Summary
Dengue is a common, sometimes life-threatening disease throughout tropical Asia and the Americas. We developed a clinically intuitive algorithm, the Early Severe Dengue Identifier, to assist in the prediction of severe, life-threatening presentations.
Keywords: dengue, diagnosis, tropical infectious diseases.
Abstract
Background.
Early prediction of severe dengue could significantly assist patient triage and case management.
Methods.
We prospectively investigated 7563 children with ≤3 days of fever recruited in the outpatient departments of 6 hospitals in southern Vietnam between 2010 and 2013. The primary endpoint of interest was severe dengue (2009 World Health Organization Guidelines), and predefined risk variables were collected at the time of enrollment to enable prognostic model development.
Results.
The analysis population comprised 7544 patients, of whom 2060 (27.3%) had laboratory-confirmed dengue; nested among these were 117 (1.5%) severe cases. In the multivariate logistic model, a history of vomiting, lower platelet count, elevated aspartate aminotransferase (AST) level, positivity in the nonstructural protein 1 (NS1) rapid test, and viremia magnitude were all independently associated with severe dengue. The final prognostic model (Early Severe Dengue Identifier [ESDI]) included history of vomiting, platelet count, AST level. and NS1 rapid test status.
Conclusions.
The ESDI had acceptable performance features (area under the curve = 0.95, sensitivity 87% (95% confidence interval [CI], 80%–92%), specificity 88% (95% CI, 87%–89%), positive predictive value 10% (95% CI, 9%–12%), and negative predictive value of 99% (95% CI, 98%–100%) in the population of all 7563 enrolled children. A score chart, for routine clinical use, was derived from the prognostic model and could improve triage and management of children presenting with fever in dengue-endemic areas.
(See the Editorial Commentary by Low and Ooi on pages 664–5.)
Dengue is the most common arboviral infection of humans and a public health burden in much of tropical Asia and Latin America [1]. Seasonal epidemics, with children and young adults most affected, place enormous stress on healthcare systems in endemic countries. The burden of dengue in 10 endemic countries was revealed with precision in phase 3 vaccine trials of the now licensed Dengvaxia vaccine, in which approximately 10% of all febrile episodes in the control groups were confirmed to be dengue cases [2]. Among these dengue cases, the percentage requiring hospitalization was 19.1% in the Asian cohort and 11.1% in the Latin American cohort [2].
The pathophysiological hallmarks of severe dengue are increased capillary permeability, coagulopathy, and a hemorrhagic diathesis. For some patients this can lead to hypovolemic shock, called dengue shock syndrome (DSS), with or without clinically severe bleeding. Other rare complications can include hepatic dysfunction, central nervous system involvement, and myocarditis [1, 3]. Progress in clinical management, particularly fluid resuscitation and monitoring in intensive care, has led to a decline in the dengue case fatality rate over the last 2 decades; it is now <1% of hospitalized cases in many settings [1], but can exceed 20% if treatment is inadequate [4]. There are no specific antiviral or disease-modifying treatments for dengue despite recent efforts [5–8].
DSS typically manifests between the fourth and sixth day of illness, during the so-called critical phase [1, 9, 10]. Features such as abdominal pain, lethargy, rapidly rising hematocrit level, and persistent vomiting have anecdotally been associated with progression to DSS or other clinically severe manifestations. However, the prognostic value of these signs and symptoms, which usually occur near the time of shock, have not been formally evaluated. Thus emergency department physicians seeing a patient with 3 days of fever in whom dengue is suspected are faced with a dilemma; to send the patient home with a request that they return for follow-up or to admit them for observation as they approach the critical phase. The hospitalization of a large number of dengue cases for observation inevitably places a burden on healthcare resources and has direct and indirect costs to patients and their families.
Previous studies have explored early prognostic factors and algorithms for predicting moderate or severe dengue. However, heterogeneity in the case populations (adults vs children, hospitalized vs outpatient) and clinical definitions of “severe” dengue [11–15] makes it difficult to make broad conclusions [1, 16–18]. Additionally, most earlier prognostic studies did not report the positive and negative predictive values of the prognostic tool [11, 19, 20]. Against this backdrop, the aim of this current study was to develop a practical, prognostic tool to enable the early prognosis of severe dengue in Vietnamese children.
METHODS
Human Research Ethics
The scientific and ethical committees of all participating hospitals and the Oxford University Tropical Research Ethical Committee (OXTREC 35-10) approved the study. Parents or guardians provided written informed consent for the child to participate. The study was conducted in compliance with good clinical practice guidelines.
Study Population
This was a prospective study of children with fever presenting to the outpatient departments of collaborating hospitals in Vietnam including the Hospital for Tropical Diseases, Children’s Hospital No. 1, Children’s Hospital No. 2, Tien Giang Provincial Hospital, Dong Nai Children’s Hospital, Binh Duong Provincial Hospital, and Long An Provincial Hospital. The predefined sample size was to enroll a sufficient number of patients to detect 160 severe dengue cases (and thereby evaluate up to 16 predefined variables for their association with severe dengue), or 13500 patients in total, whichever occurred first. For funding reasons, the study stopped after enrollment of 7563 patients (and 117 severe dengue cases). The consequences of not recruiting the targeted number of severe cases was 2-fold. First, we downsized the number of predictor variables for testing so as to be compliant with the rule of thumb of having a minimum of 10 outcome events per predictor variable in the regression procedures. A relatively smaller sample size will also have generated a larger confidence interval for the estimation statistics than would otherwise have been the case. Study enrollment commenced on 1 October 2010 and finished on 31 December 2013. Children 1–15 years of age, accompanied by a parent/guardian with a mobile phone, who presented to the designated outpatient clinic with ≤72 hours of fever and in whom the attending physician believes dengue was a possible diagnosis were eligible for enrollment. Exclusion criteria were as follows: (1) the attending physician believed they were unlikely to be able to attend follow-up or (2) the attending physician believed another (nondengue) diagnosis was more likely.
