Interferon-γ–inducible protein-10 is an affordable and easily quantifiable biomarker that can be used to accurately screen individuals on antiretroviral treatment (ART) for detectable viremia, optimizing the use of costly viral load determinations required to monitor ART in low-income countries.
Keywords: global health, cytokines, implementation research, scale-up viral load, sub-Saharan Africa
Abstract
Background
Achieving effective antiretroviral treatment (ART) monitoring is a key determinant to ensure viral suppression and reach the UNAIDS 90-90-90 targets. The gold standard for detecting virological failure is plasma human immunodeficiency virus (HIV) RNA (viral load [VL]) testing; however, its availability is very limited in low-income countries due to cost and operational constraints.
Methods
HIV-1–infected adults on first-line ART attending routine visits at the Manhiça District Hospital, Mozambique, were previously evaluated for virologic failure. Plasma levels of interferon-γ–inducible protein 10 (IP-10) were quantified by enzyme-linked immunosorbent assay. Logistic regression was used to build an IP-10–based model able to identify individuals with VL >150 copies/mL. From the 316 individuals analyzed, 253 (80%) were used for model training and 63 (20%) for validation. Receiver operating characteristic curves were employed to evaluate model prediction.
Results
From the individuals included in the training set, 34% had detectable VL. Mean age was 41 years, 70% were females, and median time on ART was 3.4 years. IP-10 levels were significantly higher in subjects with detectable VL (108.2 pg/mL) as compared to those with undetectable VL (38.0 pg/mL) (P < .0001, U test). IP-10 univariate model demonstrated high classification performance (area under the curve = 0.85 [95% confidence interval {CI}, .80–.90]). Using a cutoff value of IP-10 ≥44.2 pg/mL, the model identified detectable VL with 91.9% sensitivity (95% CI, 83.9%–96.7%) and 59.9% specificity (95% CI, 52.0%–67.4%), values confirmed in the validation set.
Conclusions
IP-10 is an accurate biomarker to screen individuals on ART for detectable viremia. Further studies should evaluate the benefits of IP-10 as a triage approach to monitor ART in resource-limited settings.
In 2014, the Joint United Nations Programme on HIV/AIDS (UNAIDS) announced bold new targets for the global response to human immunodeficiency virus (HIV), commonly known as the 90-90-90 strategy, consisting of 90% of people living with HIV aware of their status, 90% of people diagnosed with HIV on treatment, and 90% of people on treatment attaining virologic suppression, by 2020 [1]. Although considerable international efforts have resulted in a dramatic increase in antiretroviral therapy (ART) coverage in the last years, relatively little progress has been achieved in the development of simple, accurate, and affordable tools that allow proper surveillance of ART efficacy [2, 3]. In 2013, the World Health Organization (WHO) recommended routine HIV RNA (viral load [VL]) testing at 6 months after ART initiation and every 12 months as the preferred monitoring approach to supervise treatment adherence and minimize failure [4]. According to the last WHO guidelines, virologic failure (VF) is defined by a persistently detectable VL >1000 copies/mL after at least 6 months of starting a new ART regimen [5]. VL monitoring is important for timely diagnosis of VF to allow early adherence interventions, prevent further transmissions, and avoid delays in regimen switches that could lead to disease progression or emergence of drug resistances [6]. However, despite the international recommendations, VL testing is still not widely available in many low- and middle-income countries (LMICs) due to high cost and implementation constraints [3, 7]. The alternative to VL has often been clinical and/or immunological monitoring, which frequently results in patients remaining on failing ART as well as unnecessary regimen switches [8, 9].
Expression of several inflammatory and immune response cytokines is increased during HIV replication [10]. Previous studies have shown that plasma levels of interferon-γ–inducible protein 10 (IP-10) correlated with VL [11, 12] and VL set-point [13] during untreated primary HIV infection and decline after ART initiation both in early [14] and chronically HIV-infected individuals [15]. Similarly, we have seen that from a total of 42 inflammatory biomarkers, IP-10 shows the strongest association with VL and the best predictive power to identify acute HIV infection among febrile seronegative patients (Pastor et al, manuscript in preparation). We hypothesized that, because of its strong association with VL, plasma IP-10 level could be a surrogate marker of detectable viremia in ART-treated individuals, providing a simple and affordable screening tool to detect individuals with VF in LMICs.
