Abstract
Typically, animal mitochondria have very compact genomes, with few short intergenic regions, and no introns. Hence, it may seem that there is little space for unknown functions in mitochondrial DNA (mtDNA). However, mtDNA can also operate through RNA interference, as small non coding RNAs (sncRNAs) produced by mtDNA have already been proposed for humans. We sequenced sncRNA libraries from isolated mitochondria of Ruditapes philippinarum (Mollusca Bivalvia) gonads, a species with doubly uniparental inheritance of mitochondria, and identified several putative sncRNAs of mitochondrial origin. Some sncRNAs are transcribed by intergenic regions that form stable stem-hairpin structures, which makes them good miRNA-like candidates. We decided to name them small mitochondrial highly-transcribed RNAs (smithRNAs). Many concurrent data support that we have recovered sncRNAs of mitochondrial origin that might be involved in gonad formation and able to affect nuclear gene expression. This possibility has been never suggested before. If mtDNA can affect nuclear gene expression through RNA interference, this opens a plethora of new possibilities for it to interact with the nucleus, and makes metazoan mtDNA a much more complex genome than previously thought.
Keywords: mitochondria, RNA interference, sncRNAs, smithRNAs, doubly uniparental inheritance
Introduction
Since its origin from an α-proteobacterial ancestor, the mitochondrial DNA (mtDNA) has underwent a massive process of reductive genome evolution (Andersson and Kurland 1998; Khachane et al. 2007) becoming a bioenergetically specialized genetic system. Animal mitochondria usually have very compact gene-rich genomes, few short intergenic regions (except the Control Region) and no introns. Hence, there should be little space for more functions in animal mtDNA, other than coding the core subunits of oxidative phosphorylation (OXPHOS). However, additional mitochondrial genes involved in other biological functions have been described (see Breton et al. 2014 for a review). For instance, novel mtDNA-encoded proteins are known to act on pollen formation in angiosperms exhibiting cytoplasmic male sterility (CMS; Chase 2007). Another interesting case is found in octocorals, in which an additional gene (mtMutS) was proposed to be acquired by horizontal gene transfer (Bilewitch and Degnan 2011). ORFans (i.e., open reading frames having no detectable homology with known proteins; Fischer and Eisenberg 1999) were detected in mitochondrial genomes of some bivalves as well (Breton et al. 2011, Milani et al. 2013, Plazzi et al. 2013), and some of them were proposed to be involved in sex-linked functions in freshwater mussels (Breton et al. 2011), and in the Manila clam Ruditapes philippinarum (Milani et al. 2014a, 2014b, 2016), the subject of this study.
The Manila clam has an unusual mitochondrial inheritance system named doubly uniparental inheritance (DUI; Skibinski et al. 1994; Zouros et al. 1994), so far detected in about 50 bivalve species belonging to seven families. In DUI species, two mitochondrial lineages are found in gametes, one transmitted through eggs (the F-type, for female-inherited), the other through sperm (the M-type, for male-inherited). Their mtDNAs show up to 43% nucleotide divergence (Doucet-Beaupré et al. 2010). Although adult females are usually homoplasmic for the F-type, adult males are heteroplasmic: in fact, along with the F-type, the M-type is present in variable amounts in somatic tissues of males (Ghiselli et al. 2011), but M-type is the only mtDNA present in male germ line. The M-type mitochondrion, when present, has a transcriptionally functional genome (Milani et al. 2014a), and both M and F-type mitochondria are energetically active (i.e., they show the presence of membrane potential) in both soma and germ line (Milani and Ghiselli 2015).
The spatial distribution of sperm mitochondria in developing embryos has been studied in Mytilus ssp. and in R. philippinarum (both showing DUI) and a shared feature has been observed: sperm mitochondria follow two different patterns, the “aggregated” or “dispersed” patterns. In the aggregated pattern, sperm mitochondria remain clustered together after fertilization and are transported to the first cleavage furrow of the embryo, in the area where germ plasm is segregated; conversely, in the dispersed pattern, they are scattered across the embryo blastomeres (Cao et al. 2004; Obata and Komaru 2005; Cogswell et al. 2006; Milani et al. 2012). The two patters have been linked to sex: female embryos show the dispersed pattern, whereas male embryos show the aggregated pattern. The M-type will be the only mitochondrial type present in mature spermatozoa. Conversely, in females, sperm mitochondria are diluted or degraded, so that mature eggs, as well as female somatic tissues, are commonly homoplasmic for the F-type (Ghiselli et al. 2011). The above-mentioned observations led to the hypothesis that M-type mitochondria may have an active role in gonad masculinization, achieved through a series of specific signals between mitochondria and nucleus (Passamonti and Ghiselli 2009). Hence, this unique pattern of sex determination suggests novel systems of signaling between mitochondria and nucleus, which may be found in bivalves with DUI.
DUI has also been linked to the shift from hermaphroditism to gonochorism in bivalves (see Milani et al. 2016 for details), because all DUI species known so far are gonochoric, whereas most bivalves are hermaphroditic (Breton et al. 2011). Therefore, bivalves may be an interesting system to search for new mtDNA genes and functions related to sex. Moreover, in bivalves there is no evidence for sex chromosomes, and both gonads are anatomically very similar (Mackie 1984). It is unknown whether gonochoric bivalves have sex-determining nuclear loci or not, as genomic data are still very scarce. However, a model for sex determination in DUI species has been proposed and refined by several authors (Saavedra et al. 1997; Zouros 2000; Kenchington et al. 2002; Cao et al. 2004; Cogswell et al. 2006; Breton et al. 2007; Ghiselli et al. 2012; Diz et al. 2013; Zouros 2013; Milani et al. 2013), and some candidate nuclear genes have been proposed for R. philippinarum (Milani et al. 2013). Since homologs of such genes are involved in reproduction and ubiquitination, they may be also involved in the maintenance/degradation of sperm mitochondria during embryo development.
How did DUI evolve, and why is it maintained? It has been proposed that DUI arose because of genomic conflicts triggered by selfish elements that have invaded mtDNAs, resolved by the evolution of a segregation mechanism of sex-specific mtDNAs, which would have restored the uniparental inheritance of each mitochondrial type. Good candidates for mtDNA selfish elements are the lineage-specific ORFans, which have been proposed to derive from the endogenization of viral sequences (Milani et al. 2013). In this conception, DUI may have evolved multiple times, as different ORFans are found in DUI mtDNAs. In the above scenario, the previously mentioned nuclear genes are likely the restorer genes, which allow a strict segregation of the selfish elements assuring the uniparentality of mitochondrial inheritance (see Passamonti and Ghiselli 2009; and Milani et al. 2016 for details).
Notwithstanding its possible multiple origins, DUI is an evolutionary stable mechanism, which, as in the case of Unionidae, has persisted for at least 200 million years (Doucet-Beaupré et al. 2010). The establishment of two lineage-specific mitochondrial genomes represents a good prerequisite for mtDNAs to evolve new sex-related functions. Therefore, we hypothesized that, once nuclear genes have segregated M and F mitochondria into germ plasm, both M and F mtDNAs might have taken charge for subsequent gonad development by somehow driving sperm or egg formation, respectively (Passamonti and Ghiselli 2009). A role of mtDNA in germ line formation is at least conceivable, given that mitochondria are sister group of Rickettsiales, an order of α-proteobacteria able to distort host sex-ratio (see e.g., Wolbachia; Rigaud et al. 1999; Jiggin et al. 2000; Wang and Wu 2015). Moreover, cytoplasmic male sterility (CMS) is another good example of mitochondrial DNAs acting on germ line development (Chase 2007), and finally, the M-type ORFan of R. philippinarum is translated in the developing testis, and it was hypothesized that it might act as germ line distorter as well (Milani et al. 2015, 2016).
MtDNAs may also act on germ line development through RNA interference. Accordingly, we decided to explore small non coding RNAs (sncRNA) as a possible way for mitochondria to influence germ line gene expression and fate. SncRNAs are indeed good candidates as fast evolving functional elements, because they are known to originate through different processes and different kind of sequences (Shabalina and Koonin 2008).
Among sncRNAs, one of the best characterized categories are miRNAs, in which transcription profiles and target predictions have been better studied in model and nonmodel organisms. MiRNAs are small molecules with a length of usually about 21–22 nucleotides that are transcribed in the nucleus as pri-miRNAs of various lengths, then transported and matured by DROSHA and DGCR8 in the cytoplasm. The product is a pre-miRNA about 70 nt long that, once processed by the Dicer protein and the Argonaute Protein 2 (Ago2), becomes the mature miRNA (Ha and Narry Kim 2014). MiRNAs downregulate the translation of a specific mRNA by binding its UTR, usually with a perfect complementary seed sequence (Ambros 2004). The seed has been defined as nucleotides 2–8 of the miRNA, and usually it pairs perfectly with the 3′ UTR of the target mRNA (Lewis et al. 2003). Even if this is one of the features that best characterizes miRNA targets, the mechanism of action of miRNAs seems to be more complex than this, because of noncanonical seed-target pairing, different outcomes of miRNA binding, and different biogenesis. For example, some miRNAs enhancing mRNA transcription have been described (Ha and Narry Kim 2014; Cloonan 2015). Moreover, miRNA biogenesis does not always start from a pri-miRNA, but pre-tRNAs (Cloonan 2015) and pre-rRNAs (Son et al. 2013) can generate functional miRNAs as well. Accordingly, there are many conceivable ways in which the compact mitochondrial genome can originate miRNA-like products.
