For 94.3% of migrant injectors, the origin of their HIV-1 infection was assumed to be in Greece (postmigration). For injectors infected in Greece, transmissions for subtype A and CRF14_BG occurred more frequently among migrants than would be expected by chance.
Keywords: HIV, migrants, injectors, origin of infection, molecular epidemiology
Abstract
Background
High numbers of human immunodeficiency virus type 1 (HIV-1) infections among people who inject drugs (PWID) have been diagnosed in Athens, Greece, since 2011. We aimed to trace the geographic origin of HIV-1 infection for migrants who inject drugs and to investigate whether transmissions occur more frequently among migrants than among Greek nationals.
Methods
Multiple cross-sectional studies were pooled to assemble all persons diagnosed with HIV-1 in Greece between 1 January 2011 and 31 October 2014. Phylogenetic analyses used maximum likelihood estimation. The hypothesis of ethnic compartmentalization was tested by reconstructing ancestral states of characters at the tips using the criterion of parsimony over a set of bootstrap trees.
Results
Of 2274 persons, 38.4% were PWID. Phylogenetic analyses showed the existence of 4 major PWID-specific local transmission networks (LTNs): CRF14_BG (437 [58.6%]), CRF35_AD (139 [18.6%]), subtype B (116 [15.6%]), and subtype A (54 [7.2%]). Of 184 non-Greek PWID, 78.3% had been infected within the PWID-LTNs. For 173 (94.3%), the origin of their infection was assumed to be in Greece (postmigration). For PWID infected within LTNs, transmissions for subtype A and CRF14_BG occurred more frequently among migrants than would be expected by chance (phyloethnic study).
Conclusions
Our analysis showed that the majority of infections among migrants occurred postmigration. The existence of significant transmission networking among migrants highlights that this population is a priority for HIV prevention. As molecular analysis can estimate the probable country of HIV infection, it can help to inform the design of public health strategies.
Over the last several years, a considerable proportion of the newly diagnosed human immunodeficiency virus type 1 (HIV-1) infections in the European Union and the European Economic area (EU/EEA) has occurred in migrants [1]. In 2013, migrants accounted for 35% of all new HIV-1 diagnoses in the EU/EEA [1]. Heterosexual transmission accounts for the majority of these new infections among migrants, while injecting drug use represents a relatively low proportion, with the exception of migrants from Eastern Europe and Western Asia [2, 3]. Migrants are also more likely to be diagnosed late [1]. Furthermore, HIV prevention programs and HIV testing and treatment services have been described as inadequate for these populations [4]. Multiple barriers prevent migrants from accessing HIV services despite what the policy on paper suggests. In 2014, for instance, 20 of 30 countries in the EU/EEA and half of non-EU/EEA countries in Europe and Central Asia reported that migrants were a priority population [1]. Álvarez-del Arco et al for the Advancing Migrant Access to Health Services in Europe (aMASE) study reported that between half and two-thirds of HIV-infected migrants living in 9 European countries became infected after migration, irrespective of their origin and transmission category [5]. This sample included persons who inject drugs (PWID), but the small sample size in this study did not enable authors to draw subgroup-specific conclusions.
In 2011–2012, a large increase in the number of HIV-1 diagnoses among PWID was reported in the Athens metropolitan area [6, 7]. The response to this outbreak included an increase of needle/syringe programs; expanded access to opioid substitution therapy; the implementation of ARISTOTLE, a “seek, test, treat, and retain” (STTR) program [8, 9]; and the Transmission Reduction Intervention Project (TRIP), a program that intervened in the networks of recently infected PWID in Athens [10]. TRIP took place in 3 urban centers that have large HIV epidemics: Odessa, Ukraine; Athens, Greece; and Chicago, Illinois. Molecular investigation of the outbreak revealed that the majority of PWID in Athens had been infected by clades belonging to CRF14_BG, CRF35_AD, and subtypes A and B [3, 6]. Early findings showed that, for 2 of the most common local transmission networks (LTNs), the respective origins of the virus were Romania (CRF14_BG) and Afghanistan/Iran (CRF35_AD) [3, 6]. These findings, along with the higher prevalence of HIV-1 infection among male PWID from Afghanistan/Iran and Balkan Peninsula/Eastern Europe [8, 9], prompted us to investigate in the present article how the HIV-1 epidemic spread among migrant PWID. Our aims were to estimate the proportion of postmigration HIV-1 transmissions for migrant PWID, and to infer whether postmigration transmissions occurred more frequently through contacts with other migrant PWID or through contacts with Greek PWID.