METHODS
Study Population
The present analysis is a substudy of a cross-sectional cohort for detecting drug resistance in ART-treated adults enrolled between February and March 2013 at the Manhiça District Hospital (MDH), Maputo, southern Mozambique [16]. At the time of the study, current HIV national guidelines recommended ART initiation in patients with a CD4 T-cell count ≤350 cells/μL, and no routine VL monitoring was provided after ART initiation. The study protocol was approved by the institutional review boards and ethics committees of the Barcelona Clinic Hospital, Badalona Germans Trias i Pujol Hospital, Spain, and the National Committee on Health Bioethics, Mozambique. All study participants provided signed informed consent.
In brief, adults >18 years of age attending routine scheduled outpatient visits for clinical management of HIV/AIDS at the MDH were enrolled in the study. All patients had documented HIV infection, documented ART initiation ≥12 months earlier, and provided written informed consent. The sample and data collection procedures have been previously described [16] and include a single blood sample and sociodemographic and clinical data collected in a specific questionnaire.
Laboratory Procedures
HIV RNA levels were determined in plasma samples by reverse-transcription polymerase chain reaction (Abbott m2000 RealTime System with a detection limit of 150 copies/mL), and CD4+ T-cell counts were determined in whole blood by flow cytometry using FACSCalibur (BD Biosciences) as previously described [16]. IP-10 level was measured in plasma samples by enzyme-linked immunosorbent assay (Human Duo-Set ELISA, R&D Systems, Minneapolis, Minnesota) according to the manufacturer’s instructions; 0.05% Tween-20 (Sigma-Aldrich, St Louis, Missouri) 1% bovine serum albumin (Sigma-Aldrich) in phosphate-buffered saline was used as blocking solution, 3, 3′, 5, 5′-tetramethylbenzidine (Sigma-Aldrich) as substrate, and 4N sulfuric acid as stop solution. Optical density was measured at 492 and 620 nm. Values assigned to data falling outside quantification limits were the double and the half of the upper and lower quantification limits, respectively.
Statistical Analysis
Data were double-entered using Fox Pro version 2.6 (Microsoft Corporation, Redmond, Washington) and analyzed using R-3.2.2 software and Stata version 14 software (StataCorp, College Station, Texas).
Proportions and continuous variables were compared using χ2 and nonparametric Mann-Whitney U test, respectively. IP-10 values were log transformed for a better adjustment of skewed data. Spearman test was used to assess correlation coefficients for continuous variables.
To assess the capacity of IP-10 levels and clinical variables to correctly identify the cases, logistic regression with penalized likelihood was performed [17]. According to random selection of the 316 individuals included in this analysis, 80% were used for data analysis and model construction (n = 253) and 20% for model validation (n = 63). A multivariate logistic regression model was built applying a stepwise selection to the set of variables: IP-10, sex, age, CD4 T-cell count, body mass index (BMI), days on ART, and presence of symptoms. In the selection, variables with P values <.05 could enter into the model whereas a P value <.10 was required to be retained. Outcomes tested were detectable VL (defined as HIV RNA >150 copies/mL) and VF (defined as HIV RNA >1000 copies/mL). Diagnostic capacity was determined using receiver operating characteristic (ROC) analyses. ROC curves from univariate and multivariate models were compared for the best prediction.
RESULTS
Population Characteristics
Of the 332 individuals included in the cross-sectional analysis for drug resistance [16], IP-10 was determined in 316 (95.2%). We thus trained our model on 253 (80%) of the individuals with available IP-10 data. The mean age of the 253 individuals included in the training set was 41 years (standard deviation [SD], 10 years) and 70% were females. Median time on ART was 3.4 years (interquartile range [IQR], 2.1–5.3 years) and 89% were receiving zidovudine/lamivudine/nevirapine at the time of the survey. Mean BMI was 23.2 kg/m2 (SD, 3.9 kg/m2), median CD4 T-cell count was 439 cells/μL (IQR, 273–593 cells/μL), and 38% presented any type of symptoms at the time of the survey. Thirty-four percent had detectable VL (86/253) and 25% (64/253) met criteria for the standard definition of VF (VL >1000 copies/mL) [4]. In contrast, when clinical and immunological criteria were used, only 12% (29/245) of subjects were suspected to have ART failure. Population characteristics did not significantly differ from those of individuals included in the cross-sectional analysis of resistance [16] nor of those included in the validation set (P > .1).