Another kind of sncRNAs is the piRNA class: piRNAs are associated with PIWI proteins and in some way they interact with these proteins in order to silence transposons and foreign DNA (Czech and Hannon 2016). The maturation of piRNAs has only recently been described in silkworms (Izumi et al. 2016) and C. elegans (Tang et al. 2016). Typically, large, noncoding RNAs are transcribed and processed into piRNA intermediates by a complex chain of reactions, involving the cleavage of the original transcript by the Zucchini endonuclease (Czech and Hannon 2016).
The existence of mitochondrially-encoded sncRNAs was firstly confirmed by Mercer et al. (2011) for the human mitochondrial genome. Most sncRNAs detected in this pioneering study were found to be associated with tRNAs, which would be in agreement with the observation that nuclear tRNAs may enter Dicer-/RNaseZ-dependent pathways and generate RNA fragments connected with RNA interference (RNAi) (Lee et al. 2009; Haussecker et al. 2010).
Mercer et al. (2011) focused on sncRNAs expressed more than the median expression level of miRNAs in human 143B cells. However, a later study was able to identify a larger population of mitochondrially-encoded sncRNAs in both humans and mice, which were called mitosRNAs (Ro et al. 2013). Interestingly, most of them are encoded 1) on the H strand, where most mammalian genes are also located, 2) within the known 37 coding genes, and 3) in a sense orientation with the harboring gene.
The bidirectional transcription of mitochondrial genome necessarily generates sense and antisense RNAs that may interfere with each other (Barber 2001; Chakrabarti et al. 2011). Therefore, this wide array of sense sncRNAs was initially interpreted as a way for mtDNA to prevent the sense–antisense pairing of primary transcripts (as sense sncRNAs would bind antisense mRNAs): indeed, it was demonstrated that the overexpression of sense mitosRNAs enhances the expression of host genes (Ro et al. 2013). Notably, although RNAi machinery (see Agrawal et al. 2003) was found in mitochondria by Bandiera et al. (2011), the same was not found by Ro et al. (2013). Therefore, it is possible that either mitosRNAs do not act at a posttranscriptional stage, or that mitosRNA-mediated regulation in mitochondria is not a typical RNAi. Ro et al. (2013) also suggested that mitosRNAs may act on mitochondrial gene methylation, an epigenetic mechanism which is known from mtDNA too (see Shock et al. 2011).
Given the discovery of sncRNAs in the mt genomes of humans, and the possible existence of unusual mito-nuclear signaling in DUI species, we searched for sncRNAs in the mtDNA of the Manila clam R. philippinarum, by using the above-mentioned knowledge, and target prediction methods available for nonmodel organisms. Since we were interested in interactions between mitochondria and germ line formation, we performed a mitochondrial enrichment protocol on gonad samples of both sexes, we prepared and sequenced small RNA libraries, and we analyzed putative sncRNAs based on their sequence features, location on mtDNA, thermodynamic stability, and possible nuclear targets. Our analysis provided a good number of putative sncRNAs coded by the M or F mitochondrial genomes, some of them represented thousands of times in our libraries. We think that these sncRNAs, or at least some of them, may have a biological function, and, differently from mitosRNAs, may act on nuclear targets. For this reason, we decided to name them small mitochondrial highly-transcribed RNAs (smithRNAs).
Results
smithRNA Characterization
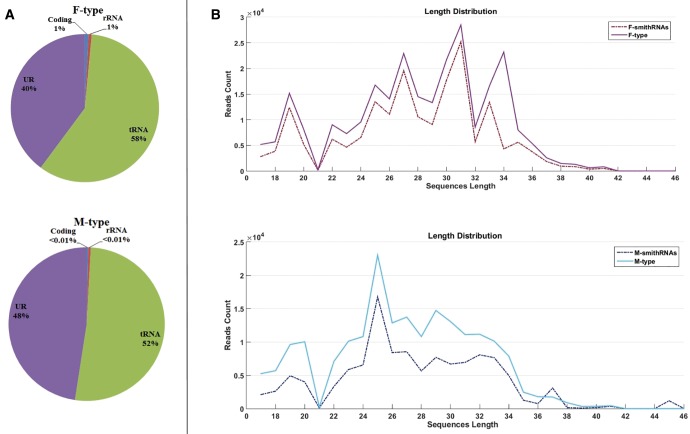
From six individuals of Ruditapes philippinarum, we obtained 386,135 filtered reads mapping to the F-type mtDNA, and 195,538 ones mapping to the M-type mtDNA. As shown in figure 1A, the reads maps almost exclusively to Unassigned Regions (URs) or tRNA genes.
Fig. 1.
The pie charts in panel (A) show the proportion of reads mapping to different regions of F- and M-mtDNA: URs (violet), tRNAs (green), rRNAs (red), and coding genes (blue). The length distribution of reads is shown in panel (B). The dash-dot line represents the 50 selected smithRNAs (see main text for details), and the solid line represents all the reads mapping to the mitochondrial genomes (F-type above, M-type below).
Among the above-mentioned putative sncRNAs, we selected 50 smithRNAs (see details about the rationale of this selection in the Material and Methods section), and their length distribution is reported in figure 1B, and compared with that of the total reads mapping on mtDNAs. The two length distributions are quite similar. On the other hand, length distribution for the total sncRNA library (which includes nuclear sncRNAs as well) is quite different (see Supplementary material S1, Supplementary Material online).
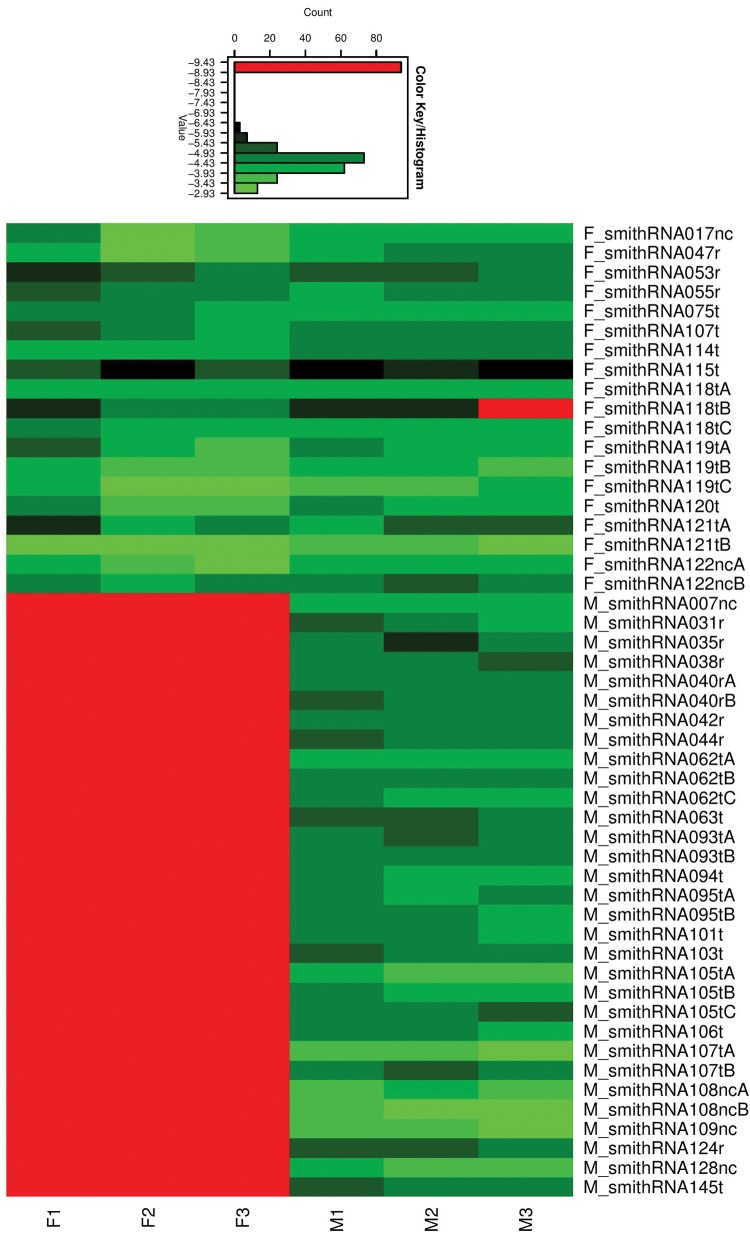
The log10 transcription level of the 50 M- and F-smithRNAs (from now on, M_ and F_smithRNAs, respectively) is reported in figure 2. The heatmap shows that M_smithRNAs are highly transcribed (black/green) in males, and absent (red) in females (as expected, because M-type mtDNA is absent in females): in general, all the 195,538 reads mapping to the male mtDNA came from the three male specimens. On the other hand, F_smithRNAs are homogeneously transcribed at high rate (black to light green) in female samples, whereas males show a heterogeneous presence of these RNAs (light red to light green): in general, the 386,135 reads mapping to the female genome came from both male and female individuals. Further information about the transcriptional levels are reported in Supplementary material S2, Supplementary Material online. As we will discuss in the next section, this distribution is what is expected in a DUI system.
