Abstract
Haemosporidians are a diverse group of vector-borne parasitic protozoa that includes the agents of human malaria; however, most of the described species are found in birds and reptiles. Although our understanding of these parasites’ diversity has expanded by analyses of their mitochondrial genes, there is limited information on these genes’ evolutionary rates. Here, 114 mitochondrial genomes (mtDNA) were studied from species belonging to four genera: Leucocytozoon, Haemoproteus, Hepatocystis, and Plasmodium. Contrary to previous assertions, the mtDNA is phylogenetically informative. The inferred phylogeny showed that, like the genus Plasmodium, the Leucocytozoon and Haemoproteus genera are not monophyletic groups. Although sensitive to the assumptions of the molecular dating method used, the estimated times indicate that the diversification of the avian haemosporidian subgenera/genera took place after the Cretaceous–Paleogene boundary following the radiation of modern birds. Furthermore, parasite clade differences in mtDNA substitution rates and strength of negative selection were detected. These differences may affect the biological interpretation of mtDNA gene lineages used as a proxy to species in ecological and parasitological investigations. Given that the mitochondria are critically important in the parasite life cycle stages that take place in the vector and that the transmission of parasites belonging to particular clades has been linked to specific insect families/subfamilies, this study suggests that differences in vectors have affected the mode of evolution of haemosporidian mtDNA genes. The observed patterns also suggest that the radiation of haemosporidian parasites may be the result of community-level evolutionary processes between their vertebrate and invertebrate hosts.
Keywords: Haemoproteus, Leucocytozoon, mitochondrial genome, Plasmodium, phylogeny, substitution rates, time tree
Introduction
Haemosporida is a highly diverse monophyletic group of vector-borne hemoparasites that includes species of medical importance such as the agents of human malaria (Cavalier-Smith 2014). Although these parasitic protozoa are found in many vertebrate hosts, including primates, the majority of species have been described in birds and reptiles (Valkiūnas 2005; Telford 2009). The use of molecular tools has propelled phylogenetic and ecological studies of haemosporidian parasites; however, most of the data on these parasites come from mitochondrial genes. The haemosporidian mtDNA genome is the smallest known among all eukaryotes, consisting of an ∼6-kb linear molecule arranged in tandem arrays. It has highly fragmented mitochondrion large subunit (LSU) and small subunit (SSU) ribosomal ribonucleic acid (rRNA) genes, with three open reading frames (ORF) encoding oxidative phosphorylation (OxPhos) proteins for electron transport and ATP generation: cytochrome c oxidase subunit 1 (cox1), cytochrome c oxidase subunit 3 (cox3), and cytochrome b (cytb) (Gray et al. 2004). Although the data on nonmammalian parasite species are still limited, the same gene content has been found in mtDNA from species of different haemosporidian genera (Perkins 2008; Pacheco, Battistuzzi, Junge, et al. 2011; Feagin et al. 2012; Hikosaka et al. 2013; Muehlenbein et al. 2015).
The mtDNA conserved gene content and copy number have facilitated its adoption in many evolutionary and ecological studies (Escalante et al. 1998; Martinsen et al. 2008; Pacheco, Battistuzzi, Junge, et al. 2011; Schaer et al. 2013). In particular, mtDNA genes have been used to describe and identify species, separate cryptic species, to infer phylogenetic relationships, and to study historical and biogeographical patterns (Bensch et al. 2000; Pacheco, Battistuzzi, Junge, et al. 2011; Levin et al. 2012; Pacheco et al. 2012; González et al. 2015; Lotta et al. 2016; Nilsson et al. 2016). Unfortunately, most studies utilize partial sequences of one gene, the cytb. The reconstructed gene lineages are then used as a proxy for species lineages (Bensch et al. 2009; Outlaw and Ricklefs 2014); however, little is known on how to interpret the observed divergence between lineages because of a limited understanding of the rates of evolution of the mtDNA genes. In particular, given that vertebrate host switches are relatively common, there is a paucity of potential events that can be used to inform calibration priors resulting in only a few molecular timing studies (Hayakawa et al. 2008; Ricklefs and Outlaw 2010; Pacheco, Battistuzzi, Junge, et al. 2011; Muehlenbein et al. 2015). Furthermore, it has been hypothesized that the phylogenetic signal of the haemosporidian mtDNA may be saturated, casting doubts on its value for phylogenetic and molecular dating analyses (Perkins 2008; Silva et al. 2015). This conjecture, however, has no empirical support (Pacheco, Battistuzzi, Junge, et al. 2011; Outlaw and Ricklefs 2014; Boundenga et al. 2016).
Beyond its use as a molecular marker, there are limited studies on how natural selection may have affected the mtDNA mode of evolution. Like in other eukaryotes, the haemosporidian mtDNA genes encode proteins involved in critical metabolic and energetic functions such as oxidative phosphorylation. However, these species may adjust their carbon source usage depending on their developmental stage (Ke et al. 2015; Jacot et al. 2016). In the case of Plasmodium species, these parasites switch their energy metabolism from glycolysis in the vertebrate host to oxidative phosphorylation in the insect vector where glucose is not readily available for ATP production (Hall et al. 2005; Hino et al. 2012). Thus, the mtDNA in these parasites may not be crucial for the survival of their asexual blood stages (vertebrate host), but it is likely to be indispensable for the development of insect (vector) stages, allowing its transmission (Hino et al. 2012; Ke et al. 2015; Jacot et al. 2016) and completion of its life cycle.
Given its vital function in the parasite transmission and the extensive use of mtDNA genes in ecological and evolutionary studies, this investigation has focused on assessing the mtDNA rate of evolution in avian haemosporidian parasites (order Haemosporida) and the factors that may affect it. In particular, an extensive data set comprised of 114 mtDNA genomes from species belonging to four genera: Leucocytozoon, Haemoproteus (subgenera Haemoproteus and Parahaemoproteus), Plasmodium, and Hepatocystis has been used to estimate the phylogeny and divergence times for the radiation of major avian haemosporidian species groups. This information is then used to estimate rates of substitution per mtDNA gene and to ascertain how natural selection has acted on different mitochondrial genes and lineages. Overall, this study shows that the mtDNA genome is suitable for phylogenetic inferences in haemosporidian parasites. Although requiring further investigation, the time estimates from the mtDNA are consistent with a scenario where the avian haemosporidian parasites may have diversified during the radiation of their vertebrate host orders. Finally, the emerging patterns show rate heterogeneity in the mtDNA among parasite clades that are explained, at least in part, by differences in natural selection regimes.
Results and Discussion
Organization of the Haemosporidian Mitochondrial Genome (mtDNA)
In this investigation, 65 new almost complete haemosporidian parasite mtDNA genomes belonging to 4 genera are reported: 3 Leucocytozoon spp., 7 Haemoproteus (Haemoproteus) spp., 24 Haemoproteus (Parahaemoproteus) spp., 29 Plasmodium spp., and 2 Hepatocystis spp. (supplementary table S1, Supplementary Material online). Thus, this study is a major expansion of the available data on avian haemosporidian parasites. Haemosporidian mitochondrial genes are translated, but the actual mechanism is unclear (Vaidya and Mather 2009; Habib et al. 2016; Vaishya et al. 2016). Although this extended data set allowed documentation of putative initiation codons on mtDNA genes, those are still uncertain in cox1 and cox3. Only the cytb in Leucocytozoon, H. (Haemoproteus) spp., and Plasmodium spp. has an ATG triplet at the 5′ terminus that could be its initiation codon. However, having an alternative initiation codon is not uncommon (Rehkopf et al. 2000). Indeed, the putative start codons of the cytb gene were ATT (Ile) or ACT (Thr) in H. (Parahaemoproteus) spp. and AGT (Ser) in Hepatocystis spp.
Even though an unusual mtDNA genome organization was recently observed in two species from the Nycteria genus (Nycteria medusiformis and Nycteria sp.) found in bats (Karadjian et al. 2016), no evidence of this type of genome rearrangements was found in this study. Indeed, a highly conserved synteny was found across the 114 mtDNA sequences. Based on the annotation proposed by Feagin et al. (2012), the small subunit ribosomal RNAs (12 SSU rRNA), large subunit ribosomal RNAs (15 LSU rRNA), and the three ORFs were found in all sequences. Importantly, the extreme fragmentation of the rRNA described for Plasmodium falciparum mtDNA (Feagin et al. 2012) was also found in all the genomes indicating that this unique organization of SSU RNAs originated early in the evolution of this group. Nevertheless, there were some differences in length between sequences mostly due to indels located between the fragmented SSU rRNA, LSU rRNA, and the ORFs. In particular, the mtDNA reported here from Hepatocystis parasites isolated from macaques exhibited indels that were longer than those found in a species of the same genus isolated from bats (Perkins 2008).
Use of mtDNA Genomes for Phylogenetic Inferences in Haemosporidian Parasites
High A/T content is among the most distinctive mtDNA features when compared with nuclear DNA, and the haemosporidian parasites are not an exception (Escalante et al. 1998; Perkins 2008). The mtDNA genomes sampled had approximately the same A/T content across species with an average of 68.7% (alignment of 114 sequences with 5,125 bp) that exceeded 80% at third codon positions for each gene (supplementary table S2, Supplementary Material online). Thus, there is no foreseeable risk of model misspecification in phylogenetic analyses. Although transitions are usually more common than transversions in mitochondrial genomes, an excess of transversional over transitional substitutions was observed in the total haemosporidian mtDNA genome (ts/tv = 0.68) and in each gene individually (ts/tv ≈ 0.66–0.73) (supplementary fig. S1, Supplementary Material online). This phenomenon has been reported also in mitochondrial genes from plants (Laroche et al. 1997; Sloan and Wu 2014). Determining whether this unusual ts/tv is the result of mutational bias or other processes requires further investigation.
The high A/T content and unusual ts/tv ratios, however, could make the mtDNA prone to saturation at the evolutionary timescale expected for the diversification of haemosporidian parasites. Thus, the mtDNA phylogenetic signal was evaluated using the entropy-based index of substitution saturation (Iss, Xia et al. 2003). Contrary to previous statements (Silva et al. 2015), no clear evidence of saturation was found in the sequences of mitochondrial genes or nonprotein coding regions among the species included in this study (supplementary fig. S1 and table S3, Supplementary Material online). Yet, when an asymmetric tree was assumed, third codon positions in cox3, cox1, and cytb were not phylogenetically informative in some groups. Nevertheless, these genes retain enough phylogenetic information when all positions were included (supplementary table S3, Supplementary Material online). Although usually insufficient for accurate phylogenetic reconstructions because of its limited number of sites, the cytb fragment (478 bp), widely used to identify lineages in avian malaria parasites and other haemosporidians (Bensch et al. 2009; Outlaw and Ricklefs 2014), showed phylogenetic signal and allowed for the correct identification of the morphologically distinct species available in our sample. Thus, this cytb fragment provides phylogenetic information that might be sufficient in ecological and parasitological investigations that do not require complex phylogenetic inferences (Outlaw and Ricklefs 2014). It is worth noting that the extensive data available on this fragment has made it a de facto barcode for avian haemosporidians (Bensch et al. 2009; Outlaw and Ricklefs 2014). However, the concordance of differentiation between morphology and mtDNA genes (including cytb) is by no means always perfect. There are species with clear morphological differences (Haemoproteus jenniae/Haemoproteus iwa, Levin et al. 2012) showing limited divergence of mtDNA genes. The data included here also showed evidence consistent with complexes of cryptic species like Leucocytozoon fringillinarum (Lotta et al. 2016) and Haemoproteus coatneyi (González et al. 2015). Thus, whereas mtDNA genes seem to be good candidates for barcoding approaches, their actual link to phenotypic and other biological criteria used to delimit parasite species (e.g., morphology) requires improving the parasite taxonomic sampling and exploration of additional characters, particularly of exoerythrocytic and vector stages.