Evaluation of an Interferon-γ–Inducible Protein 10–Based Model to Identify Detectable Viremia
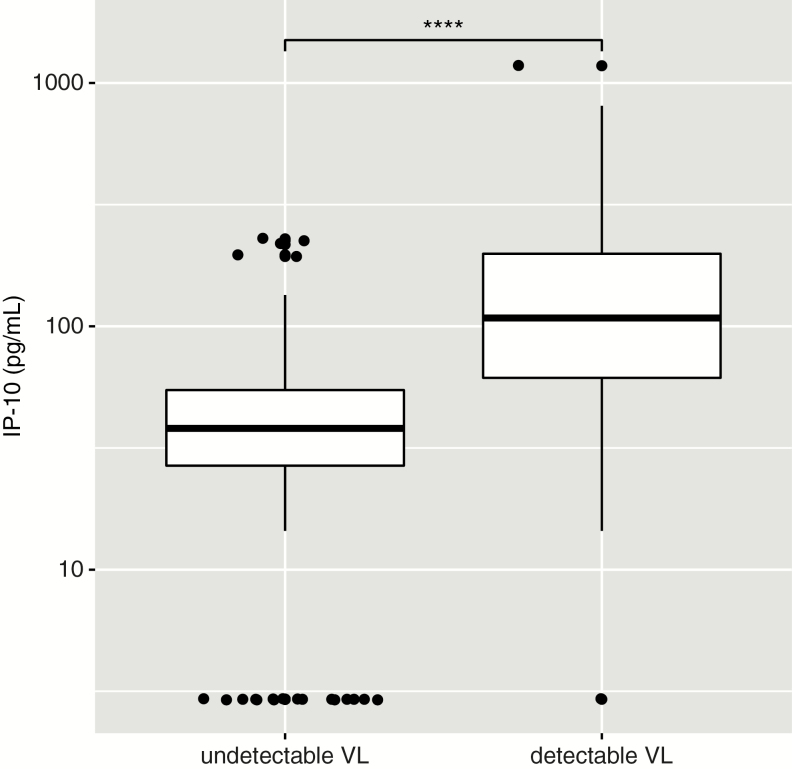
Median IP-10 levels were significantly higher among individuals with detectable VL compared to those with undetectable VL (108.2 pg/mL vs 38.0 pg/mL, respectively; U test P < .0001; Figure 1). IP-10 levels did not significantly differ from those of individuals included in the validation set (P > .1). Among those individuals with detectable VL, IP-10 levels were significantly correlated with VL (ρ = 0.33, P = .002).
Figure 1.
Comparison of interferon-γ–inducible protein 10 (IP-10) levels in antiretroviral therapy–treated individuals with and without detectable viral load (VL >150 copies/mL). Box as interquartile range (IQR), middle line as median, whiskers as Tukey values (1.5 × IQR) and dots as outliers. Nonparametric U test significance is indicated as “****” for P < .0001.