MATERIALS AND METHODS
Study Population, Setting, and Time Period
We derived our epidemiological and HIV-1 pol sequence data (n = 2274) from different sources and study designs: (1) a registry of all HIV-1–infected individuals tested for genotypic drug resistance at the National Retrovirus Reference Center in Athens from 1 January 2011 till 31 October 2014; (2) STTR program using respondent-driven sampling (ARISTOTLE) conducted from August 2012 to December 2013 in the Athens Metropolitan Area; and (3) TRIP in Athens [11], a multisite network-based intervention from June 2013 to July 2015. All patients’ samples were registered with a unique patient ID used to exclude duplicates. Our operative definition of migrants will be individuals who have non-Greek nationalities based on surveillance report.
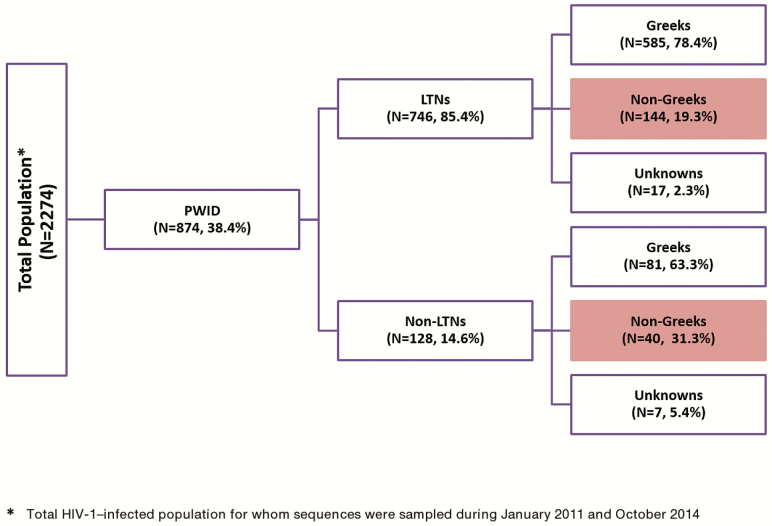
Sequences were available for 874 PWID. Of these, 184 had non-Greek nationality (Figure 1). These 874 sequences account for approximately 74.1% of all HIV-1–infected PWID diagnosed in the same time period in Greece (Hellenic Center for Disease Control & Prevention [HCDCP]; http://www.keelpno.gr).
Figure 1.
Study population. Highlighted boxes correspond to the target group of non-Greek PWID with HIV-1 infection. Abbreviations: HIV-1, human immunodeficiency virus type 1; LTN, local transmission networks; PWID, people who inject drugs.
The study was approved by the ethical committees of the National and Kapodistrian University of Athens and of the National Development and Research Institutes in New York.
Strategy to Assign Pre- or Postmigration Acquisition of Human Immunodeficiency Virus Type 1
To identify the probable country where infections were acquired, we performed the following analyses: first, we identified HIV-1 subtypes and found which sequences fell within the 4 major PWID-LTNs, as described previously [6, 12]. For sequences classified within HIV-1 clades and for recombinants that did not originate from the major LTNs, we performed phylogenetic analysis for each clade using all HIV-1 sequences available from the Athens epidemic, as well as globally sampled sequences for reference.
Below we describe the process and criteria for identifying the probable region of infection. The 4 LTNs represent transmissions which occurred during the outbreak in the Athens metropolitan area. Therefore, sequences from PWID with a non-Greek nationality within the LTNs were assumed to have been acquired locally, and thus, postmigration. This is due to the following reasons: (1) the LTNs were monophyletic clades which included sequences from PWID sampled in Athens during the outbreak, therefore suggesting that for all individuals within LTNs, HIV-1 infections were due to onward transmissions; and (2) sequences from migrants clustered as nested within the LTNs suggested that they were infected with viral strains circulated in Athens during the outbreak. Only the sequences from 2 cases in CRF14_BG and CRF35_AD LTNs clustered close to the root of the tree, suggesting that their origin might have been from a different area. Although we cannot be confident that this is true, we decided not to assign a “Greek” origin for their HIV-1 infection (ie, we assumed a more conservative scenario).
Similarly, we assumed that unique recombinant forms (URFs), which included partial genomic sequences from any of the 4 major LTNs, also corresponded to local infections. For the rest of the sequences, the geographic origin was estimated as follows: if sequences fell within monophyletic clades (including sequences from a diverse geographic area at proportions >75%), we assumed that the origin was from that area and thus, premigration. If sequences under study did not cluster with any other group(s) from a specific geographic area, we assigned no origin to this case.
