To tackle antimicrobial resistance, we need first to quantify and map its drivers. In this viewpoint, we propose a systems mapping approach to do this, supported by comprehensive data collection and novel quantification analysis.
Keywords: antimicrobial resistance, quantification, mathematical modeling, mapping
Abstract
The global threat of antimicrobial resistance (AMR) has arisen through a network of complex interacting factors. Many different sources and transmission pathways contribute to the ever-growing burden of AMR in our clinical settings. The lack of data on these mechanisms and the relative importance of different factors causing the emergence and spread of AMR hampers our global efforts to effectively manage the risks. Importantly, we have little quantitative knowledge on the relative contributions of these sources and are likely to be targeting our interventions suboptimally as a result. Here we propose a systems mapping approach to address the urgent need for reliable and timely data to strengthen the response to AMR.
Antimicrobial resistance (AMR) “poses a profound threat to human health” [1]. Policy makers, researchers, and funders have stressed the importance of developing new diagnostics and medicines, improving surveillance, and defining appropriate antimicrobial use to counter this global threat [2, 3]. Concerns on AMR are now a regular feature in the popular media, creating an impetus for politicians and policy makers to decisively address the risks. Yet, an important ingredient of an effective response has been largely overlooked: reliable and timely data to map and determine the relative contributions of AMR sources and transmission routes to overall AMR risk. Importantly, for example, we do not know what proportion of patients with infections with resistant pathogens acquired that pathogen from direct person-to-person transmission vs through the consumption of contaminated meat.
The primary drivers of AMR are thought to include suboptimal use of antimicrobial agents in hospitals, the community, and agriculture, as well as background exposure in waste water, soils, and other environmental reservoirs [3–8]. However, the extent to which these sources contribute to the development, emergence, and spread of AMR is not yet quantified [9]. Without this critical, system-wide knowledge, it is impossible to effectively optimize and target interventions.
The selection process that produces AMR occurs through exposure to antimicrobials. However, the relationship between the extent of antimicrobial exposure and the rate of AMR selection has not been quantified [10, 11]. The appearance of AMR strains in clinical environments will also be dependent on their transmission from source environments. Hence, both the sources and transmission pathways of AMR need to be identified and mapped to understand the flow of AMR to frontline clinical interfaces. For example, high antimicrobial use occurs in the agricultural environment, but we do not know how frequently this use leads to selection of AMR, if there is a dose-response relationship, or the nature and magnitude of AMR transmission from this environment into clinical settings [12]. The overall contribution of agricultural antimicrobial use to clinical AMR risk thus remains unknown. This lack of a quantified risk means that interventions to reduce antibiotic prescribing in agriculture, although logical, will have an undetermined impact on the levels of infection with AMR pathogens in clinical settings. Moreover, the lack of knowledge also hampers advocacy for any intervention in this setting.
Emerging infectious disease outbreaks, such as Ebola, and more recently the Zika virus in Brazil, demonstrate how even rare events can have catastrophic consequences for public health by overwhelming health systems that are typically designed to manage endemic, consistent, or predictable health pressures. AMR poses similar risks to health systems [1]. While multiply resistant microbial strains are likely to be rare in comparison to resistant strains that remain treatable by available and alternative compounds, the consequences of an untreatable strain overwhelming our last lines of defence would be great. As any new resistance could ultimately be the last one required for a pan-resistant strain, identifying AMR selection hotspots is critical for stemming AMR risks at the most relevant sources, while quantitative knowledge on transmission networks is central to interrupting AMR spread. With ever-limited resources, a systems approach to rank both the importance of these hotspots and the transmission pathways is required for prioritization of action or control method.
The hotspots and their relative contributions to selection and transmission are likely to vary by setting [7]. For example, countries will have different levels of direct antimicrobial exposure due to varying degrees of use of antimicrobials in agriculture [13]. Indirect factors will also vary, such as levels of sanitation, density of antimicrobial-producing pharmaceutical companies, and political will to tackle AMR (eg, with the formation of national action plans [14, 15]). Until this systems variation (both between and within countries) and then the fundamental information on the relative contribution of each of these factors are known, it will not be possible to develop policies or efficiently allocate resources to develop targeted and context-specific interventions for multiple settings.