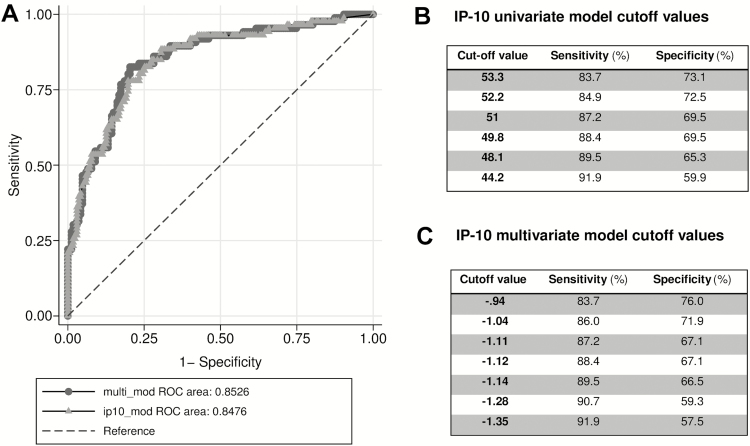
Univariate analysis showed that IP-10 was significantly and positively associated with detectable VL (odds ratio, 1.47 per 10% IP-10 pg/mL increase; P < .0001). The ROC curve demonstrated high predictive power for classification of individuals with detectable VL with an area under the curve (AUC) = 0.85 (95% confidence interval [CI], .80–.90). Sex, age, BMI, CD4 T-cell count, days on ART, and presence of any symptoms at the visit day were considered for inclusion in a multivariate analysis together with IP-10. However, only IP-10 and CD4 T-cell count were retained in the model and the resulting multivariate model did not increase the classification performance (AUC = 0.85 [95% CI, .80–.90]; Figure 2).
Figure 2.
Performance of univariate and multivariate interferon-γ–inducible protein 10 (IP-10) models in predicting detectable viral load. A, Comparison between receiver operating characteristic (ROC) curves for univariate IP-10 model (line with circles) and multivariate model (line with triangles) including IP-10 and CD4 T-cell count. IP-10 univariate model cutoff points (pg/mL) (B) and multivariate model score cutoff points (C) with their respective sensitivity and specificity values.
Then, ROC curve for the univariate IP-10 model was used to evaluate several cutoff values prioritizing the highest sensitivity. A cutoff of IP-10 ≥44.2 pg/mL was selected, providing a sensitivity of 91.9% (95% CI, 83.9%–96.7%) and a specificity of 59.9% (95% CI, 52.0%–67.4%) for predicting detectable viremia (Figure 2B).
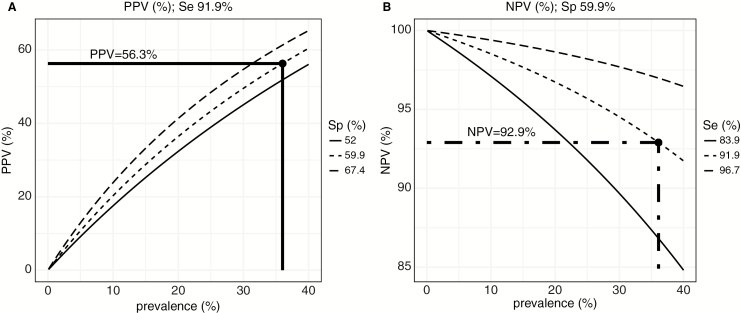
The IP-10 model with a cutoff of ≥44.2 pg/mL was assessed for predictive accuracy to identify individuals with detectable VL. We calculated positive and negative predictive values (PPV and NPV, respectively) using model sensitivity and specificity and their respective 95% CIs (Figure 3). Applying the prevalence of detectable VL of 36% observed in the cross-sectional resistance study [16] and the IP-10 model sensitivity and specificity, the PPV would be 56.3% (Figure 3A) and the NPV would be 92.9% (Figure 3B).
Figure 3.
Accuracy of the interferon-γ–inducible protein 10 model for identifying patients with detectable viral load according to the observed prevalence of individuals on antiretroviral therapy with detectable viremia. A, Positive predictive value (PPV) estimated for sensitivity (Se) = 91.9% and 3 different specificity (Sp) scenarios according to the estimated confidence interval (Sp = 59.9% [95% confidence interval, 52.0%–67.4%]). B, Negative predictive value (NPV) estimated for specificity = 59.9% and 3 different sensitivity scenarios according to the estimated confidence interval (Se = 91.9% [95% CI, 83.9%–96.7%]). Dashed/dotted line indicates PPV and NPV at the prevalence of detectable VL observed in the cross-sectional resistance study [16].
Validation of the Model Accuracy
When we applied the IP-10 model to the validation panel of samples, 80% of the 20 individuals with detectable VL and 58% of the 43 individuals with undetectable VL were correctly classified. The sensitivity and specificity derived from the validation panel were not statistically different from those obtained with the training set (equality of proportions test P = .115 and .953, respectively), thus confirming the predictive power of the univariate IP-10 model for identification of ART-treated individuals with detectable VL.
