supplementary table S10, Supplementary Material online) using the Middle Eastern populations as the outgroup.
Approximate Bayesian Computation
Based on the inferences from PCA, NJ tree, and STRUCTURE (K = 5; supplementary figs. S13, S14, and S16, Supplementary Material online), Asian sheep populations could be divided into five main genetic clusters, which were generally consistent with their geographic distributions (i.e., SME, SSA, SNC, SQT, and SYK; fig. 1A and supplementary table S1, Supplementary Material online). Of the clusters, northern Chinese sheep (SNC) exhibited multiple genetic components (supplementary fig. S16, Supplementary Material online) and were detected to have undergone admixture from two or multiple sources by the f3-statistic and MixMapper analyses (fig. 2C and supplementary tables S11, S13, and S14, Supplementary Material online). This observation indicated that northern Chinese sheep have experienced a complex demographic history. To further rigorously clarify the history of Chinese sheep, we used the ABC method (Beaumont et al. 2002; Wegmann et al. 2010) and Ovine SNP50K BeadChip data to test various scenarios of admixture and differentiation (supplementary information, Supplementary Material online). In particular, we compared four alternative demographic models to investigate whether northern Chinese sheep, which comprised multiple genetic components, was formed by admixture events. In Model-1, which corresponded to the best-supported Model 3 in Yang et al. (2016b), we assumed that modern northern Chinese sheep was derived from the Middle Eastern ancestral population, which first spread to China at c. 4,800 BP (1,600 generations; generation interval = 3 years/generation; supplementary fig. S20 and supplementary information, Supplementary Material online; Kijas et al. 2012; Lv et al. 2015; Yang et al. 2016b). In Model-2, Model-3, and Model-4, which were different from each other with the early divergence patterns of three Chinese sheep groups (i.e., northern Chinese sheep, Qinghai-Tibetan sheep, and Yunnan-Kweichow sheep), we considered that modern northern Chinese sheep was formed through an admixture between the Middle Eastern sheep and early northern Chinese sheep populations (supplementary fig. S20 and supplementary information, Supplementary Material online). The relevant samples, SNPs data, the prior distributions of model parameters, the data simulation, the model choice, and the parameter estimates were detailed in supplementary information, Supplementary Material online.
Selection Tests
We performed phenotype specific tests for selection to identify the genomic regions and SNPs associated with particular phenotypes. Based on the FST estimate (Weir and Cockerham 1984), the selection tests involved grouping populations to test for specific phenotypic traits such as tail type, horn type, fertility, coat color, wool, and fur characteristics (supplementary table S26, Supplementary Material online). FST values for individual SNPs were calculated across groups of populations with different phenotypes using Genepop v.4.2 (Rousset 2008). The SNPs with significant high FST values (top 0.1% of the total SNPs) were considered to be putative signals under strong artificial selection. Functional candidate genes located within 150 kb upstream and downstream of the physical positions of these selected SNPs were identified based on the annotation of sheep reference genome assembly version 4.0 (http://www.ncbi.nlm.nih.gov/assembly/GCF_000298735.2; last accessed March 31, 2016).
For the trait of tail type, the FST-values of the SNPs on chromosomes 13 and 15 were smoothed with a local variable bandwidth kernel estimator (Flori et al. 2009). The smoothed FST-values were then calculated using the lokern package (Gasser et al. 1991) in the R software package for three pairs of comparisons: fat tail versus thin tail, thin tail versus thin tail and fat tail versus fat tail, respectively. The populations used for the fat tail versus thin tail comparison were grouped according to those described in supplementary table S26, Supplementary Material online, whereas the populations involved in the other two comparisons were randomly grouped. The top 1% of SNPs (the 392 SNPs with the top 1% highest FST values; supplementary table S22, Supplementary Material online) across the fat tail versus thin tail comparison was considered as indicative makers that could differentiate different tail types. Based on these SNPs, population genetic structure of Chinese native sheep populations with recorded tail type (supplementary table S26, Supplementary Material online) was examined using the Bayesian clustering approach as implemented in the STRUCTURE program. The assumed number of genetic clusters K was tested from 2 to 4, and other parameters were kept the same as aforementioned STRUCTURE analysis. To visualize the pattern of geographic distribution and the genetic component of Chinese native sheep with different tail types, each K value was represented as a pie chart with the tool ArcMap in the ArcGIS program v.10.1 (ESRI, Redlands, CA). When K = 4, we interpolated the proportional values of the two genetic clusters that represented short fat-tailed and long fat-tailed/fat-rumped populations into geographic maps.
