Abstract
In one patient over time, we found that concentration of Ebola virus RNA in semen during recovery is remarkably higher than blood at peak illness. Virus in semen is replication-competent with no change in viral genome over time. Presence of sense RNA suggests replication in cells present in semen.
Keywords: Ebola, persistence, semen, replication, sexual transmission
During the 2014–2016 West Africa Ebola virus disease (EVD) outbreak, late cases occurred due to sexual transmission from male EVD survivors [1]. While Ebola virus (EBOV) is known to persist in immune-privileged sites [1–4], replication kinetics and evolutionary dynamics in these sites are not well characterized. We utilized reverse-transcription quantitative polymerase chain reaction (RT-qPCR) and deep sequencing to determine concentration of viral RNA, replicative capacity, and viral evolution in blood and semen of a single EVD patient over 110 days of illness. We show that concentration of viral RNA in semen during recovery is 4 logs higher than in blood during acute illness. Virus in semen is replication-competent yet maintains a lower than expected substitution rate. Detection of sense viral RNA by strand-specific RT-qPCR suggests replication in cells present in semen.
CASE REPORT
In March 2015, a 34-year-old male healthcare worker was evacuated from Sierra Leone on day (D) 7 of documented EVD symptoms to the US National Institutes of Health Clinical Research Center. We have previously described the clinical course of severe EVD and immunological responses for this patient [5–7]. We collected serum and peripheral blood leukocyte (PBL) samples daily from D7 to D30 (all days are post–symptom onset). Semen collections followed clinical recovery from EVD beginning on D32 and continued through D244. Viral RNA in blood peaked at D8 (cycle threshold [Ct] = 23.21) and became undetectable at D24 using the EZ1 RT-qPCR assay [8]. Viral RNA in semen was detected by EZ1 RT-qPCR assay on D32 (Ct = 18.47), D66 (Ct = 29.71), and D110 (Ct = 30.07) and was at or below the limit of detection on D180 and D244. Serum samples from these days tested negative.
HIGH CONCENTRATION OF REPLICATION-COMPETENT VIRUS IN SEMEN
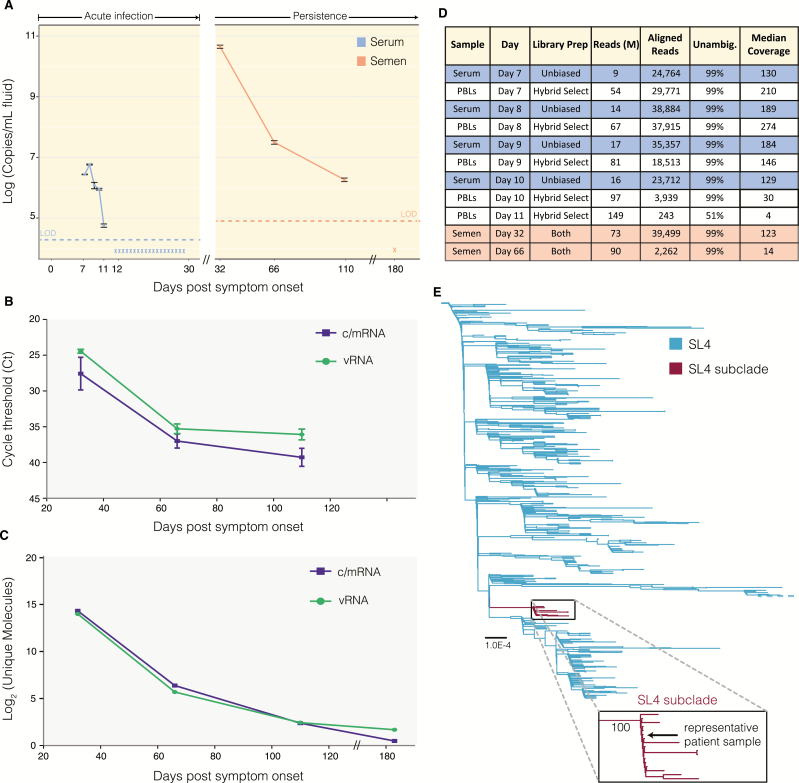
To precisely compare EBOV load, we extracted total RNA from stored serum, PBL, and semen using a modified version of the QIAgen viral RNA mini kit (see Supplementary Methods). We quantified viral copy number for each sample using the Kulesh-NP RT-qPCR assay with a standard curve, and adjusted for differences in extraction and dilution protocols between sample types (see Supplementary Methods). Notably, viral copy number in semen at D32 is 4.5 × 1010 copies/mL, while concentration in serum on D8 is 5.8 × 106 copies/mL (Figure 1A). High viral load in semen until 110 days is in line with new estimates of time to viral clearance for survivors experiencing persistence in semen (median, 158 days) [9].