Given that all the three genes and nonprotein coding regions have phylogenetic signal (supplementary table S3, Supplementary Material online), the mtDNA phylogeny was estimated using all the sites, including the third position of the three genes and the nonprotein coding regions of the mtDNA genome (fig. 1, alignment comprised of 114 sequences with 5,125 bp excluding gaps). Separately, a phylogeny was also estimated using only the three coding sequences (CDS, 107 sequences and 3,273 bp excluding gaps) (supplementary fig. S2, Supplementary Material online). Bayesian and maximum likelihood (ML) methods yield similar tree topologies, so only the Bayesian phylogenetic inferences are shown (fig. 1 and supplementary fig. S2, Supplementary Material online). Nevertheless, in addition to Bayesian posterior probabilities (pp) to the clades, the bootstrap values (b) from the ML phylogeny are also provided for comparison. The mtDNA phylogeny recovered well-supported clades providing additional information on the evolution of avian haemosporidian parasites. However, the CDS phylogeny failed to resolve the position of the most recent common ancestor for primate/rodent Plasmodium and avian/Squamata Plasmodium species (supplementary fig. S2, Supplementary Material online). These particular branches are relatively short compared with the rest of the tree, so they are more difficult to resolve. Iss values indicated that although each individual mtDNA gene is phylogenetically informative, third codon positions in cox3, cox1, and cytb of primate/rodent Plasmodium spp. are not (supplementary table S3, Supplementary Material online). This factor may have affected the support of that particular clade. Overall, this result highlights the value of adding nonprotein coding mtDNA regions and the fast-evolving third codon positions if those have phylogenetic signal when considered with the first and second codon positions (supplementary table S3, Supplementary Material online).
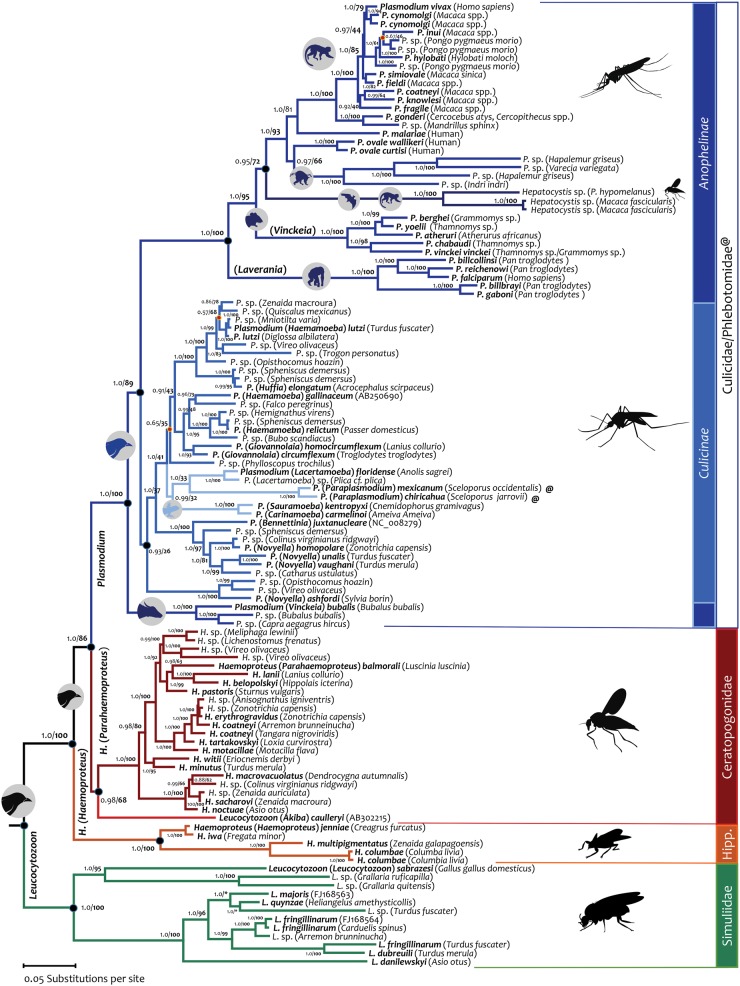
Fig. 1.
Bayesian phylogenetic hypothesis of haemosporidian parasites based on complete mitochondrial genomes (114 sequences and 5,125 bp excluding gaps). The values at the nodes are posterior probabilities together with bootstrap values (in bold) as a percentage obtained for 1,000 pseudoreplicates from a maximum likelihood tree with identical topology (see Materials and Methods). Branch colors indicate the genus–vector host species relationship. The minor inconsistencies between Bayesian and ML trees are indicated with orange dots. Species names are indicated in bold. The character @ stands for Phlebotominae and “Hipp.” corresponds to Hippoboscidae vector.
Even though reassessing the taxonomy is not the focus of this investigation, the phylogenetic analyses showed that three out of four genera (Leucocytozoon, Haemoproteus, and Plasmodium) were not recovered as monophyletic groups (fig. 1 and supplementary fig. S2, Supplementary Material online). Thus, these phylogenies not only confirm that genus Plasmodium is paraphyletic (Escalante et al. 1998; Martinsen et al. 2008, 2016; Schaer et al. 2013, 2015; Borner et al. 2016) but also highlight similar issues within avian haemosporidian parasites. In particular, Leucocytozoon (Akiba) caulleryi, the etiologic agent of chicken leucocytozoonosis (Omori et al. 2008), is not part of a monophyletic group (pp = 1.0 and b = 100%) comprised of the other Leucocytozoon species reported in this investigation and appeared within a well-supported clade that includes Haemoproteus and Plasmodium species (pp = 1.0 and b = 100%). As a result, the mtDNA phylogeny indicates that the genus Leucocytozoon is polyphyletic. Likewise, Haemoproteus is paraphyletic with respect to Leucocytozoon. However, these phylogenetic analyses support the division of Haemoproteus into two subgenera, H. (Haemoproteus) and H. (Parahaemoproteus) (Valkiūnas 2005; Martinsen et al. 2008). These Haemoproteus subgenera are distinct monophyletic groups (fig. 1 and supplementary fig. S2, Supplementary Material online) with similar morphology in the vertebrate blood but differences in their vector stages (sporogonic development), particularly in the oocyst stage in the insect hosts (Valkiūnas 2005). Furthermore, Haemoproteus (Haemoproteus) spp. are transmitted by hippoboscid flies (Hippoboscidae) and H. (Parahaemoproteus) spp. by biting midges (Ceratopogonidae) (Valkiūnas 2005). During this investigation, a potential new clade based on a single species identified as Haemoproteus antigonis was discovered (Bertram et al. 2017). Unfortunately, the low quality of the data (consensus inferred from diverse partial cytb and cox1 sequences obtained from mixed infections) does not allow its incorporation in this investigation.
The phylogenies reported here seem to be consistent with the observed association of groups of vectors at the family/subfamily level with specific clades of parasites (Valkiūnas 2005; Martinsen et al. 2008; Lotta et al. 2016). In particular, several Leucocytozoon spp., where most species are transmitted by simuliid flies (Simuliidae), form a monophyletic group. However, L. (Akiba) caulleryi is not part of the same clade and, unlike other Leucocytozoon spp., it is transmitted by biting midges. Furthermore, L. (Akiba) caulleryi appears to share a recent common ancestor with the Parahaemoproteus subgenus species (fig. 1 and supplementary fig. S2, Supplementary Material online), parasites that are transmitted by the same family of vectors (Valkiūnas 2005). This relationship is recovered using both ML and Bayesian methods; however, it has limited support (pp = 0.98 and b = 68%). In addition, while mosquitoes (Culicinae) (Valkiūnas 2005) transmit most of Plasmodium species infecting birds and lizards, P. (Paraplasmodium) mexicanum and P. (Paraplasmodium) chiricahua are transmitted by sand flies (Phlebotominae) (Telford 2009). These two species appear as sister taxa in these mtDNA phylogenies. Finally, the parasites from ungulates (Plasmodium) are also a monophyletic group and their only reported vectors have been Anopheles spp. mosquitoes (Templeton, Asada, et al. 2016; Templeton, Martinsen, et al. 2016), like the other mammalian clade (primate-rodent parasites). Although the data linking parasite clades and particular vectors seems compelling (Valkiūnas 2005; Martinsen et al. 2008; Lotta et al. 2016), it is worth mentioning that Haemoproteus parasite DNA has been found in different mosquito genera (Anopheles, Aedes, Verrallina, Culex, Coquillettidia) (Ishtiaq et al. 2008; Njabo et al. 2011). Whereas detecting parasite DNA by PCR does not prove that Haemoproteus spp. can complete the cycle in such vectors (Bukauskaitė et al. 2015), the proposed link of vector-family with distinct haemosporidian groups deserves additional exploration, and it may not necessarily be a perfect association.
In the mtDNA phylogeny, Plasmodium species infecting birds and lizards appear as a monophyletic group that contains five distinct clades, at least four of which were well supported (pp ∼ 1, b > 95%). The six lizard Plasmodium species form a monophyletic group (pp = 0.99, b = 32%); however, this result is not conclusive given the low support for this clade and the small number of species included here (fig. 1) from over 101 species and subspecies of Plasmodium described in lizards (Telford 2009). Regarding the phylogeny of haemosporidian parasites from mammals, it is worth noting that the mtDNA phylogenies were consistent with previous findings. In the case of the genus Plasmodium (fig. 1 and supplementary fig. S2, Supplementary Material online), the phylogenies obtained here supported that it is paraphyletic with respect to Hepatocystis and Nycteria genera as previously indicated from single gene or multiple-locus phylogenetic analyses (Escalante et al. 1998; Pacheco et al. 2013; Schaer et al. 2013; Borner et al. 2016; Lutz et al. 2016; Martinsen et al. 2016). The relative position of genus Hepatocystis reported here, however, is slightly different to the one described elsewhere (Borner et al. 2016; Lutz et al. 2016); this may reflect the effect of including lemur malarial parasites (Pacheco, Battistuzzi, Junge, et al. 2011) in the present analyses.
Plasmodium species from the Bovidae family appeared as a monophyletic group sharing a common ancestor with the clade that contains, in addition to parasites of Hepatocystis (fig. 1) and Nycteria genera (supplementary fig. S2, Supplementary Material online), the avian, Squamata, and mammalian Plasmodium spp. (Templeton, Asada, et al. 2016). This position in the phylogeny is consistent with the proposed host switch from mammals back to birds (Templeton, Asada, et al. 2016; Templeton, Martinsen, et al. 2016). The well-documented symbiotic relationship between ungulates and birds (Nunn et al. 2011) could have favored such event. In the context of this investigation, the ungulate malaria clade offers a novel calibration point that can be used in molecular dating analyses (see below).
Overall, this part of the study supports that whole mtDNA genomes are suitable for inferring phylogenetic relationships in haemosporidian parasites. In particular, the results presented so far can be summarized as follows: 1) the mtDNA genomes have approximately the same A/T content across species, reducing the risk of model misspecification; 2) there is no evidence indicating that the mtDNA genes are saturated at the scale of the taxonomic sampling included in this investigation; 3) similar to previous reports on parasites from mammals, the avian haemosporidian genera are paraphyletic; and 4) these phylogenies seem to be consistent with the proposed association between parasite clades and specific dipteran families/subfamilies.
Timing the Radiation of Avian Haemosporidian Genera Using the mtDNA
Divergence times were estimated from 102 different parasite species selected from the mtDNA alignment used to estimate the phylogeny (duplicate haplotypes were not included; see Materials and Methods). Although molecular timing methods differ on how to model evolutionary rate variation among lineages (Battistuzzi et al. 2010; Tamura et al. 2012), empirical studies usually rely on a single method without examining the effect of its assumptions on their time estimates. Here, a suite of relaxed molecular dating methods was used to explore the effect that their assumptions may have on the time estimates of the origin of the major clades of haemosporidian parasites. These molecular dating methods were compared under different calibration scenarios, each with at least three calibration constraints used as priors (see Materials and Methods). In particular, posterior time estimates from two Bayesian methods, namely BEAST v2.4.1 (Bouckaert et al. 2014) and MCMCTree v.4.8 (Yang 2007), were compared under different calibration scenarios (fig. 2 and supplementary figs. S3–S5, Supplementary Material online). The model implemented in BEAST assumes that evolutionary rates are independent among alignment partitions and lineages. MCMCTree allows comparing time estimates assuming autocorrelation or independence of rates across lineages. However, unlike BEAST, MCMCTree requires a time calibration at the root. Thus, in order to incorporate both models of evolutionary rate variation implemented in MCMCtree, a new calibration constraint for the root that emerged as part of this investigation was used (see Materials and Methods). In addition, a novel calibration constraint at the base of the ungulate clade was also explored (see Materials and Methods). These analyses were compared against a non-Bayesian dating method that estimates relative divergence times, RelTime (Tamura et al. 2012), which also computes absolute time estimates when calibration information is provided. The results from the three methods were compared (supplementary fig. S6, Supplementary Material online). Table 1 and supplementary table S4, Supplementary Material online, show the node ages and the estimated credibility intervals (CrI) under different scenarios using BEAST and MCMCTree, respectively (see fig. 2 and supplementary fig. S3, Supplementary Material online, for the node numbers). In particular, supplementary table S4, Supplementary Material online, clearly shows the difference between correlated and independent models.