Human Immunodeficiency Virus Type 1 Subtyping and Characterization of People Who Inject Drugs Local Transmission Networks
Details about HIV-1 subtyping and identification of the HIV-1 PWID LTNs are described in the Supplementary Materials. In brief, HIV-1 subtypes were determined by the automated HIV-1 subtyping tool, COMET version 0.2 [13], and confirmed by phylogenetic analysis. The level of agreement between phylogenetic analysis and COMET was 91.4% (2079/2274).
Investigation of Transmission Patterns Within the People Who Inject Drugs Local Transmission Networks
To investigate the postmigration transmission patterns across individuals with different nationalities, we selected those infected locally and explored whether transmissions between migrants occurred at significantly higher proportions than between migrants and Greeks as follows:
Phylogenetic analysis was carried out by maximum likelihood as implemented in FastTree version 2.1 [14] and RAxML version 8.1.15 [15] programs using the GTR+G as nucleotide substitution model.
The hypothesis that transmissions occurred more frequently between migrants in a significant manner was tested by reconstruction of ancestral states of characters at the tips (nationality) using the criterion of parsimony [16]. In this case, character changes correspond to infections between different nationalities. Past character changes were estimated over a high number of bootstrap-reconstructed phylogenetic trees (subtype A: 408; subtype B: 708; CRF14_BG: 300; CRF35_AD: 408) using PAUP*4.0 program [17].
We tested whether the estimated number of character change events differed significantly from the expected number of these events, under the null hypothesis of complete mixing. In a random mixing population, an infected individual would have the same probability to transmit the virus to any other individual, regardless of nationality. Thus, a random shuffling of taxa at the tips of any phylogenetic tree would simulate a tree inferred from such a population. We call these events “expected cross-nationality group transmissions,” as they are expected to occur under the assumption of random mixing, in contrast to the “observed cross-ethnic group transmissions” inferred from the bootstrap trees before shuffling.
We then compared the distribution of the events inferred from the 2 sets of trees.
We assumed that transmissions occurred more frequently between migrants in a significant manner (compartmentalization according to nationality) within our target group when we observed a significantly lower number of cross-nationality group transmissions than expected by chance (Supplementary Materials).
Statistical Analysis
Demographic data were summarized using frequencies and percentages. The nonparametric, 1-sided Mann-Whitney test was used to compare the distribution of the cross-ethnic group transmissions between the original bootstrap trees and the trees that were randomly reshuffled at tips (Stata 12 software, StataCorp LP).
RESULTS
Human Immunodeficiency Virus Subtyping Results
We analyzed 874 PWID. Of these, 184 (21.1%) were of non-Greek origin (Figure 1). Phylogenetic analysis revealed the existence of 4 major PWID-LTNs. PWID infected with strains of the 4 major LTNs were distributed as follows: CRF14_BG (437 [58.6%]), CRF35_AD (139 [18.6%]), subtype B (116 [15.6%]), and subtype A (54 [7.2%]).
Most of the PWID infected within the LTNs (78.4%) were of Greek origin (Figure 1 and Table 1). The HIV-1 subtype distribution in this population is shown in Table 2. CRF14_BG was the most prevalent clade in both Greeks (n = 328 [49.3%]) and non-Greeks (n = 101 [54.9%]) (Table 2).
Table 1.
Distribution of Human Immunodeficiency Virus Type 1–Infected People Who Inject Drugs Sampled Between 2011 and 2014 by Nationality
| Country/Region of Origin | PWID, No. (%) |
|---|---|
| America | 2 (0.2) |
| Central Asia | 11 (1.3) |
| Central Europe | 29 (3.3) |
| Eastern Europe | 41 (4.7) |
| Greece | 666 (76.2) |
| North Africa/Middle East | 46 (5.3) |
| South Asia | 16 (1.8) |
| Southeast Asia | 15 (1.7) |
| South and Southeast Asia | 14 (1.6) |
| Sub-Saharan Africa | 6 (0.7) |
| Western Europe | 4 (0.5) |
| Unknown | 24 (2.7) |
| Total | 874 (100) |
Abbreviation: PWID, people who inject drugs.
Table 2.
