Summary
A whole-genome sequencing approach predicted phenotypic resistance to extended spectrum β-lactams for 4 leading causes of gram-negative bacteremia in neutropenic cancer patients at a level equal or superior to data generated in the clinical microbiology laboratory.
Keywords: antimicrobial resistance, whole-genome sequencing, gram-negative bacteria, bacteremia, neutropenic fever
Abstract
Background
There is marked interest in using DNA-based methods to detect antimicrobial resistance (AMR), with targeted polymerase chain reaction (PCR) approaches increasingly being incorporated into clinical care. Whole-genome sequencing (WGS) could offer significant advantages over targeted PCR for AMR detection, particularly for species where mutations are major drivers of AMR.
Methods
Illumina MiSeq WGS and broth microdilution (BMD) assays were performed on 90 bloodstream isolates of the 4 most common gram-negative bacteria causing bloodstream infections in neutropenic patients. The WGS data, including both gene presence/absence and detection of mutations in an array of AMR-relevant genes, were used to predict resistance to 4 β-lactams commonly used in the empiric treatment of neutropenic fever. The genotypic predictions were then compared to phenotypic resistance as determined by BMD and by commercial methods during routine patient care.
Results
Of 133 putative instances of resistance to the β-lactams of interest identified by WGS, only 87 (65%) would have been detected by a typical PCR-based approach. The sensitivity, specificity, and positive and negative predictive values for WGS in predicting AMR were 0.87, 0.98, 0.97, and 0.91, respectively. Using BMD as the gold standard, our genotypic resistance prediction approach had a significantly higher positive predictive value compared to minimum inhibitory concentrations generated by commercial methods (0.97 vs 0.92; P = .025).
Conclusions
These data demonstrate the potential feasibility of using WGS to guide antibiotic treatment decisions for patients with life-threatening infections for an array of medically important pathogens.
The ever-increasing impact of antimicrobial resistance (AMR) has resulted in a broad array of efforts to improve antibiotic utilization [1–4]. Currently, most clinical microbiology laboratories require 48–72 hours to perform traditional phenotypic assays to detect AMR [5]. Thus, rapid molecular diagnostics (RMDs) of AMR have been proposed as a means to implement timely escalation or de-escalation of antimicrobial therapy [1, 6, 7]. Several studies have shown that various RMDs, including whole-genome sequencing (WGS) approaches, have good predictive values for detecting AMR in Escherichia coli and Klebsiella pneumoniae [1, 8–10]. However, to move RMDs into clinical practice, it is important to study these approaches in situations where a broad variety of species that account for a large proportion of actual infections are analyzed [1, 6, 8–11].
We have chosen to study RMDs for AMR detection in neutropenic cancer patients, as such patients are particularly prone to lethal bacterial infections, and timely initiation of active antibiotics is critical [12]. The major goal in the empiric treatment of the febrile, neutropenic patient with suspected infection is to provide adequate therapy for gram-negative bacteria [12]. Thus, the intravenous antibiotics recommended for empiric treatment of high-risk, febrile neutropenic patients are either piperacillin-tazobactam (P/T), a carbapenem (meropenem or imipenem-cilastatin), cefepime, or ceftazidime, all of which are β-lactams and possess broad activity against a range of major gram-negative pathogens, including Pseudomonas aeruginosa [12]. However, the increasing level of β-lactam resistance among bacteria causing infections in neutropenic patients threatens the ability to provide timely, effective antimicrobial therapy to these seriously ill patients [13].
DNA-based approaches, such as multiplex polymerase chain reaction (PCR) assays to detect the presence of β-lactamase genes, are considered the most likely method of increasing the speed of AMR detection [1, 6]. However, reliance on the presence or absence of β-lactamase genes to categorize AMR patterns will fail to detect resistance associated with porin mutations, efflux pump systems, or de-repression of chromosomal β-lactamase genes [8, 14]. As these are the predominant mechanisms of β-lactam resistance in P. aeruginosa and contribute significantly to AMR in Enterobacteriaceae, detection of these resistance determinants is likely to be critically important to ensure widespread clinical applicability of any RMD approach [15, 16]. Herein, we sought to determine whether WGS of gram-negative bacteria isolated from the bloodstream of neutropenic patients could be useful in classifying organisms in terms of resistance to the β-lactams used in the empiric treatment of neutropenic fever.
METHODS
Specimen Selection and Antimicrobial Susceptibility Testing
Glycerol stocks of bacterial strains causing bloodstream infection at MD Anderson Cancer Center (MDACC) in Houston, Texas, are routinely stored at –80°C for future analysis under an MDACC Institutional Review Board (IRB)–approved protocol (PA13-0334). Bacterial strains used in this study were isolated from patients with fever (maximum temperature ≥38.3°C at least 1 time or ≥38.1°C for at least 1 hour) and neutropenia (neutrophil count < 500/µL), and when the patients were considered high-risk by Infectious Diseases Society of America criteria (ie, expected duration of neutropenia at least 7 days, were clinically unstable, and/or had medical comorbidities) [12]. Isolates obtained between August 2013 and December 2014 were analyzed in this study. A waiver of informed consent to collect clinical data and analyze the isolates was provided by the MDACC IRB (PA14-0645).
