Abstract
Recent RNA-seq technology revealed thousands of splicing events that are under rapid evolution in primates, whereas the reliability of these events, as well as their combination on the isoform level, have not been adequately addressed due to its limited sequencing length. Here, we performed comparative transcriptome analyses in human and rhesus macaque cerebellum using single molecule long-read sequencing (Iso-seq) and matched RNA-seq. Besides 359 million RNA-seq reads, 4,165,527 Iso-seq reads were generated with a mean length of 14,875 bp, covering 11,466 human genes, and 10,159 macaque genes. With Iso-seq data, we substantially expanded the repertoire of alternative RNA processing events in primates, and found that intron retention and alternative polyadenylation are surprisingly more prevalent in primates than previously estimated. We then investigated the combinatorial mode of these alternative events at the whole-transcript level, and found that the combination of these events is largely independent along the transcript, leading to thousands of novel isoforms missed by current annotations. Notably, these novel isoforms are selectively constrained in general, and 1,119 isoforms have even higher expression than the previously annotated major isoforms in human, indicating that the complexity of the human transcriptome is still significantly underestimated. Comparative transcriptome analysis further revealed 502 genes encoding selectively constrained, lineage-specific isoforms in human but not in rhesus macaque, linking them to some lineage-specific functions. Overall, we propose that the independent combination of alternative RNA processing events has contributed to complex isoform evolution in primates, which provides a new foundation for the study of phenotypic difference among primates.
Keywords: alternative RNA processing event, comparative transcriptome, independent combination, isoform evolution, primate evolution, PacBio sequencing
Introduction
Alternative RNA processing events, including alternative splicing (AS), alternative cleavage, and polyadenylation (APA), and alternative promoter usage, are widely engaged in transcript processing. By generating multiple variant isoforms from one gene, these regulatory mechanisms dramatically expand the diversity and complexity of the transcriptome (Pan etal. 2008; Pal etal. 2011). Recently, the study of alternative RNA processing events has been accelerated by the development of the next-generation sequencing technology. Novel alternative events identified in many species, such as humans (Pan etal. 2008; Djebali etal. 2012; Mercer etal. 2012; Eswaran etal. 2013; Halvardson etal. 2013; Hu etal. 2015; Tilgner etal. 2015; Yan etal. 2015), chimpanzees (Wetterbom etal. 2010), and mice (Ameur etal. 2010; Pal etal. 2011; Hong etal. 2014), have provided new opportunities to investigate the complexity and evolution of the transcriptomes in mammals.
However, due to the limited sequencing length of Illumina RNA-seq, it is still technically challenging to assess the reliability of the alternative RNA processing events detected by short reads, especially the intron retention (IR) and APA. Briefly, owing to the limited scope of the information provided by short RNA-seq reads, the retained introns detected by short reads cannot be clearly distinguished from the contamination of DNA (Wang etal. 2008; Braunschweig etal. 2014; Li etal. 2015). It is also difficult to differentiate authentic PA sites from internal priming events (Beaudoing etal. 2000; Nam etal. 2002; Fu etal. 2011; Shepard etal. 2011; Derti etal. 2012; Wilkening etal. 2013).
In addition, although in some cases correlated inclusive or mutually exclusive splicing events have been found (Ubby etal. 2013; Schreiner etal. 2014; Tilgner etal. 2015), it remains difficult to investigate the combination of distant alternative RNA processing events at the whole-isoform level, which is the de facto functional entity in translation and other important molecular processes. Notably, while RNA-seq with paired-end design could be used to study pairs of nearby alternative RNA processing events, the distance between two alternative events typically exceed the insert size of the paired-end sequencing libraries. Difficulties in defining and positioning the complex alternative RNA processing events such as IR and APA hinders accurate identification of even low-order coordination. Moreover, novel alternative RNA processing events as well as the combination of these events at the isoform level may lead to the generation of thousands of novel isoforms. It is still unclear whether these novel isoforms are conserved across species, and whether they have functional implications underpinning primate evolution.
Results
Accurate and Comprehensive Cerebellum Transcriptomes in Human and Rhesus Macaque
To investigate the combinatorial mode of alternative RNA processing events and the isoform diversity in human and rhesus macaque, we first performed comparative transcriptome analyses across matched tissue in the two species, using both single molecule long-read sequencing (Iso-seq) (Eid etal. 2009; Au etal. 2013; Sharon etal. 2013; Treutlein etal. 2014) and Illumina RNA-seq. Fifteen PacBio cells were conducted on human cerebellum, producing 1,267,610 raw reads with a mean length of 15,255 bp. As the circular mode of PacBio sequencing was used, the whole length of short or intermediate transcripts was actually sequenced several times from the 5′ end to the poly(A) tail. We therefore split these raw reads into 10,255,721 subreads and self-aligned them to assemble Reads-Of-Insert (ROI) reads. Finally, 1,179,556 ROI reads were identified, with an average of seven read-through passes (see Materials and Methods and table 1). Stringent computational pipelines were then developed to sequentially remove the ROI reads without poly(A) tails, trim poly(A) tails, align the reads to the reference genome, address the alignment errors due to the intrinsic high error rate in PacBio sequencing, and differentiate authentic PA-containing reads from internal priming or DNA contamination (see Materials and Methods). Finally, a high-quality set consisting of 452,441 PacBio processed alignments from 11,466 human genes was generated (table 1).
Table 1.
Statistics of PacBio Iso-seq and Illumina RNA-seq in Human and Rhesus Macaque.
| Species | Human | Macaque 1 | Macaque 2 | Macaque (Total) |
|---|---|---|---|---|
| PacBio Iso-seq | ||||
| SMRT cells | 15 | 21 | 15 | 36 |
| Raw reads | ||||
| Count | 1,267,610 | 1,312,104 | 1,585,813 | 2,897,917 |
| Base | 19,337,965,044 | 14,848,126,759 | 27,779,716,697 | 42,627,843,456 |
| Mean length | 15,255 | 11,316 | 17,517 | 14,709 |
| Subreads | ||||
| Count | 10,255,721 | 8,534,738 | 11,572,355 | 20,107,093 |
| ROI reads | ||||
| Count | 1,179,556 | 765,100 | 1,128,117 | 1,893,217 |
| Mean length | 1,971 | 1,509 | 2,432 | 2,059 |
| Mean pass | 7 | 6 | 7 | 7 |
| PA-trimmed reads | ||||
| Count | 729,053 | 329,492 | 620,787 | 950,279 |
| Processed alignments | ||||
| Count | 452,441 | 184,712 | 404,992 | 589,704 |
| Genes | ||||
| Count | 11,466 | 7,218 | 9,300 | 10,159 |
| RNA-seq | ||||
| Reads | ||||
| Count (M) | 123 | 129 | 107 | 236 |
| Length | 100 | 90 | 150 | 90 or 150 |
| Junctions | ||||
| Count | 266,990 | 132,234 | 241,627 | 256,578 |
Similar PacBio transcriptome studies with a total of 36 PacBio cells were performed on two rhesus macaque cerebellum samples (see Materials and Methods), with 184,712 and 404,992 processed alignments generated, respectively (table 1). For comparison, we also performed Illumina poly(A)-positive, strand-specific RNA-seq on the three tissue samples with a high sequencing depth (table 1).
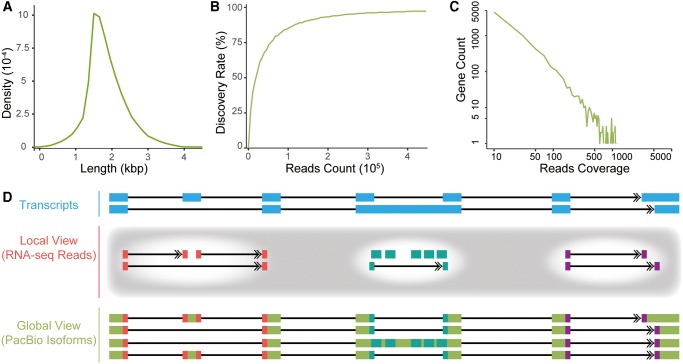
Several lines of evidence showed that the PacBio Iso-seq experiments were performed with high specificity and sensitivity. First, the lengths of isoforms generated by the PacBio Iso-seq were relatively long and, even comparable with those of the full-length human transcripts annotated in RefSeq, suggesting that the data set could be used to investigate the transcription regulation on whole-transcript level (fig. 1A). Second, the PacBio Iso-seq is representative in capturing the majority of the transcriptome of interest. Given that transcripts with >1 kbp were sequenced in the PacBio Iso-seq assays, 6,982 human genes were selected as the positive control, with the length of their longest transcripts >1 Kbp, as well as significant gene expression (RPKM >5) as estimated by the Illumina RNA-seq data from the same sample. Of these genes, 97.4% were identified by the PacBio Iso-seq at the current sequencing depth; and even at half of the current sequencing depth, 94.2% of these genes were identified (fig. 1B). Overall, 90.9% of these human genes were covered by more than one Iso-seq read (fig. 1C). Additional evaluations, such as the evaluations on sequence GC content, base quality, sequencing error, and DNA or pre-mRNA contamination rate, uniformly verified that the PacBio Iso-seq study was performed with high specificity (supplementary fig. S1, Supplementary Material online). Notably, we also compared the gene expression levels estimated by Iso-seq data and RNA-seq data, and found that they are significantly correlated (Spearman correlation coefficients = 0.8, supplementary fig. S2, Supplementary Material online), suggesting the potential applications of Iso-seq in estimating gene expression levels.
Fig. 1.