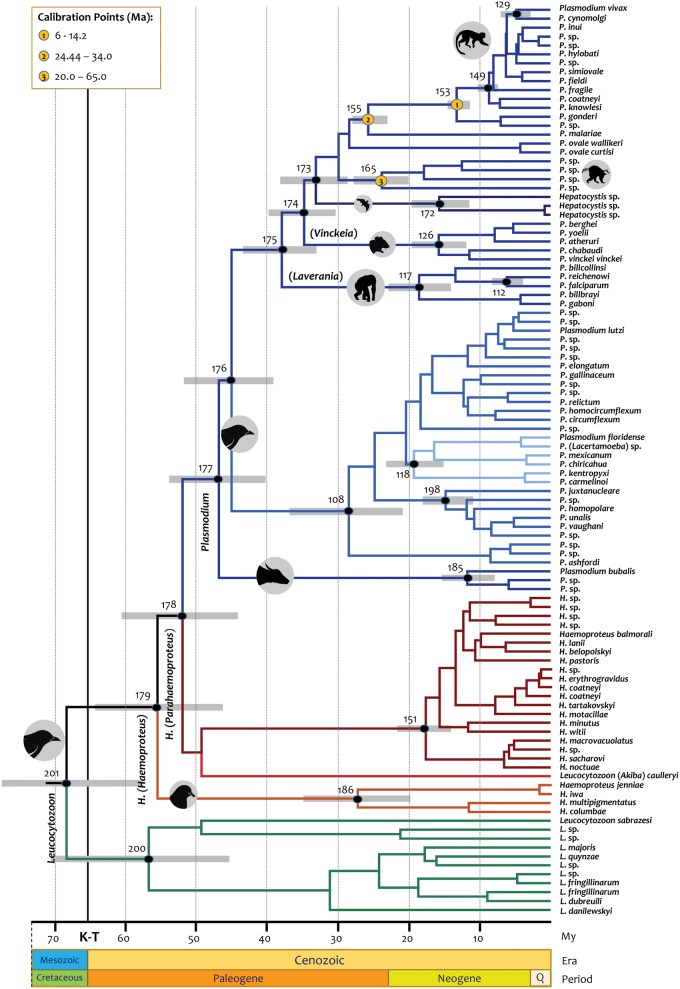
Fig. 2.
Timetree of the divergence of the major clade of haemosporidian parasites. Divergence times estimated using BEAST under calibration scenario 1 based on the minimum divergence of Macaca/Papio using fossils (6–14.2 Ma), with a maximum of 24.44 to a minimum of 34.0 Ma for the human/Macaca split, and a range of 20–65 Ma for the origin of the lemur parasites. Times are shown in My. The numbers of the nodes used in table 1 are provided. Branch colors indicate the genus–vector host species relationship.
Table 1.
Divergence Times of Major Splits in the Haemosporidian Parasite Phylogeny as Estimated by BEAST under Three Scenarios.
| Scenario 1 |
Scenario 2 |
Scenario 3 |
||||||
|---|---|---|---|---|---|---|---|---|
| Calibrations (Ma) | Node 153: 6–14.2 | Node 153: 6–14.2 | Node 153: 6–14.2 | |||||
| Node 155: 24.44–34.0 | Node 155: 24.44–34.0 | Node 155: 24.44–34.0 | ||||||
| Node 165: 20–65 | Node 165: 20–65 | Node 165: 20–65 | ||||||
| Node 201: 56.8–86.8 | Node 185: 16–28.1 | |||||||
| Node 201: 56.8–86.8 | ||||||||
| Divergence | Node | Node Age | 95% CrI | Node Age | 95% CrI | Node Age | 95% CrI | |
| Origin of haemosporidian parasites | 201 | 68.84 | 57.93–81.55 | 69.24 | 58.36–80.47 | 73.67 | 63.23–84.88 | |
| Origin of Leucocytozoon species | 200 | 57.12 | 46.18–70.97 | 57.47 | 46.71–70.18 | 61.37 | 49.64–73.65 | |
| Split H. (Haemoproteus)–H. (Parahaemoproteus)/Plasmodium | 179 | 55.86 | 47.53–64.73 | 56.14 | 47.75–64.07 | 59.55 | 51.10–67.76 | |
| Origin of Haemoproteus (Haemoproteus) species | 186 | 27.31 | 19.81–35.19 | 27.22 | 19.84–35.01 | 29.35 | 21.46–37.44 | |
| Origin of Haemoproteus (Parahaemoproteus) species | 151 | 17.63 | 14.27–21.40 | 17.73 | 14.50–21.28 | 18.91 | 15.31–22.47 | |
| Split H. (Parahaemoproteus)–Plasmodium species | 178 | 52.28 | 44.64–60.53 | 52.52 | 45.71–60.56 | 55.69 | 48.70–64.03 | |
| Origin of Plasmodium genus (+Hepatocystis) | 177 | 47.12 | 40.51–54.36 | 47.33 | 40.63–53.80 | 50.13 | 43.55–56.92 | |
| Origin of Bovidae Plasmodium species | 185 | 11.70 | 8.07–15.46 | 11.90 | 8.37–16.09 | 17.07 | 16.00–19.17 | |
| American Squamata Plasmodium species | 118 | 19.31 | 15.48–23.16 | 19.28 | 15.44–22.94 | 20.72 | 16.79–24.60 | |
| Radiation of avian Plasmodium species | 108 | 28.60 | 21.45–36.77 | 28.59 | 22.17–35.95 | 31.07 | 24.51–39.89 | |
| Split avian Plasmodium–primate/rodent Plasmodium | 176 | 45.31 | 38.95–51.96 | 45.51 | 39.69–52.27 | 48.18 | 41.98–54.62 | |
| Radiation of primate/rodent Plasmodium species | 175 | 38.15 | 33.05–43.82 | 38.26 | 33.51–43.77 | 40.42 | 34.79–45.60 | |
| Radiation of primate Plasmodium (Laverania) species | 117 | 18.56 | 14.15–22.76 | 18.66 | 14.63–19.59 | 19.78 | 15.70–24.08 | |
| Split P. falciparum–P. reichenowi | 112 | 6.06 | 4.03–8.39 | 6.10 | 3.88–8.21 | 6.47 | 4.36–8.93 | |
| Radiation of rodent Plasmodium species | 126 | 15.75 | 12.29–19.82 | 15.80 | 12.34–22.93 | 16.78 | 13.05–20.71 | |
| Radiation of Asian primate Plasmodium species | 149 | 8.57 | 6.96–10.18 | 8.54 | 6.92–10.12 | 9.02 | 7.40–10.53 | |
| Split P. vivax–P. cynomolgi | 129 | 4.67 | 3.06–6.32 | 4.65 | 3.07–6.20 | 4.97 | 3.21–6.69 | |
| Origin of Hepatocystis parasites | 172 | 15.70 | 11.53–19.74 | 15.85 | 11.80–19.91 | 16.54 | 12.45–20.91 | |
Note.—Calibration constraints, time estimates, and their associated 95% credibility intervals (CrIs) are shown in Ma. Clades and node numbers are shown in figure 2. See Materials and Methods for a description of the calibration constraints.
According to BEAST analyses under the calibration scenario 1 (fig. 2), the origin of avian haemosporidian parasites was estimated between 57.93 and 81.55 Ma (mean= 68.84 Ma, table 1) using mtDNA. This time interval clearly overlaps with a calibration point proposed for the origin of palaeognathous birds (56.8–86.8 Ma, Benton et al. 2015). Furthermore, this timeframe is consistent with molecular estimates for the origin of modern birds using avian mitochondrial genomes and targeted next-generation DNA sequencing (Pacheco, Battistuzzi, Lentino, et al. 2011; Prum et al. 2015). This event was then used to compare BEAST and MCMCTree time estimates (see Materials and Methods). Interestingly, very similar time estimates were obtained by BEAST without noticeable effects of adding this calibration constraint at the root (scenario 2) (table 1; supplementary table S4 and fig. S6A, Supplementary Material online). Although timing the origin of the modern birds is controversial with some studies reporting relatively older times (Jarvis et al. 2014; Claramunt and Cracraft 2015), all scenarios explored here indicate that the major avian haemosporidian parasite groups seem to have diversified concomitantly with the radiation of avian orders after the Cretaceous–Paleogene (K–Pg) boundary (66 Ma).
The times estimated by BEAST were similar across distinct calibration scenarios (table 1 and fig. 2;supplementary figs. S4, S5, and S6D–F, Supplementary Material online). As expected, these estimates were almost perfectly linearly related to those obtained using the MCMCTree independent rates model (coefficient of determination, R2=1) (supplementary table S4 and fig. S6A, Supplementary Material online) as evidenced by the extensive overlap of their credibility intervals (table 1 and supplementary table S4, Supplementary Material online). However, the autocorrelated rates model (Rannala and Yang 2007) yielded clearly different results when compared with the independent rates model (R2∼0.74) (supplementary table S4 and fig. S6B and G, Supplementary Material online). For example, under scenario 2 (see Materials and Methods), divergence times were ∼40% older for Haemoproteus (Parahaemoproteus) species and avian/Squamata Plasmodium, and slightly younger for primate/rodent malaria parasites and Leucocytozoon species than those obtained by the independent rates models (supplementary fig. S6B and G, Supplementary Material online). Thus, the statistical model used for evolutionary rate variation across lineages, independent or autocorrelated, has an important effect in some of our time estimates (table 1 and supplementary table S4, Supplementary Material online).
The relative divergence time estimates using RelTime method were similar to those obtained with the autocorrelated rates model (supplementary fig. S6H–J, Supplementary Material online). Although RelTime is not free from assumptions (see Materials and Methods), this observation may suggest that rates of evolution of the haemosporidian mtDNA were autocorrelated since the RelTime algorithm does not assume any particular statistical model for the variation in rates of evolution across lineages as the Bayesian methods do (Tamura et al. 2012, Tamura K, Tao Q, Kumar S, unpublished data, https://www.biorxiv.org/content/early/2017/08/24/180182; last accessed November 13, 2017). Thus, the autocorrelated and the independent rates models were compared by using Bayes factor statistics in Tracer (Rambaut et al. 2014; Ho et al. 2015). The independent rates model was a better fit to the data. However, autocorrelation can be difficult to detect (Ho et al. 2015). An alternative way to compare these different models is to validate their time estimates using a calibration constraint that was not included in the original analyses (Pacheco, Battistuzzi, Junge, et al. 2011). Unfortunately, the only fossil available does not provide information supporting one molecular clock method over the other. In particular, under the independent rates model (BEAST), the time origin of Plasmodium juxtanucleare is dated as 14.80 Ma (11.28–18.15 Ma, fig. 2). This is consistent with one of the scenarios proposed for the fossil Plasmodium dominicana (found in a Dominican amber sample), that is considered closely related to P. juxtanucleare (Poinar 2005). Although commonly dated at 15–20 Ma based on foraminifera (Iturralde-Vinent and MacPhee 1996), Dominican amber has been also dated 30–45 Ma based on coccoliths (Schlee 1990). This older age coincides with the posterior time estimates under the autocorrelated model, 26.9 Ma (20.0–34.41 Ma, Supplementary fig. S3). Since there is uncertainty about whether independent rates model is the best for the haemosporidia mtDNA, the time estimates from the autocorrelated model are also provided (supplementary table S4 and fig. S3, Supplementary Material online).
In the case of Plasmodium parasites infecting species of the Bovidae family (ungulates), this study yielded posterior time estimates for its origin between 8.07 and 15.46 Ma (11.70 Ma, fig. 2) under the independent rates model; this credibility interval overlaps with the origin of crown Bovidae reported between 15.1 and 17.3 Ma (Bibi 2013). Furthermore, this interval is close to the fossil calibration point recently proposed for the origin of Bovinae-Antilopinae (16–28.1 Ma, Benton et al. 2015). Although the time estimates for ungulate parasites based on the independent rates model were younger (supplementary fig. S4, Supplementary Material online), the estimates derived from the autocorrelated rates model overlapped with the fossil record (Benton et al. 2015) (Supplementary fig. S3, Supplementary Material online). Thus, the origin of the ungulate parasites seems to be consistent with the origin of Bovinae-Antilopinae.