To date, most AMR research has focused on evaluation of interventions (aimed at infection control for prevention and for antimicrobial stewardship) [16], surveillance, risk factor analysis, and strain characterisation (including identification of mechanisms of resistance and genetic determinants of AMR). Research on surveillance of resistance patterns suggest strong spatial variation in AMR and in the use of antimicrobials [17–19]. For instance, the majority of antibiotic prescriptions in the United Kingdom are in the community and yet the most clinically serious AMR infections are often hospital acquired [20]. Does this mean that reducing prescriptions in primary care would have a smaller effect on levels of infection with resistant pathogens than reducing prescriptions in hospitals? Or is it the key that drives colonization with and selection of AMR, with subsequent opportunities for endogenous infection once a host becomes hospitalized? Although links have been found across environments, for example, between outpatient prescribing and hospital resistance levels [21–23], few studies have explored their relative contributions and no studies, to our knowledge, have established which transmission routes contribute the most to the most serious infections with resistant pathogens in clinical settings. For example, although a link between travel and AMR spread has been established [24], and studies have revealed key genetic factors underlying transmission, no studies have quantified the relative contribution of travel to AMR in comparison to other factors.
FUTURE ACTION AREAS
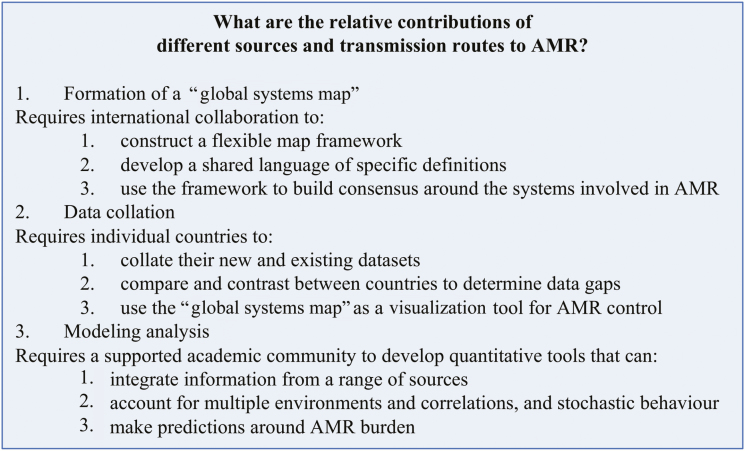
Based on these observations, we believe that there is a major gap in our understanding of AMR that requires a revolution in the analysis and quantification of the sources and transmission routes of AMR. To tackle this, we propose that a comprehensive systems mapping approach is needed, with the support of data collection and modeling. The key action points are summarized in Figure 1.
Figure 1.
Example factors influencing antimicrobial resistance selection and transmission pathways that require quantification for a more effective and efficient global response.
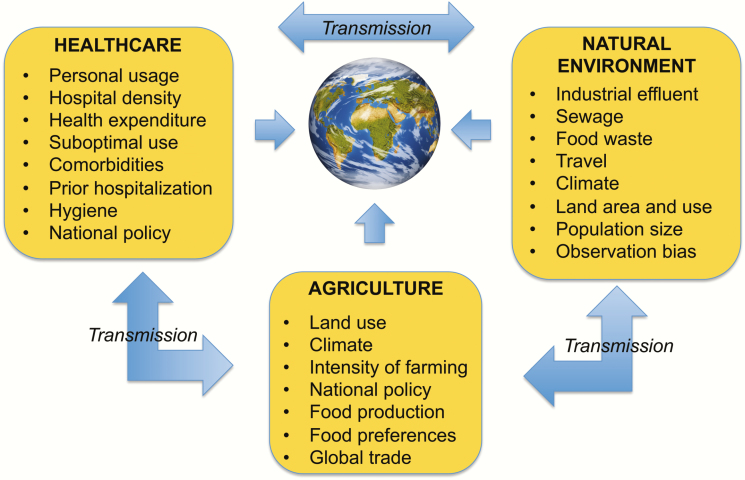
First, there is a need to establish a “global systems map” of AMR selection sources and transmission routes. Collaborative action by the global public health community is necessary to determine the relative contributions of sources and transmission routes to AMR [25, 26], including the most relevant environments and drivers at local, national, and global levels (Figure 2). While there are current efforts to identify drivers of AMR in different environments [1, 27], a comprehensive approach is lacking [3]. Research is needed to systematically map the complex network of environments and locations of selection, as well as quantifying the interplay of pathways that affect transmission (Figure 2). The formation of a “global systems map” requires international collaboration to: (1) construct a flexible map framework, perhaps within a specifically designed web-based platform, that allows for easy comparison and modification by individual countries; (2) develop a shared language of specific definitions for AMR “drivers,” “risk factors,” and “transmission pathways,” as well as for labeling environments (“sewage” or “waste water”); (3) use the framework to build consensus around the systems involved in AMR, how they differ by setting, and to continually update the systems map through conversations with all stakeholders, from patient groups to international health organizations.