The prevalence of standard VF (VL >1000 copies/mL) observed in the study population was 25% (64/253). Both univariate and multivariate models were also tested for their ability to detect individuals with standard VF, showing lower specificity for a given sensitivity than the model designed to identify individuals with detectable VL (VL >150 copies/mL; data not shown).
Comparison of Plasma Interferon-γ–Inducible Protein 10 Versus Human Immunodeficiency Virus-RNA Quantification to Predict Detectable Viral Load
We then combined the training and validation panels to estimate the number of VL assays required for ART monitoring in this population using either the IP-10–based algorithm or standard VL testing (Table 1). Whereas using VL monitoring alone would require 316 VL determinations to detect 106 individuals harboring detectable VL, the use of an IP-10 screening test followed by VL confirmation would only require 180 VL determinations. The IP-10 screening test would thus require 43% fewer VL determinations and identify 89.6% of those with detectable VL in this sample population.
Table 1.
Interferon-γ–Inducible Protein 10 Classification Performance for Predicting Viremia Compared to Gold Standard Viral Load
| VL Result (Gold Standard) | |||
|---|---|---|---|
| IP-10 Classification | Undetectable VL | Detectable VL | Total |
| No case | 125 (59.5) | 11 (10.4) | 136 (43.0) |
| Potential VF | 85 (40.5) | 95 (89.6) | 180 (57.0) |
| Total | 210 (100) | 106 (100) | 316 (100) |
Data are presented as No. (%). IP-10 model with a cutoff of ≥44.2 pg/mL was compared to gold standard VL for classification performance of individuals with detectable VL (>150 copies/mL). Note that both training and validation sets were included to simulate a hypothetical classification in a cross-sectional cohort in Mozambique.
Abbreviations: IP-10, interferon-γ–inducible protein 10; VF, virologic failure; VL, viral load.
An analysis from the Global Fund’s Price and Quality Reporting Tool found that VL reagent costs alone varied from US $13.13 to US $43.34 between countries [3, 18]. Considering US$28 as the average reagent cost for HIV-VL testing, translated to dollar values, the VL testing of these 316 treated individuals would imply around US$8850 per year to the health system. On the other hand, considering a unit cost of US$1.50 based on costs for other similar cytokines [19], the IP-10 screening, together with the 180 VL determinations required in this cohort, would imply a cost of around US$5510 per year. This means that introducing an IP-10–based screening for ART monitoring would save US$3340 a year for this cross-sectional study cohort, resulting in savings of 38% in VL-associated cost.
DISCUSSION
We have demonstrated that a cutoff value for IP-10 of ≥44.2 pg/mL gave a sensitivity of 91.9% (95% CI, 83.9%–96.7%) and a specificity of 59.9% (95% CI, 52.0%–67.4%) for predicting detectable viremia in individuals on ART for more than a year. Thus, we have shown that IP-10 is a simple biomarker that can be used to screen individuals on ART for VF, reducing the number of costly VL determinations required to monitor ART in LMICs.
The most recent WHO guidelines recommend routine tracking of ART effectiveness using VL testing at 6 and 12 months after treatment initiation and every 12 months in stable patients to minimize treatment failure [5]. However, the high cost and technical complexity of VL testing has hampered scale-up in resource-limited settings. In absence of routine VL testing, the use of clinical and CD4 monitoring in many sub-Saharan African countries has been shown to favor the emergence of VF and drug resistances [8, 16]. In Mozambique, recent cross-sectional surveys reported that 23% [7] and 36% [16] of individuals on ART, in Maputo and Manhiça District, respectively, had detectable HIV viremia.
IP-10 is an inflammatory cytokine produced as part of the innate immune response to different pathogens [20]. IP-10 has been explored for its use as both a diagnostic and prognostic marker for several infectious diseases, such as malaria, hepatitis C, or tuberculosis [20–22]. In the case of HIV infection, previous data suggested that IP-10 levels were predictive of disease progression [11, 23] and significantly decreased between 6 months [14] and 2 years [15] after ART initiation; however, to our knowledge, the use of IP-10 as a biomarker for VL levels has not been assessed for its accuracy to detect VF in individuals on ART.