Minimum inhibitory concentrations (MICs) were determined for ceftazidime, cefepime, P/T, and meropenem in duplicate using reference Clinical and Laboratory Standards Institute (CLSI) broth microdilution (BMD) methods. Quality control was performed with E. coli ATCC 25922 and P. aeruginosa ATCC 27853; all quality control results were within the specified ranges. Additionally, MIC data for each isolate generated in the MDACC clinical microbiology laboratory for the 4 β-lactams being studied were obtained from the electronic medical record. During the time period of this study, P. aeruginosa was routinely tested using Etest (bioMérieux, Marcy-L’Étoile, France) while Enterobacteriaceae were tested via the automated Vitek 2 system (bioMérieux). For both BMD and clinical microbiology data, MICs were interpreted using current CLSI recommendations with isolates classified as either as susceptible (including susceptible dose-dependent) or resistant (including intermediate).
Whole-Genome Sequencing and Analysis
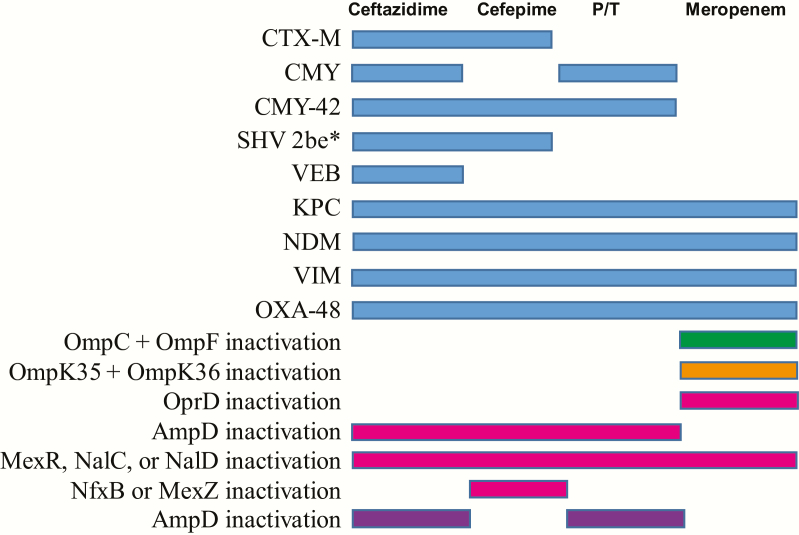
Strains of selected species of gram-negative bacteria (details on strain selection are given in the Results) were chosen for WGS based on having a broad array of antimicrobial phenotypic resistance patterns. A custom database of AMR protein sequences was built by merging the data of antibiotic resistance database (ARDB) and comprehensive antibiotic resistance database (CARD), including β-lactamase alleles or mutations leading to β-lactam resistance [1, 8–10, 14, 15, 17–28]. Further details on sequencing and predictions of genotypic resistance are provided in the Supplementary Materials and Figure 1. If any of the AMR mechanisms shown in Figure 1 for ceftazidime, cefepime, P/T, and meropenem were identified, then the isolate was predicted to be resistant to the specified antibiotic, whereas if no mechanisms were identified then the isolate was predicted to be susceptible. Predictions of AMR were performed without reference to the phenotypic data. Mechanisms of resistance were considered PCR detectable when conferred by exogenous β-lactamases. The sequences reported in this article have been deposited in the National Center for Biotechnology BioProject database PRJNA388450.
Figure 1.
Schematic for how genotypic detection of antimicrobial resistance (AMR) mechanisms was translated into phenotypic predictions. On the left are various AMR mechanisms. When detected, phenotypic resistance to the antibiotic listed at the top was predicted when a shaded bar is present. A blue bar indicates AMR was predicted for all 4 species examined. A green bar indicates a mechanism specific to Escherichia coli. An orange bar indicates a mechanism specific to Klebsiella pneumoniae. A magenta bar indicates a mechanism specific to Pseudomonas aeruginosa. A purple bar indicates a mechanism specific to Enterobacter cloacae. AmpD is separated out for P. aeruginosa and E. cloacae because of the differential effects of AmpC depression on cefepime susceptibility in these 2 organisms. Only AMR mechanisms detected in our cohort are depicted here, although we searched for all mechanisms to the indicated antibiotics found in the antibiotic resistance database (ARDB) and comprehensive antibiotic resistance database (CARD) databases. *2 be indicates extended spectrum β-lactamase SHV variant with G238S and E240K mutations [40]. Abbreviation: P/T, piperacillin-tazobactam.
Statistical Analyses
The sensitivity, specificity, and positive and negative predictive values for the genotypic prediction were calculated for each β-lactam and for each species relative to the phenotypic assignment. Interrater agreement of the various methods was determined using Cohen kappa (κ). Comparison of the sensitivity and specificity of the WGS method relative to clinical microbiology data using BMD as the gold standard was performed using McNemar test, and comparison of the positive and negative predictive values was performed using a marginal regression model [29]. Statistical significance was assigned at a P value of ≤.05. All analyses were performed in Stata software version 14 (StataCorp, College Station, Texas).