Figure 2.
Stages required for determination of the relative contributions of different sources and transmission routes to antimicrobial resistance (AMR).
Using the above map, there is then, second, a need to collect and collate data to quantify relative contributions to AMR and to populate the “global systems map” with quantitative information. Currently, there is no global database that collates information on the occurrence of antimicrobial use or AMR [25]. However, building on the first AMR global surveillance report (published in 2014 [1]) as well as existing national-level clinical datasets [18, 28], the WHO has now launched the Global Antimicrobial Resistance Surveillance System (GLASS) to fulfill 1 of the 5 strategic objectives of the WHO action plan on AMR [29]. This will collect and then report AMR rates aggregated at the national level, giving information on level of resistance within clinical isolates. This global endeavor is supported by government and nongovernmental initiatives such as the Fleming Fund in the United Kingdom and the Bill & Melinda Gates Foundation.
To populate the “global systems map” critically requires countries to support these actions, but also requires further resistance data—for example, resistance levels within samples from agriculture, water, and soil. For the identification and quantification of transmission pathways, a comparison of isolates between settings using genetic distance can help identify overlapping sources [19, 30, 31]. The map also requires systems-level information on the places where antimicrobials are prescribed and transmission pathways—for example, the amount of intensive farming (such as has been mapped globally in [32]) and what amount of antimicrobials are used where (for some drugs, this has been done globally at the national level [17]). A comparison of the existing resistance environment, using, for example, composite measures such as the Drug Resistance Index [33], can then be complemented by a comparison of underlying AMR drivers and transmission routes. This stage requires national organizations to (1) collate their new and existing datasets to inform all stages of the “global systems map” for AMR; (2) compare and contrast between countries to determine data gaps and potential ways for data collection to be effectively performed, perhaps with the inclusion of sentinel sites; and (3) use the “global systems map” as a visualization tool to identify new potential areas of AMR emergence and areas where effective control has been achieved.
Third, quantification of selection sources and transmission routes will require novel analytic approaches to measure source contributions, to establish relative importance of transmission pathways, and to predict the likely impacts of interventions. These analytic approaches will need to combine cutting-edge statistical methods as well as mathematical and systems dynamics modeling, the potential contribution of which to global health is outlined in [34]. For example, mathematical models are needed that capture the movement of AMR pathogens between environments rather than only the dynamics within a single setting (such as a hospital ward). Currently, many mathematical models are only of the transmission of resistant pathogens between individuals within a hospital [35], with some, often fixed, incoming rate of precolonization with resistant pathogens. Only by allowing the latter rate to vary, by including a dynamic modeling of the processes in external settings, can our understanding of the relative contribution of selection and transmission in different settings be determined.
Statistical methods, such as multilevel modeling, will need to be adapted to consider the complexities of time-dependent bias in AMR acquisition and different risk factor profiles. The interacting nature of selection and transmission requires adjustment for correlations between statistical hierarchies that may require novel statistical formulations. This is important, as to reveal the relative contributions of different settings, correction for interaction relationships are needed to remove bias from risk profiles.
The resulting models should holistically map and integrate complex pathways and transmission systems, and account for stochastic or random behavior of AMR spread, such as outbreaks and introductions of AMR strains or genetic determinants. This would enable the models to test for the effects of potential interventions on AMR emergence and control by considering the system as a whole. Importantly, this would allow for a “One Health” approach to AMR understanding and intervention optimization. This stage requires the academic community, supported by the public health and policy community alongside cross-sectoral agencies, to work with the “global systems map” to develop new quantitative tools that can (1) integrate information from a range of sources; (2) account for multiple environments, complex correlations, and stochastic behavior; and (3) predict the impact and hence compare interventions.
With these systems modeling tools, and given sufficient data, the relative contribution of each source and transmission pathway to AMR can then be quantified (Figure 1). Only from such quantification can come the mathematical modeling predictions as to where to optimally target interventions for control.
CONCLUSIONS
A systems approach that enables comprehensive mapping of selection sources and transmission pathways in settings at a subnational, national, and global level will enable more holistic exploration and optimization of policies and interventions designed to control AMR. Collation of data and targeted generation of hypotheses, underpinned by systems modeling approaches will help identify more effective combinations of interventions across multiple settings (eg, countries, sectors) that could efficiently combat the profound global threat that AMR poses to human health and welfare.
Notes
Disclaimer. The views expressed are those of the author(s) and not necessarily those of the National Health Service, the National Institute for Health Research (NIHR), the Department of Health, or Public Health England.
Financial support. The work was supported by the NIHR Health Protection Research Unit (HPRU) in Healthcare Associated Infection and Antimicrobial Resistance at Imperial College London in partnership with Public Health England. The authors also acknowledge support from the NIHR Imperial Biomedical Research Centre provided to A. H. H.
Potential conflicts of interest. A. H. H. is Director of the NIHR HPRU in Healthcare Associated Infection and Antimicrobial Resistance at Imperial College London. She has received an honorarium for presenting at a conference sponsored by Merck (MSD Hoddesdon). All other authors report no potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. World Health Organization. Antimicrobial resistance: global report on surveillance. Geneva, Switzerland: WHO, 2014. [Google Scholar]
- 2. Berendonk TU, Manaia CM, Merlin C et al. Tackling antibiotic resistance: the environmental framework. Nat Rev Microbiol 2015; 13:310–7. [DOI] [PubMed] [Google Scholar]
- 3. Laxminarayan R, Duse A, Wattal C et al. Antibiotic resistance—the need for global solutions. Lancet Infect Dis 2013; 13:1057–98. [DOI] [PubMed] [Google Scholar]
- 4. Cantas L, Shah SQ, Cavaco LM et al. A brief multi-disciplinary review on antimicrobial resistance in medicine and its linkage to the global environmental microbiota. Front Microbiol 2013; 4:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tello A, Austin B, Telfer TC. Selective pressure of antibiotic pollution on bacteria of importance to public health. Environ Health Perspect 2012; 120:1100–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bouki C, Venieri D, Diamadopoulos E. Detection and fate of antibiotic resistant bacteria in wastewater treatment plants: a review. Ecotoxicol Environ Saf 2013; 91:1–9. [DOI] [PubMed] [Google Scholar]
- 7. Robinson TP, Bu DP, Carrique-Mas J et al. Antibiotic resistance is the quintessential One Health issue. Trans R Soc Trop Med Hyg 2016; 110:377–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Meek RW, Vyas H, Piddock LJ. Nonmedical uses of antibiotics: time to restrict their use?PLoS Biol 2015; 13:e1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. You Y, Silbergeld EK. Learning from agriculture: understanding low-dose antimicrobials as drivers of resistome expansion. Front Microbiol 2014; 5:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Austin DJ, Kristinsson KG, Anderson RM. The relationship between the volume of antimicrobial consumption in human communities and the frequency of resistance. Proc Natl Acad Sci U S A 1999; 96:1152–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Long H, Miller SF, Strauss C et al. Antibiotic treatment enhances the genome-wide mutation rate of target cells. Proc Natl Acad Sci U S A 2016; 113:E2498–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. O’Neill J. The review on antimicrobial resistance: antimicrobials in agriculture and the environment: reducing unnecessary use and waste, 2015. Available at: https://amr-review.org/Publications.html. Accessed 26 September 2017. [Google Scholar]
- 13. Van Boeckel TP, Brower C, Gilbert M et al. Global trends in antimicrobial use in food animals. Proc Natl Acad Sci U S A 2015; 112:5649–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. World Health Organization. Antimicrobial resistance: national action plans Available at: http://www.who.int/antimicrobial-resistance/national-action-plans/en/. Accessed 15 July 2017.
- 15. World Health Organization. Worldwide country situation analysis: response to antimicrobial resistance Available at: http://apps.who.int/iris/bitstream/10665/163468/1/9789241564946_eng.pdf?ua=1&ua=1. Accessed 15 July 2017.
- 16. Holmes AH, Moore LS, Sundsfjord A et al. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 2016; 387:176–87. [DOI] [PubMed] [Google Scholar]
- 17. Van Boeckel TP, Gandra S, Ashok A et al. Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. Lancet Infect Dis 2014; 14:742–50. [DOI] [PubMed] [Google Scholar]
- 18. Center for Disease Dynamics Economics and Policy. ResistanceMap Available at: http://resistancemap.cddep.org/. Accessed 15 July 2017.
- 19. European Food Safety Authority, European Centre for Disease Prevention and Control. Antimicrobial resistance in Europe Available at: http://www.efsa.europa.eu/en/interactive_pages/AMR_Report_2015. Accessed 15 July 2017.
- 20. European Centre for Disease Prevention and Control. Antimicrobial resistance and healthcare-associated infections programme Available at: https://ecdc.europa.eu/en/about-us/who-we-are/disease-programmes/antimicrobial-resistance-and-healthcare-associated. Accessed 15 July 2017.
- 21. Vernaz N, Huttner B, Muscionico D et al. Modelling the impact of antibiotic use on antibiotic-resistant Escherichia coli using population-based data from a large hospital and its surrounding community. J Antimicrob Chemother 2011; 66:928–35. [DOI] [PubMed] [Google Scholar]
- 22. Gallini A, Degris E, Desplas M et al. Influence of fluoroquinolone consumption in inpatients and outpatients on ciprofloxacin-resistant Escherichia coli in a university hospital. J Antimicrob Chemother 2010; 65:2650–7. [DOI] [PubMed] [Google Scholar]
- 23. Hicks LA, Chien YW, Taylor TH Jr, Haber M, Klugman KP. Active Bacterial Core Surveillance Team. Outpatient antibiotic prescribing and nonsusceptible Streptococcus pneumoniae in the United States, 1996–2003. Clin Infect Dis 2011; 53:631–9. [DOI] [PubMed] [Google Scholar]
- 24. MacPherson DW, Gushulak BD, Baine WB et al. Population mobility, globalization, and antimicrobial drug resistance. Emerg Infect Dis 2009; 15:1727–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Årdal C, Outterson K, Hoffman SJ et al. International cooperation to improve access to and sustain effectiveness of antimicrobials. Lancet 2016; 387:296–307. [DOI] [PubMed] [Google Scholar]
- 26. Harbarth S, Balkhy HH, Goossens H et al. Antimicrobial resistance: one world, one fight! Antimicrob Resist Infect Control 2015; 4. [Google Scholar]
- 27. UK Department of Health. Antimicrobial resistance (AMR) systems map. 2014. Available at: https://www.gov.uk/government/publications/antimicrobial-resistance-amr-systems-map. Accessed 26 September 2017. [Google Scholar]
- 28. European Centre for Disease Prevention and Control. Annual epidemiological report 2014: antimicrobial resistance and healthcare-associated infections. Stockholm: ECDC, 2015. [Google Scholar]
- 29. World Health Organization. Global antimicrobial resistance surveillance system (GLASS) Available at: http://www.who.int/antimicrobial-resistance/global-action-plan/surveillance/glass/en/. Accessed 15 July 2017.
- 30. Valentin L, Sharp H, Hille K et al. Subgrouping of ESBL-producing Escherichia coli from animal and human sources: an approach to quantify the distribution of ESBL types between different reservoirs. Int J Med Microbiol 2014; 304:805–16. [DOI] [PubMed] [Google Scholar]
- 31. Hao H, Sander P, Iqbal Z, Wang Y, Cheng G, Yuan Z. The risk of some veterinary antimicrobial agents on public health associated with antimicrobial resistance and their molecular basis. Front Microbiol 2016; 7:1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gilbert M, Conchedda G, Van Boeckel TP et al. Income disparities and the global distribution of intensively farmed chicken and pigs. PLoS One 2015; 10:e0133381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Laxminarayan R, Klugman KP. Communicating trends in resistance using a drug resistance index. BMJ Open 2011; 1:e000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Heesterbeek H, Anderson RM, Andreasen V et al. Modeling infectious disease dynamics in the complex landscape of global health. Science 2015; 347:aaa4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Opatowski L, Guillemot D, Boëlle PY, Temime L. Contribution of mathematical modeling to the fight against bacterial antibiotic resistance. Curr Opin Infect Dis 2011; 24:279–87. [DOI] [PubMed] [Google Scholar]