The inclusion of the origin of Bovinae-Antilopinae (16–28.1 Ma, Benton et al. 2015) as a calibration constraint (scenario 3 using BEAST), yielded similar but slightly older (∼6%) posterior time estimates for all haemosporidian clades (table 1 and supplementary fig. S5, Supplementary Material online). Under the calibration scenario 3, the posterior time estimates for the origin of the genus Plasmodium (including the avian-Squamata clade) were not particularly dissimilar between the independent rate and the autocorrelated model when the Bovine–Antilopinae calibration prior was included (10% difference, supplementary table S4, Supplementary Material online). Thus, the effect of the methods’ assumptions on the posterior time estimates diminishes with the inclusion of additional calibrations (dos Reis et al. 2016). This result highlights the need for new calibration information in Haemosporida, particularly in avian parasites. Indeed, the lack of suitable calibration information within the genera Leucocytozoon and Haemoproteus may explain, at least in part, the differences in time estimates between the dating models. In the case of the genus Plasmodium, this study suggests that time estimates could be susceptible to the inclusion of additional ungulate lineages that are already being detected in biodiversity surveys (Boundenga et al. 2016). Given that the inclusion of the origin of Bovinae-Antilopinae as calibration had a minor effect on the time estimates in both models, the proposed calibration priors were further explored by calculating divergence times with and without incorporating calibrations using RelTime (Battistuzzi et al. 2015). This comparison showed that the calibration constraints used here did not affect substantially the relative time estimates yielded by the mtDNA (supplementary fig. S6C, Supplementary Material online). Thus, the proposed calibrations (including the Bovine–Antilopine calibration constraint) are internally consistent. This is a desirable attribute given the lack of information that can be used as calibrations in many haemosporidian clades.
Although there are no previous molecular dating studies with an extensive taxonomic sampling like the one included here, it is worth noting that the times and mtDNA rates obtained in this investigation are consistent with other studies that used different calibration constraints on both avian (Ricklefs and Outlaw 2010) and primate malarial parasites (Sutherland et al. 2010). Furthermore, they are similar to those found in studies carried out in ungulates and rodent parasites (Ramiro et al. 2012; Martinsen et al. 2016). However, the times estimated here clearly differ from other investigations. In particular, Silva et al. (2015) obtained older time estimates (30–300% older depending on the silent substation rate used) for the origin of Plasmodium in primates and rodents using relative rates. However, unlike the method implemented in RelTime, Silva et al. (2015) estimated pairwise divergences on proteins relative to a pair of closely related species (Plasmodium vivax and Plasmodium knowlesi). Those pairwise estimates assumed a constant rate of evolution for each protein without explicitly considering the phylogeny. Then, the relationship between relative divergences among species pairs was inferred from the slope of a linear regression (e.g., the split between P. falciparum and Plasmodium yoelii was 6.1 times older than the split of P. falciparum–Plasmodium reichenowi, see Silva et al. 2015). Finally, absolute times were calculated assuming a constant rate of silent site substitution for invertebrates and vertebrates (Silva et al. 2015). All of these are very strong propositions that rely on assuming constant rates of evolution at different levels (Graur and Martin 2004). Given the differences in methods and assumptions, their findings are difficult to compare with those reported here. Moreover, their method is not widely used, so its performance has not been properly evaluated.
This study also differs from a time estimate using genomic data for the origin of the two Plasmodium ovale subspecies (both parasitic to humans) that was clearly older than those presented here (Rutledge et al. 2017). The molecular dating method used, however, is designed to estimate the time of divergence between populations (not species) and involves very strong assumptions such as a universal rate of evolution (constant rate) and that all malarial parasites, regardless of their differences in life cycle, have the same generation time as P. falciparum (Rutledge et al. 2017). In contrast, a study on the same issue using mitochondrial and nuclear loci but with a molecular dating method suitable for species divergence yielded time estimates for primate malarias, including P. ovale subspecies, that were comparable with the results from this investigation (Sutherland et al. 2010).
Finally, the time estimates presented here do not support the classic hypothesis that haemosporidian parasites radiated with the origin of their insect vectors (Huff 1938; Escalante and Ayala 1995; Bensch et al. 2013), an event that took place 200–240 Ma. In particular, using such event as a calibration would require a series of ad hoc considerations in order to make particularly slow rates of the mtDNA compatible with what is known about the demography of human malarial parasites (Joy et al. 2003; Mu et al. 2005; Taylor et al. 2013). It would also imply an ancient divergence of avian and mammalian Plasmodium, ∼145–200 Ma (Krzywinski et al. 2006) making many observed biogeographic patterns in malarial parasites difficult to account for (Ricklefs and Outlaw 2010; Pacheco, Battistuzzi, Junge, et al. 2011; Muehlenbein et al. 2015; Martinsen et al. 2016). Furthermore, since most parasite clades would be older than the extant groups of hosts, selective extinctions (or failure to colonize particular group of hosts) are required in order to account for the lack of related parasite lineages in a broader group of vertebrate hosts and outside some geographic areas. Examples of such parasite clades are the Laverania subgenus that includes the human malaria parasite P. falciparum and its related species in African apes, the primate malaria from Southeast Asia, and the malarial parasites in ungulates (Pacheco, Battistuzzi, Junge, et al. 2011; Pacheco et al. 2012; Muehlenbein et al. 2015; Templeton, Martinsen, et al. 2016). Although these considerations do not demonstrate that this early origin is impossible, they make it less parsimonious.
In summary, time estimates from the mtDNA of haemosporidian parasites showed clear discrepancies between models assuming autocorrelated and independent evolutionary rates. As shown in this investigation, such discrepancies could be mitigated whenever new calibration information becomes available (dos Reis et al. 2016), particularly from avian lineages. Nevertheless, the mtDNA data using different methods indicate that avian haemosporidian parasites diversified during the radiation of their vertebrate host orders after the K–Pg boundary (66 Ma). This scenario needs to be evaluated as additional data (species and loci), and calibration information becomes available. However, increasing the data (loci and species sampled) without additional information that can inform calibration priors will not reduce the uncertainty in time estimates (dos Reis et al. 2016). Although each individual calibration constraint used here is an assumption that, on its own, cannot disprove others, the observation that these time priors are internally consistent (supplementary fig. S6C, Supplementary Material online) provides the most biologically plausible framework for timing haemosporidian parasites thus far.
Estimating the Rate of Evolution of the Avian Haemosporidian mtDNA Genomes
The discordant time estimates obtained from different methods could indicate disparate evolutionary rates in the mtDNA between clades of haemosporidian parasites. If differences in rates are confirmed, it might affect the biological interpretation of divergences of the mtDNA genes among haemosporidian clades. In order to explore this matter, the nucleotide substitution rates (substitutions/site/My) per partitions (three nonprotein coding regions plus the three ORFs) and per major clades were estimated using Bayesian methods under the three scenarios (table 2 and supplementary table S5, Supplementary Material online). Overall, heterogeneous rates were recovered between partitions, with coding sequences evolving faster than the nonprotein coding regions (which include the fragmented SSU rRNA and LSU rRNA). Furthermore, coding sequences evolved at distinct rates, with cox3 having the highest substitution rate (0.00474 substitutions per site per My), followed by cytb (0.00419) and cox1 (0.00371). In the case of nonprotein coding regions, nucleotide substitutions per site per My varied between 0.0016 and 0.0025 depending on their location in the genome (see table 2 for BEAST results). The same tendency was also found in MCMCTree rate estimates (supplementary table S5, Supplementary Material online). BEAST results are consistent with previous studies using similar methodologies in Plasmodium spp. infecting mammals, where the mean of the posterior distribution of the substitution rate, averaged over the entire mammal Plasmodium spp. tree, was 0.00383 (0.00189–0.00600) substitutions per site per My (Taylor et al. 2013). These rates are also similar to some previous estimates for Plasmodium using different methods and/or calibrations (Mu et al. 2005: 0.0045–0.0056; Jongwutiwes et al. 2005: 0.0026–0.0036; Cornejo and Escalante 2006: 0.0032–0.0043 substitutions per site per My).
Table 2.
Nucleotide Substitution Rates Per Site for Six Partitions (Three Nonprotein Coding Regions plus the Three Mitochondrial Genes) Estimated Using BEAST under Different Calibration Scenarios.
| Scenarios 1 |
Scenario 2 |
Scenario 3 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Node 153: 6–14.2 | Node 153: 6–14.2 | Node 153: 6–14.2 | ||||||||||
| Node 155: 24.44–34.0 | Node 155: 24.44–34.0 | Node 155: 24.44–34.0 | ||||||||||
| Calibrations (Ma) | Node 165: 20–65 | Node 165: 20–65 | Node 165: 20–65 | |||||||||
| Node 201: 56.8–86.8 | Node 185: 16–28.1 | |||||||||||
| Node 201: 56.8–86.8 | ||||||||||||
| NSR/Ma | Mean | SEM | Median | 95% CrI | Mean | SEM | Median | 95% CrI | Mean | SEM | Median | 95% CrI |
| Nonprotein coding region (884 bp) | 1.83E-03 | 2.59E-06 | 1.83E-03 | [1.49E-3–2.17E-3] | 1.83E-03 | 2.35E-06 | 1.83E-03 | [1.52E-3–2.19E-3] | 1.71E-03 | 2.12E-06 | 1.70E-03 | [1.41E-3–2.03E-3] |
| cox3 (756bp) | 4.74E-03 | 6.23E-06 | 4.73E-03 | [3.96E-3–5.53E-3] | 4.73E-03 | 5.62E-06 | 4.73E-03 | [4.01E-3–5.50E-3] | 4.41E-03 | 4.97E-06 | 4.39E-03 | [3.76E-3–5.12E-3] |
| Nonprotein coding region (848 bp) | 2.51E-03 | 3.63E-06 | 2.49E-03 | [1.99E-3–3.01E-3] | 2.50E-03 | 3.75E-06 | 2.49E-03 | [2.03E-3–3.04E-3] | 2.34E-03 | 3.04E-06 | 2.33E-03 | [1.89E-3–2.80E-3] |
| cox1 (1,434bp) | 3.71E-03 | 4.62E-06 | 3.70E-03 | [3.12E-3–4.29E-3] | 3.71E-03 | 4.55E-06 | 3.70E-03 | [3.17E-3–4.27E-3] | 3.45E-03 | 3.79E-06 | 3.44E-03 | [2.96E-3–3.96E-3] |
| cytb (1,134bp) | 4.19E-03 | 5.72E-06 | 4.18E-03 | [3.51E-3–4.91E-3] | 4.19E-03 | 5.09E-06 | 4.19E-03 | [3.52E-3–4.87E-3] | 3.90E-03 | 4.67E-06 | 3.88E-03 | [3.34E-3–4.55E-3] |
| Nonprotein coding region (1,628 bp) | 1.60E-03 | 2.15E-06 | 1.60E-03 | [1.31E-3–1.88E-3] | 1.60E-03 | 2.08E-06 | 1.60E-03 | [1.33E-3–1.87E-3] | 1.49E-03 | 1.78E-06 | 1.49E-03 | [1.26E-3–1.74E-3] |
Note.—Mean, SEM, median, and 95% credibility intervals (CrIs) are shown in Ma. Node numbers are shown in figure 2. See Materials and Methods for a description of the calibration constraints used as priors.