RESULTS
Isolate Identification and Characterization
During the period of study, there were 737 unique cases of monomicrobial gram-negative rod bacteremia. The top 4 organisms were E. coli (n = 280 cases [38%]), P. aeruginosa (n = 138 [19%]), K. pneumoniae (n = 102 [14%]), and Enterobacter cloacae (n = 39 [5%]), which together accounted for 76% of all the gram-negative bacteremia cases. Thus, these were the 4 species chosen for further analysis. A representative sample composed of 31 E. coli, 25 K. pneumoniae, 22 P. aeruginosa, and 13 E. cloacae isolates demonstrating a range of susceptibility patterns was sent for WGS. The sample was enriched for K. pneumoniae because of the broad range of MICs for each antibiotic observed for this species (Table 1). One K. pneumoniae strain was removed due to insufficient sequencing coverage depth, leaving 90 total isolates. By multilocus sequence typing (MLST) and whole-genome phylogeny, isolates were genetically diverse (Supplementary Table 1; Supplementary Figures 1–4), with the exception of 12 E. coli strains that grouped within sequence type 131, the most common extended-spectrum β-lactamase–producing E. coli genetic lineage worldwide [30].
Table 1.
Summary of Antimicrobial Susceptibility Data for Various Strain/Antibiotic Combinations
| Antibiotic | ||||
|---|---|---|---|---|
| Species | Ceftazidime | Cefepime | Piperacillin- Tazobactam | Meropenem |
| Escherichia coli (n = 31) | ||||
| Resistanta, No. (%) | 20 (64.5) | 18 (58.1) | 8 (25.8) | 1 (3.2) |
| MIC range, mg/L | <0.25 to >256 | <0.5 to >256 | 2 to >256 | <0.06 to >64 |
| Klebsiella pneumoniae (n = 24) | ||||
| Resistanta, No. (%) | 16 (66.7) | 14 (58.3) | 10 (41.7) | 6 (25.0) |
| MIC range, mg/L | <0.5 to >256 | <0.25 to >256 | 2 to >256 | <0.06 to >64 |
| Pseudomonas aeruginosa (n = 22) | ||||
| Resistanta, No. (%) | 6 (27.3) | 10 (45.5) | 9 (40.9) | 14 (63.6) |
| MIC range, mg/L | 1 to >256 | 2 to >256 | 2 to >256 | 0.12 to >64 |
| Enterobacter cloacae (n = 13) | ||||
| Resistanta, No. (%) | 7 (53.8) | 3 (23.1) | 7 (53.8) | 0 (0.0) |
| MIC range, mg/L | <0.5 to >256 | <0.5 to 32 | 0.75 to >256 | 0.25 to 1 |
Abbreviation: MIC, minimum inhibitory concentration.
aResistance as determined by Clinical and Laboratory Standards Institute guidelines using broth microdilution (reference method).
Phenotypic Analysis of β-Lactam Resistance
Phenotypic susceptibility and MIC ranges are presented in Table 1. With the exception of meropenem and E. cloacae, for which no resistant isolates were detected, we observed susceptible and nonsusceptible strains for each combination of β-lactam and species. Additionally, a wide range of MICs were detected across β-lactams for each species (Table 1). Combined with the lack of clonality observed in our WGS phylogeny data, these findings indicate that our cohort was composed of heterogeneous bacteria useful for testing the ability of WGS to predict AMR.
WGS Characterization of Predicted Antimicrobial Resistance Determinants
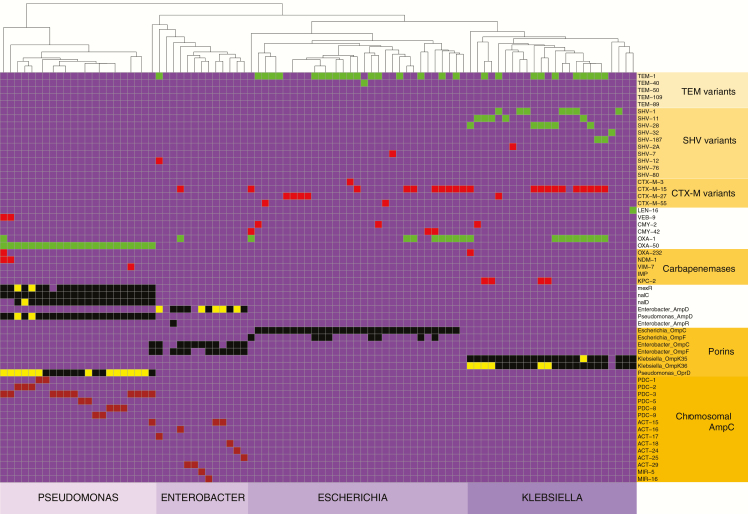
Our WGS approach identified a broad array of AMR elements relevant to a diverse range of antimicrobials (Supplementary Figure 5). Hierarchical clustering of the strains by presence/absence of AMR elements showed 100% concordance with species designation, indicating that broad inclusion of all the AMR data is sufficient to resolve the 4 species studied here (Supplementary Figure 5). For the 360 combinations of the 90 strains with 4 antimicrobials, our WGS approach identified 133 predicted instances of AMR to the 4 β-lactams of interest (Figure 2). Broadly speaking, there were 2 major categories of predicted AMR. The first resulted from acquisition of exogenous β-lactamases, such as genes encoding CTX-M or NDM enzymes. The second was due to mutations or other mechanisms resulting in inactivation of a chromosomal gene, such as insertion in the OprD porin-encoding gene in P. aeruginosa or mutation in ampD leading to hyperproduction of the chromosomally encoded AmpC protein in E. cloacae [8, 21]. The breakdown of identified AMR mechanisms by species is presented in Supplementary Figure 6. Exogenous β-lactamase acquisition predominated as a predicted AMR mechanism among E. coli and K. pneumoniae strains, whereas chromosomal gene inactivation events were the predominant AMR mechanism for P. aeruginosa and E. cloacae isolates (Table 2). In total, only 87 of the 133 (65%) of the predicted AMR events were mediated by genes encoding exogenous β-lactamases readily detected with a typical PCR-based approach.