Furthermore, to investigate genomic differentiation between argali and domestic sheep, 50 replicates of 15 randomly chosen individuals from Chinese native sheep were used in the comparisons. Pairwise FST values for individual SNPs were calculated between 15 argali and each replicate of 15 randomly chosen domestic sheep using Genepop v.4.2 (Rousset 2008). The extremely highly and lowly differentiated genomic regions and genes were scanned using the top and bottom 0.1% SNPs based on the average FST value over the 50 comparisons, and were annotated with the method described above. For the candidate genes identified, functional classification analyses were performed to discover enriched gene-function groups using the DAVID (the Database for Annotation, Visualization, and Integrated Discovery) v.6.7 (Huang et al. 2007; Huang et al. 2009).
Compilation of Archeological Records
To collect archeological information of domestic sheep from China and other regions of Asia, we conducted a literature review by searching online websites such as http://www.cnki.net, http://scholar.google.com and the online database Web of Science, using a number of different key words including “Chinese sheep and archeology”, “Chinese sheep and remains”, “Asian sheep and archeology”, and “Asian sheep and remains.” These queries were deemed sufficient to find the majority of sheep archeological information in Asia. We then compiled all the archeological records in terms of time and location.
Ethnohistorical Demographics
Regarding the historical demographics of the nomads in East Asia, we collected the information about the origin and migration of the main nomadic ethnic groups associated with sheep, i.e., “Tibetan”, “ethnic groups of Southwest China” (represented by “Yi” and “Lahu”), “Mongol”, “Jurchen,” and “Hui”, from historical literatures such as Shi Ji (Si 1131), Hou Han Shu (Fan 1131), Xin Tang Shu (Ou 1936), Yuan Shi (Song and Wang 1983), Xin Yuan Shi (Ke 1988), etc. Both the population history (divergence and admixture) of Chinese sheep as revealed by the genetic studies and the historical records of aforementioned nomadic ethnics were used to delineate the migration scenarios and peopling patterns of associated nomadic societies in East Asia, particularly China.
Data Access
The SNPs, mtDNA, and Y-chromosomal data reported in this paper are available upon request for research purpose.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank a number of persons who participated in the sampling. We also thank Dr. Ze-Hui Chen for her help in drawing the picture of sheep tail type in supplementary fig. S25, Supplementary Material online. This work was financially supported by the National Natural Science Foundation of China (grants Nos. 31272413 and 3161101336), the National Transgenic Breeding Project of China (2014ZX0800952B), the External Cooperation Program of Chinese Academy of Sciences (152111KYSB20150010), the Taishan Scholars Program of Shandong Province (No. ts201511085), the Academy of Finland (grants Nos. 250633 and 256077), and CGIAR Research Program on Livestock led by ILRI.
Author Contributions
M.-H.L. designed and supervised the project. Y.-X.Z., F.-H.L., and X.-J.H. analyzed the data. X.-L.X. and M.Z. participated in part of the laboratory work. W.-R.L., M.-J.L., Y.-T.W., J.-Q.L., Y.-G.L., Y.-L.R., F.W., E.H., J.K., and J.-L.H. contributed to the sample collections. J.Y., Y.-X.Z., and M.-H.L. wrote the paper with contribution from J.A.L., J.K., and J.-L.H. All the authors reviewed and approved the final manuscript.