Fig. 2.
The heatmap shows the transcription level of 50 putative smithRNAs in six individuals of R. philippinarum. The color scale is shown above the heatmap: for each transcriptional level (expressed as log10RPM) the associated color is shown, as well as the respective number of cells in the heatmap plotted on the “Count” axis. F1–F3: female samples; M1–M3: male samples; F_ prefix: smithRNAs mapping to F-type mtDNA; M_ prefix: smithRNAs mapping to M-type mtDNA.
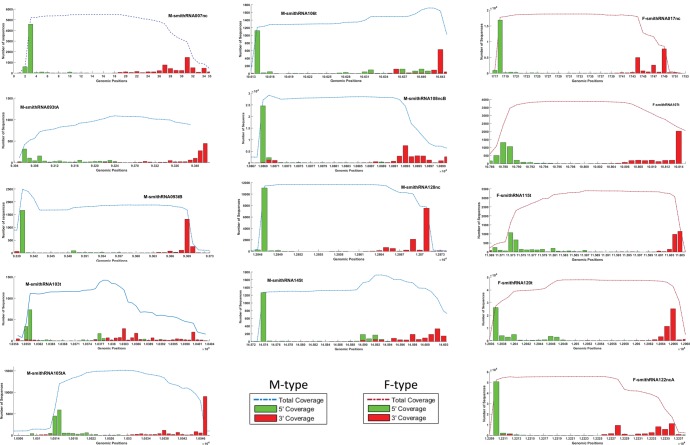
We also tried to infer if smithRNAs could be functional by analyzing the conservation of their 5′ ends. Because we do not know their biogenesis, we used both miRNA and piRNA as a model, the latter having a less conserved 3′ end (Ha and Narry Kim 2014). This step led to the restriction of the initially selected 50 smithRNAs to 14 molecules that have a very clear-cut 5′ end (fig. 3). This pattern is unlikely to be obtained by chance. Their sequences are available on figshare at https://figshare.com/s/e5db6cd8be1b0ec68087. Plots for the other 36 excluded putative smithRNAs are shown in Supplementary material S3, Supplementary Material online.
Fig. 3.
Coverage plots of the 14 smithRNAs showing the most conserved 5′ ends (i.e., almost all reads map at the very same base, or the following one, in mtDNA. Dash-dot lines represent the total coverage distribution of each position on the mitochondrial genome in the smithRNA range. Green bars show the number of reads starting at that position (5′end), and the red bars show the number of reads ending at that position (3′ end). The M_smithRNA093tA and B are plotted together in the M_smithRNA093t cluster due their overlapping starts/ends.
Target Prediction
By seed pairing, we identified at least one target nuclear gene for each of the selected 14 smithRNAs. Unfortunately, statistical tests for enrichment of putative functions of the predicted targets could not be performed due to the low sample of smithRNAs. The smithRNAs, their putative targets, and the corresponding Uniprot entry number are reported in Supplementary material S4, Supplementary Material online. No smithRNA targets mtDNA genes. Both M_ and F_smithRNAs putative targets include a wide range of biological processes, but there is some noticeable patterns. For instance, F_smithRNAs target three transcripts involved in microtubule interactions: 1) Ankyrin-3-like, which is able to link membranes to cytoskeleton; 2) Dynein heavy chain 2-like, axonemal, which is involved in the flagellar assembly; and 3) CLIP-associating protein 1, which is involved in microtubule stabilization at the plus end. Among the other transcripts targeted by F_smithRNAs, one is particularly interesting for a DUI system: the Nuclear receptor subfamily 0 group B member 1 (NR0B1). This protein has been reported to be crucial for sex determination in several species, including one bivalve (Crassostrea gigas; Vogeler et al. 2014, 2016). M_smithRNAs target three transcripts related to microtubule motors, four involved in chromatin remodeling, and nine in other biological processes. The motor proteins are: 1) Dynein heavy chain 1, which transports vesicles and organelles; 2) Dynein heavy chain 2-like, axonemal, described above; and 3) Kinesin 2-like, which is involved in vesicle and organelle transport, as well. Proteins involved in chromatin remodeling are: 1) the DNA polymerase subunit epsilon, a DNA polymerase able to affect the histone H3 acetylation; 2) the Histone-lysine N-methyltransferase SETD8, involved in histone monomethylation; 3) the Histone-lysine N-methyltransferase SETD2, which is able to trimethylate the H3 histone; and 4) Elongator complex protein 5, which acetylates the H3 histone. Among other M_smithRNA targets, U3 small nucleolar RNA-associated protein 6 is involved in translation, and the Enhancer of mRNA-decapping protein 4 in RNA degradation. Finally, even if both M_ and F_smithRNAs target the protein Dynein heavy chain 2-like, axonemal, they are predicted to act on different transcripts. The target prediction scores for each smithRNA are shown in the Supplementary material S4, Supplementary Material online.
Precursor Prediction
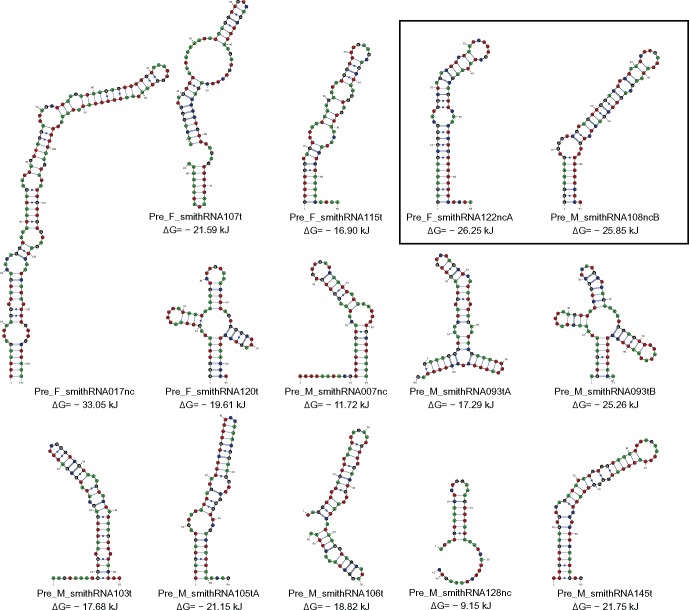
Following the hypothesis of a similar biogenesis between known sncRNAs and smithRNAs, we looked for putative precursor structures in the regions of mtDNA from which they originate. Figure 4 shows such structures. On the right side of the figure there are the two smithRNAs having the highest transcription levels (F_smithRNA122ncA, and M_smithRNA108ncB). Pre-smithRNAs showed scores comparable with those expected for miRNAs (Supplementary fig. S5A, Supplementary Material online), and formed hairpin structures that might be processed, for instance, by Dicer. Furthermore, in most cases, predicted smithRNA precursors have secondary structure ΔG values similar to or lower than random fragments of the same length extracted from different regions of the respective genome (Supplementary fig. S5B, Supplementary Material online), including rRNA and tRNA regions. However, for the male genome, tRNA scores were surprisingly comparable to the scores of random fragments of the same length extracted from other M-type regions, perhaps implying that ΔG analyses are not very well performing for this unusual experimental system.
Fig. 4.
Putative premiRNA-like precursor structures. In the box, the best candidates for each sex are shown: F_smithRNA122ncA and M_smithRNA108ncB. The variation of Gibbs free energy (ΔG) is reported below each structure, as calculated by RNAfold.
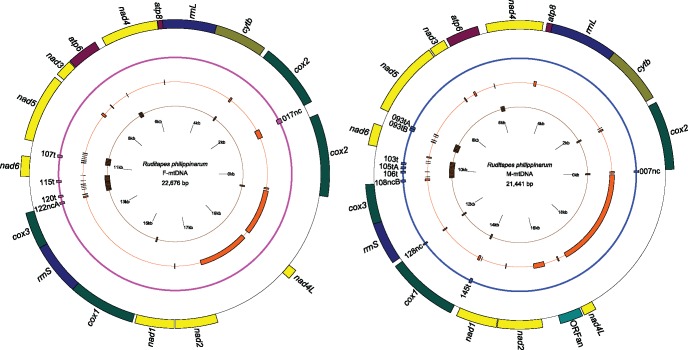
Figure 5 shows R. philippinarum M- and F-type mitochondrial genomes on which we annotated the pre-smithRNAs, URs, tRNAs, rRNAs, and coding genes. The precursors are named using the first letter for the sex identification (F/M) and the numbers and letters of the smithRNA name (i.e., F017nc for the pre_F_smithRNA017nc). Again, all the 14 smithRNAs for which we were able to predict a target nuclear gene map either to URs or tRNAs.
Fig. 5.
Maps of the F- and M-type mtDNAs. From the outer circle to the center: genes, presmithRNAs, URs, and tRNAs. Colors in the gene circle: NADH dehydrogenase complex (yellow); Cytochrome bc1 complex (olive green); Cytochrome c oxidase (dark green); ORFan (pale green); ATP synthase (purple); rRNAs (blue); presmithRNAs mapping to the F-type mtDNA (pink); presmithRNAs mapping to the M-type mtDNA (blue); URs (orange); and tRNAs (brown).
Discussion
The results we obtained are congruent with the hypothesis that mt genomes in the Manila clam Ruditapes philippinarum encode highly expressed sncRNAs (smithRNAs) that may have functional relevance. The distribution and the abundance of smithRNAs is consistent with predictions for a DUI system. Male gonads showed a high level of M_smithRNAs—as expected, since sperm is homplasmic for M—and a lower level of F_smithRNAs—as expected, because gonad tissue dissection can retain somatic parts containing F-type mtDNA. On the other hand, in ovaries only F_smithRNAs are found, as females are usually homoplasmic for F-type mtDNA. It is important to notice that female germ line may be considered as a physiological M-type Rho-0 cell lineage (sensuHashiguchi and Akiyama 2009), due to the absence of the M-type mitochondria. It is also worth noting the total absence of smithRNAs from the mtDNA lagging strand. This is also true for almost all the raw reads of our libraries (data available from the authors). As no other gene is present on the lagging strand as well (a feature that is typical of most bivalves, see Plazzi et al. 2016), these concurring observations can be taken as an indication that smithRNAs are processed as a part of the whole mitochondrial polycistronic RNA. In fact, several presmithRNAs are also spacers between genes in the URs, and others are in tRNAs. Previously characterized mitochondrial sncRNAs (from Mercer et al. 2011) are also made from URs and tRNAs. Finally, the selected 14 smithRNAs show: 1) very conserved 5′ ends, 2) 5′ phospate and 3′ hydroxil groups, 3) accessibility to their target UTRs, and 4) a ΔG pairing value on their UTR targets smaller than the suggested thresholds (see Materials and Methods). These concurring observations make the possibility that these RNAs lack any biological function very unlikely, in our opinion.
Finally, ΔGs of putative precursors are comparable with, or slightly worse than ΔG of well-characterized miRNAs (Supplementary material S5A, Supplementary Material online), and are generally lower or equal to ΔGs of fragments of similar lengths randomly extracted from both M and F mitochondrial genomes (Supplementary material S5B, Supplementary Material online), including rRNA and tRNA sequences that are known to have highly complex secondary structures. However, the observation that ΔGs of tRNAs are not significantly lower than ΔGs of other mtDNA regions (e.g., coding regions) suggests some caution, and ΔG computations must be taken only as grossly indicative in our opinion.
Because a sequenced genome for R. philippinarum is not available yet, we cannot exclude that at least some smithRNAs may originate from nuclear mitochondrial DNA segments (NUMTs). However, this hypothesis seems unlikely for many reasons: 1) NUMTs have not been reported yet for molluscs (Bensasson et al. 2001), 2) our previous molecular studies on R. philippinarum had never retrieved indirect evidence of NUMTs, and 3) M_smithRNAs could only be NUMTs if the inserted mtDNA would be cotranscribed with other male-specific nuclear genes, which we think is unlikely, because R. philippinarum has no obvious sex chromosomes, and nuclear-encoding sex loci have not been found so far in this species.
SmithRNAs Transport and Biogenesis
We reckon that smithRNAs must be transported to the cell cytoplasm in order to function through RNA interference, and specific in situ experiments are already planned for the next R. philippinarum reproductive season to confirm whether smithRNAs can migrate outside the mitochondria or not. However, the transport of mitochondrial RNA outside mitochondria has been already observed before: for example, Maniataki and Mourelatos (2005) observed the presence of several mitochondrially encoded tRNAs in the cytoplasm of human cells, and one of them was associated to the Ago2 protein, which is required for RNA-mediated gene silencing. Moreover, ribosomes of mitochondrial origin, along with extruded cristae and matrix material, are released in the germ plasm of Drosophila (Amikura et al. 2001). The inhibition of such ribosomes blocks germ line formation. A similar release of mitochondrial material has been observed in R. philippinarum as well (Milani et al. 2011), and this may be the mechanism by which some Manila clam smithRNAs can invade germ plasm. In conclusion, active transportation in the cytoplasm, and gross release after mitochondrial lyses in the germ plasm, are both suitable mechanisms to transport smithRNAs outside the mitochondrion.
At this stage we have no information about smithRNA biogenesis, but a role of Dicer is reported in mitosRNAs in human and mice cells (Ro et al. 2013), so smithRNAs might follow a similar process. However, we may propose some other possibilities, which can account for the observed length heterogeneity of smithRNAs, as reported in figure 3. For instance, Liu et al. (2015) show a Dicer-independent pathway in which Ago2 process pre-miRNAs, and produces a shorter version of a known miRNA. Interestingly, Bandiera et al. (2011) showed that Ago2 is colocalized with mitochondria, and this may suggest a role of this protein in the biogenesis of mitochondrially derived sncRNAs. We also found smithRNAs longer than 30 nucleotides that could have a maturation similar to piRNAs, in which 3′ editing is common (Izumi et al. 2016). Indeed piRNAs are usually longer than miRNAs, because of the 3′ editing.
MiRNAs are known to have a 5′ phosphorylation (Rana 2007), which is necessary for their maturation (Park et al. 2011). On the contrary, RNAse A, which is involved in RNA degradation, leaves the 5′ end hydroxilated and the 3′ end phosphorylated (Houseley and Tollervey 2009), the opposite to what we observed for smithRNAs. Our hypothesis is that the polycistronic transcript from the mitochondria is used as pri-miRNA, then a precursor similar to a pre-miRNA is produced, and it is eventually processed to obtain a mature smithRNA.
Are smithRNAs Involved in DUI?
In the present study, we used only in silico analyses, and of course in vitro and in vivo experiments are needed to confirm that the predicted smithRNAs are functional. R. philippinarum is in due course of characterization as a model species for mitochondrial biology and inheritance, and in vivo analyses will be only possible when we will develop appropriate protocols for this species.
Among all the predicted smithRNAs, M_smithRNA108ncB and F_smithRNA122ncA (see fig. 4) are the most promising candidates for functionality. They: 1) act on a single target, 2) they have the highest expression amongst the examined smithRNAs (see fig. 2), 3) are predicted to form an hairpin structure with optimal ΔG, and 4) are transcribed from formerly unassigned mtDNA regions.
The F_smithRNA122ncA was predicted to most likely target a transcript producing the NR0B1 protein, and a role of the nuclear receptor gene family in sex determination has been reported in C. gigas (Vogeler et al. 2014; Vogeler et al. 2016). The GO terms associated with this protein describe it as a masculinizing factor, and its mammalian homolog, DAX1, is transcribed at the highest level during spermatogenesis in mouse (Tamai et al. 1996). If this protein has a similar masculinizing effect on R. philippinarum gonad development, its repression by F_smithRNA122ncA (alone, or with other concurring factors) might shift the developmental cascade towards the production of an ovary, instead of a testis. The absence of any sex-specific structure in the gonad of R. philippinarum would simplify the feminizing effect of F_smithRNA122ncA, because the only target of regulation would be the gametogenic pathway. Interestingly, the same rationale—and an opposite effect—has been already hypothesized for rphm21, the ORFan of putative viral origin located in the R. philippinarum M-mtDNA at a different locus, whose product has been proposed to have a role in germ line masculinization (Milani et al. 2014b).