Human Immunodeficiency Virus Type 1 Subtype Distribution for Sequences of People Who Inject Drugs (Greeks/Non-Greeks) Sampled Between 2011 and 2014
| HIV-1 Subtype/CRF | PWID, Ethnicity, No. (%) | Total | |
|---|---|---|---|
| Greek | Non-Greek | ||
| A (LTN) | 47 (7.1) | 6 (3.3) | 53 (6.2) |
| B (LTN) | 96 (14.3) | 18 (9.8) | 114 (13.4) |
| CRF14_BG (LTN) | 328 (49.3) | 101 (54.9) | 429 (50.5) |
| CRF35_AD (LTN) | 114 (17.1) | 19 (10.3) | 133 (15.7) |
| URFs including LTN | 38 (5.7) | 24 (13.0) | 62 (7.3) |
| Subtype A | 11 (1.6) | 5 (2.8) | 16 (1.9) |
| Subtype B | 13 (2.0) | 1 (0.5) | 14 (1.7) |
| Subtype F | 0 (0.0) | 1 (0.5) | 1 (0.1) |
| CRF14_BG | 1 (0.2) | 1 (0.5) | 2 (0.2) |
| CRF35_AD | 1 (0.2) | 0 (0.0) | 1 (0.1) |
| Other CRFs and recombinant formsa | 4 (0.5) | 2 (1.1) | 6 (0.7) |
| URFs | 0 (0.0) | 1 (0.5) | 1 (0.1) |
| Unclassified | 13 (2.0) | 5 (2.8) | 18 (2.1) |
| Total | 666 (100) | 184 (100) | 850 (100) |
Abbreviations: CRF, circulating recombinant form; HIV-1, human immunodeficiency virus type 1; LTN, local transmission networks; PWID, people who inject drugs; URF, unique recombinant form.
aIncluding CRF01_AE.
Probable Countries of Infection for Migrants
Phylogenetic analysis suggested that of the PWID infected within the LTNs (n = 746), 144 (19.6%) had a non-Greek nationality and were assumed to have become infected postmigration, in the Athens Metropolitan area. However, for CRF14_BG (Figure 2B, Supplementary Figure 1B) and CRF35_AD, sequences from 2 migrants clustered very close to the root of the tree, suggesting that they might have been infected from a different area, outside of Greece. For this reason, the final conservative estimate of the number of PWID to have been infected postmigration is 142 (Table 3).
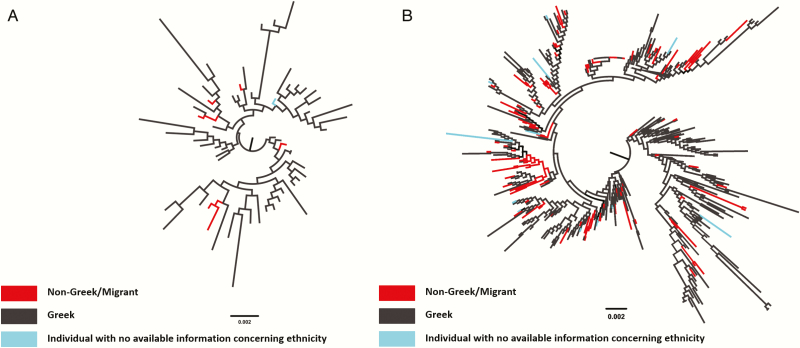
Figure 2.
Midpoint rooted maximum likelihood phylogenetic trees of HIV-1, human immunodeficiency virus type 1 outbreak sequences for the local transmission networks (phylogenetic clusters) for: (A) subtype A and (B), CRF14_BG. Viral sequences are marked in different colors according to patient’s nationality: black (Greek), red (non-Greek/migrant), and light blue (patient with no available information concerning nationality).
Table 3.
Distribution of Human Immunodeficiency Virus Type 1–Infected, Non-Greek People Who Inject Drugs Sampled Between 2011 and 2014 by Probable Region of Infection
| Geographic Origin of Infectiona | HIV-1 Subtype/CRF | No. (%) |
|---|---|---|
| Greece | A (LTN) | 6 (3.3) |
| B (LTN) | 18 (9.8) | |
| CRF14_BG (LTN) | 100 (54.4) | |
| CRF35_AD (LTN) | 18 (9.9) | |
| URFs including LTN | 24 (13.1) | |
| Subtype A (non-LTN) | 5 (2.7) | |
| Subtype B (non-LTN) | 1 (0.5) | |
| CRF01_AE (non-LTN) | 1 (0.5) | |
| Subtotal | 173 (94.3) | |
| Not in Greece | ||
| Putative origin | ||
| Greece/Israel | CRF03_AB | 1 (0.5) |
| Romania | CRF14_BG (non-LTN) | 1 (0.5) |
| Romania | Subtype F | 1 (0.5) |
| Turkey/Bulgaria | URF | 1 (0.5) |
| Turkey | Unclassified | 4 (2.4) |
| Unspecified | CRF14_BG (LTN) | 1 (0.5) |
| Unspecified | CRF35_AD (LTN) | 1 (0.5) |
| Unspecified | Unclassified | 1 (0.5) |
| Subtotal | 11 (5.7) | |
| Total | 184 (100) |
Abbreviations: CRF, circulating recombinant form; HIV-1, human immunodeficiency virus type 1; LTN, local transmission networks; URF, unique recombinant form.
aThe number of viruses with a probable region of infection outside Greece is different from the highlighted boxes shown in Figure 1 that correspond to the total number of non-Greek people who inject drugs with HIV-1 infection.