Data Collection at Enrollment
Demographic details, clinical history and findings, and blood samples for hematology, biochemistry, nonstructural protein 1 (NS1) rapid test (NS1 Ag STRIP, Bio-Rad), and quantitative reverse-transcription polymerase chain reaction (qRT-PCR) were collected at the time of patient enrollment. The attending physician was provided the routine hematology results only in order not to bias the decision on the patient’s management (eg, hospitalization vs ambulatory follow-up).
Patient Follow-up and Case Definitions
All ambulatory patients were followed up by daily phone call to determine whether they were still at home or had been hospitalized since the previous call. Daily phone calls were made until the fever had resolved and the patient had returned to daily activities. In 10% of all ambulatory patients, we collected a convalescent blood sample (2 mL) on or after day 6 of illness for the purposes of diagnostic (immunoglobulin M [IgM]) serology. The selection of cases for follow-up and collection of a convalescent blood sample were by random assignment using a computer-generated randomization list. For patients who were hospitalized at any time during the 6 days after enrollment, a second blood sample was collected at the time of discharge from hospital and a case report form completed to capture data on whether the patient evolved to severe dengue according to the World Health Organization (WHO) 2009 classification scheme.
Laboratory-confirmed dengue was defined as a positive result with either one of the following tests: (1) validated qRT-PCR assay [21]; (2) NS1 enzyme-linked immunosorbent assay (ELISA) (Platelia Dengue NS1 Ag ELISA, Bio-Rad); or (3) IgM seroconversion in paired blood samples (Panbio, Brisbane, Australia). IgM seroconversion was defined as a change in test result from negative to positive in paired plasma samples with the second sample collected ≥6 days after illness onset and >2 days after the first sample. A stepwise approach to diagnostic testing was performed. First, all enrollment plasma samples were tested with a validated, qRT-PCR assay to detect dengue virus (DENV) RNA. Next, any enrollment plasma samples that were negative in the qRT-PCR assay were tested using the Platelia Dengue NS1 Ag ELISA assay and scored according to the manufacturer’s instructions. Samples with equivalent results were repeated and, if still equivocal, they were scored as negative. Next, IgM ELISA serology (Panbio) was performed according to the manufacturer’s instructions for patients who had paired plasma samples (100% of hospitalized cases and 10% of ambulatory cases) and who were negative in both the DENV qRT-PCR assay and Platelia Dengue NS1 ELISA. Patients who had negative test results for DENV qRT-PCR and NS1 ELISA and no IgM seroconversion in paired blood samples were classified as “not dengue.” Patients who had negative test results for DENV qRT-PCR and NS1 ELISA at the time of enrollment, but did not have paired samples available for serology, were classified as a “presumptive not dengue” case. For analysis, data from “not dengue” and “presumptive not dengue” cases were pooled and were categorized as “other febrile illness.”
Statistical Methods
To develop the prognostic models for severe dengue, we used logistic regression procedures with predefined clinical, hematological, and biochemical variables and NS1 rapid test status. Bayesian information criteria were used to downselect from the original full models to the most parsimonious model for practical clinical application. Model validation was examined by 2 approaches: (1) “leave-one-site-out cross-validation”—that is, repeatedly developing the algorithm on all but 1 study site and validation on the left-out study site; and (2) temporal validation with patients recruited before 15 June 2012 as the training set and patients recruited thereafter as the evaluation set [22]. The final logistic regression model was converted to a nomogram for ease of use in clinical practice. All analyses were performed with the statistical software R version 3.1.1 (R Foundation for Statistical Computing, Vienna, Austria). Significance was assigned at P < .05 for all parameters and was 2-sided.
RESULTS
Baseline Characteristics of Study Participants
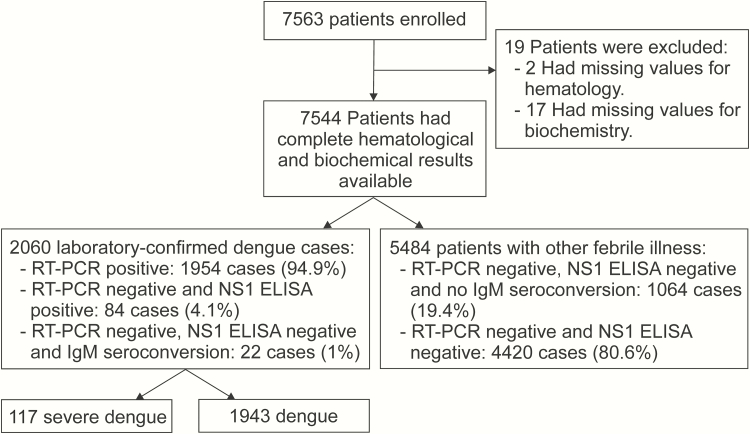
Between October 2010 and December 2013, we enrolled 7563 children with fever history of <72 hours at the outpatient departments of 7 hospitals in southern Vietnam. A description of the study profile is shown in Figure 1. There were 7544 patients with complete hematological and biochemical data, of whom 2060 (27.3%) had laboratory-confirmed dengue and 1954 (25.9%) were viremic as determined by qRT-PCR. Amongst the 2060 laboratory confirmed dengue cases, there were 117 (5.6%) cases of severe dengue (WHO 2009 Guidelines) but no deaths. Table 1 describes the baseline characteristics of the study population.