The predicted target of M_smithRNA108ncB is Dynein heavy chain 1, which is involved in the transport of molecules and organelles across the cell (Devine et al. 2016). Such movements are unidirectional, since dynein “walks” only towards the minus end of the microtubules. In DUI males, we know that sperm mitochondria migrate to the first cleavage furrow—towards the microtubule midbody—in two blastomere embryos (Milani et al. 2011). We do not know the molecular details on the processes involved in the migration of sperm mitochondria during male embryonic development, but it is at least conceivable that M_smithRNA108ncB might have some role in facilitating sperm mitochondria aggregate to move towards the first cleavage furrow. In this case, it would be transcribed during spermatogenesis and stored in the spermatozoon, reaching the zygote at fertilization; its function may be achieved by suppressing (perhaps locally) the translation of Dynein heavy chain 1 mRNAs.
Besides the two above mentioned smithRNAs, others may be tentatively linked to feminizing or masculinizing effects, in line with the DUI phenomenon. Generally speaking, the predicted targets of many smithRNAs are related to cytoskeleton organization and microtubule motors. For instance, M_smithRNA103t, which targets Kinesin 2 (KIF21A) may have a role on sperm mitochondria movements. Interestingly, in vitro knock-out of genes belonging to the KIF superfamily produces a mitochondrial aggregation (Hirokawa et al. 2009), which is similar to that observed in DUI male embryos. Another intriguing observation is that 4 M_smithRNAs are involved in chromatin remodeling, which could represent an indirect way for smithRNAs to affect gene expression of many genes, and drive gonad development towards spermatogenesis.
Are smithRNAs an Odd Feature of an Odd System?
Are smithRNAs a peculiar feature of DUI systems? The literature seems to point to a broad distribution of small mitochondrial RNAs, since they have been already found in mice and humans (Mercer et al. 2011; Ro et al. 2013). Ro et al. (2013) reported that two mitosRNAs are able to enhance the transcription of mitochondrial genes in vitro, but we cannot find any mitochondrial target gene for R. philippinarum smithRNAs.
Compared with mitosRNAs, smithRNAs are highly transcribed and may also have putative nuclear targets, so they may be a new form of retrograde signaling (i.e., the mitochondria-to-nucleus signaling). If confirmed by functional experiments, this new signaling system should be added to the known mitonuclear interactions (see f.i. Monaghan and Whitmarsh 2015). Conversely, nuclear regulators of organelle gene expression (ROGEs), which are proteins involved in the anterograde regulation, influence mitochondrial transcription (Woodson and Chory 2008), so it has to be determined whether smithRNAs would only be a case of retrograde signaling, or there is some sort of feedback regulation of smithRNA transcription through ROGEs.
In any case, smithRNAs are novel sncRNAs, originating from mtDNA, that have not been predicted (or even hypothesized) before to influence nuclear gene expression. This functional aspect of smithRNAs may be a unique feature of DUI systems, or could be more widespread, but not recognized yet. Although focused researches are needed, we would not expect that smithRNAs are limited to DUI species only, but would rather be present also in other eukaryotes. In fact: 1) mitochondria have been implicated in the evolution of sex under several hypotheses (see for example Lane 2005, Havird et al. 2015, Garg and Martin 2016, Radzvilavicius 2016, Radzvilavicius et al. 2016), so mtDNA may act as a sex-determining factor or be involved in sexual differentiation; 2) in animals the presence of intergenic regions could represent DNA with previously unrecognized functions (e.g., small RNAs), rather than “junk” DNA, and looking at transcription of these regions in other animal lineages could be a fruitful avenue for future research; 3) SmithRNAs may be primarily involved in sex determination in DUI species only, but they could have other functions (affecting other nuclear genes) in other systems; 4) a role of smithRNAs on creating reproductive isolation between closely related species may be hypothesized; and 5) SmithRNAs might also play a role in androgenesis, a peculiar reproductive mode observed also in some bivalves (e.g., Hedtke et al. 2008).
In any case, DUI represents a suitable experimental system to study mitochondrially derived sncRNAs, because its natural heteroplasmy allows for internal controls without artificial modifications. Since DUI probably evolved as a modification of the more common strictly maternal inheritance of mitochondria, its study would be particularly important to understand mitochondrial inheritance in all metazoans. Finally, this would be an important step to understand the role, when present, of mtDNA intergenic regions, which are commonly found in several animal groups, including bivalves (Mercer et al. 2011; Ghiselli et al. 2013; Ross et al. 2016; Wang et al. 2016).
Materials and Methods
Sample Preparation
Ruditapes philippinarum individuals were collected in Goro (Italy) in a single sampling campaign during the reproductive season (July/August 2016). Single clams were placed in beakers with artificial seawater—reverse osmosis water with Red Sea Coral Pro aquariology sea salt (Red Sea Europe, Verneuil-sur-Avre, France) added—that was changed every 12 h. Sex was assessed by microscopic examination of needle biopsy of gonadal tissue; three female and three male gonad samples were collected.
Half of each gonad sample was removed and placed in 1.5 ml of ice-cold isolation buffer [IB (pH 7.5): 400 mM sucrose, 100 mM KCl, 50 mM NaCl, 16 mM EGTA, 30 mM Hepes; modified after precedent works (Sokolova 2004; Munro et al. 2013)]. Samples were minced with scissors and homogenized with three passes (200 rpm) of a Potter-Elvenhjem homogenizer using a loosely fitting Teflon pestle, keeping on ice. Homogenates were then centrifuged at 2 °C at 2,500 × g for 10 min, and supernatants were centrifuged at 11,000 × g for 12 min. The obtained pellets, enriched in mitochondria, were processed for RNA extraction.
RNA was extracted using TRIzol (Thermo Fisher Scientific) following the protocol of Rio et al. (2010), and its quality/quantity was assessed with Bioanalyzer (Agilent) and NanoDrop 1000 (Thermo Fisher Scientific). Libraries were prepared by Macrogen, Inc. (Seoul, Korea) using TruSeq Small RNA Library Preparation Kits (Illumina). To select only products of RNA processing—and not from RNA turnover—the library was enriched in small RNAs (15–51 nt) whose 5′ phosphate and 3′ hydroxyl termini were preserved (Nottingham et al. 2016; Sun et al. 2016). These chemical modifications are common from RNAs that come from specific maturation pathways, such as miRNA or piRNA (Rana 2007; see also the TruSeq Small RNA Library Preparation Kits Manual); on the contrary, RNA processing by exonucleases is the most common pathway for RNA degradation (Houseley and Tollervey 2009) and usually leaves a hydroxyl group at the 5′ terminus.
Sequencing and Filtering
Sequencing was performed on an Illumina HiSeq 2000 instrument. We obtained about 23 million 51 bp single-end reads for each sample, for a total of 141 million reads. All raw Illumina sequences are available for download from the NCBI Short Read Archive (SRA) under the accession number SRP076494. Reads were filtered with a phred score threshold of 20, with a 4 bp-sliding window, using Trimmomatic0.35 (Bolger et al. 2014). Reads were also filtered out if they were similar to known PCR primers and Illumina adapter sequences. The sequence length distribution was checked using Avalanche NextGen Workbench (http://www.dnabaser.com/download/Avalanche_NextGen/index.html).
Reads were then aligned to R. philippinarum F- and M-type mitochondrial genomes (GenBank Accession Numbers: AB065375.1 and AB065374.1, respectively) using Bowtie2 (Langmead and Salzberg 2012) as implemented in the UGENE (Okonechnikov et al. 2012) bioinformatics platform. The end-to-end option was chosen with a one-nucleotide mismatch allowed. The reads aligned to each genome were imported in R (R Development Core Team 2008), and their length distribution analyzed using the seqinr package (Charif and Lobry 2007). Aligned reads were visualized using the SeqMonk program (see http://www.bioinformatics.babraham.ac.uk/projects/seqmonk) by choosing the quantification pipeline “wiggle”, setting 1 as a probe. We used this quantification to obtain the coverage that was used to draw the pie charts in figure 1A. The coverage of the starting nucleotide (5′) and the ending nucleotide (3′) of each read was extracted from the alignment using the bedtools task genomecov (Quinlan and Hall 2010).
To reduce chances of bioinformatics artifacts and false positives, we selected only smithRNAs that had a single or two close starting points. All the reads were clustered through the UCLUST algorithm, performed by USEARCH (Edgar 2010) using -id 0.99 as identity filter and storing both centroids sequences (CS) and cluster sequences (ClS). The -id option sets the identity percentage between the centroid sequence and all the sequences in the cluster. Every cluster is formed by sequences with 100% identity, all mapped to mtDNA. These settings were chosen to avoid several isoforms of the same sequence grouped in the same cluster (i.e., isoMIRs, Morin et al. 2008): indeed the variation of one or two nucleotides in the miRNA seed is sufficient to undermine its binding to the target UTR (Tan et al. 2014).
smithRNA Nomenclature
The definition “highly transcribed” we used for smithRNAs may seem arbitrary; however, a way to avoid sncRNAs found in a low copy number to be classified as smithRNAs is mandatory. The estimation of a minimal threshold to predict a biological meaning of a given miRNA is still an open question; Mercer et al. (2011) defined as highly transcribed every small RNA originated from the mitochondria that was expressed as much as the median of the endogenous miRNAs in 143B cells but, of course, we still do not have such information in our system. To reduce the chance of false positives, we arbitrarily decided not to include in the present analyses all clusters that had a size ≤200 sequences; as a result, analyzed centroids (smithRNAs) are represented by at least 200 sequences obtained by several overlapping reads (e.g., the 50 smithRNAs reported in fig. 1 were each based on 8,943 reads on average; see Supplementary material S2, Supplementary Material online). Finally, to avoid sequencing artifacts, we only considered sequences to be putative smithRNAs if they were highly transcribed in almost two out of three samples per sex.