Phylogenetic analysis showed that 40 of 184 sequences (21.7%) from migrants did not fall within the LTNs. Twenty-four (60%) were classified as recombinant forms and at least 1 subtype identified in the LTNs (subtype A, subtype B, CRF14_BG, and CRF35_AD) (Table 2). For these recombinants, the putative origin of infection was the Athens metropolitan area, because they contained partial genomic sequences from HIV-1 clades of the LTNs and were thus assumed to have been transmitted postmigration (Table 3). The subtype distribution of the remaining 16 (40%) nonclustered sequences from migrants is shown in Table 2. The exact recombination pattern for URFs including LTN is described in Supplementary Table 1.
For all sequences classified as subtypes A, B, and CRF01_AE, (Table 3; n = 7 [43.8%]), we assumed that their putative origin of infection was in Greece (Table 2). This conclusion was based on their clustering within monophyletic clades where >75% of sequences were sampled from individuals in Greece. For sequences classified as subtype F and CRF14_BG (n = 2 [12.5%]), we found that they clustered with others from Romania. For a URF (A/U), phylogenetic analysis revealed that it fell within a group of sequences from Turkey and Bulgaria, suggesting that infection may have originated from either country and thus, premigration. Greece or Israel was the most probable origin for a single CRF03_AB sequence (n = 1 [6.3%]) (Table 3). Finally, 4 unclassified sequences clustered within a monophyletic cluster from Turkey, whereas a probable origin could not be identified for a single remaining unclassified sequence.
Transmission Patterns of Postmigration Transmissions Between Individuals With Different Nationalities
Phyloethnic Study
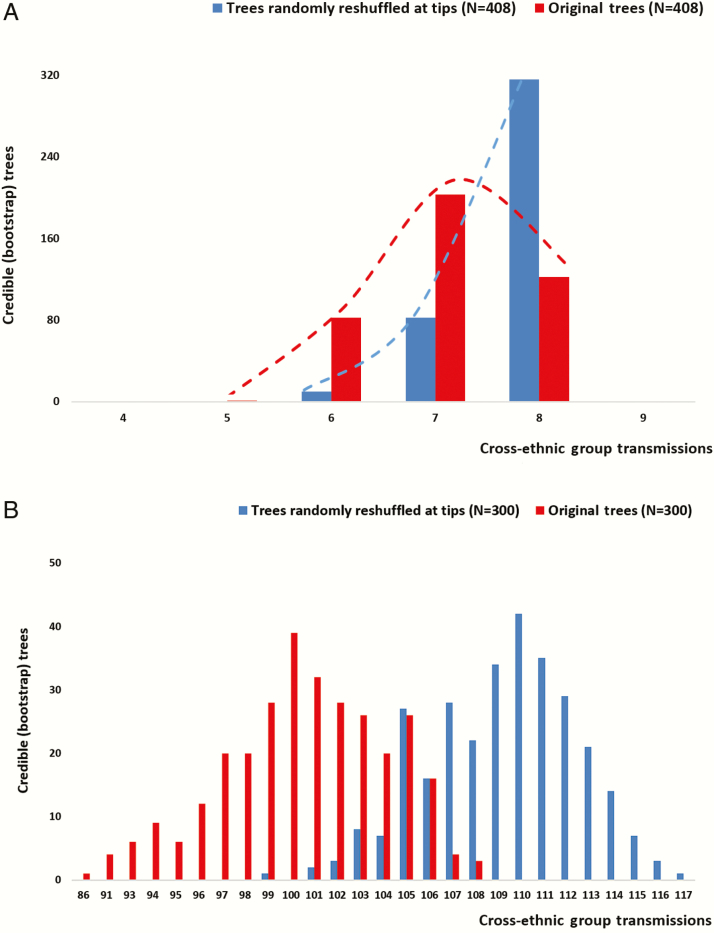
To estimate the transmission patterns of postmigration transmissions, we performed a statistical phyloethnic study, where character states at the tips of the tree correspond to nationality (Figure 2A and 2B and Supplementary Figure 1A and 1B). Interestingly, our analysis revealed evidence for significant compartmentalization according to nationality for subtype A and CRF14_BG, whereas we did not find significant differences for subtype B and CRF35_AD (Figure 3A and 3B and Supplementary Figure 2A and 2B). (For CRF35_AD, significant differences between the observed and the expected cross-ethnic group transmissions were found only for the bootstrap trees and not for the ML tree. Therefore we assume no significant compartmentalization.) Our findings imply that transmissions occur at higher proportions between migrants infected with subtype A and migrants infected with CRF14_BG than between migrants and Greeks. Notably, CRF14_BG was the most frequent clade among PWID with a non-Greek nationality (Table 2), thus suggesting that the former pattern was dominant among the injectors.