Our results show that IP-10 can indeed be used as a surrogate marker of VL with high accuracy to screen ART-treated individuals and identify patients most likely to have VL levels >150 copies/mL. This is particularly relevant in countries with scarce resources where the scale-up of ART often leads to delays and even failure to return VL results due to congested and/or centralized health facilities, which compromises both quality health services and patient retention in care [3]. Health and laboratory system strengthening together with reductions in costs and decentralization of services are required to implement the effective VL monitoring necessary to reach UNAIDS 90-90-90 targets in low-income settings [3]. Here we suggest a new algorithm to reduce the cost of ART monitoring by using IP-10 as a screening tool to target the individuals on ART most likely to require VL testing. Thus, allocation of resources to VL would be prioritized without jeopardizing quality of care. In a limited-resource setting, an IP-10 assay would halve the number of VL tests required to monitor the same number of patients and could be combined with pooled VL testing to further reduce the number of VL assays [7].. Despite the difficulty of adding an extra test to a clinical algorithm, IP-10 can be quantified by an inexpensive commercially available enzyme immunoassay with a 4-hour turnaround time and could be developed into a point-of-care test, as have other similar cytokines [19]. Therefore, WHO recommendations would be followed but VL testing would be requested only if IP-10 screening indicates potential VF. Standard recommendations for counseling, adherence support, and repeating VL within a 3-month interval would continue to be fulfilled to determine VF and subsequent antiretroviral regimen switching following WHO guidelines [5]. Pilot studies could be designed to evaluate the feasibility of integrating this test into existing laboratory facilities.
IP-10 is an affordable and easily quantifiable biomarker; however, its expression level is affected by a number of inflammatory responses [20]. Hence, its potential use to identify VF would require a second step of confirmation through VL. In this subanalysis of the cross-sectional study carried out in adults on ART at the MDH in southern Mozambique [16], to detect the 34% of individuals with detectable VL, 316 VL tests were necessary. However, using IP-10 to screen out those individuals not likely to have VF, only 180 VL tests would have had to be conducted, thus reducing by 43% the VL determinations required for ART monitoring in these settings. Although there are major pricing discrepancies across countries and policies globally [3], and disregarding costs of instrument maintenance, human resources, sample transportation, and indirect cost associated to late VF detection, according to data from our cohort, this approach could potentially save 38% of VL-derived costs to the health system. Nevertheless, this IP-10–based screening is not only presented as a cost-saving strategy, but also, and maybe more importantly, as a way to decongest the overloaded core health and laboratory facilities. Even though most of the cases with detectable viremia were identified by employing IP-10 testing, up to 10.4% were misclassified as not requiring VL confirmation. This represents an important population whose VF would remain undiagnosed. However, depending on the setting in LMICs, the consequences of this misclassification need to be weighed against the risks of a higher proportion of individuals not receiving VL monitoring due to overburdened or isolated health services.
As IP-10 plasma level is influenced by immune recovery [24], we assessed a multivariate model including clinical variables; however, predictive power was not significantly increased. Further studies should evaluate whether a combination of IP-10 with another inflammatory cytokine involved in virological responses or any other surrogate of immune activation could improve the accuracy for predicting detectable viremia and reduce misclassification. Moreover, optimization of model parameters across HIV subtypes and comorbidities present in different populations as well as sample types including dried blood spots (DBSs) could render this approach generalizable. Indeed, recent studies have shown accurate VL measurements by using DBS to monitor patients on ART in low-income settings [25]. IP-10 levels have also been demonstrated to be easily and precisely quantifiable in DBSs [26], and this approach has been recently validated for the diagnosis of tuberculosis [27] and monitoring antituberculosis treatment response [28]. Further studies should evaluate the predictive power of IP-10 levels quantified in DBSs to identified cases with detectable viremia among ART-treated individuals. This strategy would offer decentralized VF diagnosis, facilitate sample storage and transportation, and reduce test unit cost [29].