Figure 1.
High viral load and identical genomes within a single patient with Ebola virus disease. A, Viral load in semen and serum. We measured viral load from extracted RNA using primers in Supplementary Table 2 (Trombley et al, 2010). Samples were quantified against a standard curve (assay limit of detection [LOD]: 1 copy/µL) to determine copies per microliter of extracted RNA. Based on the different extraction methods, we converted these values into copies per milliliter of indicated fluid (blue: serum, n = 6 replicates per sample; orange: semen, n = 3). Asterisk indicates below-assay LOD. B, Quantification of Ebola virus (EBOV) antisense (vRNA) and sense (c/mRNA) RNA in semen samples using strand-specific quantitative reverse-transcription polymerase chain reaction (RT-qPCR). We designed RT primers for a 2-step RT-qPCR to detect antisense or sense viral RNA separately. Day 180 semen sample was not tested. C, Quantification of EBOV vRNA and c/mRNA in semen samples using single-molecule tagged amplicon sequencing. We uniquely tagged and amplified RNA molecules with random barcodes in a strand-specific manner. We deduplicated reads and counted unique numbers of barcodes. Molecules at day 180 likely represent background amplification. See Supplementary Figure 1C for quantification in peripheral blood leukocyte (PBL) samples. D, Deep sequencing metrics. We performed RNA-seq either without (unbiased) or with hybrid selection using EBOV-specific baits to reduce host RNA background. Lower viral genome coverage is observed in PBL and semen samples due to abundant cellular RNA. E, Phylogeny of the SL4 clade. We combined 1489 publicly available genomes (Diehl et al 2016) with this patient’s viral genome and generated a phylogenetic tree with 1000 bootstrap replicates. The patient sample falls within a SL4 subclade (maroon; 100% bootstrap support) of samples collected near Freetown, Sierra Leone, from late January 2015 through March 2015, representing a likely transmission network. See Supplementary Figure 2 for full tree.
To determine whether infectious EBOV was present in these samples, we attempted viral isolation by tissue culture. Because some EBOV isolates do not cause discernable plaques in tissue culture [9], we verified viral amplification by RT-qPCR. We inoculated Vero E6 cells with the D8 PBL or D32 semen samples, and saved an aliquot of each inoculum as baseline. Cytopathic effects (CPEs) were observed in semen-inoculated cells 6 days postinfection (dpi), and culture supernatant was harvested 13 dpi. CPEs in blood-inoculated cells were not observed until 14 dpi, and culture supernatant was harvested 17 dpi. We quantified viral RNA in the inoculum (baseline) and harvested supernatants. Decreased Ct values in semen harvest (Ct = 17.38) vs inoculum (Ct = 25.28), and blood harvest (Ct = 25.44) vs inoculum (Ct = 29.32), are consistent with viral growth in both samples and suggest higher concentration of infectious EBOV in semen D32 than blood D8.
Given the strikingly high viral load in semen at D32 (Figure 1A), we sought to determine if replication actively occurred within cells present in semen, or whether EBOV persisted only as extracellular virions. We utilized strand-specific RNA methods to compare levels of genomic viral RNA (vRNA), vs viral antigenomes (cRNA) and messenger RNA (mRNA) that exist in the sense coding orientation and are therefore hallmarks of active replication inside cells.
To quantify the relative abundance of vRNA vs c/mRNA, we developed a strand-specific 2-step RT-qPCR assay targeting the NP gene, adapted from previous methods [10]. Similar levels of vRNA and c/mRNA were detected in the D32, D66, and D110 semen samples, indicative of active viral replication (Figure 1B), consistent with acute blood vRNA:c/mRNA [7]. Single molecule tagged amplicon sequencing analyses supported these results. We tagged each cDNA amplicon with a unique random dodecamer barcode, allowing us to remove PCR duplicates and quantify unique amplicon copy number for each strand separately. We observed high levels of both vRNA and c/mRNA in semen (Figure 1C), at a similar ratio to that in acute phase serum or PBLs (Supplementary Figure 1A).