PacBio Iso-seq in human. (A) Distribution of the lengths of PA-trimmed PacBio reads. (B) Percentages of detectable genes (Discovery Rate) increased as the PacBio sequencing depth increased. (C) Count of genes at different read coverages. (D) Diagram illustrating the advantage of PacBio Iso-seq in deciphering transcript structure. With Illumina RNA-seq reads, alternative RNA processing events can only be inspected individually in a local view, whereas the association of distant events is indefinable. Iso-seq with single molecule, long-read sequencing provides a global view to investigate the combinatorial mode of distant alternative RNA processing events at the whole transcript level.
The PacBio Iso-seq of the two matched samples in rhesus macaque was also performed with good quality as revealed by similar evaluations (supplementary figs. S3 and S4, Supplementary Material online). Such full-length, accurate, and comprehensive PacBio Iso-seq data and the matched Illumina RNA-seq data in the two species thus provided the opportunity to investigate the alternative RNA processing events from a comparative, whole-transcript perspective (fig. 1D).
Genome-Wide Identification of Alternative RNA Processing Events in Primates
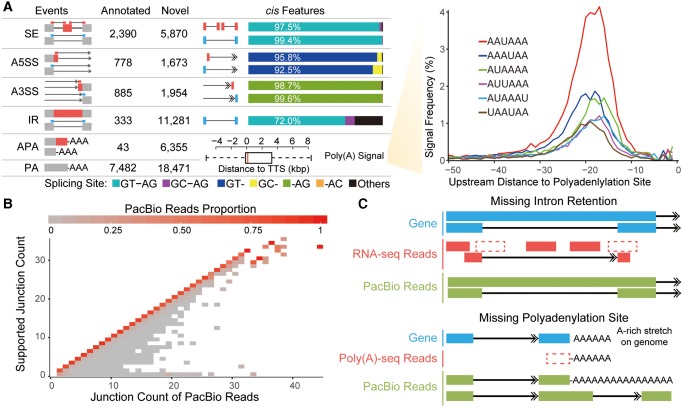
On the basis of the PacBio sequencing and the matched Illumina RNA-seq, we then identified the alternative RNA processing events in human and rhesus macaque, such as AS events including skipped exons (SE), alternative 5′ or 3′ splicing sites (A5SS or A3SS), IR, as well as alternative PA (see Materials and Methods). In total, 25,164 AS events and 6,398 APA events were identified in the human sample (fig. 2A). Among these, 20,778 (82.6%) of AS events and 6,355 (99.3%) of APA events were novel according to RefSeq annotations (release 78) (supplementary table S1, Supplementary Material online), whereas 14,854 (60.4%) of AS events and 5,767 (91.2%) of APA events were novel according to GENCODE annotations (Comprehensive version19) (supplementary table S1, Supplementary Material online).
Fig. 2.
Genome-wide identification of alternative RNA processing events in human with PacBio Iso-seq. (A) Left panel: statistics for the five categories of alternative RNA processing events (SE, A5SS, A3SS, IR, and APA) and meta-data for PA sites contributing to the APA events. Annotated: alternative RNA processing events annotated by RefSeq; Novel: alternative RNA processing events not annotated by RefSeq. For SE, A5SS, A3SS, and IR, the frequency of different splicing motifs are shown in different colors according to the legend below. For APA events, the distance of the PA site from the transcription termination site (TTS) annotated in RefSeq is summarized in the boxplot. Right panel: frequencies of the top six Poly(A) signals located upstream of PA sites. (B) For each PacBio Iso-seq read, the numbers of splicing junctions were counted (PacBio Junction Count). The numbers of junctions supported by RNA-seq reads were also counted (Junction Count Supported by RNA-seq). The density distribution of reads were then summarized and shown in tile in the figure (PacBio Junction Count on X axis, Junction Count Supported by RNA-seq on Y axis), with the density indicated by color ranging from gray to red. (C) Two schemes to demonstrate the principles of PacBio Iso-seq in the identification of intron retention and alternative polyadenylation events.
In strong support of the reliability of these alternative RNA processing events, the cis features near these events agreed with a priori knowledge of known regulatory mechanisms. Briefly, the splicing junctions for >92% of the SE, A5SS, and A3SS events were associated with a canonical GT–AG splicing site motif (fig. 2A). For IR events, 72.0% of splicing sites conformed to the canonical motif, and 7.1% were associated with GC–AG, a finding consistent with previous reports on the different mechanisms of splicing regulation in IR (Galante etal. 2004; Sakabe and de Souza 2007). To further characterize the small proportion of IR events without canonical splice site, we classified the novel IR events into two subgroups—IRs that spliced out the annotated exon (exonic IR) and IRs that retained the annotated intron (intronic IR, supplementary fig. S5B, Supplementary Material online). We found that the exonic IRs, which contributed dominantly (70.5%) to the novel IR events without canonical splice site, show unusual distribution of the splice-site motif (supplementary fig. S5C, Supplementary Material online). Although stringent criteria have been used to decrease the false-positives in here, some false-positives may still exist given the difficulties in IR identification. More verifications are thus needed to confirm these events, especially for those with nonstandard cis- features. To evaluate the reliability of PA identification, we further calculated the distances of candidate PA sites to the RefSeq-annotated transcription termination site of the nearest gene. A distribution of distances with a median score near 0 and a modest variance indicated that most of the identified PA sites were reliable. In line with this verification, the frequency of the top six poly(A) signals near these PA sites was consistent with the pattern for known PA sites (fig. 2A). Finally, as the Illumina RNA-seq has been proven efficient in defining the exon–intron structure at a local view, we used it as an independent control to evaluate whether the Iso-seq assay could also accurately define these events. We found that the splicing junctions defined by PacBio sequencing are largely supported by the RNA-seq data from the same sample (fig. 2B), indicating consistency between the two platforms in defining single splicing event. Similar procedures were conducted on the macaque data and similar patterns were observed (supplementary fig. S6 and table S1, Supplementary Material online).
Interestingly, PacBio sequencing with single molecule, long-read revealed that IR and APA events are surprisingly more prevalent than previously estimated in primates. As for the 11,466 human genes covered by Iso-seq reads, 3,590 (31.3%) were identified with IRs, the majority of which were unannotated by RefSeq and GENCODE. In addition, of the 18,471 novel PA sites detected, 15,029 were located upstream of annotated transcription termination sites. Although PacBio sequencing may have a bias towards sequencing shorter RNA molecules, our data still evidence a prevalence of unknown shorter isoforms of the transcripts in human (fig. 2A). Notably, an IR event was identifiable in Illumina RNA-seq only when junction reads were detected spanning the splicing site, and when genomic reads were uniformly distributed across the whole intronic region. Given the long length of IR introns relative to that of RNA-seq reads, as well as the typically uneven distribution of RNA-seq reads across the transcripts, it remained technically challenging to accurately identify these events with RNA-seq data only (fig. 2C). Similarly, it was challenging to identify PA sites even using an optimized RNA-seq approach, Poly(A)-seq, largely due to the false negatives introduced by the direct removal of genomic PA alignments to avoid internal priming or DNA contamination (Tian etal. 2005; Derti etal. 2012). In this regard, PacBio sequencing provided a direct and efficient approach to identify the two types of alternative RNA processing events, as the exon–intron structure on one transcript was clearly distinguishable with single molecule and long reads (fig. 2C). Briefly, IR events were accurately identified with PacBio reads that completely covered the whole intronic region (fig. 2C), and the false negatives in Poly(A)-seq were adequately addressed owing to the notion that the PA tails on PacBio reads were typically longer than the genomic A-rich stretch (fig. 2C, see Materials and Methods).
Independent Combination of Alternative RNA Processing Events throughout a Gene
On the basis of the accurate catalog of alternative RNA processing events in human and rhesus macaque, we then investigated the combinatorial mode of these events at the whole-transcript level. For SE, A5SS, and A3SS events verified by both PacBio Iso-seq and Illumina RNA-seq, we calculated the inclusion ratio of each AS event on the basis of the RNA-seq data. For IR and APA events, we calculated the inclusion ratio or the frequency of PA events on the basis of the PacBio Iso-seq data. With the inclusion ratio of local events as inputs, we then conducted 10,000 iterations of Monte Carlo simulations to test whether we could reconstruct the global profile of the real transcripts as detected by Iso-seq. Briefly, these values of inclusion ratio were used to reconstruct putative isoforms at the current depth of PacBio sequencing, under the assumption that the combination of these alternative RNA processing events is independent. The distribution of the number of putative isoforms was then compared with the real isoform number obtained by PacBio sequencing (see Materials and Methods). If alternative RNA processing events were coordinately combined (the alternative hypothesis), the observed isoform number would be significantly lower than that of the expected number (see Materials and Methods), and the frequency distribution of these putative isoforms would also be dissimilar to that of the real isoforms in PacBio sequencing.