We thus propose a novel IP-10 screening algorithm for detection of VF. At a time when UNAIDS estimated that 18.2 million people were receiving ART in 2016 and nearly 12.7 million people will initiate ART in the next few years [30], further studies are warranted to assess the impact of implementing this simple triage approach to reduce the volume of VL determinations required for monitoring viral suppression.
Notes
Author contributions. L. P., J. C., R. P., J. B., and D. N. were responsible for conceptualization and study design. M. R. and S. M. recruited subjects and collected and validated clinical data. M. R. and C. J. coordinated sample collection and processing at the field. L. P. performed biomarker quantification at the laboratory and validation of the data. L. P. and A. C. performed statistical analyses. L. P., A. C., J. B., and D. N. interpreted the data. L. P. drafted the paper. J. B. and D. N. performed critical data review and revision of manuscript writing. All authors read and approved the final version of the manuscript.
Acknowledgments. The authors are grateful for the continued support of the clinical staff at the Manhiça District Hospital, as well as the study staff working exhaustively at the field and laboratory at the Centro de Investigaçao de Saúde de Manhiça (CISM). The authors thank Victor Urrea and Llorenç Quintó for their statistical advice and support, and Laura Puyol and Helder Bulo for their contribution to study and laboratory coordination between partner institutions. The authors are particularly grateful to all study participants. ISGlobal, IrsiCaixa, and IGTP are members of the CERCA Programme, Generalitat de Catalunya.
Financial support. This work was supported by the Spanish Ministry of Health through the Institute of Health Carlos III (grant number FI12/00096 to L. P.); the Spanish Ministry of Health through Rio Hortega (grant number CM11/00278 to M. R.); the Spanish Agency for International Cooperation to CISM; and the Agencia Catalana de Cooperació al Desenvolupament to Manhiça District Hospital. The study which provided samples was supported by Gilead Sciences.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Joint United Nations Programme on HIV/AIDS (UNAIDS). 90-90-90: an ambitious treatment target to help end the AIDS epidemic 2014. Available at: http://www.unaids.org/Sites/Default/Files/Media_Asset/90-90-90_En_0.Pdf. Accessed 24 April 2017. [PubMed]
- 2. Lynen L, Van Griensven J, Elliott J. Monitoring for treatment failure in patients on first-line antiretroviral treatment in resource-constrained settings. Curr Opin HIV AIDS 2010; 5:1–5. [DOI] [PubMed] [Google Scholar]
- 3. Roberts T, Cohn J, Bonner K, Hargreaves S. Scale-up of routine viral load testing in resource-poor settings: current and future implementation challenges. Clin Infect Dis 2016; 62:1043–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. World Health Organization. Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection: recommendations for a public health approach. Geneva, Switzerland, WHO: 2013. [PubMed] [Google Scholar]
- 5. World Health Organization. Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection: recommendations for a public health approach. Geneva, Switzerland, WHO: 2016. [PubMed] [Google Scholar]
- 6. Keiser O, Chi BH, Gsponer T et al. ; IeDEA Southern Africa Collaboration Outcomes of antiretroviral treatment in programmes with and without routine viral load monitoring in Southern Africa. AIDS 2011; 25:1761–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tilghman M, Tsai D, Buene TP et al. . Pooled nucleic acid testing to detect antiretroviral treatment failure in HIV-infected patients in Mozambique. J Acquir Immune Defic Syndr 2015; 70:256–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sigaloff KC, Hamers RL, Wallis CL et al. ; PharmAccess African Studies to Evaluate Resistance (PASER) Unnecessary antiretroviral treatment switches and accumulation of HIV resistance mutations; two arguments for viral load monitoring in Africa. J Acquir Immune Defic Syndr 2011; 58:23–31. [DOI] [PubMed] [Google Scholar]