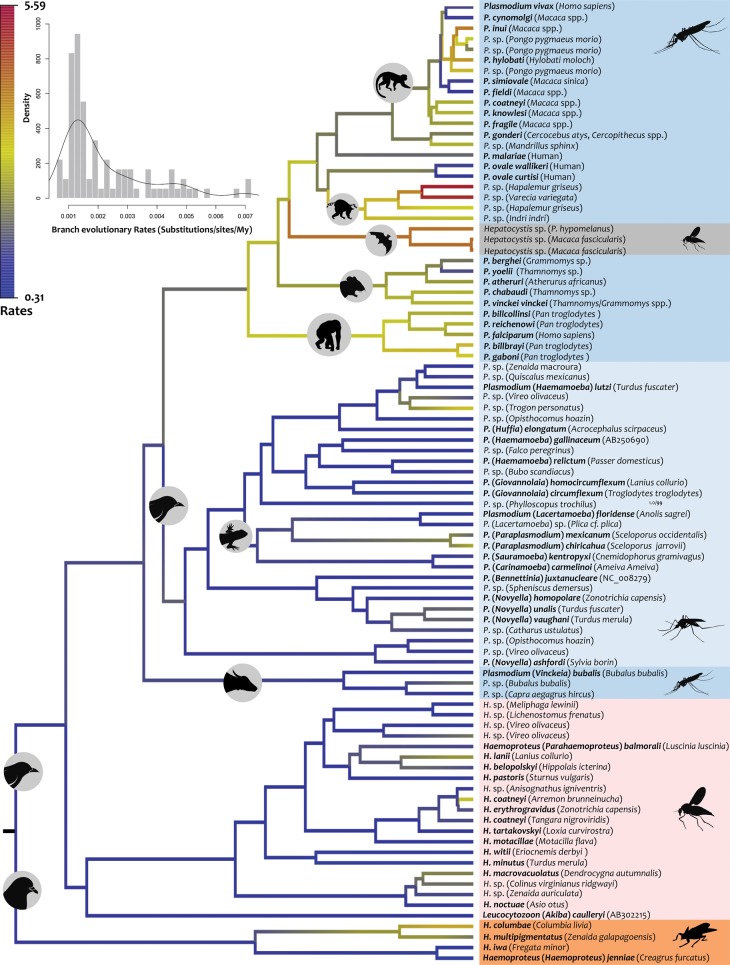
Heterogeneous nucleotide substitution rates were also found in the most recent common ancestor nodes for each haemosporidian major monophyletic group, with Plasmodium parasites from rodent and primate species evolving at a faster rate than avian haemosporidian parasites (supplementary table S6, Supplementary Material online). This pattern was subjected to further testing by estimating relative evolutionary rates per branch by using RelTime (fig. 3), and a pattern of a faster mtDNA rate in rodent-primate parasites was found. These differences in rate may account for early suggestions of an explosive species radiation of Plasmodium species (Hayakawa et al. 2008). Changes in rates between clades could be the result of factors such as differences in parasite generation times; those processes will affect rates whether those were synonymous or nonsynonymous, so they are expected to be correlated under those scenarios (Seo et al. 2004) in all genes. Thus, mtDNA rate differences between clades for each gene were also examined by estimating synonymous (nonsynonymous) rates per codon per My using a codon model as implemented in CodonRates v1.0 (supplementary table S7, Supplementary Material online) (Seo et al. 2004). As expected, synonymous rates were higher on an average than nonsynonymous rates. However, the concordance test found more variation in nonsynonymous than synonymous rates over time in cox3 (P < 0.05) and cytb (P < 0.05) but not in cox1 (P = 0.05). These patterns could be the result of differences in the strength of natural selection rather than other processes such as differences in generation time (see analyses below). Although the credibility intervals per gene overlapped across clades, there is a trend indicating that the mtDNA genes in the Parahaemoproteus subgenus are evolving at a slower rate when compared with other avian parasites. This pattern of disparate rates suggests that the use of barcoding approaches using mtDNA genes needs additional scrutiny by comparing well-characterized species in different haemosporidian clades.
Fig. 3.
Relative evolutionary rates of haemosporidian parasites. Branches are colored from high (red) to low (blue) according to their relative rates respect to the root rate (that is set to one) estimated from RelTime without calibration constraints. The histogram shows branch absolute evolutionary rates (substitutions/site/My) estimated with respect to the root rate under calibration scenario 1. Differences in vectors are indicated using a color code.
The findings reported here can be summarized as follows: 1) discrepancies in time estimates from different methods could be explained by the observed differences in evolutionary rates between haemosporidian clades; 2) compared with other avian parasites, the mtDNA genes in Parahaemoproteus subgenus appear to be evolving at a slower rate; and 3) there is a faster mtDNA rate of evolution in parasites of the rodent/primate parasite clade. This is the first report of nucleotide substitution rates estimated for each individual portion of the haemosporidian mitochondrial genome (genes and fragmented SSU rRNA/LSU rRNA) and avian haemosporidian major clades. The rates reported here provide relevant information that can be used to interpret mtDNA gene variation in avian haemosporidian parasites.
Is the Haemosporidian mtDNA Genome under Positive Selection?
In order to understand how selection acted on the mtDNA genes, methods implemented in HyPhy package were applied (Kosakovsky Pond et al. 2005, see Materials and Methods). First, a branch-site unrestricted test for episodic diversification (BUSTED, Murrell et al. 2015) was used without specifying forward branches (see Materials and Methods). BUSTED detected evidence of a low proportion of sites (1% or less, see table 3) under positive diversifying selection in all mtDNA genes. Since BUSTED is designed to aggregate information from multiple sites (losing site-level information due to necessary statistical smoothing), a test that examines each individual site independently for evidence of episodic diversification was used (MEME, Murrell et al. 2012). Importantly, MEME is capable of handling site-to-site heterogeneity in synonymous substitutions, a phenomenon suggested by the rates estimated using CodonRates v1.0. Thus, few sites under positive selection were detected in each gene using the MEME method (P < 0.05, table 3).
Table 3.
Parameter Estimates for Haemosporidian Parasite mtDNA Genes Using Models Implemented in HyPhy.
| Gene | HyPhy Test | |||||
|---|---|---|---|---|---|---|
| cox3 | ||||||
| MEME | 7 sites with evidence of episodic diversifying selection (EDS): 68, 75, 178, 192, 196, 204, 243 (P<0.05) | |||||
| BUSTED |
There is evidence of episodic diversifying selection, with LRT P of 0.000003423. |
|||||
|
Model |
log L |
AICc |
ω1 |
ω2 |
ω3 |
|
| Unconstrained model | −16,134.27 | 32,712.36 | 0.0392 (93%) | 0.565 (6.4%) | 14.6 (0.27%) | |
| Constrained model |
−16,146.85 |
32,735.5 |
0.0364 (95%) |
1.00 (0.27%) |
1.00 (4.9%) |
|
| RELAX |
Clade |
Parameter (k) |
log L |
P |
||
| H. (Parahaemoproteus) | k=1.03 (relaxation) | Null model (all k=1): −16,075.11, Alternative model = −16,059.58 | <0.001 | |||
| avian/Squamata Plasmodium spp. | k=1.21 (intensification) | |||||
| primate/rodent Plasmodium | k=1 (fixed) | |||||
| Leucocytozoon spp. | k=0.87 (relaxation) | |||||
| cox1 | ||||||
| MEME | 5 sites with evidence of EDS: 116, 169, 192, 402, 469 (P <0.05) | |||||
| BUSTED |
There is evidence of episodic diversifying selection, with LRT P of 0.001179. |
|||||
|
Model |
log L |
AICc |
ω1 |
ω2 |
ω3 |
|
| Unconstrained model | −26,321.66 | 53,085.33 | 0.0243 (97%) | 0.0463 (2.0%) | 3.48 (1.1%) | |
|
Constrained model |
−26,328.4 |
53,096.79 |
0.00181 (87%) |
0.0210 (8.1%) |
1.00 (5.1%) |
|
| RELAX |
Clade |
Parameter (k) |
log L |
P |
||
| H. (Parahaemoproteus) | k=0.56 (relaxation) | Null model (all k=1): −26,293.75, Alternative model = −26,219.05 | <0.001 | |||
| avian/Squamata Plasmodium spp. | k=4.67 (intensification) | |||||
| primate/rodent Plasmodium | k=1 (fixed) | |||||
| Leucocytozoon spp. | k=0.53 (relaxation) | |||||
| cytb | ||||||
| MEME | 4 sites with evidence of EDS: 234, 334, 343, 374 | |||||
| BUSTED |
There is evidence of episodic diversifying selection, with LRT P value of 0.003114. |
|||||
| Model | log L | AICc | ω1 | ω2 | ω3 | |
| Unconstrained model | −19,979.89 | 40,402.32 | 0.0247 (92%) | 0.516 (8.0%) | 19.7 (0.095%) | |
|
Constrained model |
−19,985.66 |
40,411.84 |
0.0317 (93%) |
0.0370 (2.8%) |
1.00 (4.0%) |
|
| RELAX |
Clade |
Parameter (k) |
log L |
P |
||
| H. (Parahaemoproteus) | k=0.82 (relaxation) | Null model (all k=1): −19,873.69, Alternative model = −19,822.87, | <0.001 | |||
| avian/Squamata Plasmodium spp. | k=1.33 (intensification) | |||||
| primate/rodent Plasmodium | k=1 (fixed) | |||||
| Leucocytozoon spp. | k=0.47 (relaxation) | |||||
Note.—Specific models, their parameters, and P are shown for each mtDNA genes. In the case of RELAX, a significant result of k < 1 indicates that selection strength has been relaxed along the test branches and a significant result k > 1 indicates that selection strength has been intensified along the test branches. An intensification on the strength of negative selection on the avian Plasmodium spp. is indicated in bold.
Lastly, in order to perform a rigorous joint test for differences in selective regimes between clades (with some branches in the phylogeny possibly designated as “nuisance”), the RELAX method (Wertheim et al. 2015) was modified (see Materials and Methods). For all three mtDNA genes, this method found strong evidence of both per site synonymous rate variation and relative relaxation/intensification of selection among clades of avian parasites. Although ω estimates indicate that strong negative selection is acting on all these genes (as is almost universally found in mtDNA selection analyses, regardless of the taxonomic range), there is clear evidence of intensification of purifying (negative) selection acting on the avian/Squamata Plasmodium spp. (table 3 and fig. 4; supplementary table S8, Supplementary Material online). In contrast, evidence of relaxation was found in all three genes for both Leucocytozoon and H. (Parahaemoproteus) spp. clades. Taken together, these findings indicate that there are differences in the strength of negative selection acting on different parasite clades, but there is no clear indication that positive selection is acting on the divergence on any particular mtDNA gene, with the exception of transient events affecting only a few sites in each gene (detected by MEME).
Fig. 4.
Branch-specific relaxation parameter inferred for each mitochondrial gene on the mtDNA phylogeny under the General Descriptive model (RELAX). Branches are colored based on relative value of the selection intensity as measured by the parameter k (red for large values and blue for small values): k > 1 indicates that selection strength has been intensified along the test branches, and a k < 1 indicates that selection strength has been relaxed along the test branches. Clades/vector relationships are identified by vertical colored bars. The scales at the top are substitutions per site.
These findings contrast with previous investigations proposing that positive selection acted on the divergence on the mtDNA cytb as a result of the switch to mammals from avian hosts in Plasmodium spp. (Outlaw and Ricklefs 2010; Outlaw et al. 2015). These earlier investigations analyzed only partial sequences of the mtDNA genes (cytb and cox1) from a handful of taxa. Nevertheless, those findings could not be reproduced using complete gene sequences even when the same tests were applied (no evidence for positive selection was found using methods implemented in PAML, see supplementary table S9, Supplementary Material online). Importantly, the new tests proposed here make fewer assumptions and are generally more powerful.
Why are there differences in selective regimes between clades? There is limited phenotypic information related to the mtDNA. However, given that the available evidence indicates that the mitochondria from haemosporidians are not metabolically important in the parasite asexual vertebrate stage (Ke et al. 2015; Jacot et al. 2016), it is unlikely that natural selection (positive or negative) could be acting differentially on the parasite mtDNA genes as a result of changes in their vertebrate host. On the other hand, since the mtDNA genes are essential in the vector stages, the proposed differences in vector families between the clades seem more biologically relevant to explain the observed differences in functional constraints (purifying selection) that led to clade rate variation in the mtDNA genes. It is worth emphasizing that these patterns of different selective regimes acting on the mtDNA emerged without assuming the proposed vector/host association. Thus, the emerging patterns in terms of differences in rates, selective regimes, and synonymous rate variation among parasite clades seem to be associated with differences in vectors at the family/subfamily levels. These patterns suggest a potential role for vectors in driving the mode and rate of evolution of the haemosporidia mtDNA.
Evolution of Haemosporidian Parasites: An Example of Evolutionary Processes at the Community Level?
Although requiring further investigation, the patterns found here suggest a more general hypothesis: the rate and mode of evolution of genes that are functionally important in the parasite sexual stages (vector stages), such as the genes in the mtDNA, could be affected by major vector switches at the family/subfamily levels. This hypothesis, contingent on the proposed association of parasite clades to a particular group of vectors, can be tested as more genomic and transcriptomic data become available. Ascertaining how major switches in vectors affected the evolution of genes expressed in the parasite sexual (vector) stages would provide new insights into the molecular basis driving the diversification/establishment of competent vectors, a critical aspect when designing interventions targeting the transmission of parasites of medical or economic importance.