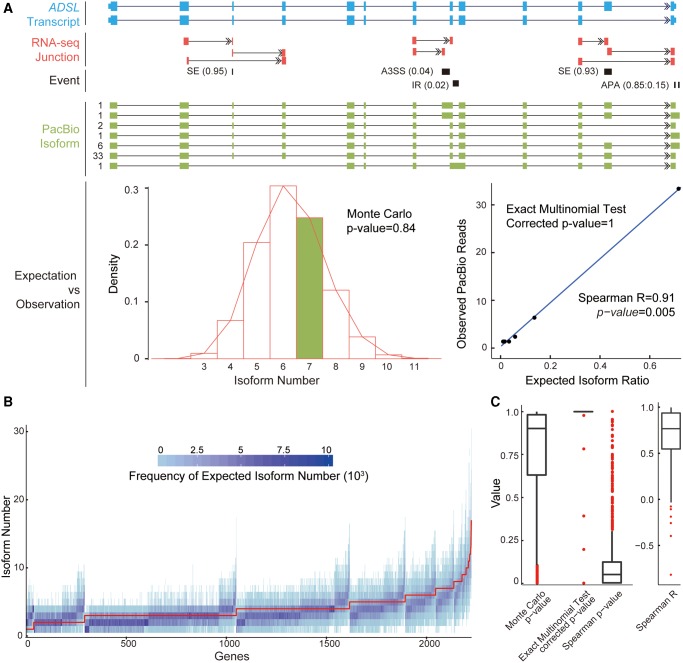
As proof of concept, one example with five alternative RNA processing events is shown in figure 3A, in which seven isoforms were found by PacBio sequencing, a number not significantly lower than the expectation according to 10,000 iterations of a Monte Carlo simulation (Monte Carlo P value = 0.84, fig. 3A). The frequency of the putative isoforms was also similar to that of the real isoforms in PacBio sequencing (Exact Multinomial Test, corrected P value = 1; Spearman correlation coefficient = 0.91, P = 0.005, fig. 3A). Similar to this case, of 2,242 human genes with at least two alternative RNA processing events and covered by at least five PacBio reads (fig. 3B), 2,163 (96.5%) showed a pattern with the observed isoform number not significantly lower than expected (Monte Carlo P value ≥0.05) (supplementary table S2, Supplementary Material online). For 1,964 of the 2,242 genes (87.6%), the frequency of the putative isoforms was consistent with that of the real isoforms in PacBio sequencing (Exact Multinomial Test, corrected P value ≥0.05; fig. 3C), with Spearman correlation coefficients indicating significant positive correlations between the observed and the expected frequencies (fig. 3C and supplementary table S2, Supplementary Material online). In other words, the global profile of the real transcripts as detected by Iso-seq could be reconstructed on the basis of the inclusion ratios as estimated by the local view, under the null hypothesis of independent combination, suggesting that nonrandom associations between different transcription events throughout a gene are rare as compared with a random mode of associations in primates. Similar patterns were identified in rhesus macaque, with 96.4% and 87.1% of genes showing the mode of independent combination in terms of the isoform number and the frequency of isoforms, respectively (supplementary fig. S7 and table S2, Supplementary Material online).
Fig. 3.
Independent combination of alternative RNA processing events in human. (A) Upper panel: as proof-of-concept, the ADSL gene is shown to demonstrate the procedures used to study the combination of alternative RNA processing events. Five alternative RNA processing events were located on the ADSL gene: four AS events and one APA. The inclusion ratio for each AS event, as well as the PA frequency ratio for APA, are shown in brackets. Middle panel: structures of PacBio isoforms with PacBio read coverage highlighted on the left of the isoform structure. Lower left panel: distribution of expected isoform numbers generated by 10,000 iterations of Monte Carlo simulations; green bar indicates the number of isoforms observed in the real PacBio Iso-seq data. Lower right panel: frequency of expected isoforms versus real isoforms in PacBio Iso-seq. (B) Distribution of isoform numbers generated by 10,000 iterations of Monte Carlo simulation shown as a heat-map across each gene (X axis). Red curve: the observed isoform numbers from PacBio Iso-seq. (C) Distribution of Monte Carlo P values for the 2,242 human genes showing whether the observed isoform number was significantly lower than expectation. The distributions of corrected P values from the Exact Multinomial Test, Spearman correlation coefficients, and the Spearman correlation P values are also shown to indicate whether the frequencies of the expected isoforms are consistent with those for the observed isoforms in PacBio sequencing.
To exclude the possibility that the general lack of coordination among alternative RNA processing events along the transcript is mainly due to splicing noise, we provided several lines of analytical evidence: First, we divided genes into different subgroups according to the expression levels in RPKM and calculated the ratio of genes with independent combination in each subgroup. We found that the percentage of genes with independent combination was not correlated with the gene expression level (supplementary fig. S8A, Supplementary Material online). Second, when only abundant isoforms supported by at least two PacBio reads were used in the calculation, the extent of absence of coordination remains unchanged. Third, the lack of coordination remained evident when we focused on annotated isoforms or isoforms without exonic IRs in the calculation (supplementary fig. S8B, Supplementary Material online). Finally, we also performed the coordination test using only alternative RNA processing events with moderate inclusion ratio (between 0.3 and 0.7), and reached the same conclusion (supplementary fig. S8B, Supplementary Material online). These analyses indicate that the general lack of coordination among alternative RNA processing events along the transcript might not be a false-positive signal attributable to splicing noise.
Prevalence of Novel Isoforms in Primates
Considering that >90% of human multi-exon genes are alternatively spliced (Pan etal. 2008; Wang etal. 2008), if these events correspond to independent combination at the isoform level, the number of isoforms would be strikingly greater than previously estimated. As expected, according to the PacBio sequencing data, for 27,383 full-length isoforms transcribed from 7,197 multi-exon genes in the human cerebellum, 76.8% (21,038) were not annotated in the latest version (release78) of RefSeq (defined as novel isoforms). Considering that the RefSeq annotation may underestimate alternative RNA processing events, we further used GENCODE (Comprehensive version19, the merged set of manual annotations from HAVANA with automatic annotations from the Ensembl) as an alternative reference to identify potential novel isoforms missed by the annotations. For the 21,038 isoforms identified as novel according to human RefSeq annotation, 16,124 isoforms (76.6%) were also missed by GENCODE annotation (supplementary table S3, Supplementary Material online). These findings indicate that the complexity of the human transcriptome is still significantly underestimated.
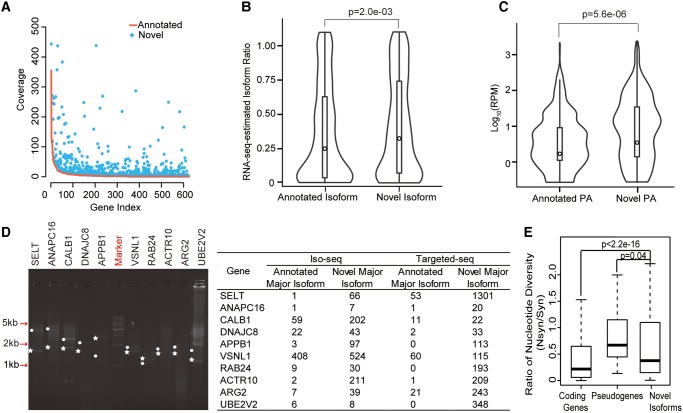
Interestingly, when comparing the expression levels between novel isoforms and annotated isoforms in RefSeq, we identified 1,119 novel major isoforms (corresponding to 624 genes), whose expression levels in the human cerebellum were even higher than those of the annotated major isoforms (fig. 4A). The patterns were verified by Illumina RNA-seq, as both the AS events (fig. 4B) and the APA events (fig. 4C) were specifically associated with the novel isoforms showing significantly higher frequencies than those of the annotated major isoforms (Wilcoxon signed-rank test, P <0.01; see Materials and Methods). Ten randomly selected cases were verified by reverse transcription PCR (RT-PCR) enrichment and PacBio targeted sequencing (Targeted-seq). In all of these genes, the novel isoforms were verified to have higher expression levels than the previously annotated major isoforms, indicating that for a substantial proportion of the human genes, the dominant transcription products remain elusive (fig. 4D, see Materials and Methods).
Fig. 4.
Validation of newly identified major novel isoforms. (A) 624 major isoforms annotated by RefSeq. Red curve, coverage of these isoforms from the PacBio Iso-seq data; blue points, coverage of 1,119 newly identified isoforms from the PacBio Iso-seq data. (B, C) Violin plots of the proportions of the newly identified isoforms, as well as the major isoforms as annotated by RefSeq. The proportions of isoforms were estimated on the basis of the inclusion ratio of alternative splicing events (B), or the sequencing coverage of PA sites (C). P values were calculated on the basis of the Wilcoxon signed-rank test. (D) Left: example of agarose electrophoresis showing ten newly identified major isoforms (dots) and ten RefSeq-annotated major isoforms (stars). Right: the coverage of these isoforms in the initial genome-wide Iso-seq and the targeted PacBio sequencing are summarized in the table. (E) The ratio of nucleotide diversity between nonsynonymous (Nsyn) sites and synonymous (Syn) sites. The ratios were calculated and shown for human annotated coding genes (Coding Genes), pseudogenes (Pseudogenes), as well as coding regions unique to novel isoforms (Novel Isoforms), as indicated.
Next, we sought to assess the functional implications of these novel isoforms missed by the “gold standard” annotations by examining whether they are selectively constrained in general. To this end, we performed population genetics analyses to compare the nucleotide diversity between nonsynonymous sites and synonymous sites, on the basis of the polymorphism data in the population of 103 human individuals (see Materials and Methods). For coding regions unique to these novel isoforms, we found that the nucleotide diversity for nonsynonymous sites is significantly lower than that of the synonymous sites, indicating these novel isoforms are selectively constrained in human population (Wilcoxon one-tail test, P value <2.2e-16). As for the ratio of nucleotide diversity between nonsynonymous (Nsyn) sites and synonymous (Syn) sites, human novel isoforms showed a higher ratio than that of annotated coding genes, but significantly lower than that of human pseudogenes (Wilcoxon one-tail test, P <0.05, fig. 4E, see Materials and Methods). When inspecting the distribution of nucleotide diversity, we found that the weak purifying selection signal was mainly from the constrained regions with a large magnitude difference in silent versus nonsynonymous diversity (supplementary fig. S9, Supplementary Material online), supporting the model that most constrained regions contributed to the signal, instead of a mixture of many unconstrained and a small number of constrained regions. As for the novel isoforms shared by human and rhesus macaque, we further performed the McDonald–Kreitman test on coding regions specific to the novel isoforms. The McDonald–Kreitman test indicated a statistically significant difference between the nonsynonymous and synonymous sites (NI: 1.31, P = 0.03), suggesting that a proportion of these novel isoforms presumably has biological functions and is selectively constrained in general.