- 9. Rawizza HE, Chaplin B, Meloni ST et al. ; APIN PEPFAR Team Immunologic criteria are poor predictors of virologic outcome: implications for HIV treatment monitoring in resource-limited settings. Clin Infect Dis 2011; 53:1283–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stacey AR, Norris PJ, Qin L et al. . Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J Virol 2009; 83:3719–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ploquin MJ, Madec Y, Casrouge A et al. . Elevated basal pre-infection CXCL10 in plasma and in the small intestine after infection are associated with more rapid HIV/SIV disease onset. PLoS Pathog 2016; 12:e1005774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Roberts L, Passmore JA, Williamson C et al. . Plasma cytokine levels during acute HIV-1 infection predict HIV disease progression. AIDS 2010; 24:819–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiao Y, Zhang T, Wang R et al. . Plasma IP-10 is associated with rapid disease progression in early HIV-1 infection. Viral Immunol 2012; 25:333–7. [DOI] [PubMed] [Google Scholar]
- 14. Gay C, Dibben O, Anderson JA et al. . Cross-sectional detection of acute HIV infection: timing of transmission, inflammation and antiretroviral therapy. PLoS One 2011; 6:e19617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hattab S, Guihot A, Guiguet M et al. . Comparative impact of antiretroviral drugs on markers of inflammation and immune activation during the first two years of effective therapy for HIV-1 infection: an observational study. BMC Infect Dis 2014; 14:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rupérez M, Pou C, Maculuve S et al. . Determinants of virological failure and antiretroviral drug resistance in Mozambique. J Antimicrob Chemother 2015; 70:2639–47. [DOI] [PubMed] [Google Scholar]
- 17. Firth D. Bias reduction of maximum likelihood estimates. Biometrika 1993; 80:27–38. [Google Scholar]
- 18. Médecins Sans Frontières (MSF). Achieving undetectable: what questions remain in scaling-up HIV. MSF Issue Br 2015; 6:1–20. [Google Scholar]
- 19. Lubell Y, Althaus T, Blacksell SD et al. . Modelling the impact and cost-effectiveness of biomarker tests as compared with pathogen-specific diagnostics in the management of undifferentiated fever in remote tropical settings. PLoS One 2016; 11:e0152420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu M, Guo S, Hibbert JM et al. . CXCL10/IP-10 in infectious diseases pathogenesis and potential therapeutic implications. Cytokine Growth Factor Rev 2011; 22:121–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lagging M, Romero AI, Westin J et al. ; DITTO-HCV Study Group IP-10 predicts viral response and therapeutic outcome in difficult-to-treat patients with HCV genotype 1 infection. Hepatology 2006; 44:1617–25. [DOI] [PubMed] [Google Scholar]
- 22. Wergeland I, Pullar N, Assmus J et al. . IP-10 differentiates between active and latent tuberculosis irrespective of HIV status and declines during therapy. J Infect 2015; 70:381–91. [DOI] [PubMed] [Google Scholar]
- 23. Liovat AS, Rey-Cuillé MA, Lécuroux C et al. . Acute plasma biomarkers of T cell activation set-point levels and of disease progression in HIV-1 infection. PLoS One 2012; 7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Massanella M, Ouchi D, Marfil S et al. . Different plasma markers of inflammation are influenced by immune recovery and cART composition or intensification in treated HIV infected individuals. PLoS One 2014; 9:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mavedzenge SN, Davey C, Chirenje T et al. . Finger prick dried blood spots for HIV viral load measurement in field conditions in Zimbabwe. PLoS One 2015; 10:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Aabye MG, Eugen-Olse J, Werlinrud AM et al. . A simple method to quantitate IP-10 in dried blood and plasma spots. PLoS One 2012; 7:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Blauenfeldt T, Heyckendorf J, Jensen SG et al. . Development of a one-step probe based molecular assay for rapid immunodiagnosis of infection with M. tuberculosis using dried blood spots. PLoS One 2014; 9e105628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vu DH, Bolhuis MS, Koster RA et al. . Dried blood spot analysis for therapeutic drug monitoring of linezolid in patients with multidrug-resistant tuberculosis. Antimicrob Agents Chemother 2012; 56:5758–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pannus P, Fajardo E, Metcalf C et al. . Pooled HIV-1 viral load testing using dried blood spots to reduce the cost of monitoring antiretroviral treatment in a resource-limited setting. J Acquir Immune Defic Syndr 2013; 64:134–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Joint United Nations Programme on HIV/AIDS (UNAIDS). Prevention gap report 2016. Geneva, Switzerland: WHO, 2016. [Google Scholar]