Beyond the evolution of the haemosporidian mtDNA, the patterns reported here suggest that the radiation of the major parasite clades cannot be described solely by assuming parasite cospeciation with their vertebrate or invertebrate hosts. The emerging picture from the mtDNA (phylogenetic analyses and timetrees) can be better explained by considering ecological and evolutionary processes at the community level (Brooks et al. 2006; Weber et al. 2017). In particular, it can be hypothesized that groups of invertebrate (hematophagous insects) and vertebrate hosts interacted during their evolution providing diverse (and even transient) biocenoses where haemosporidian parasites radiated. Indeed, the host ranges of particular parasite lineages may have undergone historical changes as a result of demographic history of each host species (vertebrate and invertebrate) that affected their population densities and spatial distributions. Given that the radiation of vector species clearly predates the origin of the vertebrate host clades (e.g., the plethora of Anopheles mosquitoes transmitting human malaria originated well before there were hominids, see Krzywinski et al. 2006), efforts should be made to understand how vertebrate–vector biocenoses emerged as a result of community-level evolutionary dynamics. Such studies will provide valuable information about the origin and diversity of haemosporidian parasites.
Materials and Methods
Samples, DNA Extraction, and Parasite Mitochondrial Genome (mtDNA) Amplification
Almost complete parasite mitochondrial genomes (mtDNA) were obtained for 63 samples from different hosts and countries (57 birds, 4 lizards, and 2 macaques). The samples were positive for haemosporidian parasites belonging to four genera: Leucocytozoon, Haemoproteus (subgenera Haemoproteus and Parahaemoproteus), Plasmodium, and Hepatocystis. Each sample was previously screened for haemosporidian parasites by microscopy (Valkiūnas 2005) and/or polymerase chain reaction (PCR), using primers targeting the cytb gene that have been used in previous studies (Bensch et al. 2000; Hellgren et al. 2004; Waldenström et al. 2004; Pacheco, Battistuzzi, Junge, et al. 2011; Levin et al. 2013; Matta et al. 2014).
Genomic DNA was extracted from whole blood or tissues (liver) using DNeasy Blood & Tissue Kit (Qiagen, GmbH, Hilden, Germany) and, the mtDNA was amplified using the oligos forward 5′ GAGGATTCTCTCCACACTT CAATTCGTACTTC 3′/reverse 5′ CAGGAAAATWA TAGACCGAACCTTGGACTC 3′ with TaKaRa LA Taq Polymerase (TaKaRa Mirus Bio Inc.) following manufacturers’ directions. In the specific case of Haemoproteus (Haemoproteus) species, oligo forward 5′ GAGGATTCTCT CCACACTTMAATTCGTAMTTCC 3′ was used with the same oligo reverse. Wherever the parasitemia was low, a nested PCR was performed using the internal oligos forward 5′ TTTCATCCTTAAATCTCGTAAC 3′/reverse 5′ GACCGAA CCTTGGACTCTT 3′. Using the mtDNA of P. falciparum (M76611) as a reference (see figure in Feagin et al. 2012), the inner forward primers anneal the fragment number 8 of the large subunit ribosomal RNA (LSU rRNA), and the inner reverse primer anneals the fragment number 1 of the LSU rRNA. The amplified mtDNA region for all species comprises at least 5,451 bp (excluding primers after nested amplification) out of 5,967 bp of the P. falciparum mtDNA genome, so only three tRNAs (7, 11, and 14) and two fragments of small subunit ribosomal RNAs (5 and 7) are missing. All PCR reactions were carried out in 50 μl, and negative controls (dH2O), and positive controls (samples from infected macaques) were included. Amplification conditions for all PCR were: a partial denaturation at 94 °C for 1 min and 30 cycles with 30 s at 94 °C and 7 min at 68 °C, followed by a final extension of 10 min at 72 °C.
At least two or three independent PCR products (50 μl) were excised from the gel (bands of ∼6 kb), purified using QIAquick Gel extraction kit (Qiagen, GmbH, Hilden, Germany), and cloned into the pGEM-T Easy Vector systems (Promega). A minimum of three clones and both strands were sequenced per sample using an Applied Biosystems 3730 capillary sequencer. Inconsistencies between the clones were not found, and only four samples with mixed infections (two distinct parasite species) were detected by PCR (sample UN234: coinfection with Plasmodium/Haemoproteus spp., and samples 01.14 C, 76.13 C, and 345.13 C: coinfection with Haemoproteus/Leucocytozoon spp.). Using BLAST against the Malavi database (Bensch et al. 2009), no discrepancies were found between the cytb sequences from the complete mtDNA and the partial cytb lineages previously reported for many of the well-characterized parasites included here (supplementary table S1, Supplementary Material online). A total of 65 new, almost complete mtDNA genomes from the 63 samples were obtained in this investigation. This data set included parasite species that had been well-identified using morphology. The sequences are available in GenBank under accession numbers KY653752 to KY653816. Supplementary table S1, Supplementary Material online, provides a complete list of the parasites (with their host species) used in the analyses reported here, including the sequences previously available in GenBank (June 2016).
Alignment Construction, Index of Substitution Saturation, and Phylogenetic Analyses
Three different nucleotide alignments were produced by using ClustalX v2.0.12 and Muscle as implemented in SeaView v4.3.5 (Gouy et al. 2010) with manual editing. The first alignment was constructed with a total of 114 mtDNA genome sequences (5,125 bp excluding gaps) belonging to four genera (Leucocytozoon, Haemoproteus, Plasmodium, and Hepatocystis). This alignment included all the sequences obtained in this study, those available from GenBank, as well as different haplotypes for some well-identified parasite species using morphology (e.g., Plasmodium lutzi, Haemoproteus columbae, and L. fringillinarum). Then, the alignment was annotated following the gene models proposed by Feagin et al. (2012). This annotation separates the mtDNA genome in fragmented small subunit ribosomal RNAs (SSU rRNA), large subunit ribosomal RNAs (LSU rRNA), and the three protein-coding genes. Subsequently, the alignment was divided into six partitions corresponding to the three nonprotein coding regions between the ORFs (fragmented SSU rRNA and LSU rRNA) and the three protein-coding genes, keeping their order in the mtDNA genome (nonprotein coding, cox3, nonprotein coding, cox1, cytb, nonprotein coding). Given that substitution saturation decreases phylogenetic information contained in the sequences, the entropy-based index of substitution saturation (Xia et al. 2003) was estimated using Dambe v6.4.81 (Xia 2017) for each gene and nonprotein coding regions. In particular, the evaluation of the phylogenetic information using this method was performed separately for the first, second, and third codon positions of the genes, and for the joined sites (1st + 2nd + 3rd). In addition, the observed numbers of transitions (s) and transversions (v) for complete mitochondrial genome and each gene were plotted against GTR distance as implemented in DAMBE v6.4.81 (Xia 2017).
A second alignment was produced using only the three protein-coding genes (CDS, 3,414 bp excluding gaps). In this alignment, each gene was considered as a partition, and only 102 sequences of different parasite species were selected from the first alignment (duplicate haplotypes were not included). Although this study focuses on avian haemosporidian parasites, the gene sequences of Nycteria gabonensis (KX090647), and Nycteria heischi (KX090648) were also included in this CDS alignment for a total of 104 species. Given the difference in mtDNA genome organization between some Nycteria parasites (e.g., N. medusiformis [KX090645] and Nycteria sp. [KX090646]), and the other haemosporidian species, neither the complete Nycteria mtDNA genomes nor genes from these two particularly divergent lineages were included in this study.
Finally, in order to estimate divergence times for these major haemosporidian genera, a third alignment (5,136 bp excluding gaps) was produced based on 102 sequences of mtDNA genomes selected from different species of the first alignment (duplicate haplotypes were not included). This alignment was also divided into six partitions including the three nonprotein coding regions between the genes (fragmented SSU rRNA and LSU rRNA) and the three genes. The complete mtDNA genome sequences for Nycteria species were not included here since neither the order nor the transcriptional directions of the three genes are conserved between Nycteria genus and the other haemosporidian parasites (Karadjian et al. 2016). The alignments are available upon request.
Phylogenetic relationships were inferred based on the first (whole mtDNA) and second (CDS) alignments using the Bayesian methods implemented in MrBayes v3.2.6 with the default priors (Ronquist and Huelsenbeck 2003) and the ML method in RAxML (Guindon et al. 2003; Stamatakis 2014). For both phylogenetic methods, a general time reversible model with gamma-distributed substitution rates and a proportion of invariant sites (GTR + Γ +I) was used for each partition; this was the model with the lowest Bayesian Information Criterion (BIC) scores for both alignments and each partition as estimated by MEGA v7.0.14 (Kumar et al. 2016). For RAxML analyses, statistical confidence was assessed with 1,000 bootstrap replicates. To better explore the likelihood surface, RAxML was run with 1,000 distinct maximum parsimony starting trees, and the tree with the greatest likelihood value was selected. Bayesian support for the nodes was inferred in MrBayes by sampling every 500 generations from two independent chains lasting 107 Markov Chain Monte Carlo (MCMC) steps. The chains were assumed to have converged once the average SD of the posterior probability was <0.01 and the value of the potential scale reduction factor (PSRF) was between 1.00 and 1.02 (Ronquist and Huelsenbeck 2003). As a “burn-in,” 50% of the sample was then discarded once convergence was reached.
The root of the phylogeny was estimated from the posterior distribution of the trees as implemented in BEAST v2.4.1 (Bouckaert et al. 2014). The estimated root (fig. 1 and supplementary fig. S2, Supplementary Material online) was consistent with those previously reported from different multi-locus data sets that included diverse haemosporidian taxa (Borner et al. 2016; Lutz et al. 2016).
Divergence Times, Rate of Evolution, and Natural Selection
Bayesian methods BEAST v2.4.1 (Bouckaert et al. 2014) and MCMCTREE v.4.8 (Yang 2007) were used to estimate time trees and nucleotide substitution rates/My per partition (three nonprotein coding regions plus the three genes) and per major clades. In BEAST, the relaxed clock (Drummond et al. 2006) was applied with the GTR + Γ+I evolutionary model with a Yule model as the tree prior. Two independent chains lasting 108 MCMC steps were run for each analysis until convergence. Three scenarios using a combination of calibration constraints were explored using BEAST. The calibration constraints used as priors include four that consider information on vertebrate hosts’ fossils (see Paleobiology Database at http://www.paleodb.org/; last accessed November 13, 2017) and one based on a biogeographical landmark for a group of vertebrate hosts. In all cases, uniform priors were used to calibrate divergences. Therefore, no particular time point within a given time interval was favored, and no punctual calibrations were used.
The first calibration constraint considers that African parasites found in Mandrillus spp. and Cercocebus spp. (Plasmodium gonderi and a Plasmodium spp., supplementary table S1, Supplementary Material online) diverged from those Plasmodium species found in Southeast Asia macaques (a monophyletic group, see node 149 in fig. 2, and also Muehlenbein et al. 2015) when Macaca branched from Papio (Mu et al. 2005; Pacheco, Battistuzzi, Junge, et al. 2011). Macaca spp. fossils indicate that such event took place 6–8 Ma as minimum and maximum boundaries, respectively (Delson 1980); however, these fossils are used as minimum times when studying primates (Perelman et al. 2011). Thus, a more inclusive alternative calibration constraint was proposed around this event assuming that the divergence at the same node took place at any time between 6 and 14.2 Ma using a uniform prior. This inclusive time interval incorporates molecular estimates for the same Papio-Macaca divergence event (Pacheco, Battistuzzi, Junge, et al. 2011); it is consistent with older fossils reported for Macaca spp. (see Paleobiology Database at http://www.paleodb.org/; last accessed November 13, 2017), and considers the fact that P. gonderi has been also reported in Chlorocebus (a Cercopithecini).
A second fossil-based calibration constraint is the minimum of 23.5 Ma for the human/Macaca split (Benton and Donoghue 2007), which is assumed to be the minimum time when the monophyletic group that includes P. malariae (a parasite found in humans) and all the Asian parasites originated. This event is used to inform a calibration prior with a maximum of 65 Ma that allows this parasite clade to be as old as the origin of primates, if the data support that (Perelman et al. 2011). The third calibration constraint considers the origin of lemur malaria, a monophyletic group of parasites that radiated in Madagascar (Pacheco, Battistuzzi, Junge, et al. 2011). This calibration assumes that these parasites were introduced to Madagascar between 20 Ma (minimum) and 65 Ma. In particular, introductions after 20 Ma are biologically unrealistic since this is the time estimated for the last colonization of terrestrial vertebrates (including mammals) into the island (Ali and Huber 2010). The maximum was set at 65 Ma allowing this clade to be as old as the origin of primates if the data support that (Perelman et al. 2011).