Figure 2.
Antimicrobial resistance mechanisms identified via whole-genome sequencing. On the x-axis are individual strains sorted by species. On the y-axis are protein forms of various β-lactam resistance mechanisms. All β-lactam resistance mechanisms identified in our cohort are included along with some additional mechanisms that were not present in any of the studied strains. Box color scheme is as follows: purple, absent; green, β-lactamase encoding gene present but not predicted to mediate resistance as not active against the β-lactams being studied; red, β-lactamase encoding gene present and predicted to mediate resistance; black, endogenous chromosomal gene present and functional (eg, ampD); yellow, endogenous chromosomal gene predicted to be inactive and to result in resistance (eg, oprD); brown, chromosomal β-lactamase encoding gene present and capable of mediating resistance if de-repressed (eg, blaPDC-1).
Table 2.
Contribution of Exogenous β-Lactamase Acquisition Versus Chromosomal Gene Mutation Driving Predicted Antimicrobial Resistance
| Species | Total Instances of Predicted AMR | No. (%) of Instances of Predicted AMR due to Exogenous β-Lactamase Acquisition | No. (%) of Instances of Predicted AMR due to Chromosomal Mutations | No. (%) of Instances of Predicted AMR due to Both Mechanisms |
|---|---|---|---|---|
| Escherichia coli | 44 | 43 (98) | 1 (2) | 0 (0) |
| Klebsiella pneumoniae | 41 | 40 (98) | 1 (2) | 0 (0) |
| Pseudomonas aeruginosa | 32 | 3 (9) | 20 (63) | 9 (28) |
| Enterobacter cloacae | 16 | 4 (25) | 11 (69) | 1 (6) |
Abbreviation: AMR, antimicrobial resistance.
Assessment of Whole-Genome Data to Predict Phenotypic AMR
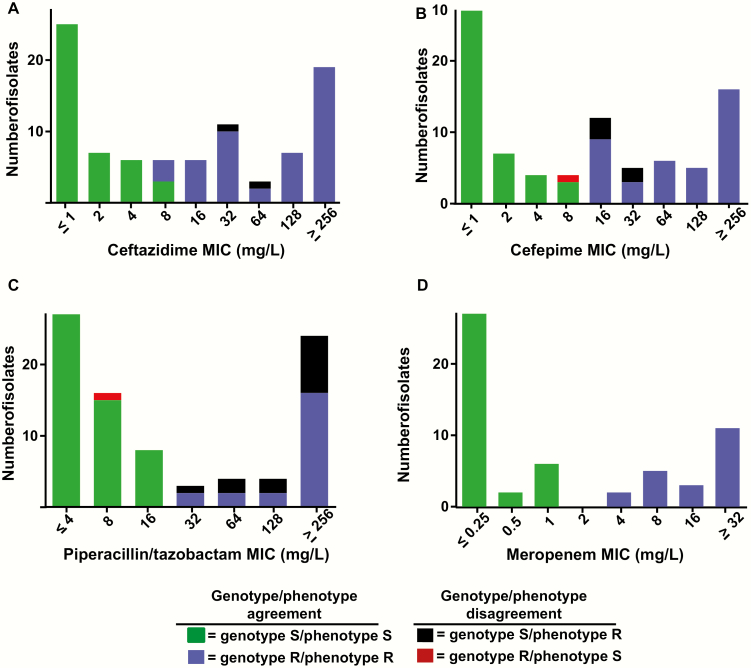
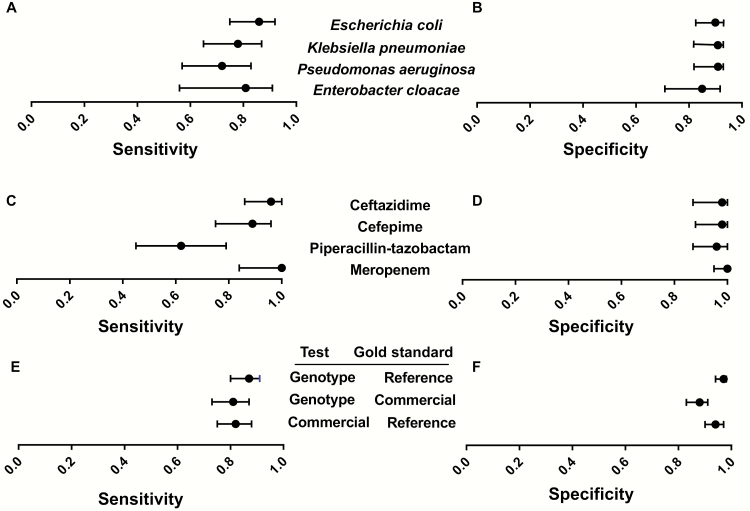
Of the 360 total determinations of AMR, there was agreement between our WGS prediction and phenotypic susceptibility as determined by BMD (hereafter called the reference method) in 336 instances (93%) and disagreement on 24 occasions (7%) (Figure 3 shows agreement/disagreement for the entire cohort whereas species specific analyses are provided in Supplementary Figures 6–10). The majority of disagreements (13 instances) were due to an inability to detect a genotypic mechanism for phenotypic resistance to P/T, which was an issue for all species except E. cloacae (Figure 3; Supplementary Table 2). The combination of cefepime and P. aeruginosa was another main source of disagreement with 4 of 10 strains that tested phenotypically resistant to cefepime lacking an identified resistance mechanism (Supplementary Figure 9). No instances of disagreement in any species were observed for meropenem. Overall, our WGS approach had a 0.87 sensitivity and 0.98 specificity vs the reference method (Figure 4). These findings translated into a positive and negative predictive value of 0.97 and 0.91, respectively.
Figure 3.
Agreement/disagreement between whole-genome sequencing and phenotypic data for antimicrobial resistance to 4 β-lactams. A–D, Minimum inhibitory concentrations for indicated β-lactams (phenotype) are shown. The colors of the bars represent various combinations of genotypic and phenotypic resistance as indicated in the legend. Abbreviations: MIC, minimum inhibitory concentration; R, resistant; S, susceptible.
Figure 4.