Figure 1.
Study profile with a description of the patient cohort and subgroups that were used to derive the prognostic models. Abbreviations: ELISA, enzyme-linked immunosorbent assay; IgM, immunoglobulin G; NS1, nonstructural protein 1; RT-PCR, reverse-transcription polymerase chain reaction.
Table 1.
Baseline Characteristics of Study Participants
| Characteristic | Severe Dengue (n = 117) |
Nonsevere Dengue (n = 1943) |
Other Febrile Illness (n = 5484) |
|---|---|---|---|
| Age, y | 9 (7–11) | 9 (6–11) | 5 (3–8) |
| BMI, kg/m2 | 16.9 (14.6–20.1) | 16.5 (14.7–19.0) | 15.6 (14.2–17.7) |
| Day of illness | |||
| 1 | 15 (12.8) | 426 (22.0) | 1659 (30.3) |
| 2 | 43 (36.8) | 789 (40.8) | 2372 (43.4) |
| 3 | 59 (50.4) | 718 (37.1) | 1437 (26.3) |
| Vomiting | 79 (67.5) | 804 (41.6) | 1902 (34.8) |
| Abdominal pain | 34 (29.1) | 385 (19.9) | 930 (17.0) |
| Mucosal bleeding | 9 (7.7) | 112 (5.8) | 134 (2.5) |
| WBC count, × 103 cells/µL | 3.7 (2.5–5.8) | 4.9 (3.6–6.9) | 9.0 (6.4–12.5) |
| Platelet count, × 103 cells/µL | 110 (86.5–147) | 182 (144–227) | 242 (201–291) |
| Hematocrit, % | 40 (37.8–42.3) | 38.6 (36.6–40.6) | 37.4 (35.3–39.7) |
| Albumin, g/L | 43.3 (40.5–45.1) | 43.8 (41.8–45.8) | 44.2 (42.2–46.2) |
| AST, U/L | 101 (61.5–155) | 50 (40–66) | 42 (35–49) |
| NS1 rapid test positive | 97 (82.9) | 1360 (70.4) | 37 (0.7) |
| Viremia concentration, log10 copies/mL | 7.5 (6.4–8.3) (n = 115) | 7.2 (6.0–8.2) (n = 1839) | … |
| Serotype | (n = 115) | (n = 1839) | |
| DENV-1 | 38 (32.5) | 725 (37.5) | |
| DENV-2 | 37 (31.6) | 404 (20.9) | |
| DENV-3 | 6 (5.1) | 181 (9.4) | |
| DENV-4 | 34 (29.1) | 519 (26.8) | |
| Unknown | 2 (1.7) | 104 (5.4) |
Values are presented as median (interquartile range) for continuous variables or frequency (%) for categorical variables. All laboratory results were acquired on the day of enrollment.Abbreviations: AST, aspartate aminotransferase; BMI, body mass index; DENV, dengue virus; NS1, nonstructural protein 1; WBC, white blood cell.
Prognostic Models for Early Identification of Severe Dengue Cases Among Febrile Patients
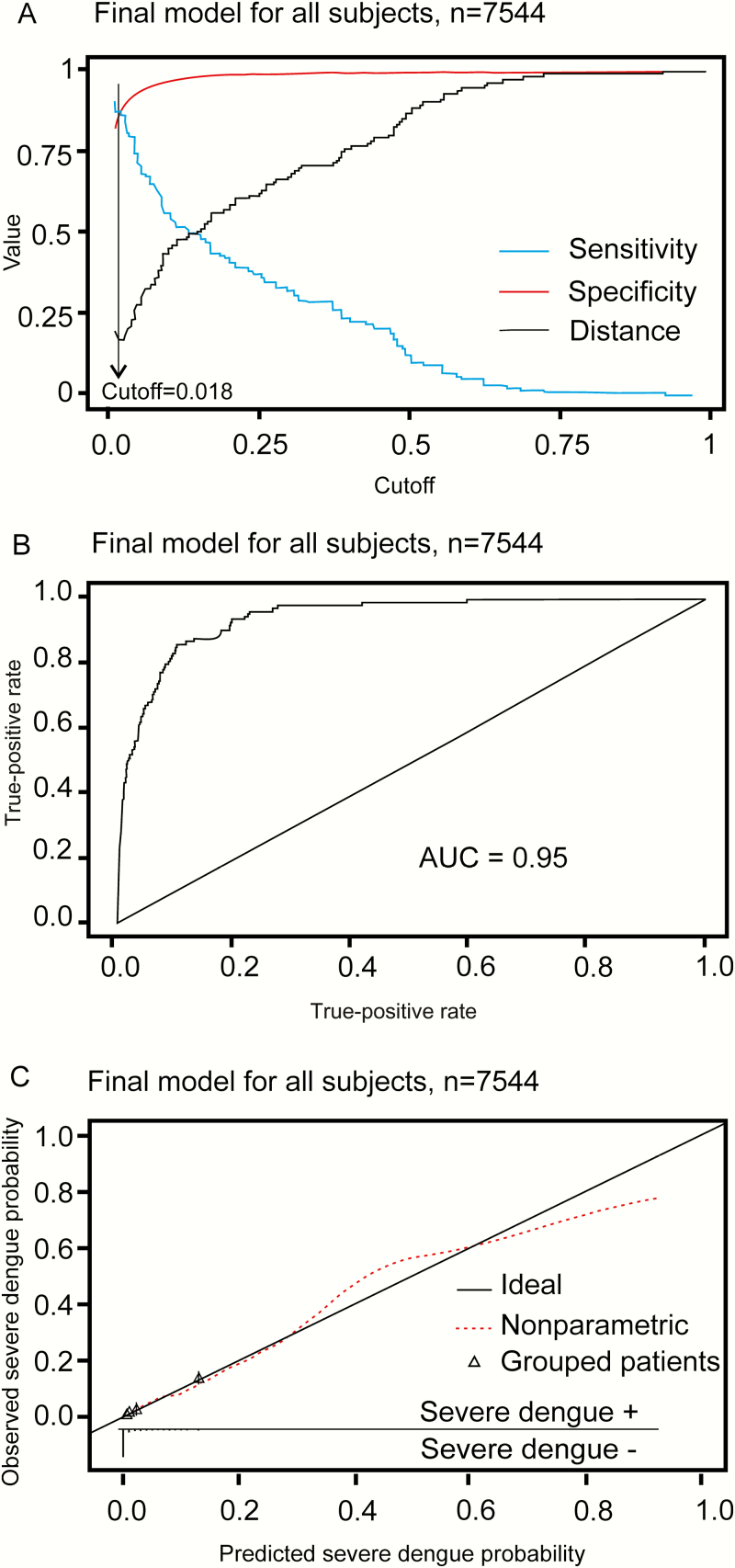
For simplicity, all continuous variables were treated as linear terms in the multivariable model development of the prognostic algorithm (Supplementary Table 1 and Supplementary Statistical Appendix). Many of the predefined clinical and laboratory variables that were collected at enrollment were associated with progression to severe dengue in the univariate analysis (Table 2). In the multivariate logistic model, a history of vomiting, lower platelet count, elevated aspartate aminotransferase (AST) level, and positivity in the NS1 rapid test were all independently associated with severe dengue within the population of febrile patients. The most practical and parsimonious