SmithRNAs were named according to the following rules: the F_ prefix is assigned to smithRNAs transcribed from F-type mtDNA; the M_ prefix is assigned to smithRNAs transcribed from M-type mtDNA; the -nc suffix is assigned to smithRNAs transcribed from URs; the -t suffix is assigned to smithRNAs transcribed from a tRNA gene; the -r suffix is assigned to smithRNAs transcribed from a rRNA gene. The numbers in the names are chosen according to their position (in hundreds of bases) on the reference mtDNA, i.e., M_smithRNA108ncB has its start in the position 10869 on the M-type genome. Furthermore, an uppercase letter suffix is added in case of smithRNA that has the same numeration: for example, there are two M_smithRNA108nc and one starts at the position 10845 and the other at the position 10869, thus they are named, respectively, M_smithRNA108ncA and M_smithRNA108ncB.
Target Prediction
We checked for the presence of putative targets searching for seed-pairing in the polyadenilated transcriptome of R. philippinarum (Ghiselli et al. 2012), which includes both nuclear and mitochondrial genes, reannotated following a pipeline for nonmodel organisms (paper in preparation, data and methods available online here: https://osf.io/cdkb9/?view_only=f0b2cde926db43719f3d705012c4eeaa).
We used two prediction tools for RNA–RNA thermodynamic interactions that take into account the accessibility of the target UTR and the binding energy. We avoided the use of machine learning algorithms, because they are trained on species that are phylogenetically very distant from R. philippinarum, and the risk of false negatives is, in our opinion, too high. In more detail, putative targets of smithRNAs were predicted with both thermodynamic and sequence-based criteria: the first criterion was assessed by checking sequence hybridization using the RNAhybrid software (Krüger and Rehmsmeier 2006), and the pairing to UTRs was evaluated with PITA (Kertesz et al. 2007). The second criterion consisted in searching for “seed-pairing” targets (Cloonan 2015), due to the absence of detailed information about the RNA interference mechanism in R. philippinarum.
Cases of seed shift (a change in the common seed location, at nucleotides 2–11 of the mature molecule) are known (Tan et al. 2014). Moreover, a central seed-pairing has been found to be more predictive in mammals (Shin et al. 2010); so miRNAs that show both seed and central pairing should be considered as more predictive. Therefore, a screening of the sequences able to do seed-pairing has been performed through blastn, using each smithRNA as query against the R. philippinarum gonadal transcriptome in which all ORFs were masked (thus with UTRs only). The searches were ran locally using the blast+ package (Camacho et al. 2009). All sequences with at least 11 nt pairing and 4–10 perfect match were kept for subsequent thermodynamic analyses on Gibbs Free Energy (ΔG) in the RNA–RNA hybrids.
We kept all the sequences with a ΔG < −20KJ and 3–10 helix constraints from RNAhybrid, and those with a ΔG < −9KJ from PITA. ΔG thresholds were chosen on the indications reported in the original publications of the tools (Krüger and Rehmsmeier 2006; Kertesz et al. 2007). Furthermore, the threshold chosen in RNAhybrid was the same used in another study dealing with miRNA target identification in a different bivalve species (Pinctada martensii; Jiao et al. 2014). The threshold of PITA is based on the prediction of known miRNA in four model species (Kertesz et al. 2007). Moreover, we have to point out that ΔGs are computed at 37 °C because of software setting restrictions, although R. philippinarum commonly lives at 12–22 °C. The ΔG is directly proportional to the temperature as described by Gibbs–Helmholtz equation (Atkins 1978), so we expect the in vivo ΔGs of R. philippinarum to be even lower than those predicted.
Precursor Prediction
Pre-smithRNA structures were computed using RNAfold (Lorenz et al. 2011), with default settings and T = 18 °C. The sequences used for prediction are usually around 70 nt long (like pre-miRNAs), and they map to the genomic portion surrounding each smithRNAs. The whole UR in which they are included was chosen for -nc smithRNAs. The -t smithRNAs precursors were obtained from the tRNA in which they were located, but in the case a small UR (3–10 nt) was next to them, this was also included.
In order to understand the reliability of the ΔG obtained in the precursor prediction of smithRNAs, we also ran simulations using sequences from other known miRNAs. To do this, we used a sample of 9,126 sequences (about 1/3 of all the sequences in miRBase) from several species, and a subsample of 192 sequences from Drosophila melanogaster, Gallus gallus, and Caenorhabditis elegans, which are species that have been thoroughly characterized for miRNAs. We computed ΔGs of their pre-miRNA using RNAfold, and then we built a probability plot using the values of the pre-miRNAs and pre-smithRNAs. The plot is shown in the Supplementary material S5A, Supplementary Material online. Although few smithRNA precursors did not have a good ΔG, the most promising ones (i.e., F_smithRNA122ncA and M_smithRNA108ncB) retrieved a value slightly higher than the median of other miRNAs. Moreover, we tested for differences in secondary structure ΔG between putative precursors and random fragments of the same size extracted from the references genomes. Custom R scripts were used to set up five sets of 5,000 fragments of the same lengths of putative precursors: different lengths of predicted smithRNA precursors were represented in the same proportions. The first set was composed by fragments randomly extracted from the reference genome; the second one was composed by sequences randomly generated using the same nucleotide compositions. The third, fourth, and fifth sets were obtained by extracting sequences only from protein coding genes, rRNA genes, and tRNA genes, respectively; whenever it was not possible to draw 5,000 different fragments from a given region, all the possible fragments were drawn. Final boxplots are shown in Supplementary figure S5B, Supplementary Material online.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
A.P., F.G., and M.P. conceived the study, and M.P., F.P., and F.G. supervised it. A.P. and L.M. performed wet lab analyses. L.M. developed the protocol of mitochondrial enrichment for R. philippinarum. A.P. performed the computational analyses and processed the data. A.P., F.G., F.P., and M.P. prepared the figures, analyzed the data and interpreted the results. The manuscript was written by A.P. and M.P. and revised by all authors.
Supplementary Material
Acknowledgments
We wish to acknowledge two anonymous referees for their valuable suggestions to improve this manuscript, and Edoardo Turolla (Istituto Delta, Ferrara, Italy) for providing the specimens for this analysis. This work was supported by the RFO grant of the University of Bologna (Italy), the Canziani bequest funded to M.P.; the Italian Ministry of Education, University and Research MIUR - FIR Programme no. RBFR13T97A funded to F.G.; the Italian Ministry of Education, University and Research MIUR - SIR Programme no. RBSI14G0P5 funded to L.M.