Sensitivity and specificity of genotypic predicted resistance. A–F, Data shown are either sensitivity or specificity (as noted in the y-axis) plus 95% confidence intervals. A–D, Data derived from predicted genotypic resistance using reference method (broth microdilution) as gold standard. Data are grouped by indicated organism (A and B) or by indicated antimicrobial (C and D). E and F, Summary data for all organism–antimicrobial combinations. Top line shows performance of genotypic prediction using reference method as gold standard. Middle line shows performance of genotypic prediction using commercial methods as gold standard. Bottom line shows performance of commercial methods using reference method as gold standard.
Comparison of BMD With Data Derived From the Clinical Microbiology Laboratory
Although BMD is considered the gold standard for determining antimicrobial susceptibility, it is a time- and resource-consuming methodology and thus rarely performed in clinical microbiology laboratories. Instead, a combination of non-BMD (ie, automated susceptibility testing and diffusion-based) approaches is frequently used [31, 32]. Therefore, we next sought to compare whether using data from the clinical microbiology laboratory (hereafter called commercial methods) as the gold standard would significantly alter the predictive capacity of our WGS approach. We observed marked variation between the commercial methods and reference method MICs, particularly for the combinations of E. coli and K. pneumoniae with ceftazidime and cefepime (Supplementary Table 2). Primarily this was due to higher MICs using the reference method in these 2 organisms for strains in which CTX-M enzymes were present, a finding that has been previously reported [33]. When all combinations of strains and antimicrobials were considered, the sensitivity and specificity for our WGS approach was higher when we used the reference method rather than the commercial methods MICs as the gold standard, and the 95% confidence intervals (CIs) were nonoverlapping for specificity but not sensitivity (Figure 4E and 4F; Supplementary Table 2).
Our finding that using different reference gold standards significantly impacted the test performance of our WGS prediction model led us to directly compare our WGS prediction model with the commercial methods data while using the reference method as the gold standard. The interrater agreement (Cohen κ) for our WGS prediction with the reference method was 0.86 (95% CI, .81–.91), whereas it was 0.78 (95% CI, .72–.85) for the commercial methods data with the reference method. Using the reference method as the gold standard, the positive predictive value of our WGS approach was significantly higher compared to the commercial methods data (0.97 vs 0.92; P = .025), and the difference in specificity showed a trend toward a statistically significant difference (0.98 vs 0.95; P = .07) (Table 3). Although the sensitivity and negative predictive values were also higher for our WGS approach compared to the commercial methods data, the differences were not statistically significant (Table 3). Thus, our WGS approach was at least equivalent, and in some characteristics significantly better, in terms of classification of AMR for the β-lactams studied here compared to the commercial methods data.
Table 3.
Diagnostic Performance of Whole-Genome Sequencing Versus Clinical Microbiology Data Using Broth Microdilution as the Gold Standard
| Diagnostic Performance | WGS | Clinical Microbiology | P Value |
|---|---|---|---|
| Sensitivity (95% CI) | 0.87 (.81–.92) | 0.82 (.76–.88) | .36a |
| Specificity (95% CI) | 0.98 (.96–.999) | 0.95 (.92–.98) | .07a |
| Positive predictive value (95% CI) | 0.97 (.94–.999) | 0.92 (.88–.97) | .025b |
| Negative predictive value (95% CI) | 0.91 (.88–.95) | 0.88 (.84–.92) | .24b |
Abbreviations: CI, confidence interval; WGS, whole-genome sequencing.
aMcNemar test.
bScore statistic derived from marginal regression model.
DISCUSSION
In light of the increasing burden of antimicrobial-resistant infections, there is significant impetus to improve the rapidity and accuracy of bacterial AMR identification [1, 6]. Although PCR-based assays are currently being incorporated into routine clinical care in many microbiology laboratories [34], there is growing recognition that a WGS approach will be needed to capture the diverse array of insertions, deletions, and single-nucleotide polymorphisms (SNPs) that contribute to AMR [35]. We chose to examine neutropenic fever as a proof of concept for WGS prediction of clinically relevant AMR because such patients are treated with a limited number of antimicrobials for which the AMR pathways are fairly well delineated [12]. Using an approach that incorporated both gene presence/absence and mutations in genes encoding an array of proteins involved in β-lactamase expression, porin function, and efflux pump expression, we achieved high rates of both positive and negative AMR prediction values for 4 β-lactams empirically used in the neutropenic fever setting.
These data add to a number of investigations showing that genetic approaches can effectively predict antimicrobial susceptibility for an array of medically important bacterial pathogens [1, 6, 8–11, 36–38]. Our study, however, provides key advances. First, research into genetic predictors of gram-negative AMR has mainly focused on E. coli and K. pneumoniae given that AMR in these organisms is primarily mediated by β-lactamase gene acquisition events that are relatively easy to detect [1, 10]. Genotypic predictions of AMR in P. aeruginosa and E. cloacae have been more problematic and scarce despite the importance of these organisms to healthcare-associated infections [8, 9]. A major reason for this difficulty is that these organisms contain genes encoding a variety of resistance mechanisms to the β-lactams studied here, such as AmpC-like enzymes or efflux pumps, but de-repression of such genes, and not simply their presence, are needed to confer a resistant phenotype [15]. Thus, accurate genotypic assessment of β-lactam AMR needs to incorporate identification of small genetic changes such as polymorphisms in AmpD leading to AmpC hyperproduction, inactivation of genes regulating efflux pump production, and porin mutations [15, 20, 21]. Importantly, these AMR mechanisms are not readily detected by most PCR-based methodologies. In our cohort, the vast majority of genetic changes mediating AMR to the β-lactams of interest in P. aeruginosa and E. cloacae were caused by mutations or insertions/deletion in genes encoding either regulatory elements or porins (Table 2). For the entire cohort, PCR methodologies capable of detecting exogenous β-lactamases would have identified less than two-thirds of all AMR mechanisms, indicating that WGS, if feasible to implement and automate, could be a more comprehensive, and clinically relevant, approach.
A second important aspect of our study was that we compared our WGS results to susceptibility data derived by reference BMD methods and to data generated by commercial methods. In clinical laboratories, susceptibility testing by automated technologies and diffusion-based methods (ie, Etest) are widely utilized [1, 6, 32]; however, such approaches are known to have shortcomings with particular species-antimicrobial combinations [33, 39]. Indeed, we found that when we compared our WGS data to MICs generated by commercial methods, we observed a significant decrease in the positive predictive value and specificity. To minimize inaccuracies in the interpretation of genotypic data, comparisons should ideally be made to susceptibility results generated by reference BMD methods. This assertion is supported by our finding that WGS data were at least as good, and possibly superior, to susceptibility testing results from commercial methods in predicting AMR. As WGS technologies evolve, the possibility of incorporating genome-wide analyses in the clinical microbiology laboratory grows. Given that automated DNA extraction, sequencing, and analysis may ultimately be significantly less resource intensive for the clinical microbiology laboratory compared with current techniques, the WGS approach could become the preferred methodology for species identification and AMR detection. For this to happen, however, the genetic basis of resistance for all clinically useful antimicrobials must be more fully characterized, and clinical trials to test whether WGS approaches can significantly improve patient care will need to be conducted.
There are several limitations to this study that bear mentioning. First, all of the isolates were from a single center at which broad-spectrum antimicrobial use is common, meaning that we do not know how our data translate into other healthcare settings. However, there was limited clonality among our isolates, the genetic bases for AMR identified in our strains are similar to those previously reported [1, 8, 10], and our MLST populations were comparable to those described elsewhere for the studied species [8, 10, 30]. Second, as all data were derived from genotypic information, we did not confirm that our genotypic prediction actually resulted in changes in transcript level or protein function, such as efflux pump upregulation or OprD inactivation. Given the high sensitivity and specificity of our genotypic predictions, however, it seems that we were able to correctly identify when genotypic changes resulted in alterations in protein function that eventually affected AMR. Third, we had to use manual sequence inspection to identify SNPs and in/dels that abrogated gene function, which was a time-consuming process that will require significant computational biology improvements to move to an automated platform. Finally, we did not extend our genotypic prediction to a broad array of antimicrobials, but rather focused solely on 4 broad spectrum β-lactams. Given that the clinical microbiology laboratory needs to provide physicians with information regarding a diverse panel of antimicrobials, it will be important to extend this line of investigation to the full range of antibiotics used in clinical practice, and we are currently pursuing such studies.
In summary, we have shown that WGS combined with single nucleotide–level analysis accurately predicts resistance and susceptibility to β-lactam antimicrobials used in the treatment of suspected infections in neutropenic cancer patients. Further efforts to routinely incorporate such an approach into the care of patients with suspected and proven infections could markedly alter how the clinical microbiology laboratory identifies and reports AMR to clinicians.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Acknowledgments. We thank the personnel of the clinical microbiology laboratory at MD Anderson Cancer Center for assistance with collecting isolates.
Financial support. Financial support for this study was provided by the Shelby Foundation (R. Lee Clark Fellow Award to S. A. S.); the National Institutes of Health (grant number K08 AI114883 to R. K. S.); and the National Cancer Institute (grant number P30-CA016672 via the Bioinformatics Shared Resource at MD Anderson Cancer Center). J. K., M. K., and B. C. are supported by the Cancer Prevention and Research Institute of Texas (grant number RP150596).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Evans SR, Hujer AM, Jiang H et al. ; Antibacterial Resistance Leadership Group Rapid molecular diagnostics, antibiotic treatment decisions, and developing approaches to inform empiric therapy: PRIMERS I and II. Clin Infect Dis 2016; 62:181–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ward C, Stocker K, Begum J, Wade P, Ebrahimsa U, Goldenberg SD. Performance evaluation of the Verigene (Nanosphere) and FilmArray (BioFire) molecular assays for identification of causative organisms in bacterial bloodstream infections. Eur J Clin Microbiol Infect Dis 2015; 34:487–96. [DOI] [PubMed] [Google Scholar]
- 3. Walker T, Dumadag S, Lee CJ et al. Clinical impact of laboratory implementation of verigene BC-GN microarray-based assay for detection of gram-negative bacteria in positive blood cultures. J Clin Microbiol 2016; 54:1789–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goodman KE, Lessler J, Cosgrove SE et al. ; Antibacterial Resistance Leadership Group A clinical decision tree to predict whether a bacteremic patient is infected with an extended-spectrum β-lactamase-producing organism. Clin Infect Dis 2016; 63:896–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Caliendo AM, Gilbert DN, Ginocchio CC et al. ; Infectious Diseases Society of America (IDSA) Better tests, better care: improved diagnostics for infectious diseases. Clin Infect Dis 2013; 57(suppl 3):S139–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Evans SR, Hujer AM, Jiang H et al. Informing antibiotic treatment decisions: evaluating rapid molecular diagnostics (RMDs) to identify susceptibility and resistance to carbapenems against Acinetobacter spp. PRIMERS III. J Clin Microbiol 2016; 55:134–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tuite N, Reddington K, Barry T, Zumla A, Enne V. Rapid nucleic acid diagnostics for the detection of antimicrobial resistance in gram-negative bacteria: is it time for a paradigm shift? J Antimicrob Chemother 2014; 69:1729–33. [DOI] [PubMed] [Google Scholar]
- 8. Kos VN, Déraspe M, McLaughlin RE et al. The resistome of Pseudomonas aeruginosa in relationship to phenotypic susceptibility. Antimicrob Agents Chemother 2015; 59:427–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kos VN, McLaughlin RE, Gardner HA. Elucidation of mechanisms of ceftazidime resistance among clinical isolates of Pseudomonas aeruginosa by using genomic data. Antimicrob Agents Chemother 2016; 60:3856–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stoesser N, Batty EM, Eyre DW et al. Predicting antimicrobial susceptibilities for Escherichia coli and Klebsiella pneumoniae isolates using whole genomic sequence data. J Antimicrob Chemother 2013; 68:2234–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pesesky MW, Hussain T, Wallace M et al. Evaluation of machine learning and rules-based approaches for predicting antimicrobial resistance profiles in gram-negative bacilli from whole genome sequence data. Front Microbiol 2016; 7:1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Freifeld AG, Bow EJ, Sepkowitz KA et al. Clinical practice guideline for the use of antimicrobial agents in neutropenic patients with cancer: 2010 update by the Infectious Diseases Society of America. Clin Infect Dis 2011; 52:e56–93. [DOI] [PubMed] [Google Scholar]
- 13. Blennow O, Ljungman P. The challenge of antibiotic resistance in haematology patients. Br J Haematol 2016; 172:497–511. [DOI] [PubMed] [Google Scholar]
- 4. Poole K. Pseudomonas aeruginosa: resistance to the max. Front Microbiol 2011; 2:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 2009; 22:582–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guh AY, Bulens SN, Mu Y et al. Epidemiology of carbapenem-resistant Enterobacteriaceae in 7 US communities, 2012–2013. JAMA 2015; 314:1479–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jia B, Raphenya AR, Alcock B et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 2017; 45:D566–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu B, Pop M. ARDB—antibiotic resistance genes database. Nucleic Acids Res 2009; 37:D443–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bush K, Jacoby GA. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother 2010; 54:969–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guérin F, Isnard C, Cattoir V, Giard JC. Complex regulation pathways of AmpC-mediated β-lactam resistance in Enterobacter cloacae complex. Antimicrob Agents Chemother 2015; 59:7753–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Babouee Flury B, Ellington MJ, Hopkins KL et al. Association of novel nonsynonymous single nucleotide polymorphisms in ampD with cephalosporin resistance and phylogenetic variations in ampC, ampR, ompF, and ompC in Enterobacter cloacae isolates that are highly resistant to carbapenems. Antimicrob Agents Chemother 2016; 60:2383–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jacoby GA. AmpC beta-lactamases. Clin Microbiol Rev 2009; 22:161–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bradford PA, Urban C, Jaiswal A et al. SHV-7, a novel cefotaxime-hydrolyzing beta-lactamase, identified in Escherichia coli isolates from hospitalized nursing home patients. Antimicrob Agents Chemother 1995; 39:899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Naas T, Poirel L, Nordmann P. Minor extended-spectrum beta-lactamases. Clin Microbiol Infect 2008; 14(suppl 1):42–52. [DOI] [PubMed] [Google Scholar]
- 25. Zhang Y, Jiang X, Wang Y et al. Contribution of β-lactamases and porin proteins OmpK35 and OmpK36 to carbapenem resistance in clinical isolates of KPC-2-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 2014; 58:1214–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goessens WH, van der Bij AK, van Boxtel R et al. Antibiotic trapping by plasmid-encoded CMY-2 β-lactamase combined with reduced outer membrane permeability as a mechanism of carbapenem resistance in Escherichia coli. Antimicrob Agents Chemother 2013; 57:3941–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hentschke M, Kotsakis SD, Wolters M, Heisig P, Miriagou V, Aepfelbacher M. CMY-42, a novel plasmid-mediated CMY-2 variant AmpC beta-lactamase. Microb Drug Resist 2011; 17:165–9. [DOI] [PubMed] [Google Scholar]
- 28. Queenan AM, Shang W, Bush K, Flamm RK. Differential selection of single-step AmpC or efflux mutants of Pseudomonas aeruginosa by using cefepime, ceftazidime, or ceftobiprole. Antimicrob Agents Chemother 2010; 54:4092–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leisenring W, Alonzo T, Pepe MS. Comparisons of predictive values of binary medical diagnostic tests for paired designs. Biometrics 2000; 56:345–51. [DOI] [PubMed] [Google Scholar]
- 30. Mathers AJ, Peirano G, Pitout JD. Escherichia coli ST131: the quintessential example of an international multiresistant high-risk clone. Adv Appl Microbiol 2015; 90:109–54. [DOI] [PubMed] [Google Scholar]
- 31. Bauer KA, Perez KK, Forrest GN, Goff DA. Review of rapid diagnostic tests used by antimicrobial stewardship programs. Clin Infect Dis 2014; 59(suppl 3):S134–45. [DOI] [PubMed] [Google Scholar]
- 32. Fournier PE, Drancourt M, Colson P, Rolain JM, La Scola B, Raoult D. Modern clinical microbiology: new challenges and solutions. Nat Rev Microbiol 2013; 11:574–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rhodes NJ, Richardson CL, Heraty R et al. Unacceptably high error rates in Vitek 2 testing of cefepime susceptibility in extended-spectrum-β-lactamase-producing Escherichia coli. Antimicrob Agents Chemother 2014; 58:3757–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Salimnia H, Fairfax MR, Lephart PR et al. Evaluation of the FilmArray blood culture identification panel: results of a multicenter controlled trial. J Clin Microbiol 2016; 54:687–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dunne WM Jr, Westblade LF, Ford B. Next-generation and whole-genome sequencing in the diagnostic clinical microbiology laboratory. Eur J Clin Microbiol Infect Dis 2012; 31:1719–26. [DOI] [PubMed] [Google Scholar]
- 36. McDermott PF, Tyson GH, Kabera C et al. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob Agents Chemother 2016; 60:5515–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhao S, Tyson GH, Chen Y et al. Whole-genome sequencing analysis accurately predicts antimicrobial resistance phenotypes in Campylobacter spp. Appl Environ Microbiol 2015; 82:459–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gordon NC, Price JR, Cole K et al. Prediction of Staphylococcus aureus antimicrobial resistance by whole-genome sequencing. J Clin Microbiol 2014; 52:1182–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thomson KS, Cornish NE, Hong SG, Hemrick K, Herdt C, Moland ES. Comparison of Phoenix and VITEK 2 extended-spectrum-beta-lactamase detection tests for analysis of Escherichia coli and Klebsiella isolates with well-characterized beta-lactamases. J Clin Microbiol 2007; 45:2380–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Howard C, van Daal A, Kelly G, Schooneveldt J, Nimmo G, Giffard PM. Identification and minisequencing-based discrimination of SHV beta-lactamases in nosocomial infection-associated Klebsiella pneumoniae in Brisbane, Australia. Antimicrob Agents Chemother 2002; 46:659–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.