prognostic model, herein called the Early Severe Dengue Identifier (ESDI), included history of vomiting, platelet count, AST (per 2-fold increase), and NS1 rapid test status at the time of enrollment (Table 3). Figure 2A illustrates the performance characteristics of the ESDI at various cutoffs in the febrile population. At the cutoff of 0.02, the ESDI had a sensitivity of 87% (95% CI, 80%–92%), specificity of 88% (95% CI, 87%–89%), positive predictive value of 10% (95% CI, 9%–12%), and negative predictive value of 99% (95% CI, 98%–100%) for correctly discriminating severe dengue from other cases within the febrile population. This cutoff was very close to the point on the receiver operating characteristic (ROC) curve closest to the upper left corner (perfect model), which was 0.018 (Figure 2A). The area under the ROC curve (AUC) of the ESDI in the analyzed population was 0.95 using the cutoff of 0.02 (Figure 2B).
Table 2.
Univariate Analysis of Candidate Predictors of Severe Dengue Among Laboratory-Confirmed Dengue and All Subjects
| Predictor | Severe Dengue vs Nonsevere Dengue Among Laboratory-Confirmed Dengue Cases | Severe Dengue vs All Other Study Participants | ||||
|---|---|---|---|---|---|---|
| OR | 95% CI | P Value | OR | 95% CI | P Value | |
| Age (+ 1 y) | 1.00 | .95–1.06 | .901 | 1.16 | 1.10–1.21 | <.001 |
| BMI (+ 1 kg/m2) | 1.03 | .98–1.09 | .223 | 1.10 | 1.04–1.15 | <.001 |
| Day of illness (+ 1 d) | 1.53 | 1.17–1.99 | .002 | 1.97 | 1.52–2.56 | <.001 |
| Vomiting: Yes | 2.92 | 1.96–4.33 | <.001 | 3.60 | 2.45–5.36 | <.001 |
| Abdominal pain: Yes | 1.65 | 1.09–2.49 | .018 | 1.89 | 1.25–2.81 | .003 |
| Mucosal bleeding: Yes | 1.35 | .67–2.74 | .405 | 2.40 | 1.12–4.54 | .027 |
| WBC (+ 1000 cells/µL) | .85 | .78–.93 | <.001 | .66 | .60–.71 | <.001 |
| Platelet count (+ 10 000 cells/µL) | .82 | .79–.85 | <.001 | .77 | .74–.79 | <.001 |
| Hematocrit (+ 1%) | 1.14 | 1.08–1.20 | <.001 | 1.20 | 1.14–1.26 | <.001 |
| Albumin (+ 1 g/L) | .92 | .87–.97 | .003 | .89 | .85–.94 | <.001 |
| AST (per 2-fold increase) | 3.74 | 3.00–4.68 | <.001 | 5.07 | 4.18–6.16 | <.001 |
| NS1 rapid test positive: Yes | 2.06 | 1.26–3.36 | .004 | 20.82 | 13.12–34.77 | <.001 |
| Viremia (+ 1 log10 copies/mL) | 1.22 | 1.07–1.39 | .003 | … | … | … |
| Serotypea | ||||||
| DENV-1 | 1.00 | … | … | … | … | … |
| DENV-2 | 1.74 | 1.09–2.79 | .020 | … | … | … |
| DENV-3 | .63 | .26–1.52 | .305 | … | … | … |
| DENV-4 | 1.25 | .78–2.02 | .355 | … | … | … |
Abbreviations: AST, aspartate aminotransferase; BMI, body mass index; CI, confidence interval; DENV, dengue virus; NS1, nonstructural protein 1; OR, odds ratio; WBC, white blood cell.aUnivariate effect of serotype on severe dengue estimated from univariate logistic regression, using DENV-1 as reference group.
Table 3.
Multivariate Analysis of Candidate Predictors for Severe Dengue in All Subjects
| Predictor | Full Model With Predefined Candidate Predictors | Reduced Model by Stepwise BIC (ESDI) | ||||
|---|---|---|---|---|---|---|
| OR | 95% CI | P Value | OR | 95% CI | P Value | |
| Age (+ 1 y) | .97 | .91–1.05 | .481 | … | … | … |
| BMI (+ 1 kg/m2) | 1.03 | .97–1.09 | .369 | … | … | … |
| Day of illness (+ 1 d) | 1.69 | .58–1.18 | .295 | … | … | … |
| Vomiting: Yes | 2.13 | 1.36–3.32 | <.001 | 2.14 | 1.4–3.3 | <.001 |
| Abdominal pain: Yes | .98 | .60–1.61 | .933 | … | … | … |
| Mucosal bleeding: Yes | 1.74 | .79–3.85 | .192 | … | … | … |
| WBC count (+ 1000 cells/µL) | 1.04 | .95–1.14 | .41 | … | … | … |
| Platelet count (+ 10 000 cells/µL) | .83 | .79–.88 | <.001 | .85 | .81–.88 | <.001 |
| Hematocrit (+ 1 %) | 1.06 | 1.00–1.14 | .059 | … | … | … |
| Albumin (+ 1 g/L) | 1.00 | .93–1.07 | .937 | … | … | … |
| AST (per 2-fold increase) | 2.67 | 2.09–3.43 | <.001 | 2.72 | 2.14–3.45 | <.001 |
| NS1 rapid test positive: Yes | 9.91 | 5.58–17.59 | <.001 | 8.61 | 5.12–14.48 | <.001 |
Full model with predefined candidate predictors: clinical, hematological, and biochemical features and NS1 rapid test status.
Abbreviations: AST, aspartate aminotransferase; BIC, Bayesian information criteria; BMI, body mass index; CI, confidence interval; ESDI, Early Severe Dengue Identifier; NS1, nonstructural protein 1; OR, odds ratio; WBC, white blood cell.
Figure 2.
Performance of the Early Severe Dengue Identifier (ESDI) in reverse-transcription polymerase chain reaction–positive dengue patients. A, Possible sensitivity/specificity trade-offs for different cutoff values of the ESDI and the distance from the corresponding points on the receiver operating characteristic (ROC) curve to the upper left corner (perfect model). B, ROC curve. C, Calibration plot displaying a scatterplot-smoother of predicted vs observed risks (dashed line), predicted vs observed risks for 10 patient strata of equal size grouped according to predicted risks (triangles), and the ideal identity line (solid line). The rugs at the bottom of the graphs characterize the distribution of predicted risks in severe dengue and nonsevere dengue cases, respectively. Abbreviation: AUC, area under the curve.
The calibration plot shows that the ESDI overestimates the risk of severe dengue in the highest decile of predicted probability (Figure 2C)—that is, patients scored as having the highest decile of risk actually have a lower true risk than the model suggests. A summary of the performance characteristics of the ESDI by temporal and leave-one-site out validation is presented in Table 4. The ESDI clearly showed good discriminative performance for both temporal and leave-one-site out validation with an AUC of at least 0.96. Leave-one-serotype-out validation results (Supplementary Table 2) demonstrated the ESDI was robust to differences in the serotypes that contributed to the dengue case population.
Table 4.
Performance of the Early Severe Dengue Identifier in All Subjects
| Parameter | Apparent Performance | Temporal Validation | Leave-One-Site-Out Validation |
|---|---|---|---|
| Calibration intercept | 0.09 | 0.11 | 0.13 (–.09 to .13) |
| Calibration slope | 1.12 | 1.20 | 1.17 (.95–1.22) |
| AUC | 0.95 (.92–.98) | 0.96 (.93–.99) | 0.96 (.91–.98) |
| Sensitivity (cutoff 0.02) | 0.87 (.80–.92) | 0.93 (.86–.97) | 0.91 (.78–.95) |
| Specificity (cutoff 0.02) | 0.88 (.87–.89) | 0.85 (.84–.86) | 0.86 (.82–.92) |
| PPV (cutoff 0.02) | 0.10 (.09–.12) | 0.10 (.09–.12) | 0.10 (.09–.12) |
| NPV (cutoff 0.02) | 0.99 (.98–1) | 0.99 (.98–1) | 0.99 (.97–1) |
Data in parentheses are 95% confidence intervals. The prognostic model for severe dengue in all patients was the reduced model by stepwise Bayesian information criteria, derived from the original full model that included all predefined clinical and hemobiochemical features and nonstructural protein 1 rapid test status.Abbreviations: AUC, area under the curve; NPV, negative predictive value; PPV, positive predictive value.
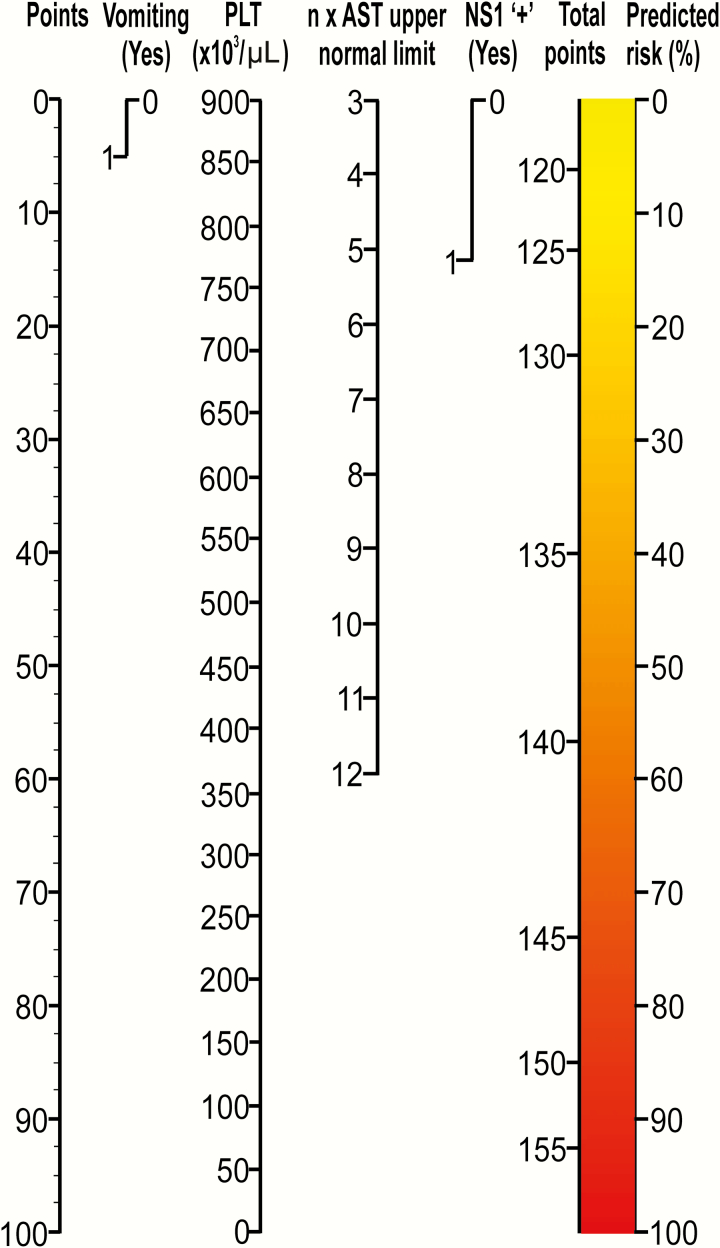
A Practical Tool for Physicians to Predict Severe Dengue
A nomogram was developed to predict severe dengue using the 4 independent parameters in the ESDI (Figure 3). The nomogram is used by totaling the points assigned on the scales for each independent parameter. For example, a patient with vomiting, a platelet count of 100000 cells/µL, positive NS1 rapid test, and an AST level of 280 U/L (7-fold increase compared with the upper normal value of 40 U/L) has a score of 5 + 89 + 14 + 26 = 134, and the corresponding risk of severe dengue is approximately 35%.
Figure 3.
Nomogram of the prognostic model to predict the risk of severe dengue. A vertical line from a predictor value to the “points” axis assigns points to the 4 required variables: vomiting, platelet count (PLT), nonstructural protein 1 (NS1) rapid test status, and aspartate aminotransferase (AST) level. The sum of these points (total points) can then be translated to the corresponding predicted risk of severe dengue.
DISCUSSION
The great majority of clinically severe complications in pediatric dengue patients occur between the fourth and sixth day of illness [1, 9, 10]. Thus there is a window of opportunity in the first few days of illness to both make a diagnosis and try to identify those patients at greatest risk of severe complications so that their management can be adapted accordingly. Yet the current standard of care in relation to diagnosis and prognosis in many endemic countries relies mostly on subjective clinical skills that are variable across different countries, across different levels of the health system, and between individual physicians. Here we demonstrate the feasibility of evidence-based early prognosis (within 72 hours of illness onset) of severe dengue using simple clinical and laboratory investigations that are available in many endemic settings. The ESDI model could be helpful to physicians looking for an evidence-based tool to improve triage and management. It could also deliver efficiencies to clinical trials (ie, enable enrollment of patients at greater risk of severe dengue).
The ability to make an early, evidence-based prognosis of severe dengue could have practical rewards to clinical care, health systems, and clinical research. First, the ESDI could assist clinical services in identifying at-risk patients for triage to more regular observations than would occur under the current standard of care; this could mean hospitalization or more regular visits in an outpatient setting. Additionally, for outpatient care, communication to the patient’s caregivers could be appropriately calibrated and with attention to WHO-nominated clinical warning signs [1]. The benefits of early prognosis, with accompanying closer clinical management, could potentially include a reduction in the frequency with which cases progress to severe disease and, hence, cost to the healthcare system and to families. However, the generally low positive predictive value of the ESDI (10% at a cutoff of 0.02) inevitably means that a large number of cases identified as being at risk of severe disease will in fact have uneventful disease evolutions. Nonetheless, the ESDI potentially offers a tool to clinicians in some circumstances because they currently work in a vacuum of evidence with respect to early prognosis of pediatric dengue cases.
In the context of the multiparameter ESDI, the inclusion of NS1 status (by rapid test) delivered a small but incremental improvement to prognostic model performance. Clinical trials of candidate dengue therapeutics [5–7] have employed NS1 rapid tests to enable early diagnosis and enrollment. However, NS1 rapid tests have no utility as stand-alone prognostic tests. For example, a clinical trial of early prednisolone therapy in 225 NS1 rapid test–positive Vietnamese children observed that only 6.7% of cases (in the placebo arm) developed DSS. Treatment trials in NS1 rapid test–positive adult dengue cases observe an even lower incidence of severe dengue [5, 7, 8]. Thus, currently, early-phase clinical trials endeavoring to use severe dengue as an endpoint would require very large patient sample sizes to meet their objectives. The ESDI could potentially enhance the efficiency of some trials by enriching the study population with cases at higher risk of developing severe dengue, compared with the NS1 rapid test used alone.
Many of the parameters used in the ESDI are collectable in primary care settings in Vietnam and other endemic countries. For example, the NS1 antigen rapid test is available in many hospital emergency department/outpatient settings in Vietnam. NS1 rapid tests are recommended by WHO as a routine screening test for patients with clinically suspected dengue in the acute febrile phase [23]. The diagnostic performance, and limitations, of this test have been extensively described in this patient cohort [24] and elsewhere [25–32]. Previous studies also demonstrated that viremia levels in the first 72 hours of illness were positively correlated with NS1 rapid test positivity [31, 33–36]. Blood hematology (platelet count) and biochemistry (AST level) were also important to the operation of the ESDI; hence, only in settings where these services are routinely available could the ESDI be utilized. Potts et al previously observed that elevated AST in the first 3 days of illness was a predictor for severe dengue [20]. Presumably, early elevations in blood AST concentrations are a signal of the severity of disseminated virus infection and tissue damage.
Not all DENV serotypes were equally associated with severe outcomes. DENV-2 in particular was overrepresented among severe cases, consistent with previous studies [37, 38]. For reasons of study design, we could not dissociate whether DENV-2 was acting as a proxy for secondary infection, itself a well-recognized risk factor for severe dengue, or was independently associated with severe dengue.
Only one previous prospective study, of 1384 febrile Thai children (including 37 with DSS), by Potts et al, is similar in study design to that reported here. Potts et al used classification and regression tree analysis to derive an algorithm from laboratory variables collected at the time of enrollment (platelet count, white blood cell count, monocyte percentage, and hematocrit). The best algorithm had 97% sensitivity and 48% specificity for the identification of patients who progressed to DSS [20]; however, positive and negative predictive values were not reported. The study described here includes several important points of difference including (1) a much larger sample size and inclusion of clinically severe cases who did not have DSS; (2) acquisition of clinical and laboratory data; and (3) validation of model performance.
Our study, and the resulting ESDI, has some inherent limitations. First, the ESDI will not be suitable in all outpatient settings because NS1 rapid tests and biochemistry are not always available. The ESDI is only applicable to observations made in the first 72 hours of illness; it is uncertain what the test performance will be outside this window of presentation. The evolution of dengue is probably impacted by clinical management, so the incidence rate of severe dengue described in this study could be context dependent; for the same patient population, other settings might observe a lower or higher incidence of severe dengue and this could impact the prognostic classifier. Finally, although the ESDI has a good discriminative ability (AUC = 0.95), the low positive predictive value (common to many algorithms seeking to classify relatively rare events) means that the number of true severe dengue cases will be overestimated. Application of the ESDI may result in excessive hospitalizations, unnecessary follow-up procedures, and the associated economic burden than would occur under the current standard of care. Further “pilot phase” research is needed to understand the benefits and disadvantages of the ESDI in routine practice and clinical research.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Acknowledgments. The authors are grateful for the collaboration of the patients and their families who participated in this research, and the many healthcare workers who made this study possible.
Financial support. This work was supported by the Australian National Health and Medical Research Council (GNT1006549) and the Wellcome Trust (GNT084368/Z/07/Z).
Potential conflicts of interest. All authors: No reported conflicts. The authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. World Health Organization. Dengue: guideline for diagnosis, treatment, prevention and control. Geneva, Switzerland: WHO, 2009. [PubMed] [Google Scholar]
- 2. L’Azou M, Moureau A, Sarti E, et al. ; CYD14 Primary Study Group; CYD15 Primary Study Group Symptomatic dengue in children in 10 Asian and Latin American countries. N Engl J Med 2016; 374:1155–66. [DOI] [PubMed] [Google Scholar]
- 3. Martinez-Torres E, Polanco-Anaya AC, Pleites-Sandoval EB. Why and how children with dengue die? Rev Cubana Med Trop 2008; 60:40–7. [Google Scholar]
- 4. Ranjit S, Kissoon N. Dengue hemorrhagic fever and shock syndromes. Pediatr Crit Care Med 2011; 12:90–100. [DOI] [PubMed] [Google Scholar]
- 5. Nguyen NM, Tran CN, Phung LK, et al. A randomized, double-blind placebo controlled trial of balapiravir, a polymerase inhibitor, in adult dengue patients. J Infect Dis 2013; 207:1442–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tam DT, Ngoc TV, Tien NT, et al. Effects of short-course oral corticosteroid therapy in early dengue infection in Vietnamese patients: a randomized, placebo-controlled trial. Clin Infect Dis 2012; 55:1216–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Low JG, Sung C, Wijaya L, et al. Efficacy and safety of celgosivir in patients with dengue fever (CELADEN): a phase 1b, randomised, double-blind, placebo-controlled, proof-of-concept trial. Lancet Infect Dis 2014; 14:706–15. [DOI] [PubMed] [Google Scholar]
- 8. Tricou V, Minh NN, Van TP, et al. A randomized controlled trial of chloroquine for the treatment of dengue in Vietnamese adults. PLoS Negl Trop Dis 2010; 4:e785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Simmons CP, Farrar JJ, Nguyen vV, Wills B. Dengue. N Engl J Med 2012; 366:1423–32. [DOI] [PubMed] [Google Scholar]
