Hepatitis C viral load testing using dried blood spots reliably identifies candidates for treatment and is easily incorporated into large-scale surveys, including those conducted in resource-limited settings such as the Democratic Republic of the Congo.
Keywords: DRC, HCV, Africa, m2000, RNA
Abstract
Background
Efficient viral load testing is needed for hepatitis C (HCV) surveillance and diagnosis. HCV viral load testing using dried blood spots (DBSs), made with a single drop of finger-prick whole blood on filter paper, is a promising alternative to traditional serum- or plasma-based approaches.
Methods
We adapted the Abbott Molecular m2000 instrument for high-throughput HCV viremia testing using DBSs with simple specimen processing and applied these methods to estimate the national burden of infection in the Democratic Republic of the Congo (DRC). We tested DBSs collected during the 2013–2014 DRC Demographic and Health Survey, including 1309 adults ≥40 years of age. HCV-positive samples underwent targeted sequencing, genotyping, and phylogenetic analyses.
Results
This high-throughput screening approach reliably identified HCV RNA extracted from DBSs prepared using whole blood, with a 95% limit of detection of 1196 (95% confidence interval [CI], 866–2280) IU/mL for individual 6-mm punches and 494 (95% CI, 372–1228) IU/mL for larger 12-mm punches. Fifteen infections were identified among samples from the DRC Demographic and Health Survey; the weighted country-wide prevalence of HCV viremia was 0.9% (95% CI, 0.3%–1.6%) among adults ≥40 years of age and 0.7% (95% CI, .6%–.8%) among human immunodeficiency virus–infected subjects. All successfully genotyped cases were due to genotype 4 infection.
Conclusions
DBS-based HCV testing represents a useful tool for the diagnosis and surveillance of HCV viremia and can easily be incorporated into specimen referral systems. Among adults ≥40 years of age in the DRC, 100000–200000 may have active infection and be eligible for treatment.
Recent evidence suggests that dried blood spots (DBSs) can be used confidently for hepatitis C (HCV) viral load testing in the laboratory, but they have not been field tested in large-scale surveys [1, 2]. DBSs are prepared with whole blood from a finger prick in the field and can be stored at room temperature for months with only minimal degradation of HCV RNA [3]. DBSs are easily incorporated into specimen referral networks and offer significant advantages over traditional venous blood–based approaches, which typically require venipuncture, centrifugation, and a cold chain [4]. Such costly sample processing requirements are not feasible in large-scale surveys, especially in resource-limited settings.
In developing countries such as the Democratic Republic of the Congo (DRC), HCV infections frequently go undiagnosed and untreated. The burden of HCV in sub-Saharan Africa has not been well characterized, despite estimates indicating that the region harbors some of the highest viral hepatitis rates in the world [5]. One study of elderly inhabitants of Kinshasa found that nearly 26% were HCV seropositive and 14% were viremic, although prevalence rates are likely lower in younger age groups [6].
DBSs are commonly collected in sub-Saharan Africa during large-scale health surveys, such as Demographic and Health Surveys (DHSs). More than 27000 subjects were enrolled in the 2013–2014 DRC DHS, which included DBS collection from a nationally representative sample for the purpose of human immunodeficiency virus (HIV) and vaccine-related serological testing and malaria molecular testing [7]. However, these DBS samples also afforded the opportunity to delineate the extent and characteristics of HCV infection throughout the country.
Using DBSs, we employed a commercially available, high-throughput testing platform that is widely deployed in African reference laboratories to detect HCV viremia. We subsequently performed targeted sequencing and phylogenetic analyses of HCV RNA extracted from these DBSs to define prevalent genotypes and explore their evolutionary history. Our results demonstrate the utility of DBS-based HCV viral load testing and genotyping in resource-limited settings such as the DRC for identifying individuals who may benefit from treatment with new, highly effective direct-acting antivirals.