Figure 3.
Distribution of viral migration events corresponding to transmissions between migrants and Greeks (cross-nationality group transmissions) estimated for trees randomly reshuffled at tips (shown in blue) and the original bootstrap trees (shown in red). Distributions were estimated for: (A) subtype A and (B) CRF14_BG.
Identification of Putative Origin Based on Genetic Distances
To further confirm our previous findings about the significant clustering of migrants, we estimated the putative closest partner for each individual found among the PWID-LTNs and examined whether the 2 partners were of similar nationality. This analysis was performed only for subtype A and CRF14_BG, for which we found evidence for significant clustering between migrants.
Notably, we found that 4 of 6 (66.6%) of the subtype A closest partners of migrants were also of non-Greek nationality. Similarly, 75.2% (76 of 101) of the CRF14_BG closest partners were of similar nationality. These findings further confirm our phylogenetic analysis findings that sequences from migrants cluster together at high proportions.
DISCUSSION
Our findings show that most of the HIV-1 infections in migrant PWID were acquired after they moved to Greece, suggesting that this population was severely affected during the Athens outbreak. Additionally, we show that HIV-1 transmissions occur more frequently through contacts between migrants. These estimates were based on diverse methods using clinical and behavioral data, as well as molecular analyses.
Tracing the most probable country of infection is complex. Additional methods are needed besides surveillance and other traditional epidemiological tools. Given concerns about the accuracy of previous methods, the European Centre for Disease Prevention and Control has taken an initiative for the estimation of the putative origin of HIV-1 transmissions based on CD4 decline modeling [1]. We herein provide evidence that molecular analyses comprise an additional method for the reliable estimation of the probable country of infection. Moreover, we used novel molecular methods to explore whether postmigration transmissions occur more frequently among migrants. To the best of our knowledge, this is one of very few studies using molecular epidemiology to address this issue [18, 19].
We investigated the patterns of the HIV-1 epidemic across groups with different nationalities, using data from all available HIV-1–infected individuals after 2010 in Greece. Notably, our results suggest that the majority (94.3%) of infections among migrant PWID originated in Greece, and specifically, within the LTNs in the Athens metropolitan area. Given that the outbreak sequences form monophyletic groups within the CRF14_BG, CRF35_AD, subtype A, and subtype B clades, this implies that with the exception of the founder strain, the rest of these infections occurred locally. Hypothetically, if any transmissions in PWID had occurred outside Greece, then the corresponding sequences would cluster outside the suboutbreak groups. The origin of the founder strains was previously estimated to be in Romania (CRF14_BG), Afghanistan/Iran (CRF35_AD), and Greece (subtypes A and B) [3, 12].
Our findings are consistent with recent evidence that shows that migrants and transmission categories become infected at disproportionate prevalence after migrating to Europe [20, 21]. This is in contrast with previous findings from European countries where the levels of postmigration transmissions were lower [22]. In studies using molecular methods, a previous analysis from a Swiss cohort suggested that approximately 20% of non–subtype B newly diagnosed transmissions among individuals with an African or Asian nationality occurred as a result of domestic transmissions in Switzerland [18]. In Italy, individuals from generalized HIV-1 epidemics were found to be less likely to belong to local clusters than Latin Americans and Italians [23]. Similarly, Yebra et al reported from Madrid, Spain, that only 9% of sub-Saharan Africans belonged to local clusters [24]. The characteristics of the epidemic (eg, HIV-1 clades, sex, and risk group distributions) differed significantly between migrants and local populations [21], suggesting that postmigration acquisition of HIV-1 occurred but was probably not dominant. This picture is consistent across Western Europe, where non–subtype B transmissions have been mostly introduced from areas with generalized HIV-1 epidemics. Greece and Portugal provide exceptions where subtypes A and G have been spread within local transmission networks [25].