As PacBio Iso-seq provided an efficient means to the fine mapping of transcript structure, the candidates identified by Iso-seq likely represent bona fide transcripts expressed in the tissues. We thus did not use read coverage thresholds (as usually done in processing RNA-seq data with short reads) to call isoforms. To eliminate the possible effects of transcription or splicing noises on the identification of novel isoforms, we removed candidate novel isoforms contributed solely by rare alternative RNA processing events (the number of reads supporting the events < 2) or exonic IRs with unusual distribution of the splice-site motif, and subsequently found that nearly 90.0% of previously defined novel isoforms remains in the list (supplementary fig. S10A, Supplementary Material online). For coding regions unique to these novel isoforms, we found that the nucleotide diversity for nonsynonymous sites is significantly lower than that of the synonymous sites (supplementary fig. S10B, Supplementary Material online). It is thus possible that although some of these transcripts may represent noise of transcription and splicing, a proportion of them presumably has biological functions and is selectively constrained in general in the population.
Similar measurements were made in the macaque cerebellum samples, with 20,819 (86.7%) of the transcripts not included in current annotations, and 770 novel major isoforms (corresponding to 433 genes) were identified whose expression levels in the macaque cerebellum were higher than those of the annotated major isoforms (supplementary figs. S9 and S11 and table S3, Supplementary Material online).
Isoform Evolution Underpins Lineage-Specific Traits in Primates
We then investigated whether isoforms identified by PacBio sequencing are conserved across closely related species, such as human and rhesus macaque. For the 3,550 orthologous genes covered by full-length Iso-seq reads in both human and rhesus macaque (Macaque 1, fig. 5A), 3,019 genes (85.0%) encode nonconserved human isoforms that could not be detected in rhesus macaque Iso-seq (see Materials and Methods and fig. 5A).
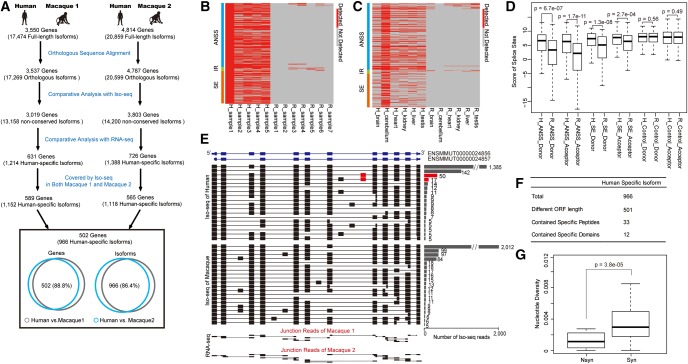
Fig. 5.
Identification and verification of genes encoding isoforms specific to human. (A) Diagram showing the comparative transcriptome study in human on the basis of both PacBio Iso-seq and RNA-seq data. (B) Heatmap showing the status of the candidate human lineage-specific events in multiple human individuals and macaque animals. (C) Heatmap showing the status of the candidate human lineage-specific events in multiple tissues between human and rhesus macaque. Red: observed events; Gray: events not detected. (D) Cis-features for lineage-specific alternative RNA processing events in human. The splice site score of each lineage-specific splice site in human and rhesus macaque was calculated and shown. For lineage-specific A5SS and A3SS events detected in human only, the splice score in human (H_ANSS_Donor and H_ANSS_Acceptor) are significantly higher than that in rhesus macaque (R_ANSS_Donor and R_ANSS_Acceptor). For lineage-specific exon skipping events detected in human only, the splice score for 5′ and 3′ splice sites in human (H_SE_Donor and H_SE_Acceptor) are significantly higher than that in rhesus macaque (R_SE_Donor and R_SE_Acceptor). For lineage nonspecific exon skipping events detected in both human and rhesus macaque, the splice scores are generally comparable in the two species. (E) Validation of isoforms specific to human in PIGU gene using ultradeep targeted Iso-seq sequencing. Isoforms with relatively high sequencing depth (≥5) were shown. For each type of isoform identified by the Iso-seq, the structure of the isoform was shown with the number of the Iso-seq reads supporting it. The human lineage-specific exon was highlighted in red. Junction reads identified by RNA-seq were also shown to indicate the splicing junctions. (F) Functional implications of isoforms specific to human. (G) Boxplots of nucleotide diversity for nonsynonymous (Nsyn) and synonymous (Syn) sites in coding regions specific to novel isoforms specific to human.
Considering the relatively low-throughput of Iso-seq at the given cost, false-positives may exist in defining lineage-specific isoforms through direct comparison of sequencing data. To control for the false-positives, we further integrated RNA-seq data from the same RNA samples as used in the Iso-seq to identify lineage-specific RNA processing events represented only in one species, further defined as lineage-specific isoforms (see Materials and Methods and fig. 5A). Overall, for the 3,019 genes that are initially identified with nonconserved human isoforms, 631 genes were confirmed with the RNA-seq data to encode isoforms specific to human but not rhesus macaque. This lineage-specific identification was not due to differential gene expression—these genes showed similar expression profile between human and rhesus macaque (Spearman correlation coefficient = 0.75, P value <2.2e-16) and are not enriched in genes differentially expressed between the two species (Fisher’s Exact test, P > 0.05, supplementary fig. S12, Supplementary Material online).
To exclude the possibility that these lineage-specific isoforms may actually represent population-level polymorphism, we replicated analysis on the basis of the Iso-seq data and matched RNA-seq data in another rhesus macaque animal (Macaque 2, fig. 5A and supplementary table S3, Supplementary Material online), and identified 726 genes encoding isoforms specific to human but not rhesus macaque. Among the 726 candidate genes, 565 were also included in the initial pipeline, and 502 (88.8%) were identified to encode isoforms specific to human in both of the identifications. To further account for population-level variability, we collected public RNA-seq data of human/macaque cerebellum to verify these lineage-specific events in larger populations of human and rhesus macaque. Overall, more than 86.6% of the lineage-specific alternative RNA processing events could be detected in at least two human individuals, in contrast to 6.7% detected in macaque samples (fig. 5B), indicating that the population-level variability may not have a strong effect in the definition of lineage-specific alternative RNA processing events. Moreover, we also performed comparable analysis of lineage-specific events in multiple tissues between human and rhesus macaque, and found that most lineage-specific alternative RNA processing events identified in human cerebellum were undetected in multiple macaque tissues, indicating that the specificity is likely mediated by cis-elements (fig. 5C). Accordingly, when calculating the strength of these lineage-specific splice sites (Yeo and Burge 2004), we found that the splice scores in human are significantly higher than that in rhesus macaque, whereas the scores are generally comparable for lineage nonspecific events in the two species, suggesting the roles of cis-element in the formation of these events specific to human but not rhesus macaque (fig. 5D).
Similar analyses were performed to identify genes encoding isoforms specific to rhesus macaque but not human, from which 90 genes were identified to encode 131 lineage-specific isoforms (see Materials and Methods and supplementary fig. S13 and table S3, Supplementary Material online).
To verify these lineage-specific isoforms detected in human but not in rhesus macaque, three candidate genes with potential lineage-specific isoforms at different Iso-seq coverages were randomly selected, and the full-length isoforms of the corresponding genes were enriched by RT-PCR and sequenced with PacBio. Overall, we obtained ultradeep coverage of these genes in both human and rhesus macaque (Iso-seq reads counts of each gene ranging from 455 to 2,731), and confirmed all of these lineage-specific isoforms (fig. 5E;supplementary fig. S14 and table S4, Supplementary Material online).
Instead of being transcription noise, these lineage-specific isoforms may have substantial impact on the proteome. Briefly, 501(51.9%) of human-specific isoforms and 49 (37.4%) of macaque-specific isoforms had ORF lengths different from the known isoforms (fig. 5F and supplementary fig. S13, Supplementary Material online). The translation of 33 human-specific isoforms was supported by MS-bank peptides (Wilhelm etal. 2014) that could be specifically assigned to the novel isoforms rather than the annotated known isoforms. Moreover, for 12 isoforms, the isoform-specific regions encoded whole or parts of known functional modules such as protein domains and signal peptides, suggesting a direct impact on the molecular functions of the protein products (fig. 5F).
To further investigate the functionality of these lineage-specific isoforms, we performed population genetics analyses to investigate whether selective constraints have acted on these isoforms specific to human but not rhesus macaque, on the basis of whole-genome sequencing data from 103 human individuals (see Materials and Methods). In the coding regions specific to these lineage-specific isoforms, we found that the nucleotide diversity of nonsynonymous sites was significantly lower than that of synonymous sites (Wilcoxon one-tail test, P value = 3.8e-05, fig. 5G), indicating that these isoforms are selectively constrained in human population. Taken together, the independent combination of alternative RNA processing events has contributed to the generation of numerous lineage-specific, selectively constrained isoforms. This new catalog of isoforms thus provides important foundation for the study of phenotypic difference among primates.
Discussion
Prevalence of IR and APA Regulation in Primates
Although the study of alternative RNA processing events has been dramatically accelerated by the next generation sequencing technology, it remains technically challenging to accurately define these events by short RNA-seq reads alone, especially for the IR and APA events. PacBio Iso-seq provided a direct and convincing approach to identify alternative RNA processing events including IR and APA, as the exon–intron structure and poly(A) signals on one transcript were clearly distinguishable with single molecule and long PacBio reads (fig. 2C). Using the platform, we substantially expanded the repertoire of alternative RNA processing events in primates, and demonstrated that IR and APA events in primates are surprisingly more prevalent than previously estimated. The data therefore constitute a more comprehensive foundation for further investigation of the combinatorial mode of these events at the whole-transcript level as well as their biological significance.