LACK OF VIRAL DIVERSIFICATION DURING INFECTION AND PERSISTENCE
We assessed the genetic diversity of EBOV over the course of illness to investigate if variants change over time. We performed unbiased deep sequencing on serum, PBLs, and semen, and subsequently performed hybrid selection on PBLs and semen to enrich for EBOV content in these cell-rich samples [11]. We assembled near-complete EBOV genome sequences (99% unambiguous bases; range, 30–274 times median coverage) from serum and PBLs collected during D7–D10 of acute illness, as well as a partial genome (51% unambiguous bases, 4 times coverage) from a D11 PBL sample (Figure 1D). High levels of host RNA decreased the fraction of EBOV sequencing reads in semen compared to serum (Supplementary Figure 1B). We assembled near-complete EBOV genomes for samples collected on D32 (123 times coverage) and D66 (14 times coverage), but were unable to assemble a genome for D110.
Consensus viral genomes were identical for all 10 of the samples sequenced using unbiased and hybrid selection approaches, and these genomes belong to the EBOV Makona Sierra Leone clade 4 (SL4) [12] (Figure 1E). This consensus genome contains 6 single-nucleotide polymorphisms (SNPs) that separate it from the ancestral clade 4: 3 synonymous SNPs in the GP, VP24, and L genes (A250A, V169V, E1837E) and 3 SNPs in the 3ʹ untranslated region of VP40 and VP24 (A5619C, T5849C, C11407T) (Supplementary Figure 1C). Four of these SNPs co-occur in only 16 patient samples collected in Sierra Leone from 18 January to 19 March 2015, which form a distinct phylogenetic cluster within the SL4 lineage, suggesting a transmission chain between these patients (Figure 1E; Supplementary Figure 1D).
Because of our unbiased and hybrid select sequencing approaches, changes in viral quasispecies were accurately quantified over time. In all samples with sufficient coverage (>30 times), we detected an intrahost single-nucleotide variant (iSNV) in NP (F90F; C739T). In the serum, PBL, and semen samples, this iSNV maintained a frequency of 2%–10% throughout infection (Supplementary Table 1). We observed 1 transient iSNV in the D32 semen sample (T1186T, T15138C) at 26% frequency in L, not detected in acute samples or the D66 semen sample.
We used our single-molecule tagged amplicon sequencing to confirm the frequency of the C739T iSNV in our unbiased sequencing. We removed amplicon duplicates with the same dodecamer barcode (PCR duplicates) before counting the C739T frequency. This approach yielded similar frequencies of 3%–8% (Supplementary Table 1) in support of our unbiased sequencing, suggesting that viral quasispecies did not change drastically over 110 days, consistent with a reduced substitution rate seen in other EVD survivors [1].
DISCUSSION
Here, we present the first case study evaluating EBOV copy number and genetic diversity in blood vs semen during acute and persistent infection of a single patient. We observed that concentration of viral RNA in the early recovery semen sample is 4 logs higher than in blood during peak infection. In cells inoculated with semen vs blood, we observed faster CPE and detected a larger increase in viral RNA after culture, indicative of higher concentration of infectious virus. We found evidence of actively replicating EBOV consistent with high levels of sense c/mRNA. Viral genome and quasispecies were largely maintained during the course of infection or persistence.
Together, our data suggest that high concentration of virus in semen is likely attributable to active EBOV replication within cells present in semen and not solely due to accumulating extracellular virions. Originally, the finding of a reduced viral substitution rate during persistence here and by others [1] raised the possibility of high EBOV load due to minimal viral replication and reduced immune clearance. However, our detection of significant amounts of c/mRNA strongly points to replicating virus [13] in cells present in semen. Because substitution rate is a function of replication/mutation rate and selective pressures, our data suggest that the lower EBOV substitution rate is related to reduced selection (eg, immunoprivilege) rather than reduced replication.
Cell type(s) that support productive EBOV infection in the male reproductive system have not been definitively determined. While inflammatory or sloughed epithelial cells might be present in semen, orchitis was not observed clinically in our patient, and delayed viral clearance from semen suggests lack of significant immune cell infiltration. Immunohistochemical analyses of human autopsy specimens suggest that seminiferous tubule cells could be infected [14]. Our detection of EBOV c/mRNA in semen samples also suggests possible EBOV replication within sperm cells.
While limited to a single case, our investigation provides evidence for high concentration of EBOV and active viral replication in cells present in semen during recovery, given detection of sense RNA. Improved understanding of the natural course and pathogenesis of EBOV in the male reproductive system, as well as immune responses to infection in this site, will further inform recommendations for control of sexual transmission and clarify the impact of EVD on reproductive health in male survivors.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Acknowledgments. GenBank accession numbers span KY366412.1 through KY366421.1.
Disclaimer. The content of this publication does not necessarily reflect the views or policies of the US Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
Financial support. This work was supported by the Intramural Research Programs of the National Institutes of Health (NIH) Clinical Center, Critical Care Medicine Department and the National Institute of Allergy and Infectious Diseases (NIAID) (grant number U19AI110818 to the Broad Institute). This work was supported by the Howard Hughes Medical Institute (to P. C. S.). This material is based on work supported by the National Science Foundation (grant number DGE 1144152 to A. E. L.).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Diallo B, Sissoko D, Loman NJ et al. Resurgence of Ebola virus disease in guinea linked to a survivor with virus persistence in seminal fluid for more than 500 days. Clin Infect Dis 2016; 63:1353–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mate SE, Kugelman JR, Nyenswah TG et al. Molecular evidence of sexual transmission of Ebola virus. N Engl J Med 2015; 373:2448–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jacobs M, Rodger A, Bell DJ et al. Late Ebola virus relapse causing meningoencephalitis: a case report. Lancet 2016; 388:498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Deen GF, Knust B, Broutet N et al. Ebola RNA persistence in semen of Ebola virus disease survivors—preliminary report. N Engl J Med 2015. doi:10.1056/NEJMoa1511410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chertow DS, Childs RW, Arai AE, Davey RT Jr. Cardiac MRI findings suggest myocarditis in severe Ebola virus disease. JACC Cardiovasc Imaging 2016. doi:10.1016/j.jcmg.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chertow DS, Nath A, Suffredini AF et al. Severe meningoencephalitis in a case of Ebola virus disease: a case report. Ann Intern Med 2016; 165:301–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kash JC, Walters KA, Kindrachuk J et al. Longitudinal peripheral blood transcriptional analysis of a patient with severe Ebola virus disease. Sci Transl Med 2017; 9. doi:10.1126/scitranslmed.aai9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. US Food and Drug Administration. Ebola Zaire (EZ1) rRT-PCR (TaqMan) assay on ABI 7500 Fast Dx, Lightcycler, and JBAIDS: instruction booklet Available at: http://wwwfdagov/downloads/MedicalDevices/Safety/EmergencySituations/UCM408334pdf. Accessed 1 February 2017.
- 9. Sissoko D, Duraffour S, Kerber R et al. Persistence and clearance of Ebola virus RNA from seminal fluid of Ebola virus disease survivors: a longitudinal analysis and modelling study. Lancet Glob Health 2017; 5:e80–8. [DOI] [PubMed] [Google Scholar]
- 10. Kawakami E, Watanabe T, Fujii K et al. Strand-specific real-time RT-PCR for distinguishing influenza vRNA, cRNA, and mRNA. J Virol Methods 2011; 173:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Matranga CB, Andersen KG, Winnicki S et al. Enhanced methods for unbiased deep sequencing of Lassa and Ebola RNA viruses from clinical and biological samples. Genome Biol 2014; 15:519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Holmes EC, Dudas G, Rambaut A, Andersen KG. The evolution of Ebola virus: insights from the 2013-2016 epidemic. Nature 2016; 538:193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoenen T, Jung S, Herwig A, Groseth A, Becker S. Both matrix proteins of Ebola virus contribute to the regulation of viral genome replication and transcription. Virology 2010; 403:56–66. [DOI] [PubMed] [Google Scholar]
- 14. Martines RB, Ng DL, Greer PW, Rollin PE, Zaki SR. Tissue and cellular tropism, pathology and pathogenesis of Ebola and Marburg viruses. J Pathol 2015; 235:153–74. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.