The recent outbreak among PWID in the Athens Metropolitan Area provides a different case where HIV-1 acquisition occurred across individuals with diverse nationalities. These findings may imply that there is a difference in nature between sexually vs parenterally transmitted epidemics, at least in the context of a new epidemic among PWID. This is of public health importance, as for an outbreak among PWID, prevention actions should target the entire injecting population, including ethnic minorities. In addition to domestic transmissions, we also found a small proportion of imported infections from abroad, and most importantly, from Turkey.
The results of our phyloethnic study suggest that for those infected postmigration, HIV-1 transmissions occur more frequently through contacts between migrants in a significant manner. In the Athens outbreak, we show that although migrants represented a small proportion of all HIV-infected PWID, transmissions occurred more frequently among them (at least for 2 of the LTNs). These findings are novel and suggest first that migrants engage in riskier behaviors with other migrants (at least from their own nationality) than with Greeks. This is consistent with extensive research that suggests similar patterns of safer practices with “strangers.” Second, these findings suggest that migrants’ risk networks are primarily with other migrants. This is also consistent with parallel findings that racial/ethnic categories of PWID tend to inject within-group and to engage in sex within-group more often than between racial/ethnic groups [26].
Our study has some limitations. The exact source for an individual cannot always be identified since there might be intermediate links that have not been sampled. In the present study, since we investigated the potential geographic origin of HIV transmissions and not the putative source, our estimations are expected to be credible, except in the case of migrants’ sequences that did not fall within phylogenetic clusters from specific geographic areas (LTNs). For such cases, the levels of confidence are lower than for sequences falling within LTNs. For those within LTNs, the acquisition might have originated from a remote area outside the region of the LTN. However, the local nature of the outbreak, the monophyletic clustering of LTNs, the fact that the PWID-specific viral strains have not been detected outside of Greece, and the nested clustering of sequences from migrants in the LTNs (except for 2 sequences that appeared close to the root) all make the non-Greek origin hypothesis quite unlikely. A final limitation of the present study is that, although we used similar criteria to estimate the geographic origin of transmissions for sequences outside of the LTNs, the nonrepresentative sampling of sequences available on public databases may have introduced bias in our estimations.
In conclusion, we show that molecular methods can be used not only to trace the most probable country of infection but also to infer additional features of an HIV epidemic. We also found evidence that postmigration transmissions dominate the PWID outbreak in Athens and demonstrated the existence of significant transmission between migrants. These findings are of public health importance, as this knowledge can be used to inform the design of prevention strategies for communicable diseases (ie, increased access to opioid substitution therapy, needle/syringe programs, testing, and treatment services) especially in the case of epidemics among PWID. Using molecular methods to inform HIV real-time prevention would be of particular importance, although certain limitations can make real-time implementation unfeasible.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Financial support. This work was supported by National Strategic Reference Framework 2007–2013 and cofounded by the European Social Fund and Greek national resources (MIS 365008). Additional financial support was provided by the Hellenic Scientific Society for the Study of AIDS and STDs and the US National Institute on Drug Abuse (“Preventing HIV Transmission by Recently-Infected Drug Users”; grant number DP1 DA034989).