References
- Agrawal N, Dasaradhi PV, Mohmmed A, Malhotra P, Bhatnagar RK, Mukherjee SK.. 2003. RNA interference: biology, mechanism, and applications. Microbiol Mol Biol Rev. 67:657–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. 2004. The functions of animal microRNAs. Nature 431:350–355. [DOI] [PubMed] [Google Scholar]
- Amikura R, Kashikawa M, Nakamura A, Kobayashi S.. 2001. Presence of mitochondria-type ribosomes outside mitochondria in germ plasm of Drosophila embryos. Proc Natl Acad Sci U S A. 98:9133–9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson SG, Kurland CG.. 1998. Reductive evolution of resident genomes. Trends Microbiol. 6:263–268. [DOI] [PubMed] [Google Scholar]
- Atkins PW. 1978. Physical chemistry Oxford: Oxford University Press. [Google Scholar]
- Bandiera S, Cagnard N, Hanein S, Chrétien D, Munnich A, Lyonnet S, Henrion-Caude A.. 2011. Nuclear outsourcing of RNA interference components to human mitochondria. PLoS One 6:e20746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber GN. 2001. Host defense, viruses and apoptosis. Cell Death Differ. 8:113–126. [DOI] [PubMed] [Google Scholar]
- Bensasson D, Zhang D, Hartl DL, Hewitt GM.. 2001. Mitochondrial pseudogenes: evolution’s misplaced witnesses. Trends Ecol Evol. 16:314–321. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilewitch JP, Degnan SM.. 2011. A unique horizontal gene transfer event has provided the octocoral mitochondrial genome with an active mismatch repair gene that has potential for an unusual self-contained function. BMC Evol Biol. 11:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton S, Doucet-Beaupré H, Stewart DT, Hoeh WR, Blier PU.. 2007. The unusual system of doubly uniparental inheritance of mtDNA: isn’t one enough? Trends Genet. 23:465–474. [DOI] [PubMed] [Google Scholar]
- Breton S, Milani L, Ghiselli F, Guerra D, Stewart DT, Passamonti M.. 2014. A resourceful genome: updating the functional repertoire and evolutionary role of animal mitochondrial DNAs. Trends Genet. 30:555–564. [DOI] [PubMed] [Google Scholar]
- Breton S, Stewart DT, Shepardson S, Trdan RJ, Bogan AE, Chapman EG, Ruminas AJ, Piontkivska H, Hoeh WR.. 2011. Novel protein genes in animal mtDNA: a new sex determination system in freshwater mussels (Bivalvia: Unionoida)? Mol Biol Evol. 28:1645–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL.. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Kenchington E, Zouros E.. 2004. Differential segregation patterns of sperm mitochondria in embryos of the blue mussel (Mytilus edulis). Genetics 166:883–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti A, Jha BK, Silverman RH.. 2011. New insights into the role of RNase L in innate immunity. J Interferon Cytokine Res. 31:49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charif D, Lobry JR.. 2007. SeqinR 1.0-2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis In: Bastolla U, Porto M, Roman HE, Vendruscolo M, editors. Structural approaches to sequence evolution: molecules, networks, populations. New York (NY: ): Springer Verlag; p. 207–232. [Google Scholar]
- Chase CD. 2007. Cytoplasmic male sterility: a window to the world of plant mitochondrial-nuclear interactions. Trends Genet. 23:81–90. [DOI] [PubMed] [Google Scholar]
- Cloonan N. 2015. Re-thinking miRNA–mRNA interactions: intertwining issues confound target discovery. Bioessays 37:379–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogswell AT, Kenchington EL, Zouros E.. 2006. Segregation of sperm mitochondria in two- and four-cell embryos of the blue mussel Mytilus edulis: implications for the mechanism of doubly uniparental inheritance of mitochondrial DNA. Genome 49:799–807. [DOI] [PubMed] [Google Scholar]
- Czech B, Hannon GJ.. 2016. A happy 3′ ending to the piRNA maturation story. Cell 164:838–840. [DOI] [PubMed] [Google Scholar]
- Devine MJ, Birsa N, Kittler JT.. 2016. Miro sculpts mitochondrial dynamics in neuronal health and disease. Neurobiol Dis. 90:27–34. [DOI] [PubMed] [Google Scholar]
- Diz AP, Dudley E, Cogswell A, MacDonald BW, Kenchington ELR, Zouros E, Skibinski DOF.. 2013. Proteomic analysis of eggs from Mytilus edulis females differing in mitochondrial DNA (mtDNA) transmission mode. Mol Cell Proteomics. 12:3068–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet-Beaupré H, Breton S, Chapman EG, Blier PU, Bogan AE, Stewart DT, Hoeh WR.. 2010. Mitochondrial phylogenomics of the Bivalvia (Mollusca): searching for the origin and mitogenomic correlates of doubly uniparental inheritance of mtDNA. BMC Evol Biol. 10:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. [DOI] [PubMed] [Google Scholar]
- Fischer D, Eisenberg D.. 1999. Finding families for genomic ORFans. Bioinformatics 15:759–762. [DOI] [PubMed] [Google Scholar]
- Garg SG, Martin WF.. 2016. Mitochondria, the cell cycle, and the origin of sex via a syncytial eukaryote common ancestor. Gen Biol Evol. 86:1950–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiselli F, Milani L, Chang PL, Hedgecock D, Davis JP, Nuzhdin SV, Passamonti M.. 2012. De novo assembly of the Manila clam Ruditapes philippinarum transcriptome provides new insights into expression bias, mitochondrial doubly uniparental inheritance and sex determination. Mol Biol Evol. 29:771–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiselli F, Milani L, Guerra D, Chang PL, Breton S, Nuzhdin SV, Passamonti M.. 2013. Structure, transcription, and variability of metazoan mitochondrial genome: perspectives from an unusual mitochondrial inheritance system. Genome Biol Evol. 5:1535–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiselli F, Milani L, Passamonti M.. 2011. Strict sex-specific mtDNA segregation in the germ line of the DUI species Venerupis philippinarum (Bivalvia: Veneridae). Mol Biol Evol. 28:949–961. [DOI] [PubMed] [Google Scholar]
- Ha M, Narry Kim V.. 2014. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 15:509–524. [DOI] [PubMed] [Google Scholar]
- Hashiguchi K, Akiyama QZ.. 2009. Establishment of human cell lines lacking mitochondrial DNA. Methods Mol Biol. 554:383–391. [DOI] [PubMed] [Google Scholar]
- Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA.. 2010. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 16:673–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havird JC, Hall MD, Dowling DK.. 2015. The evolution of sex: a new hypothesis based on mitochondrial mutational erosion: mitochondrial mutational erosion in ancestral eukaryotes would favor the evolution of sex, harnessing nuclear recombination to optimize compensatory nuclear coadaptation. Bioessays 37:951–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedtke SM, Stanger-Hall K, Baker RJ, Hillis DM.. 2008. All-male asexuality: origin and maintenance of androgenesis in the Asian Corbicula. Evolution 625:1119–1136. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Noda Y, Tanaka Y, Niwa S.. 2009. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 10:682–696. [DOI] [PubMed] [Google Scholar]
- Houseley J, Tollervey D.. 2009. The many pathways of RNA degradation. Cell 136:763–776. [DOI] [PubMed] [Google Scholar]
- Izumi N, Shoji K, Sakaguchi Y, Honda S, Kirino Y, Suzuki T, Katsuma S, Tomari Y.. 2016. Identification and functional analysis of the Pre-piRNA 3' trimmer in silkworms. Cell 164:962–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Zheng Z, Du X, Wang Q, Huang R, Deng Y, Shi S, Zhao X.. 2014. Identification and characterization of MicroRNAs in pearl oyster Pinctada martensii by Solexa deep sequencing. Mar Biotechnol. 16:54–62. [DOI] [PubMed] [Google Scholar]
- Jiggin FM, Hurst GD, Majerus ME.. 2000. Sex-ratio-distorting Wolbachia causes sex-role reversal in its butterfly host. Proc Biol Sci. 267:69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenchington E, MacDonald B, Cao L, Tsagkarakis D, Zouros E.. 2002. Genetics of mother-dependent sex ratio in blue mussels (Mytilus spp.) and implications for doubly uniparental inheritance of mitochondrial DNA. Genetics. 161:1579–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E.. 2007. The role of site accessibility in microRNA target recognition. Nat Genet. 39:1278–1284. [DOI] [PubMed] [Google Scholar]
- Khachane AN, Timmis KN, Martins dos Santos VA.. 2007. Dynamics of reductive genome evolution in mitochondria and obligate intracellular microbes. Mol Biol Evol. 24:449–456. [DOI] [PubMed] [Google Scholar]
- Krüger J, Rehmsmeier M.. 2006. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 34:W451–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N. 2005. Power, sex, suicide: mitochondria and the meaning of life Oxford: Oxford University Press. [Google Scholar]
- Langmead B, Salzberg S.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Shibata Y, Malhotra A, Dutta A.. 2009. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 23:2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB.. 2003. Prediction of mammalian microRNA targets. Cell 115:787–798. [DOI] [PubMed] [Google Scholar]
- Liu YP, Karg M, Harwig A, Herrera-Carrillo E, Jongejan A, Van Kampen A, Berkhout B.. 2015. Mechanistic insights on the dicer-independent AGO2-mediated processing of AgoshRNAs. RNA Biol. 12:92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Höner zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL.. 2011. ViennaRNA package 2.0. Algorithms Mol Biol. 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie GL. 1984. Bivalves In: Tompa AS, Verdonk NH, van den Biggelaar JAM, editors. The Mollusca vol. 7, reproduction. New York: Academic Press, Inc. [Google Scholar]
- Maniataki E, Mourelatos Z.. 2005. Human mitochondrial tRNAMet is exported to the cytoplasm and associates with the Argonaute 2 protein. RNA 11:849–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, et al. 2011. The human mitochondrial transcriptome. Cell 146:645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani L, Ghiselli F.. 2015. Mitochondrial activity in gametes and transmission of viable mtDNA. Biol Direct. 10:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani L, Ghiselli F, Guerra D, Breton S, Passamonti M.. 2013. A comparative analysis of Mitochondrial ORFans: new clues on their origin and role in species with Doubly Uniparental Inheritance of Mitochondria. Genome Biol Evol. 5:1408–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani L, Ghiselli F, Iannello M, Passamonti M.. 2014a. Evidence for somatic transcription of male-trasmitted mitochondrial genome in the DUI species Ruditapes philippinarum (Bivalvia: Veneridae). Curr Genet. 60:163–173. [DOI] [PubMed] [Google Scholar]
- Milani L, Ghiselli F, Maurizii MG, Nuzhdin SV, Passamonti M.. 2014b. Paternally transmitted mitochondria express a new gene of potential viral origin. Genome Biol Evol. 6:391–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani L, Ghiselli F, Maurizii MG, Passamonti M.. 2011. Doubly uniparental inheritance of mitochondria as a model system for studying germ line formation. PLoS One 6:e28194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani L, Ghiselli F, Passamonti M.. 2012. Sex-linked mitochondrial behavior during early embryo development in Ruditapes philippinarum (Bivalvia Veneridae) a species with the Doubly Uniparental Inhetitance (DUI) of mitochondria. J Exp Zool Part B. 318:182–189. [DOI] [PubMed] [Google Scholar]
- Milani L, Ghiselli F, Passamonti M.. 2016. Mitochondrial selfish elements and the evolution of biological novelties. Curr Zool. 62:687–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani L, Ghiselli F, Pecci A, Maurizii MG, Passamonti M.. 2015. The expression of a novel mitochondrially-encoded gene in gonadic precursors may drive paternal inheritance of mitochondria. PLoS One. 10:e0137468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan MR, Whitmarsh AJ.. 2015. Mitochondrial proteins moonlighting in the nucleus. Trends Biochem Sci. 40:728–735. [DOI] [PubMed] [Google Scholar]
- Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. 2008. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 18:610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro D, Pichaud N, Paquin F, Kemeid V, Blier PU.. 2013. Low hydrogen peroxide production in mitochondria of the long-lived Arctica islandica: underlying mechanisms for slow aging. Aging Cell 12:584–592. [DOI] [PubMed] [Google Scholar]
- Nottingham RM, Wu DC, Qin Y, Yao J, Hunicke-Smith S, Lambowitz AM.. 2016. RNA-seq of human reference RNA samples using a thermostable group II intron reverse transcriptase. RNA 22:597–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata M, Komaru A.. 2005. Specific location of sperm mitochondria in mussel Mytilus galloprovincialis zygotes stained by MitoTracker. Dev Growth Differ. 47:255–263. [DOI] [PubMed] [Google Scholar]
- Okonechnikov K, Golosova O, Fursov M, the UGENE team. 2012. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28:1166–1167. [DOI] [PubMed] [Google Scholar]
- Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel JD, Kim VN.. 2011. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 475:201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passamonti M, Ghiselli F.. 2009. Doubly uniparental inheritance: two mitochondrial genomes, one precious model for organelle DNA inheritance and evolution. DNA Cell Biol. 28:79–89. [DOI] [PubMed] [Google Scholar]
- Plazzi F, Ribani A, Passamonti M.. 2013. The complete mitochondrial genome of Solemya velum (Mollusca: Bivalvia) and its relationships with Conchifera. BMC Genomics 14:409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plazzi F, Puccio G, Passamonti M.. 2016. Comparative large-scale mitogenomics evidences clade-specific evolutionary trends in mitochondrial DNAs of Bivalvia. Genome Biol Evol. 8:2544–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. 26:383–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2008. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing.
- Radzvilavicius AL. 2016. Evolutionary dynamics of cytoplasmic segregation and fusion: mitochondrial mixing facilitated the evolution of sex at the origin of eukaryotes. J Theor Biol. 404:160–168. [DOI] [PubMed] [Google Scholar]
- Radzvilavicius AL, Hadjivasiliou Z, Pomiankowski A, Lane N.. 2016. Selection for mitochondrial quality drives evolution of the germline. PLoS Biol. 14:e2000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana TM. 2007. Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Biol. 8:23–26. [DOI] [PubMed] [Google Scholar]
- Rigaud T, Moreau J, Juchault P.. 1999. Wolbachia infection in the terrestrial isopod Oniscus asellus: sex ratio distortion and effect on fecundity. Heredity 83:469–475. [DOI] [PubMed] [Google Scholar]
- Rio DC, Ares MJ, Hannon GJ, Nilsen TW.. 2010. Purification of RNA using TRIzol (TRI Reagent). Cold Spring Harb Protoc. 2010. doi: 10.1101/pdb.prot5439. [DOI] [PubMed] [Google Scholar]
- Ro S, Ma HY, Park C, Ortogero N, Song R, Hennig GW, Zheng H, Lin YM, Moro L, Hsieh JT, et al. 2013. The mitochondrial genome encodes abundant small noncoding RNAs. Cell Res. 23:759–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross E, Blair D, Guerrero-Hernández C, Sánchez Alvarado AG.. 2016. Comparative and transcriptome analyses uncover key aspects of coding- and long non-coding RNAs in flatworm mitochondrial genomes. G3 (Bethesda) 6:1191–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra C, Reyero MI, Zouros E.. 1997. Male-dependent doubly uniparental inheritance of mitochondrial DNA and female-dependent sex-ratio in the mussel Mytilus galloprovincialis. Genetics. 145:1073–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina SA, Koonin EV.. 2008. Origins and evolution of eukaryotic RNA interference. Trends Ecol Evol. 23:578–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin C, Nam J-W, Farh KK-H, Chiang HR, Shkumatava A, Bartel DP.. 2010. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell. 38:789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shock LS, Thakkar PV, Peterson EJ, Moran RG, Taylor SM.. 2011. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc Natl Acad Sci U S A. 108:3630–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibinski DO, Gallagher C, Beynon CM.. 1994. Mitochondrial DNA inheritance. Nature 368:817–818. [DOI] [PubMed] [Google Scholar]
- Sokolova IM. 2004. Cadmium effects on mitochondrial function are enhanced by elevated temperatures in a marine poikilotherm, Crassostrea virginica Gmelin (Bivalvia: Ostreidae). J Exp Biol. 207:2639–2648. [DOI] [PubMed] [Google Scholar]
- Son DJ, Kumar S, Takabe W, Kim CW, Ni CW, Alberts-Grill N, Jang IH, Kim S, Kim W, Won Kang S, et al. 2013. The atypical mechanosensitive microRNA-712 derived from pre-ribosomal RNA induces endothelial inflammation and atherosclerosis. Nat Commun. 4:3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Cheng YW, Lai L, Huang TC, Wang J, Wu X, Wang Y, Huang Y, Wang J, Zhang K, et al. 2016. Signature miRNAs in colorectal cancers were revealed using a bias reduction small RNA deep sequencing protocol. Oncotarget 7:3857–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamai KT, Monaco L, Alastalo TP, Lalli E, Parvinen M, Sassone-Corsi P.. 1996. Hormonal and developmental regulation of DAX-1 expression in Sertoli cells. Mol Endocrinol. 10:1561–1569. [DOI] [PubMed] [Google Scholar]
- Tan GC, Chan E, Molnar A, Sarkar R, Alexieva D, Isa IM, Robinson S, Zhang S, Ellis P, Langford CF, et al. 2014. 5′ isomiR variation is of functional and evolutionary importance. Nucleic Acids Res. 42:9424–9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Tu S, Lee H-C, Weng Z, Mello CC.. 2016. The RNase PARN-1 trims piRNA 3′ ends to promote transcriptome surveillance in C. elegans. Cell 164:974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogeler S, Bean TP, Lyons BP, Galloway TS.. 2016. Dynamics of nuclear receptor gene expression during Pacific oyster development. BMC Dev Biol. 16:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogeler S, Galloway TS, Lyons BP, Bean TP.. 2014. The nuclear receptor gene family in the Pacific oyster, Crassostrea gigas, contains a novel subfamily group. BMC Genomics 15:369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CR, Lou Y, Gao JF, Qiu JH, Zhang Y, Gao Y, Chang QC.. 2016. Comparative analyses of the complete mitochondrial genomes of the two murine pinworms Aspiculuris tetraptera and Syphacia obvelata. Gene 585:71–75. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wu M.. 2015. An integrated phylogenomic approach toward pinpointing the origin of mitochondria. Sci Rep. 5:7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Chory J.. 2008. Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet. 9:383–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouros E. 2000. The exceptional mitochondrial DNA system of the mussel family Mytilidae. Genes Genet Syst. 75:313–318. [DOI] [PubMed] [Google Scholar]
- Zouros E. 2013. Biparental inheritance through uniparental transmission: the doubly uniparental inheritance (DUI) of mitochondrial DNA. Evol Biol. 40:1–31. [Google Scholar]
- Zouros E, Oberhauser Ball A, Saavedra C, Freeman KR.. 1994. An unusual type of mitochondrial DNA inheritance in the blue mussel Mytilus. Proc Natl Acad Sci USA. 91:7463–7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.