MATERIALS AND METHODS
Study Population
We randomly selected 25% of adults enrolled in the 2013–2014 DRC DHS from each of the DRC’s 11 provinces who were at least 40 years of age and provided DBS samples, with sampling stratified by province. We purposefully oversampled HIV-infected subjects, including 50% of all HIV-infected adults who were at least 40 years of age. Additionally, we subsequently performed focused regional testing on adults from Kasai Occidental and Kasai Oriental Provinces, the region with highest HCV viremia prevalence during initial testing. Because provincial borders were redrawn shortly after completion of the DHS, data analysis was conducted using the 26 newly drawn provinces adopted in 2015. DBS samples were collected in the field according to DHS protocol, transported at room temperature to the DRC’s National AIDS Control Program national reference laboratory in Kinshasa, and stored at –80°C as individual 6-mm punches until further testing [8]. This study was approved by the Kinshasa School of Public Health and the University of North Carolina institutional review boards.
Validation of RNA Extraction and Hepatitis C Viral Load Testing Methods
We used mock DBSs to determine the performance characteristics of the Abbott m2000 RealTime System (Abbott Laboratories, Abbott Park, Illinois), the mSample Preparation System (m2000sp) for RNA extraction, and the RealTime HCV Viral Load assay for quantitative reverse-transcription polymerase chain reaction (qRT-PCR) viral load testing. First, we used a HCV genotype 2 plasma sample diluted in whole blood to make DBSs containing target viral load concentrations of 250000, 25000, 5000, 2000, 1000, 500, and 250 international units (IU)/mL. Seventy-microliter aliquots of each dilution were placed onto Whatman filter paper (GE Healthcare, Little Chalfont, United Kingdom) and allowed to dry at room temperature overnight. Experiments to evaluate the reproducibility of quantitative viral load results and the assay’s limits of detection (LOD) were performed using multiple replicates.
To simulate samples available for testing through the 2013–2014 DRC DHS, we initially tested individual 6-mm punches from each DBS. After the 6-mm DBS punches were incubated in 0.5 mL Abbott mSample Preparation System DBS Buffer at 55°C for 30 minutes, 400 µL of eluate was transferred into the m2000sp reaction vessel and processed using the RealTime HCV 0.2 mL assay according to the manufacturer’s instructions for RNA extraction, amplification, and detection. In addition, we evaluated the assay’s performance using larger 12-mm punches. These larger samples were directly loaded into the m2000sp reaction vessel, incubated in 1.3 mL DBS buffer at room temperature for 30 minutes, and processed using the RealTime HCV 1 mL DBS HCV RNA open mode protocol for sample extraction, amplification, and detection. Viral load results obtained from the DBSs were compared to known viral loads in whole blood.
Field Samples: Hepatitis C Viral Load Testing and Genotyping
Individual 6-mm punches from DBSs collected during the DHS underwent elution, RNA extraction, and HCV qRT-PCR as described above. For positive samples, we attempted genotyping after manual RNA extraction from a new 6-mm punch of each positive sample using TRIzol (Thermo Fisher Scientific, Waltham, Massachusetts) and chloroform. Each punch was incubated in 400 µL of TRIzol reagent for 60 minutes at room temperature with agitation and subsequently processed according to the manufacturer’s instructions for RNA extraction. For an additional subset of samples selected from the Kasai region, only manual RNA extraction using leftover sample stored at –70°C in DBS buffer from initial viral load testing was attempted. For this subset, we incubated 250 µL of each DBS buffer sample with 750 µL of TRIzol for 5 minutes at room temperature with agitation, followed by RNA extraction according to the manufacturer’s instructions. Immediately after isolating RNA from the DBS punches or DBS buffer, we generated complementary DNA (cDNA) using random hexamer primers and SuperScript IV Reverse Transcriptase (Thermo Fisher Scientific) following the manufacturer’s protocol, with inclusion of RNaseOut. Twenty microliters of cDNA was generated for each sample and stored at –20°C prior to PCR for HCV gene amplification.
We then genotyped samples by characterizing the 5ʹ-untranslated region (5ʹ-UTR), core/E1, and NS5B regions of the HCV genome using PCR and Sanger sequencing. These regions are regularly used for HCV genotyping and represent areas of low, medium, and high divergence between genotypes, respectively [9]. Primers and reaction conditions are described in Supplementary Table 1. For nested PCR reactions, first-round products were diluted 100-fold in nuclease-free water prior to inclusion in second-round reactions. For samples that failed to amplify NS5B using 2 µL of cDNA template, assays were repeated using 5 µL of cDNA template. Amplicons were visualized using 1% agarose gel electrophoresis prior to sequencing.
Amplicons underwent bi-directional Sanger sequencing at Eton Biosciences (Research Triangle Park, North Carolina). Raw sequence reads were first processed using Sequencher 5.4 (Gene Codes Corporation, Ann Arbor, Michigan). Sequences were trimmed to yield reads in which the first and last 10 bases contained <1 base with confidence values <25. Contigs were generated by aligning forward and reverse sequences from a given sample and resolving discrepancies by visual inspection of chromatograms. Contigs were imported into MEGA 7.0, along with additional reference genomes downloaded from GenBank, for phylogenetic analysis [10, 11]. Reference genome phylogenies were generated using HCV coding regions only (aligned by codon), and sequences were aligned using MUSCLE [12]. Phylogenies were generated in MEGA using a general time reversible model with a discrete gamma distribution and assuming that a proportion of sites are evolutionarily invariable. All trees were bootstrapped 100 times. Trees were visualized in R (R Core Team, Vienna, Austria) using the APE package [13]. Accession numbers for sequences uploaded to GenBank are as follows: KY793121–KY793148.
Data Analysis
We used probit analysis to assess the limits of detection of the Abbott platform and generate confidence intervals (CIs). We calculated the national and provincial prevalence of HCV viremia using sample weights employed during the DHS to account for complex sampling design [14]. Logistic regression models were used to test hypotheses and generate Wald P values. HCV prevalence rates by province were mapped using ArcGIS version 10.3 (ESRI, Redlands, California) and analyzed for spatial clustering by DHS cluster using the Moran I statistic. Statistical analyses were performed using SAS 9.4 software (SAS Institute, Cary, North Carolina), except for probit analysis, which was conducted using version 9.3.
RESULTS
Initial testing confirmed that the m2000 system can be applied to detect HCV infection using DBS samples. During testing of thirty-nine 6-mm DBS punches made using whole blood containing RNA concentrations above the RealTime HCV Viral Load assay’s limits of quantification—250000 (5.4 log10) IU/mL, 25000 (4.4 log10) IU/mL, and 5000 (3.7 log10) IU/mL of HCV RNA—the RealTime HCV assay quantified all DBS replicates with minimal variation between replicates (mean 3.4 log10 IU/mL, 2.6 log10 IU/mL, and 1.8 log10 IU/mL; coefficient of variation 1.8%, 5.8%, and 5.4%, respectively; Supplementary Table 2). Among these samples, quantitative viral loads from individual 6-mm DBS punches were 1.8–2.0 log10 lower than the known viral load in whole blood. Testing of 122 individual 6-mm punches from DBSs made using whole blood containing 250–5000 (2.4–3.7 log10) IU/mL revealed a 95% LOD of 1196 (95% CI, 866–2280) IU/mL; Table 1). Subsequent testing of 139 larger 12-mm punches revealed a 95% LOD of 494 (95% CI, 372–1228) IU/mL (Table 1). Having confirmed that the assay reliably detected HCV infections using DBSs made from whole blood, we then applied these methods to estimate the national and provincial prevalence of HCV viremia using DBSs collected during the 2013–2014 DRC DHS.
Table 1.
Limits of Detection Testing of the RealTime Hepatitis C Virus (HCV) Viral Load Assay Using RNA Extracted From Circular Punches of Mock Dried Blood Spots Made With Whole Blood Containing Various Target Concentrations of HCV RNA
| Target Concentration, IU/mL Whole Blood | 6-mm DBS Punch | 12-mm DBS Punch | ||||
|---|---|---|---|---|---|---|
| Tested, No. | Detected, No. | % | Tested, No. | Detected, No. | % | |
| 5000 | 15 | 15 | 100 | 31 | 31 | 100 |
| 2000 | 19 | 19 | 100 | 16 | 16 | 100 |
| 1000 | 30 | 27 | 90 | 40 | 40 | 100 |
| 500 | 30 | 21 | 70 | 33 | 31 | 94 |
| 250 | 28 | 9 | 32 | 28 | 21 | 75 |
Abbreviations: DBS, dried blood spot; IU, international units.
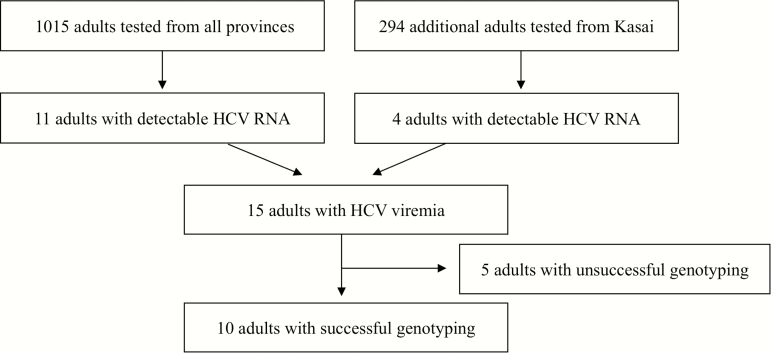
We initially selected 1015 of the 3939 adults who were at least 40 years of age and had DBSs available for testing, including 19 of 38 HIV-infected subjects (Figure 1). Subsequently, focused regional testing was also performed on 294 additional adults from Kasai Occidental and Kasai Oriental provinces, the region with highest HCV viremia prevalence during initial testing.
Figure 1.
Study population and hepatitis C virus (HCV) testing results, including subsequent testing of additional samples selected from Kasai Orientale and Kasai Occidental provinces after initial testing.
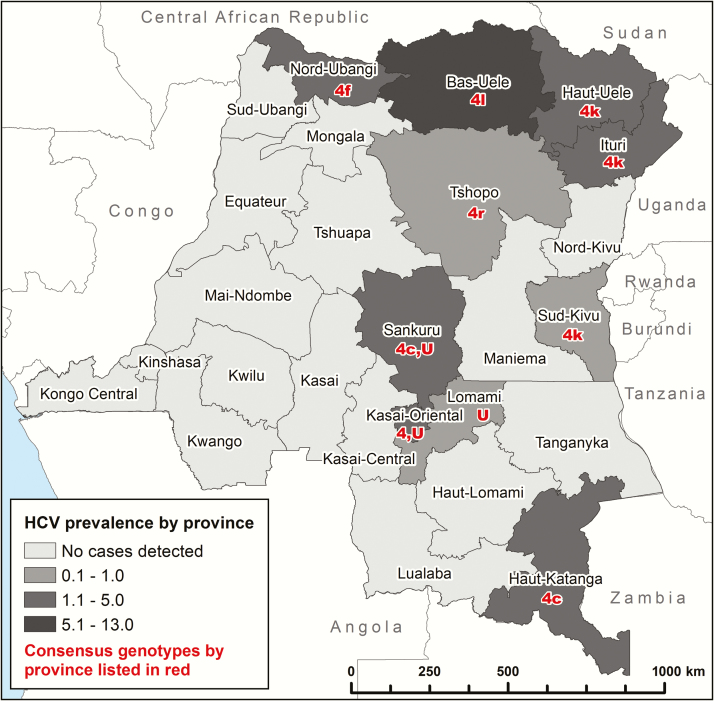
We identified 15 cases of HCV viremia among 1309 subjects tested, representing a national weighted prevalence of 0.9% (95% CI, .3%–1.6%) among adults 40–55 years of age (Table 2). HCV viremic subjects were older than HCV-negative subjects, but neither sex, hemoglobin level, HIV status, urban residence, nor wealth index were risk factors for HCV viremia. One case of HIV/HCV coinfection was observed, representing a 0.7% (95% CI, .6%–.8%) weighted national prevalence of HCV viremia among HIV-infected subjects or 4.7% of the 21 HIV-infected subjects tested. While initial results suggested the possibility of geographic clustering in the central Kasai Orientale and Kasai Occidental provinces, analysis after testing of additional samples and incorporation of sample weights to account for complex survey design did not reveal significant spatial clustering (Moran I = 0.016, P = .08; Figure 2 and Supplementary Table 3).
Table 2.
Characteristics of Adults With Hepatitis C Virus Viremia
| Characteristica | All Subjects | HCV RNA Positive | HCV RNA Negative | P Value |
|---|---|---|---|---|
| No. | 1309 | 15 | 1294 | … |
| Prevalence, % (95% CI) | … | 0.9 (.3–1.6) | 99.1 (98.4–99.7) | … |
| Age, y, median (IQR) | 44.9 (41.9–48.6) | 49.0 (43.9–55.7) | 44.9 (41.9–48.5) | .02 |
| Age strata, y, No. (%) | .01 | |||
| 40–44 | 518 (43.6) | 3 (13.7) | 515 (43.9) | |
| 45–49 | 471 (34.7) | 5 (36.2) | 466 (34.7) | |
| 50–54 | 174 (12.0) | 2 (9.2) | 172 (12.0) | |
| ≥55 | 146 (9.7) | 5 (40.9) | 141 (9.4) | |
| Male sex, No. (%) | 798 (60.4) | 12 (74.6) | 786 (60.2) | .41 |
| HIV infection, No. (% [95% CI]) | 21 (1.5) | 1 (1.1 [.8–1.4]) | 20 (1.5 [.6–2.5]) | .74 |
| Hemoglobin,b mean, g/dL (95% CI) | 13.4 | 13.5 (13.1–13.8) | 13.4 (13.3–13.5) | .88 |
| Urban place of residence, No. (% [95% CI]) | 390 (30.0) | 4 (32.6 [21.2–43.9]) | 386 (29.9 [25.5–34.3]) | .85 |
| Wealth index quintile, mean (95% CI)c | 3.0 | 3.0 (2.4–3.6) | 2.9 (2.8–3.1) | .87 |
Abbreviations: CI, confidence interval; HCV, hepatitis C virus; HIV, human immunodeficiency virus; IQR, interquartile range.
aAll summary statistics are weighted to account for complex survey design. Sample sizes are unweighted. Percentages reflect the weighted proportion of subjects (all, HCV RNA positive, or HCV RNA negative) with the specified characteristic.
bAltitude-adjusted.
cDemographic and Health Survey–calculated wealth index, where 1 = poorest and 5 = richest compared to other participants [25].
Figure 2.
Hepatitis C virus (HCV) viremia prevalence by province, stratified by the weighted percent of the population with HCV viremia. The number in red corresponds to the genotype and the letters following the number correspond to the subtype. Abbreviation: U, unknown.
We successfully genotyped 10 of the 15 subjects. The 5 samples with unknown genotype had low quantitative HCV viral loads (Supplementary Table 4), ranging from detectable (<1.48 log10) to 2.76 log10 IU/mL from a single 6-mm DBS punch. As expected, manual TRIzol/chloroform RNA extraction directly from DBSs was more effective than extraction from leftover DBS buffer. Using manual extraction followed by 2-step RT-PCR and gel electrophoresis, we visualized HCV amplicons from 9 of 11 (81.8%) DBSs and 1 of 4 (25%) leftover DBS buffer aliquots, a difference at least partially explained by the effect of sample dilution during m2000 processing.
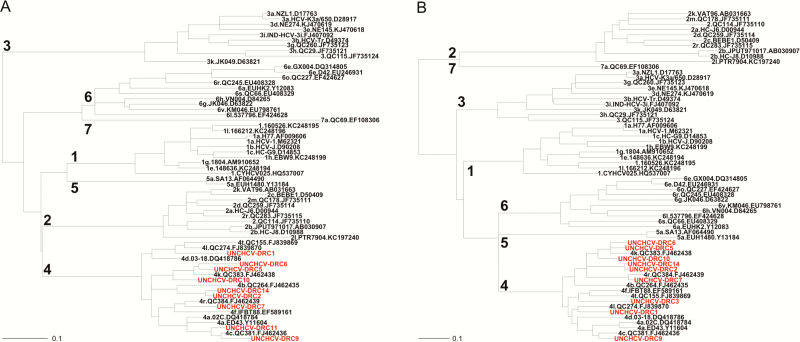
Phylogenetic analyses confirmed that genotype 4 infections are most common in the DRC and revealed what may be a novel subtype. Genotype 4k was the most common in this cohort (30%), followed by 4c (20%), 4f (10%), 4l (10%), and 4r (10%). One sample produced contradictory genotyping results—the 5ʹ-UTR amplicon clustered with a reference 4a sequence, whereas the NS5B amplicon clustered with a reference 4l. Because the NS5B region of HCV is more phylogenetically informative, we assigned it to subtype 4l but could not definitively exclude the possibility of intersubtype recombination [15]. Samples 2 and 14 show high sequence homology but fail to cluster with any known subtype. Because there was insufficient sample for sequencing of their entire coding region; however, assignment of a new subtype designation according to recently proposed definitions was not possible [16].
Sequences clustered with known HCV genotype 4 subtypes (Figure 3), with the exception of samples 2 and 14 as described above. Additionally, NS5B sequences clustered with Central African sequences (Supplementary Figure 1), suggesting an autochthonous origin of genotype 4 HCV as previously proposed [17].
Figure 3.
Phylogenetic analyses. Samples from the current study (red) clustered with reference sequences (black) from known genotype 4 subtypes for both the hepatitis C virus NS5B (A) and core/E1 (B) regions, except for samples 2 and 14. Reference sequence identifying information is separated by periods and includes the following (from left to right): genotype and subtype, isolate identifier, GenBank accession number.
DISCUSSION
We successfully detected and sequenced HCV RNA from DBSs collected during a national population-based survey, producing the first national HCV prevalence estimates from the DRC. High-throughput sample extraction and HCV qRT-PCR using the Abbott m2000 system provided an efficient means of HCV RNA screening with limited specimen processing, requiring only a brief incubation followed by direct analysis of the eluate. In addition, we successfully genotyped two-thirds of all HCV viremia cases.
This approach has broad utility throughout much of sub-Saharan Africa, as the required laboratory infrastructure is already established for HIV viral load testing. By employing a similar workflow to what is required for ongoing HIV projects, ministries of health in resource-limited settings can identify subjects eligible for HCV treatment. HCV RNA levels from untreated HCV-positive blood donors remain high and generally stable during longitudinal studies, indicating that the current assay’s limits of detection would permit identification of virtually all patients in need of antiviral therapy [18].
This approach allowed for the first national HCV viremia prevalence estimates for the DRC, indicating that 0.9% of adults aged 40–55 are actively viremic (95% CI, .3%–1.6%). The prevalence and provincial distribution of cases we observed is consistent with recent serological analyses of subjects with acute febrile jaundice who were referred for yellow fever testing in the DRC [19]. The prevalence of HCV viremia was similar to US estimates, where infected individuals are increasingly cured through treatment with direct-acting antivirals [20, 21]. While treatment is available in select settings in Africa through discounted pricing or generic licensing agreements, treatment remains largely unavailable in the DRC. Genotype 4 infections predominate in the DRC and are amenable to treatment with recently approved direct-acting antiviral combination regimens.
The association between older age and HCV viremia supports the hypothesis that HCV transmission rates were high in mid-20th-century DRC due to iatrogenic causes, including mass treatment campaigns involving injectable agents prior to the discovery of blood-borne viruses [6, 22]. Additionally, the predominance of genotype 4 and its subtypes is consistent with reports that suggest an autochthonous origin in Central Africa [17]. We did not observe any cases of HCV genotype 7, a recently described genotype that may have originated in Central Africa [11].
While the comprehensive survey design of the DHS allowed us to estimate the prevalence of HCV viremia, additional research is required to refine these estimates. Specifically, provinces without cases of HCV viremia should not be assumed to be free of HCV infections. Additionally, our genotyping methods maximized our ability to examine the genetic diversity of HCV using limited clinical samples but are not readily adapted for high-throughput use. We chose to use reflex TRIzol-chloroform RNA extraction and sequencing for phylogenetic analysis for 2 reasons. First, TRIzol-chloroform generally outperforms high-throughput RNA extraction approaches, typically achieving approximately 1 log10 greater RNA concentration than column-based kits in our hands. Second, sequencing was necessary to permit identification of genotype 7 cases that might be missed by commercially available assays for routine genotyping. While we expect that commercially available genotyping assays could be applied successfully to RNA extracted using automated methods, the recent release of pan-genotypic, direct-acting antivirals suggests that the role of genotyping may be less important in pretreatment testing algorithms, particularly in resource-limited environments. A simplified approach that links viremic patients to care, irrespective of genotype, is especially appealing in settings with limited laboratory capacity such as the DRC.
DBS-based testing is likely feasible using other commercially available HCV RNA assays using protocols optimized for this approach. Future studies could involve initial serological screening for HCV antibodies to reduce costs, followed by reflex RNA testing using an entire DBS rather than a single 6-mm punch to improve assay sensitivity [23]. This approach was not possible in the current study due to limited sample quantity. The advent of point-of-care HCV antibody testing could further streamline testing by allowing for real-time, targeted screening followed by reflex DBS-based RNA testing and linkage to care.
In conclusion, DBS-based viral load testing allowed us to determine the prevalence and characteristics of HCV viremia in the DRC. Based on crude 2016 population estimates, we expect that at least 100000–200000 adults ≥40 years of age are actively infected and eligible for treatment [24]. Without access to direct-acting HCV antivirals, many of these individuals will suffer the devastating effects of chronic HCV infection, including cirrhosis, hepatocellular carcinoma, and death. High-throughput screening for HCV RNA in resource-limited settings can be achieved using existing laboratory infrastructure, allowing for the identification of individuals eligible for curative treatment.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Author contributions. Both G. C. and V. H. played a role in the study design and data analysis.
Acknowledgments. The authors acknowledge the administrators of the 2013–2014 Democratic Republic of the Congo Demographic and Health Survey and the thousands of subjects who participated, without whom this study would not have been possible. The authors also thank Stephanie Doctor, Amy Whitesell, and Alexandra Wilcox for help in the laboratory, Jonathan Juliano for counsel on the study design, and Donald Murphy for helpful advice and for sharing primer sequences.
Financial support. This work was funded by the National Institute of Allergy and Infectious Diseases (award numbers 5T32AI007151 to J. B. P. and 5R01AI107949 to S. R. M.); the National Science Foundation (award number BCS-1339949 to C. K. and M. E.); and Abbott Laboratories, which performed the initial HCV viremia screening assays and LOD testing.
Potential conflicts of interest. G. C. and V. H. are employees and shareholders of Abbott Laboratories. S. M. L. has received research funding from Gilead Sciences and Merck, consulting fees from Merck, and royalties from AbbVie. D. R. M. has received grant support from AbbVie and Gilead Sciences. M. W. F. has received research grants from AbbVie, Bristol-Myers Squibb, Gilead Sciences, and Merck, and serves as a consultant to AbbVie, Bristol-Myers Squibb, Gilead Sciences, Merck, and TARGET PharmaSolutions. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Soulier A, Poiteau L, Rosa I et al. Dried blood spots: a tool to ensure broad access to hepatitis C screening, diagnosis, and treatment monitoring. J Infect Dis 2016; 213:1087–95. [DOI] [PubMed] [Google Scholar]
- 2. Mössner BK, Staugaard B, Jensen J, Lillevang ST, Christensen PB, Holm DK. Dried blood spots, valid screening for viral hepatitis and human immunodeficiency virus in real-life. World J Gastroenterol 2016; 22:7604–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bennett S, Gunson RN, McAllister GE et al. Detection of hepatitis C virus RNA in dried blood spots. J Clin Virol 2012; 54:106–9. [DOI] [PubMed] [Google Scholar]
- 4. Fonjungo PN, Alemnji GA, Kebede Y et al. Combatting global infectious diseases: a network effect of specimen referral systems. Clin Infect Dis 2017; 64:796–803. [DOI] [PubMed] [Google Scholar]
- 5. World Health Organization. Guidelines for the screening, care and treatment of persons with chronic hepatitis C infection. Geneva, Switzerland: WHO, 2016. [PubMed] [Google Scholar]
- 6. Hogan CA, Iles J, Frost EH et al. Epidemic history and iatrogenic transmission of blood-borne viruses in mid-20th century Kinshasa. J Infect Dis 2016; 214:353–60. [DOI] [PubMed] [Google Scholar]
- 7. Ministère du Plan et Suivi de la Mise en oeuvre de la Révolution de la Modernité (MPSMRM), Ministère de la Santé Publique (MSP), and ICF International. Enquête Démographique et de Santé en République Démocratique du Congo 2013–2014. Rockville, MD: MPSMRM, MSP and ICF International, 2014. [Google Scholar]
- 8. ICF International. MEASURE DHS biomarker field manual. Calverton, MD: ICF International, 2012. [Google Scholar]
- 9. Murphy DG, Willems B, Deschênes M, Hilzenrat N, Mousseau R, Sabbah S. Use of sequence analysis of the NS5B region for routine genotyping of hepatitis C virus with reference to C/E1 and 5’ untranslated region sequences. J Clin Microbiol 2007; 45:1102–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 2016; 33:1870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Murphy DG, Sablon E, Chamberland J, Fournier E, Dandavino R, Tremblay CL. Hepatitis C virus genotype 7, a new genotype originating from central Africa. J Clin Microbiol 2015; 53:967–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004; 32:1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 2004; 20:289–90. [DOI] [PubMed] [Google Scholar]
- 14. Rutstein SO, Rojas G.. Guide to DHS statistics. Calverton, MD: Demographic and Health Surveys, 2006. [Google Scholar]
- 15. Galli A, Bukh J. Comparative analysis of the molecular mechanisms of recombination in hepatitis C virus. Trends Microbiol 2014; 22:354–64. [DOI] [PubMed] [Google Scholar]
- 16. Smith DB, Bukh J, Kuiken C et al. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology 2014; 59:318–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iles JC, Raghwani J, Harrison GL et al. Phylogeography and epidemic history of hepatitis C virus genotype 4 in Africa. Virology 2014; 464–465:233–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kuramoto IK, Moriya T, Schoening V, Holland PV. Fluctuation of serum HCV-RNA levels in untreated blood donors with chronic hepatitis C virus infection. J Viral Hepat 2002; 9:36–42. [DOI] [PubMed] [Google Scholar]
- 19. Makiala-Mandanda S, Le Gal F, Ngwaka-Matsung N et al. High prevalence and diversity of hepatitis viruses in suspected cases of yellow fever in the Democratic Republic of Congo. J Clin Microbiol 2017; 55:1299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Centers for Disease Control and Prevention. Viral hepatitis Available at: https://www.cdc.gov/hepatitis/hcv/cfaq.htm—overview. Accessed 20 January 2017.
- 21. Klevens RM, Hu DJ, Jiles R, Holmberg SD. Evolving epidemiology of hepatitis C virus in the United States. Clin Infect Dis 2012; 55(suppl 1):S3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Iles JC, Abby Harrison GL, Lyons S et al. Hepatitis C virus infections in the Democratic Republic of Congo exhibit a cohort effect. Infect Genet Evol 2013; 19:386–94. [DOI] [PubMed] [Google Scholar]
- 23. Ross RS, Stambouli O, Grüner N et al. Detection of infections with hepatitis B virus, hepatitis C virus, and human immunodeficiency virus by analyses of dried blood spots–performance characteristics of the ARCHITECT system and two commercial assays for nucleic acid amplification. Virol J 2013; 10:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Central Intelligence Agency. The World Factbook: Democratic Republic of the Congo Available at: https://www.cia.gov/library/publications/the-world-factbook/geos/cg.html. Accessed 9 January 2017.
- 25. ICF International. Wealth index construction Available at: http://www.dhsprogram.com/topics/wealth-index/Wealth-Index-Construction.cfm. Accessed 24 July 2017.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.