It is well accepted that IR is the most common form of AS in the majority of plants, but occurs at lower frequency in drosophila and human ranging ∼5–15% (Zhang etal. 2014; Zhang etal. 2015). By applying new pipelines for IR detection with high-coverage RNA-seq data, a recent study by Braunschweig etal. reported that 53% and 51% of human and mouse introns could potentially be retained, indicating that IR could be more prevalent in mammals (Braunschweig etal. 2014). Using Iso-seq to directly detect intron–exon structure on the isoform level, our current study provided new evidence for the prevalence of IR in primates. The IR events identified in this study showed significantly higher PSI than introns without IR (Wilcoxon signed-rank test, P = 0) (supplementary fig. S5A, Supplementary Material online), and 79.1% of splicing sites conformed to the canonical GT–AG motif or the previously reported GC–AG motif, indicating that these IR events may not represent random readout of nascent transcripts that has yet to undergo splicing at a particular intron. Given that IR events have been implicated in a variety of biological processes and complex human diseases (Yap etal. 2012; Wong etal. 2013; Jung etal. 2015), their prevalence in human and macaque genes represents a new layer of molecular mechanisms underlying complex regulatory mechanisms and disease processes.
As for the PA sites prevalently distributed across the genomes of human and rhesus macaque, we first analyzed the length of the poly(A) tail with Iso-seq data. As expected, the highly expressed genes have significantly longer poly(A) tails in both human and rhesus macaque (Wilcoxon signed-rank test, P value = 9.4e-13 and 2.0e-262 for human and rhesus macaque, respectively, supplementary fig. S15, Supplementary Material online). Surprisingly, 15,029 of 18,471 newly identified PA sites were located upstream of the annotated transcription termination sites. Although PacBio sequencing may have a bias towards sequencing shorter RNA molecules, this high preponderance of upstream PA sites revealed by our sequencing data nevertheless suggests pervasiveness of shorter transcript isoforms encoded by genes in primates. More studies are thus needed to investigate whether the isoforms with shorter 3′-UTRs have distinct molecular properties such as RNA stability, as well as their potential link to development (Ji etal. 2009; Li etal. 2012) and disease (Mayr and Bartel 2009).
A General Lack of Coordination among Alternative RNA Processing Events along the Transcript
It is generally accepted that distant alternative RNA processing events may be subject to some cis- or trans- regulation, thus exhibiting coordinated representation. Although previous reports also suggested correlations between distant alternative RNA processing events on single RNA molecules (Ubby etal. 2013; Schreiner etal. 2014; Tilgner etal. 2015), there is no evidence to date at a genome-wide scale. Surprisingly, PacBio Iso-seq in our study demonstrated that independent combination of different alternative RNA processing events may be rather the more predominant means by which isoforms are generated. This notion was substantiated by the finding that the global profile of the real transcripts as detected by Iso-seq could be reconstructed on the basis of the inclusion ratios as estimated by the local view under the null hypothesis of independent combination. The finding not only extended our understanding into the formation mechanism of the complex transcriptome, but also provided theoretical support to many de novo transcript assembly softwares, such as Trinity (Grabherr etal. 2011) and Scripture (Guttman etal. 2010), which assemble a draft repertoire of full-length transcripts on the basis of short RNA-seq reads with local view.
On the other hand, despite our result that nonrandom associations between different transcription events throughout a gene are rare as compared with random associations, we identified 37 human genes with coordinated alternative RNA processing events (see Materials and Methods, supplementary table S2, Supplementary Material online). Viewed together, although we could theoretically assemble a draft repertoire of full-length transcripts on the basis of short RNA-seq reads, PacBio Iso-seq is needed to establish the fine map of the transcriptome, especially when the coordinated events play essential roles in the biological systems of interest.
Prevalence of Unknown Isoforms Missed by “Gold Standard” Annotations in Primates
RefSeq annotations with a set of canonical transcript structures have been widely used in gene functional studies as “gold standard.” The latest version of RefSeq annotation, by integrating the recently emerged RNA-seq profiles, has become a more comprehensive resource for transcript structure annotation (O’Leary etal. 2016). However, according to even the latest version, 76.8% of the multi-exon isoforms identified in Iso-seq of human cerebellum were not annotated. Considering that the RefSeq annotation may underestimate some alternative RNA processing events, we further included GENCODE with both manual annotations and Ensembl (version 89) automatic annotations as an alternative reference, yet 76.6% novel isoforms missed by human RefSeq annotation were also unannotated by GENCODE.
We further investigated whether these novel isoforms correspond to genes with relatively high expression, and found that there was no significant correlation between the density of novel isoforms and the expression of the gene. Interestingly, genes with more alternative RNA processing events showed a higher density of novel isoforms (supplementary fig. S10C, Supplementary Material online), suggesting that the current annotations underestimated the complexity of the isoforms, especially for genes with more alternative RNA processing events. Overall, the advantages of Iso-seq in convincingly delineating full-length transcript structure significantly expand the repertoire of the known transcripts even in extensively studied species such as human.
Comparative Transcriptome Study on Isoform Level
Despite its significant advance in annotating isoforms, PacBio Iso-seq is currently not good enough in comparative analyses, mainly due to the relatively low throughput of sequencing at a given cost. To define lineage-specific isoforms at the Iso-seq sequencing depth in our study, false-positives may exist in direct comparison of Iso-seq data. To control for the false-positives, we then increased the sensitivity in comparisons by combining the corresponding RNA-seq data with the same extraction of Iso-seq. Subsequently, only isoforms with lineage-specific RNA processing events defined by RNA-seq data were considered as lineage-specific isoforms. We further extended our analyses using the Iso-seq data and matched RNA-seq data in another rhesus macaque animal, and kept only isoforms supported by both of the identifications. Even using this conservative strategy, 502 genes were identified to encode isoforms specific to human but not rhesus macaque. More cross-species differences on isoform level could be expected, especially when considering the prevalence of alternative RNA processing events with significantly different inclusion ratios during primate evolution (Barbosa-Morais etal. 2012; Merkin etal. 2012), which could result in numerous species-biased isoforms through isoform switching. These differences in isoform level detected in the matched tissue of closely related species may thus underpin the complex phenotypic difference during primate evolution.
The expression of these lineage-specific transcripts may have a substantial impact on the proteome. Our results also showed that population-level selective constraints have acted on these isoforms, thus indicating their potential lineage-specific functions. Pathway functional enrichment analyses further revealed that the isoforms specific to human are enriched in pathways associated with essential biological processes, such as intracellular transport, translation, and protein folding (modified Fisher’s Exact P value <0.05). This new catalog of isoforms identified by PacBio Iso-seq thus provides a new foundation for the study of phenotypic difference among primates. Furthermore, taking into account the rapid evolution of alternative RNA processing events in mammals (Barbosa-Morais etal. 2012; Merkin etal. 2012), our data demonstrate that direct generalization of the findings of gene functions in model animals, such as mouse and rat, to human may require additional validation.
Due to the high costs of PacBio sequencing experiments, here we performed a human–macaque comparative transcriptome analysis, focusing on the brain tissues as a proof-of-concept study. Further studies with more tissue types from additional primate species are needed to extend the scope of the findings. The use of rhesus macaque as an outgroup in this study was mainly due to two reasons. First, although rhesus macaque is a distantly related model to study human biology compared with chimpanzee or bonobo, it is closely related in contrast to other widely used model animals such as rat and mouse. Moreover, we have established an AAALAC-accredited macaque facility with availability of high-quality macaque tissue samples for the transcriptome study with Iso-seq. Second, we have performed a series of functional genomics studies in rhesus macaque in previous studies (Zhang etal. 2013; Zhang etal. 2014; Zhong etal. 2016). Annotations from these studies, especially the accurate gene models and population genetics profiles, provide a comprehensive macaque genomic context for studying the diversity of the transcriptome in primates. However, our current results do not exclude the possibility that the lineage-specific transcripts specifically detected in human may represent a mixture of human-specific as well as hominoid-specific transcripts. More transcriptome data in chimpanzee and bonobo are thus needed to accurately define human-specific novel transcripts. Finally, we focused on brain samples in this pilot study as it has been reported that alternative RNA processing events are frequently lineage-specific, with relatively small cross-tissue differences (Barbosa-Morais etal. 2012; Merkin etal. 2012), whereas exploring the complexity and evolution of the brain transcriptome may provide mechanistic insights to complex diseases and human evolution (Li etal. 2010; Zhang etal. 2011; Xie etal. 2012). Similar long-read sequencing experiments involving additional tissue types are certainly needed in future studies to capture transcriptome diversity at a systems level.
Materials and Methods
Ethics Statement
The rhesus macaque cerebellum samples were obtained from the AAALAC-accredited (Association for Assessment and Accreditation of Laboratory Animal Care) animal facility at the Institute of Molecular Medicine in Peking University. The animal samples were obtained in accordance with protocols approved by the Animal Care and Use Committee of Peking University and followed good practice.
RNA Extraction, Library Preparation, and Deep Sequencing
Total RNA from human cerebellum was purchased from Clontech (Cat. No. 636535). Cerebellum samples for rhesus macaque were obtained from two macaque animals, one of them was used in previous studies (Zhang etal. 2014; Chen etal. 2015). Total RNA was extracted using TRI Reagent (Sigma, catalog # T9424), after the sample was ground in liquid nitrogen. The quality of the RNA samples was assessed using an Agilent 2100 Bioanalyzer and electrophoresis. The quantity of total RNA was measured by NanoDrop2000. First-strand cDNA was synthesized using a SMARTer PCR cDNA Synthesis Kit (Clontech, catalog #634925), subsequently quantified by real-time PCR, and subjected to PacBio sequencing. SMRT bell libraries were generated using a SMRTbell Template prep kit 1.0 (LOT: 005746, Pacific Biosciences) and the template preparation and sequencing protocol for 1-kb libraries. SMRT bell templates were bound to polymerases (P6) using the DNA Binding kit P6 V2 (LOT: 005612) and v2 primers. Polymerase-template complexes were bound to magnetic beads using the Magbead binding kit (part #100-134-800, Pacific Biosciences). Iso-seq was then carried out on a real-time sequencer (Pacific Biosciences) using C4 sequencing reagents. Poly(A)-positive, strand-specific RNA-seq for the same samples was performed on a HiSeq 2000, following the previously reported protocol (Zhang etal. 2013; Zhang etal. 2014). All sequencing data in this study are available at NCBI SRA under accession numbers SRR630492, SRR3466506, SRR3476690, SRR3476739, SRR5038768, and SRR5038792. The quality-filtered Iso-seq data are available on RhesusBase website (http://www.rhesusbase.org/download/download.jsp; last accessed July 31, 2017).
Processing and Evaluation of PacBio Data
PacBio data were processed and evaluated with several tools in SMRT Analysis (v2.2.0 and v2.3.0) and in-house pipelines (supplementary fig. S16, Supplementary Material online). Briefly, the raw reads in fastq format were extracted from h5 format by bash5tool in the pbh5tools packages. The suggested parameters (MinReadScore = 0.75, MinSRL = 50, MinRL = 50) were adopted to filter and trim the raw reads. The Circular Consensus module in ConsensusTools was then applied to extract ROI reads with the minimal predicted accuracy (minPredictedAccuracy) set at 75. Particularly, the parameter “minFullPasses = 0” was used to increase the sensitivity in ROI reads identification.
Primer sequences flanking ROI reads were trimmed by the hmmer_wrapper tool with the parameters “–primer_search_window 150 –min-score 8 –must-see-polyA –left-nosee-ok,” according to the sequences of primers used during library preparation. In this procedure, primer-trimmed reads were further processed to scan the PA tail (reads with a PA tail were referred to as PA-containing reads, otherwise as PA-free reads). PA-containing reads were trimmed to obtain PA-trimmed reads. To recover the possible PA-containing reads missed by hmmer_wrapper possibly due to the stringent parameter sets, PA-free reads defined by hmmer_wrapper were further processed using in-house scripts. Briefly, a 50-bp window was set at the tail of the read and slid toward the 5′ end. The read was trimmed off from the 3′ end of the sliding window until the A base fraction in the window was ≤0.65. 15,389 reads could be trimmed were recovered as PA-containing reads and pooled with the previous PA-trimmed reads identified by hmmer_wrapper to obtain the final PA-trimmed reads available for subsequent analysis. Reports of pre-mapping evaluation were generated with tools in SMRT Analysis, such as filter_stats, filter_subread_summary, and reads_of_insert_report (supplementary figs. S1 and S3, Supplementary Material online). GC content, base quality, sequencing error, DNA, or pre-mRNA contamination were also evaluated with in-house scripts (supplementary figs. S1 and S3, Supplementary Material online).
PA-trimmed reads were then mapped to the reference genome (hg19 and rheMac2) by GMAP (Wu and Watanabe 2005) with the parameters “–min-intronlength 70 –intronlength 1,100,000 –totallength 2,500,000 –trimendexons 9” for hg19, and “–min-intronlength 70 –intronlength 800,000 –totallength 2,000,000 –trimendexons 15” for rheMac2. The alignments generated were then filtered by in-house scripts, using the parameters (Coverage ≥67% and Identity ≥75%) consulted a previous study (Tilgner etal. 2013).
Next, several additional procedures were conducted to remove ambiguous reads. First, nonuniquely mapped reads were discarded using an alignment score defined as “Coverage × Identity.” For reads with multiple hits, those with an alignment score of the secondary hit <0.8 times of the score of the best hit were retained and defined as uniquely mapped reads. The threshold of 0.8 is actually more stringent than the 0.98 used in (Tilgner etal. 2013). Second, conflicting reads were discarded on the basis of the strand information. We first adjusted the strand information according to the sequence features of the Iso-seq reads. Briefly, due to the circular mode of Iso-seq, an Iso-seq read may be presented as the reverse complementary sequence of the transcript, and a stretch of T bases may be found at the front of the read. If a certain number of T bases could be found at the front of a read (with parameters similar to define the PA tail), the T bases were considered as PA tail and the read sequence was converted into its reverse complementary sequence accordingly. In addition, for spliced read, if the “Splicing Site Score” of its opposite strand was higher than that of the current strand by 2 scores, the strand was modified to the opposite strand. Here, “Splicing Site Score” was measured by summing the number of canonical splice site motifs (GT, GC, and AT for donor; AG and AC for acceptor). After these adjustments, the strand to which the read was mapped should be the same with the annotated transcript. Accordingly, if the strand was opposite to the annotation due to possible mis-mapping, this conflicting read was discarded. For unspliced read, more stringent criteria were used to control for false-positives, in that only reads with high degree of overlap with RefSeq transcripts (>80%) were kept.
Due to the intrinsically high error rate in PacBio sequencing, severe errors near the splice junction could introduce three types of alignment error: 1) missing a mini-exon, 2) adding an extra exon, and 3) misjudging an exon–intron boundary. For conflicting PacBio alignments, junction information from RNA-seq, as well as RefSeq Genes (release 78) for human and revised Ensembl annotations for rhesus macaque (Zhang etal. 2014) were further combined with the PacBio alignments to correct these errors. Briefly, for exons annotated by RefSeq but absent from PacBio alignments, we tried to find the missed exonic region in the two adjacent exons upstream and downstream of the candidate missed exon. We summed the lengths of extra exonic regions in the two adjacent exons in PacBio alignment relative to the RefSeq annotation. If the length of the candidate missing exon was comparable with the summed length (ratio between 50% and 200%), it was included in the PacBio alignment on the basis of RefSeq annotations. Similarly, a candidate extra exon was removed on the basis of RefSeq annotations. For conflicting exon–intron boundaries defined by PacBio, the junctions defined by PacBio alignments, RNA-seq, and RefSeq annotations were compared, and the ends of each junction in PacBio alignment were revised to the site with the most supporting evidence among these three types of junctions. The PacBio alignments were further filtered by discarding those with any exon overlapping with or adjacent to a genomic gap.
To eliminate false positives in PA identification due to DNA contamination or internal priming events, procedures similar to PA tail-trimming were conducted on the regions downstream of the 3′ end of the alignment to identify an A-rich stretch on the genome. Alignments were kept only when the PA tail was 20-bp longer than that of the A-rich stretch on the genome. In some cases, reads were aligned by soft clipping due to sequencing errors at the end of the reads, which may have some influence on the identification of PA sites. Alignments with a 3′-end clipping length >30 bp were discarded. Overall, PacBio alignments passing all the filters were defined as processed PacBio alignments and used in the subsequent analysis.
Identification and Evaluation of Alternative RNA Processing Events
To accurately identify AS events with the PacBio reads, processed PacBio alignments were further filtered. Briefly, alignments overlapping two different genes were discarded to obtain unambiguous alignments. These alignments were aggregated with RNA-seq junctions to identify SE, A5SS, and A3SS events. IR events were identified using only PacBio alignments. Two types of reads were needed to define an IR event: 1) reads completely covering the intron and the flanking exons; and 2) reads spliced at the donor and acceptor sites of the intron. The donor and acceptor sites were then used together to uniquely define an IR event. As for the definition of PA sites, the 3′ end of each processed PacBio alignment was defined as a cleavage site and clustered following a previously published approach (Derti etal. 2012). Briefly, cleavage sites within 30 bp were clustered into a 3′ cluster, and the cleavage site with the highest read coverage was defined as the PA site. If there were several cleavage sites with equivalent highest coverage, the downstream site was chosen. Alternative RNA processing events were identified in RefSeq Gene annotations using a similar approach and used as the reference to classify the alternative RNA processing events as being annotated or novel.
Combinatorial Mode of Alternative RNA Processing Events
The inclusion ratio was introduced to quantify the relative ratio of SE and A5/3SS events on the basis of RNA-seq reads, according to the previously reported approach (Wang etal. 2008). The inclusion ratios of IR events and PA frequency ratios were then calculated on the basis of the PacBio alignments. Briefly, the inclusion ratio is defined as the ratio of the number of inclusion reads to inclusion plus exclusion reads. Inclusion reads are those that support the retention of the alternative exon in SE, the alternative region in A5/3SS and the alternative intron in IR. Inversely, the exclusion reads are those that support the skipping of the alternative exon in SE, the spliced-out of the alternative region in A5/3SS and the alternative intron in IR.
To investigate the combinatorial mode of the alternative RNA processing events in one molecule, 10,000 iterations of Monte Carlo simulations were conducted with the null hypothesis of independent combination. Briefly, for a gene covered by N PacBio reads, N putative PacBio reads were simulated, with an inclusion ratio for each AS event and a PA frequency ratio for each APA event equal to the ratios as estimated using the real data (RNA-seq for SE and A5/3SS; Iso-seq for IR and APA). In such an occasion, false-positives introduced by low sequencing coverage were controlled. According to the simulation results, for each gene, the distribution of the expected number of simulated isoforms at the current sequencing depth was calculated (defined as “Expected”), and was compared with the real number of isoforms observed in Iso-seq data (defined as “Observed”). The Monte Carlo P value was then calculated as the proportion of times that “Observed” was less than “Expected.” A P value ≥0.05 indicated that “Observed” was not significantly less than “Expected,” that is, the combination is independent. In addition, the expected frequencies of isoforms were calculated according to the inclusion/exclusion ratio and PA frequencies, and compared with the observed. Exact multinomial tests were performed to determine whether the expected and the observed frequencies were consistent. A consistent comparable result (corrected P value ≥0.05) indicates that the combination is independent. The correlation of the expected and the observed frequencies were also quantified using the Spearman correlation coefficient.
Identification of Novel Isoforms
The 5′ and 3′ ends of processed PacBio alignments were clustered into 5′ and 3′ clusters, using an approach similar to the above cleavage-site clustering. The consensus split mapped molecule (CSMM) was defined from the processed PacBio alignments, with a shared 5′ cluster, 3′ cluster, and consensus splicing site at every split site. CSMMs were then compared with the RefSeq annotations (release 78), and a CSMM was assigned to a multi-exon gene if the CSMM and the gene shared at least one splice site in common. CSMMs of unclear origin (such as spanning multiple genes or partially aligning to one gene) were discarded.
The structures of CSMMs and annotated transcripts of the assigned gene were then compared, and a CSMM was defined as a novel isoform if it contained novel junctions, a novel PA site, or a novel combination of annotated junctions. To compare the CSMM coverage in annotated isoforms and novel isoforms, one novel isoform was defined as the major novel isoform when it had higher coverage than the annotated isoforms of the same gene.
Validation of Major Novel Isoforms by RNA-Seq, Poly(a)-Seq, and Targeted PacBio Sequencing
Major novel isoforms were classified into two types: 1) those only with a novel PA site, and 2) those containing novel alternative RNA processing events and/or a novel combination of annotated alternative RNA processing events. In order to evaluate whether the major novel isoforms were authentic, human brain Poly(A)-seq (GSM747470), and macaque brain Poly(A)-seq (GSM747488) data (Derti etal. 2012) were downloaded from the UCSC Genome Browser and used to evaluate the frequency of these candidate major novel isoforms. The inclusion ratio of each AS event and the frequency of each PA site were then calculated on the basis of the RNA-seq or Poly(A)-seq data as described earlier.
Ten randomly selected cases were also verified by RT-PCR enrichment and targeted PacBio sequencing. Briefly, using a SMARTer RACE 5′/3′ Kit from Clontech Laboratories, Inc., reverse transcription was carried out in three steps. First, 5× First-Strand Buffer (RNAse-free), 100 mM DTT, and 20 mM dNTPs were mixed and set aside at room temperature. Second, 1 µg of total RNA, 3′-CDS Primer A, and sterile water were mixed and incubated at 72 °C for 3 min, then cooled to 42 °C for 2 min. Finally, the buffer from the previous two steps was mixed with 20 U RNase inhibitor and 100 U SMARTScribe Reverse Transcriptase in a 20-µl reaction system. Samples were incubated at 42 °C for 90 min and at 70 °C for 10 min. The reaction product was then diluted with 90 µl Tricine-EDTA buffer. All PCR reactions were performed with the TaKaRa LA Taq kit (Takara Bio Inc.). To amplify the cDNAs, PCR reactions were performed in a 50-µl mixture containing 2.5 U of TaKaRa LA Taq, LA PCR Buffer II (Mg2+-free), 2.5 mM MgCl2, 2.5 mM dNTP mixture, 200 ng cDNA, 5 µl 10× Universal Primer Mix, and 12.5 µM Gene Specific Primer. After an initial denaturation step at 94 °C for 1 min, the reaction was performed for 30 cycles, with 30 sec at 94 °C, 30 sec at 60 °C, and 3 min at 72 °C. Final extension was carried out at 72 °C for 5 min. The PCR products were then purified by a QIAquick PCR Purification Kit for PacBio library preparation and sequencing. Primers used in the RT-PCR enrichment experiments are shown in supplementary table S5, Supplementary Material online.
Identification and Validation of Lineage-Specific Isoforms in Human and Rhesus Macaque
LiftOver (-minblocks = 0.6, -minmatch = 0.75) was used to link orthologous regions between human and rhesus macaque, and genes covered by at least one full-length PacBio read in both species were used in the comparative transcriptome study. A conserved isoform was defined as follows: 1) all splicing sites of the isoform were exactly matched in the two species, and 2) the 5′ and 3′ ends of the isoform were exactly the same, or no further apart than 30 bp, in the two species.
Considering the relatively low-throughput of Iso-seq, for nonconserved isoforms between species, we further combined the corresponding RNA-seq data with the same extraction of the Iso-seq to control for the false-positives and identify lineage-specific isoforms. Following the direct comparison of Iso-seq data between human and rhesus macaque, the matched RNA-seq data were further used to identify lineage-specific RNA processing events represented only in one species. Briefly, RNA-seq reads with deep coverage on the candidate lineage-specific isoforms were used to calculate the inclusion ratio of each event in human and rhesus macaque. Poly(A)-seq data were also used to evaluate the frequency of PA sites (Derti etal. 2012). One alternative RNA processing event was retained only when it represented specifically in one species, and not supported by RNA-seq in another species. Only isoforms with lineage-specific RNA processing events were kept and defined as lineage-specific isoforms.
To evaluate the accuracy of these isoforms specific to human but not rhesus macaque, three candidate genes with potential human-specific isoforms with different Iso-seq coverage were randomly selected, and the full-length isoforms of the corresponding genes were enriched by RT-PCR and sequenced with PacBio RS II, with a similar pipeline as described earlier. Primers used in the RT-PCR enrichment experiments were listed in supplementary table S4, Supplementary Material online.
Population Genetic Analyses in Human and Rhesus Macaque
On the basis of the polymorphism data from a population of 103 human individuals and 31 macaques animals (Chen etal. 2015; Zhong etal. 2016), we measured the nucleotide diversity (π) for the coding regions specific to the lineage-specific novel isoforms. Given that the individuals archived in 1000 Genomes project were sequenced at varied sequencing coverage (median sequencing depth of 7.1-folds) (Auton et al. 2015), genotyping and nucleotide diversity estimation for variants and ambiguous allele frequency may be problematic, particularly for low-coverage data of certain individuals. Rather than directly using previously annotated polymorphism sites in 1000 Genomes project, we thus profiled the human polymorphism data by reanalyzing whole genome sequencing data of 103 individuals from different subpopulations and archived with relatively high and comparable sequencing coverage (median sequencing depth of 12-folds; in phase-3 release of the 1000 Genomes Project) (Zhong etal. 2016). We used the same pipeline as 1000 Genomes Project to call variants, and finally identified 21,485,040 single nucleotide polymorphism sites across the whole genome, with 95.93% sites also archived in 1000 Genomes project. With whole-genome sequencing data of 31 independent macaque animals (median sequencing depth of 40-folds), we also compiled a comprehensive polymorphism map in rhesus macaque with 46,146,548 polymorphism sites (Chen etal. 2015; Zhong etal. 2016). We calculated the nonsynonymous/synonymous ratio of the nucleotide diversity for annotated coding regions, novel coding regions unique to novel isoforms, as well as regions of 18 human pseudogenes (Wang etal. 2006). The Wilcoxon one-tail tests were performed to determine whether the nucleotide diversity between synonymous and nonsynonymous sites, or the nonsynonymous/synonymous ratio of the nucleotide diversity, were significantly different, with a P value cutoff of 0.05.
We also performed codon-level alignment between human and rhesus macaque to clarify the divergent sites and polymorphic sites into synonymous and nonsynonymous groups. The divergent nonsynonymous sites (Dn), polymorphic nonsynonymous sites (Pn), divergent synonymous sites (Ds), and polymorphic synonymous sites (Ps) were then counted as described in the R package (Popgenome). Finally, the neutrality index and its associated P value were calculated by using published pipeline (Zhong etal. 2016).
Functional Annotations for Lineage-Specific Isoforms
Peptide evidence from large-scale mass spectrometry studies was extracted from PRIDE (Vizcaino etal. 2013), PeptideAtlas (Deutsch 2010), ProteomicsDB (Kim etal. 2014), and Human Proteome Map (Wilhelm etal. 2014). A peptide was considered to support the protein expression of an isoform only if 1) when performing BLAT similarity searches against all human proteins (RefSeq, BLAT settings -t = prot -q = prot -stepSize = 5), its whole sequence exactly matched the CDS region of the novel isoform, with the second-best hit in the proteome (if existing) including at least one mismatch; and 2) when performing BLAT similarity searches against the human genome, its whole sequence identically and exclusively matched the CDS region of the novel isoform (hg19 or rheMac2, BLAT settings -stepSize = 5 -stepSize = 5 -t = dnax -q = prot). Peptide sequences aligned to both novel isoforms and annotated isoforms were discarded. InterProScan5 (Jones etal. 2014) was also used with default parameters to find functional motifs for the coding regions specific to the lineage-specific novel isoforms. For functional enrichment analyses, DAVID (Huang da etal. 2009) was used to obtain a list of biological processes enriched in the genes with human-specific isoforms, with a P value cut-off of 0.05 in the Fisher’s Exact test.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
C.Y.L. conceived the idea and designed the study. S.J.Z. and C.W. performed most of the experiments. S.Y., A.F., X.L., J.Y.C., and A.H. performed part of the experiments. S.J.Z., C.W., S.Y., Y.L., Q.S., and X.Z. analyzed data and performed statistical analysis. C.Y.L., S.J.Z., C.W., B.C.M.T., and X.W. wrote the paper. All authors read and approved the final manuscript.
Supplementary Material
Acknowledgments
We thank Dr Heping Cheng at Peking University and Dr Yong E. Zhang at the Chinese Academy Of Sciences for insightful suggestions. This work was supported by grants from the National Key Basic Research Program of China (2013CB531202), the National Natural Science Foundation of China (31522032, 31471240, 31171269, and 31521062), and the National Young Top-Notch Talent Support Program of China. The funders played no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript. We acknowledge Dr IC Bruce for critical reading of the manuscript.
References
- Ameur A, Wetterbom A, Feuk L, Gyllensten U.. 2010. Global and unbiased detection of splice junctions from RNA-seq data. Genome Biol. 113: R34.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au KF, Sebastiano V, Afshar PT, Durruthy JD, Lee L, Williams BA, van Bakel H, Schadt EE, Reijo-Pera RA, Underwood JG, Wong WH.. 2013. Characterization of the human ESC transcriptome by hybrid sequencing. Proc Natl Acad Sci U S A. 11050: E4821–E4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR.. 2015. A global reference for human genetic variation. Nature 5267571: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, et al. 2012. The evolutionary landscape of alternative splicing in vertebrate species. Science 3386114: 1587–1593. [DOI] [PubMed] [Google Scholar]
- Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D.. 2000. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 107: 1001–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig U, Barbosa-Morais NL, Pan Q, Nachman EN, Alipanahi B, Gonatopoulos-Pournatzis T, Frey B, Irimia M, Blencowe BJ.. 2014. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 2411: 1774–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JY, Shen QS, Zhou WZ, Peng J, He BZ, Li Y, Liu CJ, Luan X, Ding W, Li S, et al. 2015. Emergence, retention and selection: a trilogy of origination for functional de novo proteins from ancestral LncRNAs in primates. PLoS Genet. 117: e1005391.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM, Babak T.. 2012. A quantitative atlas of polyadenylation in five mammals. Genome Res. 226: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch EW. 2010. The PeptideAtlas project. Methods Mol Biol. 604: 285–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. 2012. Landscape of transcription in human cells. Nature 4897414: 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, et al. 2009. Real-time DNA sequencing from single polymerase molecules. Science 3235910: 133–138. [DOI] [PubMed] [Google Scholar]
- Eswaran J, Horvath A, Godbole S, Reddy SD, Mudvari P, Ohshiro K, Cyanam D, Nair S, Fuqua SA, Polyak K, et al. 2013. RNA sequencing of cancer reveals novel splicing alterations. Sci Rep. 3: 1689.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sun Y, Li Y, Li J, Rao X, Chen C, Xu A.. 2011. Differential genome-wide profiling of tandem 3′ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res. 215: 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galante PA, Sakabe NJ, Kirschbaum-Slager N, de Souza SJ.. 2004. Detection and evaluation of intron retention events in the human transcriptome. RNA 105: 757–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 297: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. 2010. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 285: 503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halvardson J, Zaghlool A, Feuk L.. 2013. Exome RNA sequencing reveals rare and novel alternative transcripts. Nucleic Acids Res. 411: e6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SE, Song HK, Kim do H.. 2014. Identification of tissue-enriched novel transcripts and novel exons in mice. BMC Genomics 15: 592.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Scott HS, Qin G, Zheng G, Chu X, Xie L, Adelson DL, Oftedal BE, Venugopal P, Babic M, et al. 2015. Revealing missing human protein isoforms based on ab initio prediction, RNA-seq and proteomics. Sci Rep. 51: 10940.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 41: 44–57. [DOI] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B.. 2009. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci U S A. 10617: 7028–7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 309: 1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Lee D, Lee J, Park D, Kim YJ, Park WY, Hong D, Park PJ, Lee E.. 2015. Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat Genet. 4711: 1242–1248. [DOI] [PubMed] [Google Scholar]
- Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, et al. 2014. A draft map of the human proteome. Nature 5097502: 575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CY, Zhang Y, Wang Z, Zhang Y, Cao C, Zhang PW, Lu SJ, Li XM, Yu Q, Zheng X, et al. 2010. A human-specific de novo protein-coding gene associated with human brain functions. PLoS Comput Biol. 63: e1000734.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Rao X, Mattox WW, Amos CI, Liu B.. 2015. RNA-seq analysis of differential splice junction usage and intron retentions by DEXSeq. PLoS One 109: e0136653.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sun Y, Fu Y, Li M, Huang G, Zhang C, Liang J, Huang S, Shen G, Yuan S, et al. 2012. Dynamic landscape of tandem 3′ UTRs during zebrafish development. Genome Res. 2210: 1899–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP.. 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 1384: 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Gerhardt DJ, Dinger ME, Crawford J, Trapnell C, Jeddeloh JA, Mattick JS, Rinn JL.. 2012. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol. 301: 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkin J, Russell C, Chen P, Burge CB.. 2012. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science 3386114: 1593–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam DK, Lee S, Zhou G, Cao X, Wang C, Clark T, Chen J, Rowley JD, Wang SM.. 2002. Oligo(dT) primer generates a high frequency of truncated cDNAs through internal poly(A) priming during reverse transcription. Proc Natl Acad Sci U S A. 999: 6152–6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, et al. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44: D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Gupta R, Kim H, Wickramasinghe P, Baubet V, Showe LC, Dahmane N, Davuluri RV.. 2011. Alternative transcription exceeds alternative splicing in generating the transcriptome diversity of cerebellar development. Genome Res. 218: 1260–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ.. 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 4012: 1413–1415. [DOI] [PubMed] [Google Scholar]
- Sakabe NJ, de Souza SJ.. 2007. Sequence features responsible for intron retention in human. BMC Genomics 8: 59.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner D, Nguyen TM, Russo G, Heber S, Patrignani A, Ahrne E, Scheiffele P.. 2014. Targeted combinatorial alternative splicing generates brain region-specific repertoires of neurexins. Neuron 842: 386–398. [DOI] [PubMed] [Google Scholar]
- Sharon D, Tilgner H, Grubert F, Snyder M.. 2013. A single-molecule long-read survey of the human transcriptome. Nat Biotechnol. 3111: 1009–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y.. 2011. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA 174: 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS.. 2005. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 331: 201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Jahanbani F, Blauwkamp T, Moshrefi A, Jaeger E, Chen F, Harel I, Bustamante CD, Rasmussen M, Snyder MP.. 2015. Comprehensive transcriptome analysis using synthetic long-read sequencing reveals molecular co-association of distant splicing events. Nat Biotechnol. 337: 736–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Raha D, Habegger L, Mohiuddin M, Gerstein M, Snyder M.. 2013. Accurate identification and analysis of human mRNA isoforms using deep long read sequencing. G3 (Bethesda) 33: 387–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B, Gokce O, Quake SR, Sudhof TC.. 2014. Cartography of neurexin alternative splicing mapped by single-molecule long-read mRNA sequencing. Proc Natl Acad Sci U S A. 11113: E1291–E1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubby I, Bussani E, Colonna A, Stacul G, Locatelli M, Scudieri P, Galietta L, Pagani F.. 2013. TMEM16A alternative splicing coordination in breast cancer. Mol Cancer 12: 75.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino JA, Cote RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J, et al. 2013. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41(Database issue): D1063–D1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB.. 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 4567221: 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Grus WE, Zhang J.. 2006. Gene losses during human origins. PLoS Biol. 43: e52.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetterbom A, Ameur A, Feuk L, Gyllensten U, Cavelier L.. 2010. Identification of novel exons and transcribed regions by chimpanzee transcriptome sequencing. Genome Biol. 117: R78.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M, Schlegl J, Hahne H, Moghaddas Gholami A, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, et al. 2014. Mass-spectrometry-based draft of the human proteome. Nature 5097502: 582–587. [DOI] [PubMed] [Google Scholar]
- Wilkening S, Pelechano V, Jarvelin AI, Tekkedil MM, Anders S, Benes V, Steinmetz LM.. 2013. An efficient method for genome-wide polyadenylation site mapping and RNA quantification. Nucleic Acids Res. 415: e65.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JJ, Ritchie W, Ebner OA, Selbach M, Wong JW, Huang Y, Gao D, Pinello N, Gonzalez M, Baidya K, et al. 2013. Orchestrated intron retention regulates normal granulocyte differentiation. Cell 1543: 583–595. [DOI] [PubMed] [Google Scholar]
- Wu TD, Watanabe CK.. 2005. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 219: 1859–1875. [DOI] [PubMed] [Google Scholar]
- Xie C, Zhang YE, Chen JY, Liu CJ, Zhou WZ, Li Y, Zhang M, Zhang R, Wei L, Li CY.. 2012. Hominoid-specific de novo protein-coding genes originating from long non-coding RNAs. PLoS Genet. 89: e1002942.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Weyn-Vanhentenryck SM, Wu J, Sloan SA, Zhang Y, Chen K, Wu JQ, Barres BA, Zhang C.. 2015. Systematic discovery of regulated and conserved alternative exons in the mammalian brain reveals NMD modulating chromatin regulators. Proc Natl Acad Sci U S A. 11211: 3445–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap K, Lim ZQ, Khandelia P, Friedman B, Makeyev EV.. 2012. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes Dev. 2611: 1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Burge CB.. 2004. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 11(2-3): 377–394. [DOI] [PubMed] [Google Scholar]
- Zhang C, Gschwend AR, Ouyang Y, Long M.. 2014. Evolution of gene structural complexity: an alternative-splicing-based model accounts for intron-containing retrogenes. Plant Physiol. 1651: 412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Yang H, Yang H.. 2015. Evolutionary character of alternative splicing in plants. Bioinform Biol Insights 9(Suppl 1): 47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SJ, Liu CJ, Shi M, Kong L, Chen JY, Zhou WZ, Zhu X, Yu P, Wang J, Yang X, et al. 2013. RhesusBase: a knowledgebase for the monkey research community. Nucleic Acids Res. 41(Database issue): D892–D905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SJ, Liu CJ, Yu P, Zhong X, Chen JY, Yang X, Peng J, Yan S, Wang C, Zhu X, et al. 2014. Evolutionary interrogation of human biology in well-annotated genomic framework of rhesus macaque. Mol Biol Evol. 31: 1309–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YE, Landback P, Vibranovski MD, Long M.. 2011. Accelerated recruitment of new brain development genes into the human genome. PLoS Biol. 910: e1001179.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Peng J, Shen QS, Chen JY, Gao H, Luan X, Yan S, Huang X, Zhang SJ, Xu L, et al. 2016. RhesusBase PopGateway: genome-wide population genetics atlas in rhesus macaque. Mol Biol Evol. 335: 1370–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.