The fourth calibration constraint is that the parasites of ungulates originated at the proposed time for the origin of Bovinae-Antilopinae (16–28.1 Ma, Benton et al. 2015), where 16 Ma corresponds to Pseudoeotragus seegrabensis fossil (Bibi 2013) from the MN4 planktic foraminifera zone (Burdigalian stage) and 28.1 Ma to the soft maximum constraint at the base of the Chattian stage of the late Oligocene (28.1 Ma), encompassing many equivocal stem bovids, but lacking those from the crown (Benton et al. 2015). Finally, the proposed root calibration is the origin of Palaeognathae (56.8–86.8 Ma, Benton et al. 2015). The upper limit (56.8 Ma) corresponds to Lithornis celetius fossil (Houde 1988) from the Fort Union Formation (early Tiffanian) of Montana and Wyoming and the lower limit (86.8 Ma) to the soft maximum constraints equivalent to the age of the Niobrara Chalk Formation, dated as Santonian (86.3–83.6 ± 0.5 Ma). Like in the case of the other calibration constraints, uniform priors were used.
These calibrations were organized into three scenarios. A first scenario (scenario 1) included a combination of the relaxed 6–14.2 Ma calibration of the Papio/Macaca parasite divergence (95% quantile = 13.8) (Delson 1980; Mu et al. 2005); a maximum of 23.5 and a minimum of 34.0 Ma for the human/Macaca split (95% quantile = 33.5); and a range of 20–65 Ma (95% quantile = 62.8) for the origin of the lemur parasites (Pacheco, Battistuzzi, Junge, et al. 2011). The second scenario (scenario 2) comprised the calibrations used in the first scenario plus a calibration at the root (the origin of Palaeognathae). Lastly, a third scenario (scenario 3) considered all the calibration information of scenario 2 plus a calibration constraint proposed for the origin of Bovinae-Antilopinae (16–28.1 Ma, Benton et al. 2015).
In the case of MCMCTree, a calibration constraint is needed for the tree root, so only the second and third scenarios were considered. Estimates of the branch lengths were calculated by the approximate likelihood method (dos Reis and Yang 2011) under the HKY85+Γ+I substitution model on the MrBayes tree obtained using the third alignment. Then, MCMC algorithm was used to estimate divergence times on the constrained tree topology in two separate runs. Both independent and autocorrelated rates models (Rannala and Yang 2007) were employed with the following parameters: a diffuse gamma prior G(1, 1) for the overall substitution rate; a rate drift parameter σ2 G(1, 1); and the parameters of the birth–death process with species sampling fixed at λ = μ = 1 and ρ = 0. A total of 2×106 iterations was run, sampling every 100 with 2,000 iterations as burn-in, and then both runs were concatenated to provide the final posterior values. Although the time estimates under both Bayesian methods and distinct calibration scenarios were expected to overlap, their comparison is important since the calibrations rely on different assumptions (Pacheco, Battistuzzi, Junge, et al. 2011; Ramiro et al. 2012). The autocorrelation and the independent rates models were compared by Bayes factor statistics in Tracer (http://tree.bio.ed.ac.uk/software/tracer/; last accessed November 13, 2017.) (Ho et al. 2015).
Besides the Bayesian approaches, divergence times were also estimated in a non-Bayesian framework with RelTime method (Tamura et al. 2012). RelTime algorithm does not assume any statistical model for the variation in rates of evolution across lineages as the Bayesian methods do (Tamura K, Tao Q, Kumar S, unpublished data, https://www.biorxiv.org/content/early/2017/08/24/180182; last accessed November 13, 2017). However, RelTime computes the rate of the ancestral branch as the average of those in the two immediate descendant branches (Tamura K, Tao Q, Kumar S, unpublished data, https://www.biorxiv.org/content/early/2017/08/24/180182; last accessed November 13, 2017) and that it is not free from assumptions. In this case, calculations were carried out on the command line version of MEGA7 (Kumar et al. 2012, 2016). The substitution model was the same as that used for Bayesian analyses (GTR + Γ+I). RelTime only requires minimum and/or maximum boundaries for calibration constraints, so the same boundaries as those given to the calibration density priors in Bayesian analyses were used. The calibration located at the root of the phylogenetic tree was automatically removed as RelTime does not consider it because the assumption of equal rates of evolution between ingroup and outgroup sequences is not testable (Kumar et al. 2016). Two divergence time analyses were performed in RelTime, with and without incorporating calibrations (Battistuzzi et al. 2015). This allows exploring the effect of calibration constraints used as priors on time estimates. In particular, absolute times were estimated under two distinct calibration scenarios (scenario 1 and scenario 1 plus the calibration of the Bovinae crown node). Secondly, relative noncalibrated times were estimated in RelTime with the same procedure described earlier, except for the fact that calibration boundaries were not provided.
Absolute rates of synonymous and nonsynonymous substitutions per codon per My for each gene and per major clades were obtained using CodonRates v1.0 in a Bayesian framework (Seo et al. 2004). Because tree topology, fossil constraints, and calibration prior for the ingroup root node are required for this method, MrBayes topology with Babesia bovis (EU075182) as an outgroup, a calibration point of 71.8 ± 5 Ma for the ingroup root of the tree (56.8–86.8 Ma, Benton et al. 2015), and the calibrations used in the first scenario were employed in these analyses.
In order to elucidate selective pressures during the evolution of the haemosporidian mtDNA genomes, several methods were performed with HyPhy package (Kosakovsky Pond et al. 2005) implemented in DataMonkey Web Server (http://www.datamonkey.org/; last accessed November 13, 2017). A branch-site unrestricted test for episodic diversification (BUSTED, Murrell et al. 2015) was used to investigate whether individual genes were subject to diversifying selection anywhere in the tree and along the sequence. Then, evidence of both episodic and pervasive positive selection at the level of an individual site was identified using a mixed effects model of evolution (MEME, Murrell et al. 2012). Sites positively selected were identified at a significance level of P <0.05. In order to perform a rigorous joint test for differences in selective regimes between clades (with some branches in the phylogeny possibly designated as “nuisance”), the RELAX method (Wertheim et al. 2015) was modified. RELAX is a hypothesis testing framework that asks whether the strength of natural selection has been relaxed or intensified along a specified set of test branches (Wertheim et al. 2015). Thus, it allows identifying trends and/or shifts in the stringency of natural selection on a given gene between lineages. Specifically, the method was extended to support more than two clades in a single analysis and added a site-level synonymous rate variation component to augment branch-site level variation used in ω ratio. This test performs a likelihood ratio test (LRT) by comparing the null model in which the relation parameter (k) is constrained to 1 for all clades, to the alternative model in which K is a free, clade-specific, parameter, and one clade (primate/rodent Plasmodium clade) is designated as reference (k = 1). RELAX models each branch-site combination as a draw from a distribution of ω with three values (ω−<1, ωN=1, ω+>1). For a specific clade, these values are modified by the k parameter via ω → ωK. Hence, estimates of k > 1 imply intensification of selection, and k < 1 implies a relaxation of selection, relative to the reference clade.
Finally, in order to compare this study against previous findings (Outlaw and Ricklefs 2010; Outlaw et al. 2015), CODEML (PAML package v4.9, Yang 2007) was applied to conduct site models along with more complex branch-site models (Zhang et al. 2005). First, to estimate the proportion of sites under selection per gene, model 1 (neutral) versus model 2 (selection) and models M7 (beta; Model = 0, NSsites = 7) versus M8 (beta + ω; Model = 0, NSsites = 8) were compared. M7 and M8 estimate the variation in dN/dS using a beta distribution, with M8 specifically addressing the proportion of sites under positive selection. A LRT was conducted using the log likelihoods from each model. Significance was assigned using a chi-square distribution and 2 degrees of freedom (df; Wong et al. 2004). Then, to identify whether specific clades evolve at different rates and potentially could be under unique signatures of selection, branch-site models, implemented also in CODEML (MA and MA-null, also referred to as Branch-model M2 and M2 Null) were used, which allow ω to vary among lineages (Zhang et al. 2005). To understand if specific clades are more likely to be characterized by a ω value differing from the rest of the phylogeny, branch-model M0 (Model = 0, NSsites = 0) was compared with the branch model M2 (two-ratio model; Model = 2, NSsites = 0). In addition, signatures of positive selection for specific clades was evaluated by comparing the branch model MA (Model = 2, NSsites =2, fix_ω = 0, ω > 1), which allows ω to vary >1, with the null hypothesis MA-null (Model = 2, NSsites = 2, fix_ω = 1, ω = 1) in which ω is fixed at 1 (neutrality). These two analyses were run independently with the three major clades labeled as “foreground” lineages (Haemoproteus [Haemoproteus] spp., Plasmodium spp. from avian/Squamata, and Plasmodium spp. from rodent/primate). From the LRT, a chi-square distribution was used to assign significance using 1 df. All these methods are susceptible to site level synonymous rate variation.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported in part by the US National Institutes of Health (grant R01 GM080586 to A.A.E.). The authors thank Professor Martha L. Calderón Espinosa at “Instituto de Ciencias” (Universidad Nacional de Colombia) for her support to obtain reptile samples and their taxonomic identification; we thank all the students of the Host-Parasite Relationship Research Group at Universidad Nacional de Colombia, especially Ingrid A. Lotta. We thank the staff of the P. B. Šivickis Laboratory of Parasitology, Nature Research Centre, particularly Tatjana A. Iezhova and Vaidas Palinauskas, for participation in parasite sampling. We also thank Steven Weaver, Bui Quang Minh, Monica M. Acosta, Benjamin L. Rice, Michael Li, and Roberto Aguilar for their valuable support. We thank Georges Snounou for his comments on an early version of this manuscript. We thank the DNA laboratory at the School of Life Sciences (Arizona State University) for their technical support. Finally, the authors thank the editor and reviewers for their valuable comments.