References
- Adachi T, Ishii K, Matsumoto Y, Hayashi Y, Hamamoto H, Sekimizu K.. 2014. Niemann-Pick disease type C2 protein induces triglyceride accumulation in silkworm and mammalian cell lines. Biochem J. 459:137–147. [DOI] [PubMed] [Google Scholar]
- Aldenderfer M. 2011. Peopling the Tibetan Plateau: insights from archaeology. High Alt Med Biol. 12:141–147. [DOI] [PubMed] [Google Scholar]
- Anderson R, McEwan J, Brauning R, International Sheep Genomics Consortium, Kijas J, Dalrymple B, Worley K, Daetwyler H, Heaton M, Van Stijn T et al. 2014. Development of a high density (600K) Illumina Ovine SNP chip and its use to fine map the yellow fat locus. San Diego: Plant and Animal Genome XXII Conference. [Google Scholar]
- Arslan S, Açikkol A, Korkmaz EM.. 2011. The optimization of aDNA extraction protocol and sex determination of Bronze Age individuals from Oylum Höyük (Kilis, Turkey). Turk J Biol. 35:647–653. [Google Scholar]
- Bandelt HJ, Forster P, Röhl A.. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 16:37–48. [DOI] [PubMed] [Google Scholar]
- Barton L, Newsome SD, Chen FH, Wang H, Guilderson TP, Bettinger RL.. 2009. Agricultural origins and the isotopic identity of domestication in northern China. Proc Natl Acad Sci U S A. 106:5523–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont MA, Zhang W, Balding DJ.. 2002. Approximate Bayesian computation in population genetics. Genetics 162:2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai DW, Han L, Zhang XL, Zhou H, Zhu H.. 2007. DNA analysis of archaeological sheep remains from China. J Archaeol Sci. 34:1347–1355. [Google Scholar]
- Cai DW, Tang ZW, Yu HX, Han L, Ren XY, Zhao XB, Zhu H, Zhou H.. 2011. Early history of Chinese domestic sheep indicated by ancient DNA analysis of Bronze Age individuals. J Archaeol Sci. 38:896–902. [Google Scholar]
- Chang C, Koster HA.. 1986. Beyond bones: toward an archaeology of pastoralism. Adv Archaeol Method Theory 9:97–148. [Google Scholar]
- Chen SY, Duan ZY, Sha T, Xiangyu J, Wu SF, Zhang YP.. 2006. Origin, genetic diversity, and population structure of Chinese domestic sheep. Gene 376:216–223. [DOI] [PubMed] [Google Scholar]
- Christian D. 2000. Silk roads or steppe roads? The silk roads in world history. J World Hist. 11:1–26. [Google Scholar]
- Demirci S, Baştanlar EK, Dağtaş ND, Pişkin E, Engin A, Özer F, Yüncü E, Doğan ŞA, Togan İ.. 2013. Mitochondrial DNA diversity of modern, ancient and wild sheep (Ovis gmelinii anatolica) from Turkey: new insights on the evolutionary history of sheep. PLoS One 8:e81952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson J, Dodson E, Banati R, Li X, Atahan P, Hu S, Middleton RJ, Zhou X, Nan S.. 2014. Oldest directly dated remains of sheep in China. Sci Rep. 4:7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong GH, Jia X, Elston R, Chen F, Li SC, Wang L, Cai LH, An CB.. 2013. Spatial and temporal variety of prehistoric human settlement and its influencing factors in the upper Yellow River valley, Qinghai Province, China. J Archaeol Sci. 40:2538–2546. [Google Scholar]
- Earl DA, vonHoldt BM.. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 4:359–361. [Google Scholar]
- Excoffier L, Estoup A, Cornuet JM.. 2005. Bayesian analysis of an admixture model with mutations and arbitrarily linked markers. Genetics 169:1727–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Lischer HE.. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 10:564–567. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK.. 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Xing Y, Zhang Z, Ai HS, Ouyang Z, Ouyang J, Yang M, Li P, Chen Y, Gao J, et al. 2013. A further look at porcine chromosome 7 reveals VRTN variants associated with vertebral number in Chinese and Western pigs. PLoS One 8:e62534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y. 1131. Hou Han Shu, Section on Xi Qiang Zhuan. Nanjing: Jiangnan East Road Transport Department. [Google Scholar]
- Felsenstein J. 1989. PHYLIP - phylogeny inference package (version 3.2). Cladistics 5:164–166. [Google Scholar]
- Flori L, Fritz S, Jaffrézic F, Boussaha M, Gut I, Heath S, Foulley JL, Gautier M.. 2009. The genome response to artificial selection: a case study in dairy cattle. PLoS One 4:e6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz LA, Schraiber JG, Madsen O, Megens HJ, Cagan A, Bosse M, Paudel Y, Crooijmans RP, Larson G, Groenen MA.. 2015. Evidence of long-term gene flow and selection during domestication from analyses of Eurasian wild and domestic pig genomes. Nat Genet. 47:1141–1148. [DOI] [PubMed] [Google Scholar]
- Frantz LA, Mullin VE, Capitan MP, Lebrasseur O, Ollivier M, Perri A, Linderholm A, Mattiangeli V, Teasdale MD, Dimopoulos EA, et al. 2016. Genomic and archaeological evidence suggest a dual origin of domestic dogs. Science 352:1228–1231. [DOI] [PubMed] [Google Scholar]
- Gasser T, Kneip A, Köhler W.. 1991. A flexible and fast method for automatic smoothing. J Am Stat Assoc. 86:643–652. [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, et al. 2010. A draft sequence of the Neandertal genome. Science 328:710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grousset R. 1941. The History of the Mongol Empire. Shanghai: The Commercial Press. [Google Scholar]
- Hanotte O, Bradley DG, Ochieng JW, Verjee Y, Hill EW, Rege JEO.. 2002. African pastoralism, genetic imprints of origins and migrations. Science 296:336–339. [DOI] [PubMed] [Google Scholar]
- He GY. 2000. Di Qiang Yuan Liu Shi. Nanchang: Jiangxi Education Press. [Google Scholar]
- Hellenthal G, Busby GB, Band G, Wilson JF, Capelli C, Falush D, Myers S.. 2014. A genetic atlas of human admixture history. Science 343:747–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Robertson A.. 1968. The effects of inbreeding at loci with heterozygote advantage. Genetics 60:615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4:44–57. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, et al. 2007. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 35:W169–W175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D.. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 23:254–267. [DOI] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA.. 2007. Clumpp: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806. [DOI] [PubMed] [Google Scholar]
- Jeong C, Ozga AT, Witonsky DB, Malmström H, Edlund H, Hofman CA, Hagan RW, Jakobsson M, Lewis CM, Aldenderfer MS, et al. 2016. Long-term genetic stability and a high-altitude East Asian origin for the peoples of the high valleys of the Himalayan arc. Proc Natl Acad Sci U S A. 113:7485–7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston SE, McEwan JC, Pickering NK, Kijas JW, Beraldi D, Pilkington JG, Pemberton JM, Slate J.. 2011. Genome-wide association mapping identifies the genetic basis of discrete and quantitative variation in sexual weaponry in a wild sheep population. Mol Ecol. 20:2555–2566. [DOI] [PubMed] [Google Scholar]
- Kamp KA, Yoffee N.. 1980. Ethnicity in ancient Western Asia during the early second millennium BC: archaeological assessments and ethnoarchaeological prospectives. Bull Am Sch Orient Res. 237:85–104. [Google Scholar]
- Ke SM. 1988. Xin Yuan Shi, section on Volume Three “Tai Zu Xia”, Volume Four “Tai Zong”, Volume Nine “Shi Zu San”. Beijing: China Bookstore Press. [Google Scholar]
- Kijas JW, Lenstra JA, Hayes B, Boitard S, Neto LRP, Cristobal MS, Servin B, McCulloch R, Whan V, Gietzen K, et al. 2012. Genome-wide analysis of the world’s sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biol. 10:e1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson G, Albarella U, Dobney K, Rowley-Conwy P, Schibler J, Tresset A, Vigne JD, Edwards CJ, Schlumbaum A, Dinu A, et al. 2007. Ancient DNA, pig domestication, and the spread of the Neolithic into Europe. Proc Natl Acad Sci U S A. 104:15276–15281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson G, Burger J.. 2013. A population genetics view of animal domestication. Trends Genet. 29:197–205. [DOI] [PubMed] [Google Scholar]
- Li FB. 2008. The regional distribution and changes of Sichuan Huis in history. Chongqing: Southwest University, MA. [Google Scholar]
- Lipson M, Loh PR, Levin A, Reich D, Patterson N, Berger B.. 2013. Efficient moment-based inference of admixture parameters and sources of gene flow. Mol Biol Evol. 30:1788–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson M, Loh PR, Patterson N, Moorjani P, Ko YC, Stoneking M, Berger B, Reich D.. 2014. Reconstructing Austronesian population history in island Southeast Asia. Nat Commun. 5:4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv FH, Agha S, Kantanen J, Colli L, Stucki S, Kijas JW, Joost S, Li MH, Ajmone Marsan P.. 2014. Adaptations to climate-mediated selective pressures in sheep. Mol Biol Evol. 31:3324–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv FH, Peng WF, Yang J, Zhao YX, Li WR, Liu MJ, Ma YH, Zhao QJ, Yang GL, Wang F, et al. 2015. Mitogenomic meta-analysis identifies two phases of migration in the history of Eastern Eurasian sheep. Mol Biol Evol. 32:2515–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows JR, Hanotte O, Drögemüller C, Calvo J, Godfrey R, Coltman D, Maddox JF, Marzanov N, Kantanen J, Kijas JW.. 2006. Globally dispersed Y chromosomal haplotypes in wild and domestic sheep. Anim Genet. 37:444–453. [DOI] [PubMed] [Google Scholar]
- Meadows JR, Kijas JW.. 2009. Re-sequencing regions of the ovine Y chromosome in domestic and wild sheep reveals novel paternal haplotypes. Anim Genet. 40:119–123. [DOI] [PubMed] [Google Scholar]
- Meyer MC, Aldenderfer MS, Wang Z, Hoffmann DL, Dahl JA, Degering D, Haas WR, Schlütz F.. 2017. Permanent human occupation of the central Tibetan Plateau in the early Holocene. Science 355:64–67. [DOI] [PubMed] [Google Scholar]
- Miller JM, Poissant J, Kijas JW, Coltman DW, International Sheep Genomics Consortium. 2011. A genome-wide set of SNPs detects population substructure and long range linkage disequilibrium in wild sheep. Mol Ecol Resour. 11:314–322. [DOI] [PubMed] [Google Scholar]
- Moioli B, Pilla F, Ciani E.. 2015. Signatures of selection identify loci associated with fat tail in sheep. J Anim Sci. 93:4660–4669. [DOI] [PubMed] [Google Scholar]
- Ou YX. 1936. Xin Tang Shu, Section on Tu Bo. Taibei: The Commercial Press of Taiwan. [Google Scholar]
- Pagani L, Lawson DJ, Jagoda E, Mörseburg A, Eriksson A, Mitt M, Clemente F, Hudjashov G, DeGiorgio M, Saag L, et al. 2016. Genomic analyses inform on migration events during the peopling of Eurasia. Nature 538:238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palstra FP, Heyer E, Austerlitz F.. 2015. Statistical inference on genetic data reveals the complex demographic history of human populations in Central Asia. Mol Biol Evol. 32:1411–1424. [DOI] [PubMed] [Google Scholar]
- Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, Reich D.. 2012. Ancient admixture in human history. Genetics 192:1065–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D.. 2006. Population structure and eigenanalysis. PLoS Genet. 2:e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips DJ. 2001. Peoples on the move: introducing the nomads of the world. Carlisle: Pigquant. [Google Scholar]
- Pickrell JK, Pritchard JK.. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8:e1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P.. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Brown KT, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2015. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. [Google Scholar]
- Reich D, Thangaraj K, Patterson N, Price AL, Singh L.. 2009. Reconstructing Indian population history. Nature 461:489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds J, Weir BS, Cockerham CC.. 1983. Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics 105:767–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA. 2004. Distruct: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138. [Google Scholar]
- Rousset F. 2008. Genepop'007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour. 8:103–106. [DOI] [PubMed] [Google Scholar]
- Ryder ML. 1983. Sheep and Man. London: Duckworth. [Google Scholar]
- Sambrook J, Russell D.. 2000. Molecular cloning: a laboratory manual, 3rd edn.New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Shou WH, Qiao EF, Wei CY, Dong YL, Tan SJ, Shi H, Tang WR, Xiao CJ.. 2010. Y-chromosome distributions among populations in Northwest China identify significant contribution from Central Asian pastoralists and lesser influence of western Eurasians. J Hum Genet. 55:314–322. [DOI] [PubMed] [Google Scholar]
- Si MQ. 1131. Shi Ji, Sections on Yin Ben Ji and Qin Ben Ji. Yangzhou: Huainan Road Transport Department. [Google Scholar]
- Skoglund P, Malmström H, Raghavan M, Storå J, Hall P, Willerslev E, Gilbert MT, Götherström A, Jakobsson M.. 2012. Origins and genetic legacy of Neolithic farmers and hunter-gatherers in Europe. Science 336:466–469. [DOI] [PubMed] [Google Scholar]
- Song L, Wang H.. 1983. Yuan Shi, Volume One “Tai Zu”, Volume Three “Xian Zong”, Volume Nine “Shi Zu Liu”. Beijing: China Publishing House. [Google Scholar]
- Szpiech ZA, Jakobsson M, Rosenberg NA.. 2008. ADZE: a rarefaction approach for counting alleles private to combinations of populations. Bioinformatics 24:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang K, Thornton KR, Stoneking M.. 2007. A new approach for using genome scans to detect recent positive selection in the human genome. PLoS Biol. 5:e171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapio M, Marzanov N, Ozerov M, Cinkulov M, Gonzarenko G, Kiselyova T, Murawski M, Viinalass H, Kantanen J.. 2006. Sheep mitochondrial DNA variation in European, Caucasian, and Central Asian areas. Mol Biol Evol. 23:1776–1783. [DOI] [PubMed] [Google Scholar]
- Wagner M, Wu X, Tarasov P, Aisha A, Ramsey CB, Schultz M, Schmidt-Schultz T, Gresky J.. 2011. Radiocarbon-dated archaeological record of early first millennium B.C. mounted pastoralists in the Kunlun Mountains, China. Proc Natl Acad Sci U S A. 108:15733–15738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Węcek K, Hartmann S, Paijmans JL, Taron U, Xenikoudakis G, Cahill JA, Heintzman PD, Shapiro B, Baryshnikov G, Bunevich AN, et al. 2017. Complex admixture preceded and followed the extinction of wisent in the wild. Mol Biol Evol. 34:598–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmann D, Leuenberger C, Neuenschwander S, Excoffier L.. 2010. ABCtoolbox: a versatile toolkit for approximate Bayesian computations. BMC Bioinformatics 11:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CH, Wang HH, Liu G, Wu M, Cao J, Liu Z, Liu R, Zhao F, Zhang L, Lu J, et al. 2015. Genome-wide analysis reveals population structure and selection in Chinese indigenous sheep breeds. BMC Genomics 16:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC.. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370. [DOI] [PubMed] [Google Scholar]
- Xie ZC, Chen QY, Yang SH, Sun FY, Yu YC.. 2008. BMP2 induces adipogenic differentiation of C3H10T1/2 cells. Chin J Biochem Mol Biol. 24:142–147. [Google Scholar]
- Xu S, Gupta S, Jin L.. 2010. PEAS v1.0: a package for elementary analysis of SNP data. Mol Ecol Resour. 10:1085–1088. [DOI] [PubMed] [Google Scholar]
- Yang J, Huang L, Yang M, Fan Y, Li L, Fang S, Deng W, Cui L, Zhang Z, Ai H, et al. 2016a. Possible introgression of the VRTN mutation increasing vertebral number, carcass length and teat number from Chinese pigs into European pigs. Sci Rep. 6:19240.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Li WR, Lv FH, He SG, Tian SL, Peng WF, Sun YW, Zhao YX, Tu XL, Zhang M, et al. 2016b. Whole-genome sequencing of native sheep provides insights into rapid adaptations to extreme environments. Mol Biol Evol. 33:2576–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunusbayev B, Metspalu M, Metspalu E, Valeev A, Litvinov S, Valiev R, Akhmetova V, Balanovska E, Balanovsky O, Turdikulova S, et al. 2015. The genetic legacy of the expansion of Turkic-speaking nomads across Eurasia. PLoS Genet. 11:e1005068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Peng WF, Yang GL, Lv FH, Liu MJ, Li WR, Liu YG, Li JQ, Wang F, Shen ZQ, et al. 2014. Y chromosome haplotype diversity of domestic sheep (Ovis aries) in northern Eurasia. Anim Genet. 45:903–907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.