Potential conflicts of interest. All authors: No potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. European Centre for Disease Prevention and Control. Thematic report: migrants. Monitoring implementation of the Dublin Declaration on Partnership to Fight HIV/AIDS in Europe and Central Asia: 2014 progress report. Stockholm: ECDC, 2015. [Google Scholar]
- 2. Hernando V, Alvarez-del Arco D, Alejos B et al. HIV infection in migrant populations in the European Union and European Economic Area in 2007–2012: an epidemic on the move. J Acquir Immune Defic Syndr 2015; 70: 204–11. [DOI] [PubMed] [Google Scholar]
- 3. Paraskevis D, Nikolopoulos G, Fotiou A et al. Economic recession and emergence of an HIV-1 outbreak among drug injectors in Athens metropolitan area: a longitudinal study. PLoS One 2013; 8:e78941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Deblonde J, Sasse A, Del Amo J et al. Restricted access to antiretroviral treatment for undocumented migrants: a bottle neck to control the HIV epidemic in the EU/EEA. BMC Public Health 2015; 15:1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Álvarez-Del Arco D, Monge S, Rivero-Montesdeoca Y, Burns F, Noori T, Del Amo J. Implementing and expanding HIV testing in immigrant populations in Europe: comparing guideline’s recommendations and expert’s opinions. Enferm Infecc Microbiol Clin 2017; 35:47–51. [DOI] [PubMed] [Google Scholar]
- 6. Paraskevis D, Nikolopoulos G, Tsiara C et al. HIV-1 outbreak among injecting drug users in Greece, 2011: a preliminary report. Euro Surveill 2011; 16. [DOI] [PubMed] [Google Scholar]
- 7. Nikolopoulos GK, Sypsa V, Bonovas S et al. Big events in Greece and HIV infection among people who inject drugs. Subst Use Misuse 2015; 50:825–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hatzakis A, Sypsa V, Paraskevis D et al. Design and baseline findings of a large-scale rapid response to an HIV outbreak in people who inject drugs in Athens, Greece: the ARISTOTLE programme. Addiction 2015; 110:1453–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sypsa V, Paraskevis D, Malliori M et al. Homelessness and other risk factors for HIV infection in the current outbreak among injection drug users in Athens, Greece. Am J Public Health 2015; 105:196–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nikolopoulos GK, Pavlitina E, Muth SQ et al. A network intervention that locates and intervenes with recently HIV-infected persons: the Transmission Reduction Intervention Project (TRIP). Sci Rep 2016; 6:38100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tsang MA, Schneider JA, Sypsa V et al. Network characteristics of people who inject drugs within a new HIV epidemic following austerity in Athens, Greece. J Acquir Immune Defic Syndr 2015; 69:499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Paraskevis D, Paraschiv S, Sypsa V et al. Enhanced HIV-1 surveillance using molecular epidemiology to study and monitor HIV-1 outbreaks among intravenous drug users (IDUs) in Athens and Bucharest. Infect Genet Evol 2015; 35:109–21. [DOI] [PubMed] [Google Scholar]
- 13. Struck D, Lawyer G, Ternes AM, Schmit JC, Bercoff DP. COMET: adaptive context-based modeling for ultrafast HIV-1 subtype identification. Nucleic Acids Res 2014; 42:e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 2010; 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014; 30:1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Slatkin M, Maddison WP. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics 1989; 123:603–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wilgenbusch JC, Swofford D. Inferring evolutionary trees with PAUP. Curr Protoc Bioinformatics 2003. doi:10.1002/0471250953.bi0604s00. [DOI] [PubMed] [Google Scholar]
- 18. von Wyl V, Kouyos RD, Yerly S et al. ; Swiss HIV Cohort Study The role of migration and domestic transmission in the spread of HIV-1 non-B subtypes in Switzerland. J Infect Dis 2011; 204:1095–103. [DOI] [PubMed] [Google Scholar]
- 19. Dauwe K, Mortier V, Schauvliege M et al. Characteristics and spread to the native population of HIV-1 non-B subtypes in two European countries with high migration rate. BMC Infect Dis 2015; 15:524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chaix ML, Seng R, Frange P et al. ; ANRS PRIMO Cohort Study Group Increasing HIV-1 non-B subtype primary infections in patients in France and effect of HIV subtypes on virological and immunological responses to combined antiretroviral therapy. Clin Infect Dis 2013; 56:880–7. [DOI] [PubMed] [Google Scholar]
- 21. Abecasis AB, Wensing AM, Paraskevis D et al. HIV-1 subtype distribution and its demographic determinants in newly diagnosed patients in Europe suggest highly compartmentalized epidemics. Retrovirology 2013; 10:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fakoya I, Álvarez-del Arco D, Woode-Owusu M et al. A systematic review of post-migration acquisition of HIV among migrants from countries with generalised HIV epidemics living in Europe: implications for effectively managing HIV prevention programmes and policy. BMC Public Health 2015; 15:561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lai A, Bozzi G, Franzetti M et al. Phylogenetic analysis provides evidence of interactions between Italian heterosexual and South American homosexual males as the main source of national HIV-1 subtype C epidemics. J Med Virol 2014; 86:729–36. [DOI] [PubMed] [Google Scholar]
- 24. Yebra G, Holguín A, Pillay D, Hué S. Phylogenetic and demographic characterization of HIV-1 transmission in Madrid, Spain. Infect Genet Evol 2013; 14:232–9. [DOI] [PubMed] [Google Scholar]
- 25. Thomson MM, Najera R. Increasing HIV-1 genetic diversity in Europe. J Infect Dis 2007; 196:1120–4. [DOI] [PubMed] [Google Scholar]
- 26. Bell DC, Montoya ID, Atkinson JS. Partner concordance in reports of joint risk behaviors. J Acquir Immune Defic Syndr 2000; 25:173–81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.