- 10. Phung KL, Dong THT, Tran VD, et al. Clinical characteristics of dengue shock syndrome in Vietnamese children; a 10-year prospective study in a single hospital. Clin Infect Dis 2013; 57:1577–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tanner L, Schreiber M, Low JG, et al. Decision tree algorithms predict the diagnosis and outcome of dengue fever in the early phase of illness. PLoS Negl Trop Dis 2008; 2:e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Khan MI, Anwar E, Agha A, et al. Factors predicting severe dengue in patients with dengue fever. Mediterr J Hematol Infect Dis 2013; 5:e2013014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fox A, Le NM, Simmons CP, et al. Immunological and viral determinants of dengue severity in hospitalized adults in Ha Noi, Viet Nam. PLoS Negl Trop Dis 2011; 5:e967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chuansumrit A, Puripokai C, Butthep P, et al. Laboratory predictors of dengue shock syndrome during the febrile stage. Southeast Asian J Trop Med Public Health 2010; 41:326–32. [PubMed] [Google Scholar]
- 15. Carrasco LR, Leo YS, Cook AR, et al. Predictive tools for severe dengue conforming to World Health Organization 2009 criteria. PLoS Negl Trop Dis 2014; 8:e2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gregory CJ, Santiago LM, Arguello DF, Hunsperger E, Tomashek KM. Clinical and laboratory features that differentiate dengue from other febrile illnesses in an endemic area—Puerto Rico, 2007–2008. Am J Trop Med Hyg 2010; 82:922–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Daumas RP, Passos SR, Oliveira RV, et al. Clinical and laboratory features that discriminate dengue from other febrile illnesses: a diagnostic accuracy study in Rio de Janeiro, Brazil. BMC Infect Dis 2013; 13:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ho TS, Wang SM, Lin YS, Liu CC. Clinical and laboratory predictive markers for acute dengue infection. J Biomed Sci 2013; 20:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Falconar AK, Romero-Vivas CM. Simple prognostic criteria can definitively identify patients who develop severe versus non-severe dengue disease, or have other febrile illnesses. J Clin Med Res 2012; 4:33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Potts JA, Gibbons RV, Rothman AL, et al. Prediction of dengue disease severity among pediatric Thai patients using early clinical laboratory indicators. PLoS Negl Trop Dis 2010; 4:e769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hue KD, Tuan TV, Thi HT, et al. Validation of an internally controlled one-step real-time multiplex RT-PCR assay for the detection and quantitation of dengue virus RNA in plasma. J Virol Methods 2011; 177:168–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harrell F. Regression modeling strategies with applications to linear models, logistic regression, and survival analysis. New York: Springer, 2001. [Google Scholar]
- 23. World Health Organization. Handbook for clinical management of dengue. Geneva, Switzerland: WHO, 2012. [Google Scholar]
- 24. Tuan NM, Nhan HT, Chau NV, et al. Sensitivity and specificity of a novel classifier for the early diagnosis of dengue. PLoS Negl Trop Dis 2015; 9:e0003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hang VT, Nguyet NM, Trung DT, et al. Diagnostic accuracy of NS1 ELISA and lateral flow rapid tests for dengue sensitivity, specificity and relationship to viraemia and antibody responses. PLoS Negl Trop Dis 2009; 3:e360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guzman MG, Jaenisch T, Gaczkowski R, et al. Multi-country evaluation of the sensitivity and specificity of two commercially-available NS1 ELISA assays for dengue diagnosis. PLoS Negl Trop Dis 2010; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Duong V, Ly S, Lorn Try P, et al. Clinical and virological factors influencing the performance of a NS1 antigen-capture assay and potential use as a marker of dengue disease severity. PLoS Negl Trop Dis 2011; 5:e1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dussart P, Petit L, Labeau B, et al. Evaluation of two new commercial tests for the diagnosis of acute dengue virus infection using NS1 antigen detection in human serum. PLoS Negl Trop Dis 2008; 2:e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kumarasamy V, Chua SK, Hassan Z, et al. Evaluating the sensitivity of a commercial dengue NS1 antigen-capture ELISA for early diagnosis of acute dengue virus infection. Singapore Med J 2007; 48:669–73. [PubMed] [Google Scholar]
- 30. Lapphra K, Sangcharaswichai A, Chokephaibulkit K, et al. Evaluation of an NS1 antigen detection for diagnosis of acute dengue infection in patients with acute febrile illness. Diagn Microbiol Infect Dis 2008; 60:387–91. [DOI] [PubMed] [Google Scholar]
- 31. Tricou V, Vu HT, Quynh NV, et al. Comparison of two dengue NS1 rapid tests for sensitivity, specificity and relationship to viraemia and antibody responses. BMC Infect Dis 2010; 10:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zainah S, Wahab AH, Mariam M, et al. Performance of a commercial rapid dengue NS1 antigen immunochromatography test with reference to dengue NS1 antigen-capture ELISA. J Virol Methods 2009; 155:157–60. [DOI] [PubMed] [Google Scholar]
- 33. Thomas L, Najioullah F, Verlaeten O, et al. Relationship between nonstructural protein 1 detection and plasma virus load in dengue patients. Am J Trop Med Hyg 2010; 83:696–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chau TN, Anders KL, Lien le B, et al. Clinical and virological features of dengue in Vietnamese infants. PLoS Negl Trop Dis 2010; 4:e657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Duyen HT, Ngoc TV, Ha do T, et al. Kinetics of plasma viremia and soluble nonstructural protein 1 concentrations in dengue: differential effects according to serotype and immune status. J Infect Dis 2011; 203:1292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Libraty DH, Young PR, Pickering D, et al. High circulating levels of the dengue virus nonstructural protein NS1 early in dengue illness correlate with the development of dengue hemorrhagic fever. J Infect Dis 2002; 186:1165–8. [DOI] [PubMed] [Google Scholar]
- 37. Vaughn DW, Green S, Kalayanarooj S, et al. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J Infect Dis 2000; 181:2–9. [DOI] [PubMed] [Google Scholar]
- 38. Fried JR, Gibbons RV, Kalayanarooj S, et al. Serotype-specific differences in the risk of dengue hemorrhagic fever: an analysis of data collected in Bangkok, Thailand from 1994 to 2006. PLoS Negl Trop Dis 2010; 4: e617. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.