References
- Ali JR, Huber M.. 2010. Mammalian biodiversity on Madagascar controlled by ocean currents. Nature 4637281:653–656.http://dx.doi.org/10.1038/nature08706 [DOI] [PubMed] [Google Scholar]
- Battistuzzi FU, Billing-Ross P, Murillo O, Filipski A, Kumar S.. 2015. A protocol for diagnosing the effect of calibration priors on posterior time estimates: a case study for the Cambrian explosion of animal phyla. Mol Biol Evol. 327:1907–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battistuzzi FU, Filipski A, Hedges SB, Kumar S.. 2010. Performance of relaxed-clock methods in estimating evolutionary divergence times and their credibility intervals. Mol Biol Evol. 276:1289–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensch S, Hellgren O, Križanauskienė A, Palinauskas V, Valkiūnas G, Outlaw D, Ricklefs RE.. 2013. How can we determine the molecular clock of malaria parasites? Trends Parasitol. 298:363–369. [DOI] [PubMed] [Google Scholar]
- Bensch S, Hellgren O, Pérez-Tris J.. 2009. MalAvi: a public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol Ecol Resour. 95:1353–1358. [DOI] [PubMed] [Google Scholar]
- Bensch S, Stjernman M, Hasselquist D, Ostman O, Hansson B, Westerdahl H, Pinheiro RT.. 2000. Host specificity in avian blood parasites: a study of Plasmodium and Haemoproteus mitochondrial DNA amplified from birds. Proc Biol Sci. 2671452:1583–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton MJ, Donoghue PC.. 2007. Paleontological evidence to date the tree of life. Mol Biol Evol. 241:26–53. [DOI] [PubMed] [Google Scholar]
- Benton MJ, Donoghue PCJ, Asher RA, Friedman M, Near TJ, Vinther J.. 2015. Constraints on the timescale of animal evolutionary history. Palaeontol Electron. 18.1.1FC:1–107. [Google Scholar]
- Bertram MR, Hamer SA, Hartup BK, Snowden KF, Medeiros MC, Outlaw DC, Hamer GL.. 2017. A novel Haemosporida clade at the rank of genus in North American cranes (Aves: Gruiformes). Mol Phylogenet Evol. 109:73–79. [DOI] [PubMed] [Google Scholar]
- Bibi F. 2013. A multi-calibrated mitochondrial phylogeny of extant Bovidae (Artiodactyla, Ruminantia) and the importance of the fossil record to systematics. BMC Evol Biol. 13:166.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner J, Pick C, Thiede J, Kolawole OM, Kingsley MT, Schulze J, Cottontail VM, Wellinghausen N, Schmidt-Chanasit J, Bruchhaus I.. 2016. Phylogeny of haemosporidian blood parasites revealed by a multi-gene approach. Mol Phylogenet Evol. 94(Pt A):221–231. [DOI] [PubMed] [Google Scholar]
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ.. 2014. BEAST 2: a software platform for bayesian evolutionary analysis. PLoS Comput Biol. 104:e1003537.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boundenga L, Makanga B, Ollomo B, Gilabert A, Rougeron V, Mve-Ondo B, Arnathau C, Durand P, Moukodoum ND, Okouga AP, et al. 2016. Haemosporidian parasites of antelopes and other vertebrates from Gabon, Central Africa. PLoS One 112:e0148958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DR, León-Règagnon V, McLennan DA, Zelmer D.. 2006. Ecological fitting as a determinant of the community structure of platyhelminth parasites of anurans. Ecology 87:S76–SS8. [DOI] [PubMed] [Google Scholar]
- Bukauskaitė D, Žiegytė R, Palinauskas V, Iezhova TA, Dimitrov D, Ilgūnas M, Bernotienė R, Markovets MY, Valkiūnas G.. 2015. Biting midges (Culicoides, Diptera) transmit Haemoproteus parasites of owls: evidence from sporogony and molecular phylogeny. Parasite Vectors 8:303.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. 2014. Gregarine site-heterogeneous 18S rDNA trees, revision of gregarine higher classification, and the evolutionary diversification of Sporozoa. Eur J Protistol. 505:472–495.http://dx.doi.org/10.1016/j.ejop.2014.07.002 [DOI] [PubMed] [Google Scholar]
- Claramunt S, Cracraft J.. 2015. A new time tree reveals Earth history’s imprint on the evolution of modern birds. Sci Adv. 111:e1501005.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornejo OE, Escalante AA.. 2006. The origin and age of Plasmodium vivax. Trends Parasitol. 2212:558–563.http://dx.doi.org/10.1016/j.pt.2006.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delson E. 1980. Fossil macaques, phyletic relationships and a scenario of deployment In: Lindburg DG, editor. The macaques: studies in ecology, behavior and evolution. New York: Van Nostrand Reinhold Co; p. 10–29. [Google Scholar]
- dos Reis M, Donoghue PC, Yang Z.. 2016. Bayesian molecular clock dating of species divergences in the genomics era. Nat Rev Genet. 172:71–80. [DOI] [PubMed] [Google Scholar]
- dos Reis M, Yang Z.. 2011. Approximate likelihood calculation on a phylogeny for Bayesian estimation of divergence times. Mol Biol Evol. 287:2161–2172.http://dx.doi.org/10.1093/molbev/msr045 [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Ho SY, Phillips MJ, Rambaut A.. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 45:e88.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante AA, Ayala FJ.. 1995. Evolutionary origin of Plasmodium and other Apicomplexa based on rRNA genes. Proc Natl Acad Sci U S A. 9213:5793–5797.http://dx.doi.org/10.1073/pnas.92.13.5793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante AA, Freeland DE, Collins WE, Lal AA.. 1998. The evolution of primate malaria parasites based on the gene encoding cytochrome b from the linear mitochondrial genome. Proc Natl Acad Sci U S A. 9514:8124–8129.http://dx.doi.org/10.1073/pnas.95.14.8124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feagin JE, Harrell MI, Lee JC, Coe KJ, Sands BH, Cannone JJ, Tami G, Schnare MN, Gutell RR.. 2012. The fragmented mitochondrial ribosomal RNAs of Plasmodium falciparum. PLoS One 76:e38320.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González AD, Lotta IA, García LF, Moncada LI, Matta NE.. 2015. Avian haemosporidians from Neotropical highlands: evidence from morphological and molecular data. Parasitol Int. 644:48–59. [DOI] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O.. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 272:221–224.http://dx.doi.org/10.1093/molbev/msp259 [DOI] [PubMed] [Google Scholar]
- Graur D, Martin W.. 2004. Reading the entrails of chickens: molecular timescales of evolution and the illusion of precision. Trends Genet. 202:80–86.http://dx.doi.org/10.1016/j.tig.2003.12.003 [DOI] [PubMed] [Google Scholar]
- Gray MW, Lang BF, Burger G.. 2004. Mitochondria of protists. Annu Rev Genet. 38:477–524.http://dx.doi.org/10.1146/annurev.genet.37.110801.142526 [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O, Rannala B.. 2003. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 525:696–704.http://dx.doi.org/10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- Habib S, Vaishya S, Gupta K.. 2016. Translation in organelles of apicomplexan parasites. Trends Parasitol. 3212:939–952.http://dx.doi.org/10.1016/j.pt.2016.07.005 [DOI] [PubMed] [Google Scholar]
- Hall N, Karras M, Raine JD, Carlton JM, Kooij TWA, Berriman M, Florens L, Janssen CS, Pain A, Christophides GK, et al. 2005. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science 3075706:82–86. [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Culleton R, Otani H, Horii T, Tanabe K.. 2008. Big bang in the evolution of extant malaria parasites. Mol Biol Evol. 2510:2233–2239. [DOI] [PubMed] [Google Scholar]
- Hellgren O, Waldenström J, Bensch S.. 2004. A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium, and Haemoproteus from avian blood. J Parasitol. 904:797–802. [DOI] [PubMed] [Google Scholar]
- Hikosaka K, Kita K, Tanabe K.. 2013. Diversity of mitochondrial genome structure in the phylum Apicomplexa. Mol Biochem Parasitol. 1881:26–33. [DOI] [PubMed] [Google Scholar]
- Hino A, Hirai M, Tanaka TQ, Watanabe Y, Matsuoka H, Kita K.. 2012. Critical roles of the mitochondrial complex II in oocyst formation of rodent malaria parasite Plasmodium berghei. J Biochem. 1523:259–268. [DOI] [PubMed] [Google Scholar]
- Ho SYW, Duchêne S, Duchêne D.. 2015. Simulating and detecting autocorrelation of molecular evolutionary rates among lineages. Mol Ecol Resour. 154:688–696. [DOI] [PubMed] [Google Scholar]
- Houde PW. 1988. Paleognathous birds from the early Tertiary of the Northern Hemisphere. Publ Nuttall Ornithological Club. 22:1–148. [Google Scholar]
- Huff CG. 1938. Studies on the Evolution of Some Disease-Producing Organisms. Q Rev Biol. 132:196–206.http://dx.doi.org/10.1086/394557 [Google Scholar]
- Ishtiaq F, Guillaumot L, Clegg SM, Phillimore AB, Black RA, Owens IP, Mundy NI, Sheldon BC.. 2008. Avian haematozoan parasites and their associations with mosquitoes across Southwest Pacific Islands. Mol Ecol. 1720:4545–4555. [DOI] [PubMed] [Google Scholar]
- Iturralde-Vinent MA, MacPhee RDE.. 1996. Age and paleogeographical origin of Dominican amber. Science 2735283:1850–1852.http://dx.doi.org/10.1126/science.273.5283.1850 [Google Scholar]
- Jacot D, Waller RF, Soldati-Favre D, MacPherson DA, MacRae JI.. 2016. Apicomplexan energy metabolism: carbon source promiscuity and the quiescence hyperbole. Trends Parasitol. 321:56–70. [DOI] [PubMed] [Google Scholar]
- Jarvis ED, Mirarab S, Aberer AJ, Li B, Houde P, Li C, Ho SY, Faircloth BC, Nabholz B, Howard JT, et al. 2014. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 3466215:1320–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongwutiwes S, Putaporntip C, Iwasaki T, Ferreira MU, Kanbara H, Hughes AL.. 2005. Mitochondrial genome sequences support ancient population expansion in Plasmodium vivax. Mol Biol Evol. 228:1733–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joy DA, Feng X, Mu J, Furuya T, Chotivanich K, Krettli AU, Ho M, Wang A, White NJ, Suh E, et al. 2003. Early origin and recent expansion of Plasmodium falciparum. Science 3005617:318–321. [DOI] [PubMed] [Google Scholar]
- Karadjian G, Hassanin A, Saintpierre B, Gembu Tungaluna GC, Ariey F, Ayala FJ, Landau I, Duval L.. 2016. Highly rearranged mitochondrial genome in Nycteria parasites (Haemosporidia) from bats. Proc Natl Acad Sci U S A. 11335:9834–9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke H, Lewis IA, Morrisey JM, McLean KJ, Ganesan SM, Painter HJ, Mather MW, Jacobs-Lorena M, Llinás M, Vaidya AB.. 2015. Genetic investigation of tricarboxylic acid metabolism during the Plasmodium falciparum life cycle. Cell Rep. 111:164–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski J, Grushko OG, Besansky NJ.. 2006. Analysis of the complete mitochondrial DNA from Anopheles funestus: an improved dipteran mitochondrial genome annotation and a temporal dimension of mosquito evolution. Mol Phylogenet Evol. 392:417–423.http://dx.doi.org/10.1016/j.ympev.2006.01.006 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Peterson D, Tamura K.. 2012. MEGA-CC: computing core of molecular evolutionary genetics analysis program for automated and iterative data analysis. Bioinformatics 2820:2685–2686.http://dx.doi.org/10.1093/bioinformatics/bts507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 337:1870–1874.http://dx.doi.org/10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche J, Li P, Maggia L, Bousquet J.. 1997. Molecular evolution of angiosperm mitochondrial introns and exons. Proc Natl Acad Sci U S A. 9411:5722–5727.http://dx.doi.org/10.1073/pnas.94.11.5722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin II, Valkiūnas G, Iezhova TA, O’Brien SL, Parker PG.. 2012. Novel Haemoproteus species (Haemosporida: Haemoproteidae) from the swallow-tailed gull (Lariidae), with remarks on the host range of hippoboscid-transmitted avian hemoproteids. J Parasitol. 984:847–854. [DOI] [PubMed] [Google Scholar]
- Levin II, Zwiers P, Deem SL, Geest EA, Higashiguchi JM, Iezhova TA, Jiménez-Uzcátegui G, Kim DH, Morton JP, Perlut NG, et al. 2013. Multiple lineages of avian malaria parasites (Plasmodium) in the Galapagos Islands and evidence for arrival via migratory birds. Conserv Biol. 276:1366–1377. [DOI] [PubMed] [Google Scholar]
- Lotta IA, Pacheco MA, Escalante AA, González AD, Mantilla JS, Moncada LI, Adler PH, Matta NE.. 2016. Leucocytozoon diversity and possible vectors in the Neotropical highlands of Colombia. Protist 1672:185–204. [DOI] [PubMed] [Google Scholar]
- Lutz HL, Patterson BD, Kerbis Peterhans JC, Stanley WT, Webala PW, Gnoske TP, Hackett SJ, Stanhope MJ.. 2016. Diverse sampling of East African haemosporidians reveals chiropteran origin of malaria parasites in primates and rodents. Mol Phylogenet Evol. 99:7–15. [DOI] [PubMed] [Google Scholar]
- Martinsen ES, McInerney N, Brightman H, Ferebee K, Walsh T, McShea WJ, Forrester TD, Ware L, Joyner PH, Perkins SL, et al. 2016. Hidden in plain sight: cryptic and endemic malaria parasites in North American white-tailed deer (Odocoileus virginianus). Sci Adv. 22:e1501486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinsen ES, Perkins SL, Schall JJ.. 2008. A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): evolution of life-history traits and host switches. Mol Phylogenet Evol. 471:261–273.http://dx.doi.org/10.1016/j.ympev.2007.11.012 [DOI] [PubMed] [Google Scholar]
- Matta NE, Pacheco MA, Escalante AA, Valkiūnas G, Ayerbe-Quiñones F, Acevedo-Cendales LD.. 2014. Description and molecular characterization of Haemoproteus macrovacuolatus n. sp. (Haemosporida, Haemoproteidae), a morphologically unique blood parasite of black-bellied whistling duck (Dendrocygna autumnalis) from South America. Parasitol Res. 1138:2991–3000. [DOI] [PubMed] [Google Scholar]
- Mu J, Joy DA, Duan J, Huang Y, Carlton J, Walker J, Barnwell J, Beerli P, Charleston MA, Pybus OG, et al. 2005. Host switch leads to emergence of Plasmodium vivax malaria in humans. Mol Biol Evol. 228:1686–1693. [DOI] [PubMed] [Google Scholar]
- Muehlenbein MP, Pacheco MA, