The appropriate application of machine learning to data in healthcare has the potential to transform patient risk stratification for infectious diseases. We present an introduction to machine learning basics for the healthcare epidemiologist.
Keywords: machine learning, patient risk stratification, healthcare epidemiologist, data-driven, computation
Abstract
The increasing availability of electronic health data presents a major opportunity in healthcare for both discovery and practical applications to improve healthcare. However, for healthcare epidemiologists to best use these data, computational techniques that can handle large complex datasets are required. Machine learning (ML), the study of tools and methods for identifying patterns in data, can help. The appropriate application of ML to these data promises to transform patient risk stratification broadly in the field of medicine and especially in infectious diseases. This, in turn, could lead to targeted interventions that reduce the spread of healthcare-associated pathogens. In this review, we begin with an introduction to the basics of ML. We then move on to discuss how ML can transform healthcare epidemiology, providing examples of successful applications. Finally, we present special considerations for those healthcare epidemiologists who want to use and apply ML.
INTRODUCTION
Increasingly, healthcare epidemiologists must process and interpret large amounts of complex data [1]. As the role of healthcare epidemiologists has expanded, so too has the pervasiveness of electronic health data [2]. The availability of large quantities of high-quality patient- and facility-level data has generated new opportunities. In particular, these data could lead to an improved understanding of risk factors for development of healthcare-associated infections (HAIs), improved patient risk stratification, and identification of pathways for intra- and interfacility spread of infectious diseases—all of which would allow for targeted prevention approaches.
In the past, a large fraction of clinical data were ignored (or not collected at all). This limitation was due to both the size and complexity of the data and the absence of techniques for collecting and storing such data. These data are frequently underused and undervalued; however, new and improved methods for data collection and storage (eg, electronic health records) provide opportunities to tackle the issue of analysis. In particular, machine learning (ML) has begun to infiltrate the clinical literature broadly. The appropriate application of ML in healthcare epidemiology (HE) promises returns on the field’s investment in data collection.
In this review, we begin by describing the basics of ML and then move on to discuss how it applies to HE, providing examples of successful research applications. Finally, we describe some of the practical considerations for design and implementation of ML applied to HE.
WHAT IS MACHINE LEARNING?
The definition of ML is broad. ML is the study of tools and methods for identifying patterns in data. These patterns can then be used to either increase our understanding of the current world (eg, identify risk factors for infection) or make predictions about the future (eg, predict who will become infected). ML draws on concepts from many fields including computer science, statistics, and optimization. At their core, almost all ML problems can be formulated as an optimization problem with respect to a dataset. In such settings, the goal is to find (or “learn” in ML parlance) a model that best explains the data (Figure 1). While there are many different types of ML, most applications fall into 1 of 3 categories: supervised, unsupervised, or reinforcement learning.
Figure 1.
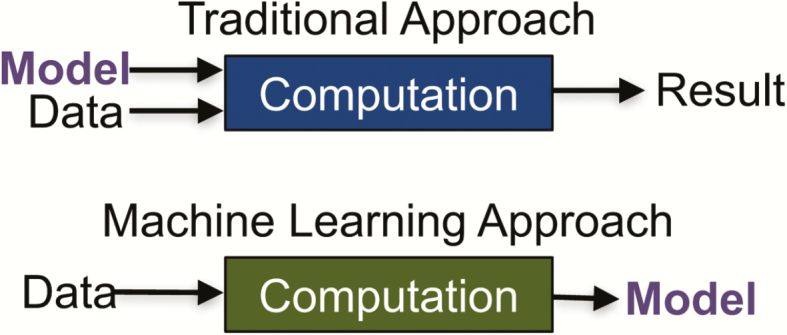
Traditional vs. machine learning (ML) approach. In a traditional approach to data analysis, one starts with the model as input to the machine. In an ML (or data-driven) approach, one starts with the data and outputs a model that can then be applied to new data.
Here, we focus on supervised learning, a setting in which the data are “labeled” according to a specific outcome of interest (eg, patients are either infected or not infected). The algorithm then learns a mapping from a set of covariates (eg, patient demographics) to the outcome. This part is performed on the training data. Once learned, this mapping can be applied to new test data either for identification or prediction tasks. For example, given a dataset of patients described by their demographics and admission details, one can try to predict the specific outcome of 30-day readmission.
Many different learning algorithms exist to accomplish this task (eg, logistic regression, decision trees, ensemble approaches, and deep neural networks). These techniques differ in their underlying objective function and constraints. While closely tied to traditional statistics, ML-based analyses often seek nonlinear relationships among hundreds or thousands of covariates. Unsurprisingly, such techniques do best when a large amount of “training” data is available (ie, when there are many examples to learn from). Here, one aims to learn a model that will generalize beyond the data one has already seen. The goal is generalization not memorization. In many cases, especially in settings with hundreds or thousands of covariates (ie, high-dimensional settings), it may be straightforward to learn a model that works well when applied to the training data but fails when applied to never-before-seen data. In such cases, the model is said to have “overfit” the training data (ie, it has simply memorized the data). Different regularization methods exist to deal with such issues and depend on the underlying learning framework. For example, in a least squares regression setting, L2 regularization is commonly applied (ie, ridge regression). These techniques push algorithms toward simpler models. The optimization loosely follows Occam’s razor, preferring simpler models over more complex ones.
As a field, ML has experienced a number of successes in recent years and continues to have an impact across several disciplines. The common thread across these disciplines is the availability of data. For example, the computer vision community (ie, the field of computer science focused on image-related tasks) has benefited tremendously from recent advances in ML. For many image recognition tasks, the performance of ML algorithms has approached or even surpassed that of humans [3, 4]. These advances have, in part, been driven by large image databases (eg, “ImageNet” consists of more than 14 million images [5]). In addition to providing training data, such databases serve as a resource against which researchers can benchmark their proposed algorithms. ML has also led to recent breakthroughs in machine translation [6, 7]. Models that take an input sequence (eg, an English sentence) and generate the target sequence (eg, the sentence translated to French). Such models are trained on tens of millions of sentence pairs. Here, the success came from training “deeper” models (ie, more complex models) capable of capturing context within sequences.
With the recent increase in availability of clinically relevant datasets, researchers have applied ML techniques to a wide range of clinical tasks [8–22], from identification/diagnostic tasks (eg, automatic classification of skin lesions or arrhythmia detection [23, 24]) to prediction tasks (eg, predicting 30-day readmissions [25]). While more research is required before we will understand the full clinical impact of this work, efforts are already underway to integrate ML tools into clinical practice [26–29]. As we continue to amass more data in HE, we will be better positioned to take advantage of these data and recent advances in ML. Below, we describe the impact ML is beginning to have on the field of infectious disease and HE more specifically.
HOW WILL ML AFFECT INFECTIOUS DISEASE (AND MORE SPECIFICALLY HE)?
The applications of ML in infectious disease are diverse and include risk stratification for specific infections (eg, specific HAIs), identifying the relative contribution of specific risk factors to overall risk, understanding pathogen–host interactions, and predicting the emergence and spread of infectious diseases. Here, we review 4 recent projects that highlight the diversity of applications in HE and infectious disease.
Predicting Risk of Nosocomial Clostridium difficile Infection (CDI) [30–33]
Despite efforts to reduce incidence, HAIs remain prevalent, in part, because we lack an effective clinical tool for accurately measuring patient risk. Along these lines, researchers have sought to develop models for predicting patient risk of CDI. ML-driven approaches can successfully leverage the entire contents of the electronic health record (EHR). These clinical data contain information regarding medications, procedures, locations, healthcare staff, lab results, vital signs, demographics, patient history, and admission details. ML techniques learn to map these data to a value that estimates the patient’s probability of CDI. Although more complex than low-dimensional tools for calculating patient risk, models that leverage the richness of the EHR can be significantly more accurate [33]. Such models, based on thousands of variables, have been extended to change over the course of an admission, capturing how risk factors change over time [34]. These time-varying models could be incorporated into an EHR system, using streaming data to generate daily risk estimates for each inpatient.
Predicting Reservoirs of Zoonotic Diseases [35]
Zoonotic diseases account for billions of human infections and millions of deaths per year globally [36]. Researchers have applied ML to datasets that contain information on rodent species that carry zoonotic pathogens [35]. Using nearly 100 predictor variables (eg, lifespan, habitat), the authors identified reservoir status with high accuracy. Furthermore, their model predicted new hyperreservoir species (ie, those identified as harboring 2 or more zoonotic pathogens). The ability to identify geographic areas with higher likelihood of harboring rodent reservoirs of new or emerging zoonoses could help direct surveillance, vector control, and research into vaccines and therapeutics.
Predicting Clinical Outcomes in Ebola Virus Disease (EVD) Infection Using Initial Clinical Symptoms [37]
Though ML is often applied to large datasets, one recent study successfully applied ML techniques to a limited clinical dataset from a small patient cohort. The authors learned a model to predict clinical outcomes in patients presenting with Ebola Virus Disease during the 2013–2016 West African epidemic. Using a publicly available de-identified dataset, they accurately predicted outcome of infection with only a few clinical symptoms and laboratory results.
Predicting Patients at Greatest Risk of Developing Septic Shock [18]
Prediction holds the promise of early intervention. In sepsis, early intervention can reduce mortality in patients who go on to develop septic shock [38]. Using publicly available data stored in the MIMIC-II Clinical Database (described more below), researchers learned to predict with high sensitivity which patients were likely to develop septic shock. Importantly, the prediction could be made at a median of more than a day prior to onset of septic shock, providing clinicians sufficient time to potentially prevent disease or mitigate its severity.
These applications of ML could, in theory, facilitate the targeting of specific interventions to high-risk groups. However, the potential positive impact of ML on the field of infectious disease goes well beyond facilitating targeted interventions. In particular, such models could be used to design more efficient clinical trials. Often, clinical trials can be underpowered and inconclusive because a small fraction of the study population experiences the outcome of interest. ML-based risk stratification models could help identify patients at several times the baseline risk, making it possible to have adequately powered efficacy results with fewer enrolled patients. ML models can also be used to help generate testable hypotheses. Although the relationships uncovered by ML models are not necessarily causal, study of the model can generate hypotheses. Further investigation of such hypotheses could then lead to new findings related to disease risk.
MACHINE LEARNING IN HE: A USER’S GUIDE—SPECIAL CONSIDERATIONS, CHALLENGES, AND PITFALLS
Technical and methodological details about performing ML analyses is beyond the scope of this review. For an in-depth introduction to ML, we refer the reader to several excellent resources [39–42]. Rather than presenting a detailed user’s guide, here, we focus on a few special considerations/requirements in the context of patient risk stratification and HE (ie, for settings in which the goal is to map patient data to a continuous value representing patient risk for a specific outcome).
It Starts with Data
Data may come from a number of sources. The examples mentioned in the section above are not specific to one particular dataset or even one data type. Researchers have successfully applied ML to clinical notes [20], physiological waveforms [10, 43], structured EHR data [44], radiologic images [45], and even unstructured data from publications [13]. Clinicians interested in using EHR data for ML may engage leadership within their institution to both obtain access to institutional data and to establish the resources to organize the data. Clinicians should not underestimate the amount of time required for this step and should also be aware that clinical insight throughout the process is essential.
Sharable Data Are Key
Shared datasets serve an important purpose by facilitating comparison of different ML approaches to specific clinical problems. Without a shared dataset, it becomes difficult to compare methods in a meaningful way.
The Data Will Be Messy
Healthcare practitioners are well aware of the extent of inconsistencies, inaccuracies, and errors present in health data, in particular, clinical notes. Often, the vast majority of such projects are dedicated to “data wrangling,” that is, data extraction and preprocessing. While the adage “garbage in garbage out” still holds, no amount of ML can identify relationships not present in the data. When the size of the data grows, in particular, the number of examples, it can still be possible to identify a signal, despite the presence of noise. Techniques like regularization (see above) and a held-out test set (see below) can help identify whether or not there’s enough of a signal to learn meaningful relationships.
Choose the Right Target
When choosing the target (ie, outcome of interest), it is important that one has access to accurate data regarding that target. For example, if predicting the development of CDI is the target, one must know which patients developed CDI in order to develop the model. Sometimes, it is impossible to obtain complete certainty (eg, not all laboratory tests are 100% accurate). ML techniques can, however, handle the presence of some uncertainty in the data. In addition, it is important to remember that the outcome used during training is the outcome that the model is learning to predict. For example, we may want to predict risk of CDI. However, since not all patients are tested, in reality we are predicting risk of a positive laboratory result for CDI. This distinction is subtle but important. In particular, if the hospital were to change its testing protocol, then the predictive performance of an existing model may change.
Keep a Held-Out Test Set
As mentioned above, because such analyses often deal with a large number of covariates, one can easily overfit, obtaining a model that works well on the training set but does not generalize. Thus, it is important to split one’s data into separate training and test sets (eg, 80% of the data is used for training and 20% for testing). Use the training set for model selection, and hold the test set aside for final model evaluation. One may split the data multiple times at random or choose a temporal split (eg, training on data from 2010–2014, testing on data from 2015). By splitting the data temporally, one can estimate how changes over time may affect predictive performance.
Beware of Data Leakage
Beware of results that are too good to be true. In cases where the discriminative power is well beyond that of humans, there is often some form of “data leakage.” For example, one of the covariates may accidentally encode the outcome (eg, receipt of empiric oral vancomycin probably indicates that a clinician has already diagnosed CDI). This type of potential pitfall makes it important to “look inside” the model to try to understand why it is making the predictions it is or test the model prospectively.
Good Accuracy Isn’t Enough
When evaluating predictive performance, it is important to keep in mind the clinical task of interest. For example, if the goal is to learn a model to predict daily risk of CDI, then the model should be applied daily to the test data, rather than just prior to the event of interest. In addition, both calibration (ie, how well the estimate risk maps to actual risk) and discriminative performance (ie, how well the model distinguishes high-risk from low-risk patients) are important to consider. Finally, the transparency (ie, interpretability) of one’s model can be as important as its accuracy. A black-box model that only tells a user who is at risk may be less actionable than a transparent model that tells a user why the patient is at risk. A model’s ability to explain its predictions can help identify “bugs” or data leakage. In addition, it can point researchers to testable hypotheses that have biological plausibility.
Hospital-specific Models Using Generalizable Methods
In the past, researchers have most often aimed to learn models that generalize across hospitals or healthcare settings. Such models may do well on average but can perform poorly when applied to specific institutions. This limitation is, in part, because of institutional differences in the way data are collected and stored [46]. Rather than seeking models that generalize across all hospitals, we should seek generalizable methods that can be used to generate institution-specific models. Such an approach allows institutions to train models specific to their data collection practices and patient populations.
It Takes a Team
Finally, and most importantly, applied ML in HE requires teams composed of experts from a variety of disciplines. Leadership of these teams is likely to comprise individuals with expertise in infectious disease, statistics, optimization, and computer science. For studies that use EHR data, individuals with expertise in clinical data architecture are essential to team success. Such work cannot take place in the isolation of a single department or discipline. ML experts are unlikely to make a meaningful clinical contribution using ML without close collaboration with a clinical expert. Conversely, although open-source ML tools exist, without a good understanding of the underlying algorithms, the misapplication of ML to clinical data can lead to misleading results and incorrect conclusions.
While the increased availability of data and ML tools holds the promise of improved patient outcomes, we should proceed cautiously. To date, ML has featured more prominently in research than in practice. At the point at which ML has proven efficacy in HE, additional questions will remain in translation to practice. These include training and education of healthcare epidemiologists and creation and maintenance of ML tools and applications. Barriers to implementation may be expected to vary by institution size, resources, and interest in the technology. More research is required before we will fully understand the good, the bad, and the unintended consequences of ML in HE [47].
CONCLUSIONS
ML has resulted in important contributions to a number of disciplines in recent years, including vision and natural language processing. In these fields, more complex models can take advantage of the large amount of existing training data (eg, images in vision or sentences in natural language). Similarly, we are on the verge of a major shift in HE. Through the appropriate application of ML to increasingly available electronic health data—including genomic data—healthcare epidemiologists will be able to better understand the underlying risk for acquisition of infectious diseases and transmission pathways, develop targeted interventions, and reduce HAIs. While powerful, it is important to remember that ML cannot identify relationships that are not present in the data. Moreover, ML does not replace the need for standard statistical analyses or randomized, control trials. Instead, ML can serve as a tool to augment HE’s current toolbox. Going forward, the greatest impact will come from interdisciplinary teams that work together to make sense of the data.
Notes
Acknowledgments. We acknowledge John Guttag, PhD, and David C. Hooper, MD, for thoughtful review of the manuscript, and Erin Ryan, MPH, CCRP, for assistance with manuscript preparation.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH; K01AI110524); the the Massachusetts General Hospital-Massachusetts Institute of Technology Grand Challenge to E. S. S.; and the National Science Foundation (IIS-1553146) and NIAID of NIH (U01AI124255) to J. W.
Potential conflict of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Kaye KS, Anderson DJ, Cook E et al. Guidance for infection prevention and healthcare epidemiology programs: healthcare epidemiologist skills and competencies. Infect Control Hosp Epidemiol 2015; 36:369–80. [DOI] [PubMed] [Google Scholar]
- 2. Bates DW, Saria S, Ohno-Machado L, Shah A, Escobar G. Big data in health care: using analytics to identify and manage high-risk and high-cost patients. Health Aff (Millwood) 2014; 33:1123–31. [DOI] [PubMed] [Google Scholar]
- 3. He K, Zhang X, Ren S, Sun J. Delving deep into rectifiers: surpassing human-level performance on imagenet classification. In IEEE International Conference on Computer Vision (ICCV) 2015 2015. [Google Scholar]
- 4. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In Computer Vision and Pattern Recognition (CVPR), 2016 IEEE Conference on 2016. [Google Scholar]
- 5. ImageNet 2016. [cited 2017; Available from: http://www.image-net.org/.
- 6. Zhou J, Cao Y, Wang X, Li P, Xu W.. Deep recurrent models with fast-forward connections for neural machine translation. arXiv preprint arXiv:1606.04199, 2016. [Google Scholar]
- 7. Lewis-Kraus G. The great AI awakening. In The New York Times Magazine. 2016. [Google Scholar]
- 8. Opmeer BC. Electronic health records as sources of research data. JAMA 2016; 315:201–2. [DOI] [PubMed] [Google Scholar]
- 9. Roski J, Bo-Linn GW, Andrews TA. Creating value in health care through big data: opportunities and policy implications. Health Aff (Millwood) 2014; 33:1115–22. [DOI] [PubMed] [Google Scholar]
- 10. Shoeb AH, Guttag J. Application of machine learning to epileptic seizure detection. Proceedings of the 27th International Conference on Machine Learning (ICML-10), 2010: p. 975–82. [Google Scholar]
- 11. Saria S, Rajani AK, Gould J, Koller D, Penn AA. Integration of early physiological responses predicts later illness severity in preterm infants. Sci Transl Med 2010; 2:48ra65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ghassemi M, et al. , A Multivariate Timeseries Modeling Approach to Severity of Illness Assessment and Forecasting in ICU with Sparse, Heterogeneous Clinical Data. Proceedings of the AAAI Conference on Artificial Intelligence. AAAI Conference on Artificial Intelligence, 2015 2015: p. 446–453. [PMC free article] [PubMed] [Google Scholar]
- 13. Wallace BC, Dahabreh IJ, Trikalinos TA, Barton Laws M, Wilson I, Charniak E. Identifying differences in physician communication styles with a log-linear transition component model. In Twenty-Eighhth AAAI Conference on Artificial Intelligence. 2014. [Google Scholar]
- 14. Visweswaran S, Cooper GF. Learning instance-specific predictive models. J Mach Learn Res 2010; 11:3333–69. [PMC free article] [PubMed] [Google Scholar]
- 15. Wang X, Sontag D, Wang F. Unsupervised learning of disease progression models. In 20th ACMSIGKDD International Conference on Knowledge Discovery and Data Mining 2014. [Google Scholar]
- 16. Kale DC, Gong D, Che Z et al. An examination of multivariate time series hashing with applications to health car. In IEEE International Conference on Data Mining (ICDM) 2014. p. 260–69. [Google Scholar]
- 17. Wiens J, Guttag J. Active learning applied to patient-adaptive heartbeat classification. Advances in Neural Information Processing Systems (NIPS), 2010: p. 2442–50. [Google Scholar]
- 18. Henry KE. A targeted real-time early warning score (TREWScore) for septic shock. Sci Transl Med 2015; 7:299ra122–299ra122. [DOI] [PubMed] [Google Scholar]
- 19. Wallace BC, Kuiper J, Sharma A, Zhu MB, Marshall IJ. Extracting PICO sentences from clinical trial reports using supervised distant supervision. J Mach Learn Res 2016; 17. [PMC free article] [PubMed] [Google Scholar]
- 20. Rumshisky A, Ghassemi M, Naumann T et al. Predicting early psychiatric readmission with natural language processing of narrative discharge summaries. Transl Psychiatry 2016; 6:e921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wallace BC, Kuiper J, Sharma A, Zhu MB, Marshall IJ. Extracting PICO sentences from clinical trial reports using supervised distant supervision. J Mach Learn Res 2016; 17:1–25. [PMC free article] [PubMed] [Google Scholar]
- 22. Rumshisky A, Ghassemi M, Naumann T et al. Predicting early psychiatric readmission with natural language processing of narrative discharge summaries. Transl Psychiatry 2016; 6:e921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Esteva A, Kuprel B, Novoa RA et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017; 542:115–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rajpurkar P, Hannun AY, Haghpanahi M, Bourn C, Ng AY. Cardiologist-level arrhythmia detection with convolutional neural networks. arXiv preprint arXiv:1707.01836, 2017.
- 25. Bayati M, Braverman M, Gillam M et al. Data-driven decisions for reducing readmissions for heart failure: general methodology and case study. PLoS One 2014; 9:e109264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. DeepMindHealth: Helping clinicians get patients from test to treatment, faster 2017; Available from: https://deepmind.com/applied/deepmind-health/.
- 27. Watson for Oncology. Oncology and Genomics 2017; Available from: https://www.ibm.com/watson/health/oncology-and-genomics/oncology/.
- 28. Caradigm: a GE Healthcare company 2017; Available from: https://www.caradigm.com/en-us/.
- 29. InnerEye - Assistive AI for Cancer Treatment 2017. [cited 2017; Available from: https://www.microsoft.com/en-us/research/project/medical-image-analysis/#.
- 30. Wiens J, Guttag J, Horvitz E. Patient risk stratification with time-varying parameters: A multitask learning approach. J Mach Learn Res 2016; 17:1–23. [Google Scholar]
- 31. Wiens J, Guttag J, Horvitz E. A study in transfer learning: leveraging data from multiple hospitals to enhance hospital-specific predictions. J Am Med Inform Assoc 2014; 21:699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wiens J, Horvitz E, Guttag JV. Patient risk stratification for hospital-associated c. diff as a time-series classification task. in Advances in Neural Information Processing Systems 2012. [Google Scholar]
- 33. Wiens J, Campbell WN, Franklin ES, Guttag JV, Horvitz E. Learning data-driven patient risk stratification models for Clostridium difficile. Open Forum Infect Dis 2014; 1:ofu045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wiens J, Guttag J, Horvitz E. Patient risk stratification with time-varying parameters: a multitask learning approach. J Mach Learn Res 2016; 17:2797–819. [Google Scholar]
- 35. Han BA, Schmidt JP, Bowden SE, Drake JM. Rodent reservoirs of future zoonotic diseases. Proc Natl Acad Sci U S A 2015; 112:7039–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bank TW. Zoonotic Disease Prevention and Control, One Health, and the Role of the World Bank. 2015. [Google Scholar]
- 37. Colubri A, Silver T, Fradet T, Retzepi K, Fry B, Sabeti P. Transforming clinical data into actionable prognosis models: machine-learning framework and field-deployable app to predict outcome of Ebola patients. PLoS Negl Trop Dis 2016; 10:e0004549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dellinger RP, Levy MM, Rhodes A et al. ; Surviving Sepsis Campaign Guidelines Committee including the Pediatric Subgroup Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock: 2012. Crit Care Med 2013; 41:580–637. [DOI] [PubMed] [Google Scholar]
- 39. An introduction to machine learning with scikit-learn 2016. [cited 2017; Available from: http://scikit-learn.org/stable/tutorial/basic/tutorial.html.
- 40. Pedregosa F, Varoquaux G, Gramfortet A et al. Scikit-learn: Machine learning in Python. J Mach Learn Res 2011; 12:2825–30. [Google Scholar]
- 41. In-depth introduction to machine learning in 15 hours of expert videos 2014. [cited 2017; Available from: http://www.dataschool.io/15-hours-of-expert-machine-learning-videos/.
- 42. Ng A. Machine learning [cited 2017; Online course]. Available from: https://www.coursera.org/learn/machine-learning.
- 43. Saria S, Rajani AK, Gould J, Koller D, Penn AA. Integration of early physiological responses predicts later illness severity in preterm infants. Sci Transl Med 2010; 2:48ra65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ghassemi M, Pimentel MAF, Naumann T, Brennan T, Clifton DA, Szolovits P, Feng M. A multivariate time series modeling approach to severity of illness assessment and forecasting in icu with sparse, heterogeneous clinical data. in Proceedings of the... AAAI Conference on Artificial Intelligence. AAAI Conference on Artificial Intelligence 2015. NIH Public Access. [PMC free article] [PubMed] [Google Scholar]
- 45. Ahmed B, Thesen T, Blackmon KE, Kuzniecky R, Devinsky O, Brodley CE. Decrypting cryptogenic epilepsy: semi-supervised hierarchical conditional random fields for detecting cortical lesions in MRI-negative patients. J Mach Learn Res 2016; 17:3885–9–14. [Google Scholar]
- 46. Wiens J, Guttag J, Horvitz E. A study in transfer learning: leveraging data from multiple hospitals to enhance hospital-specific predictions. J Am Med Inform Assoc 2014; 21:699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cabitza F, Rasoini R, Gensini G. Unintended consequences of machine learning in medicine. JAMA 2017; 318:517–8. [DOI] [PubMed] [Google Scholar]


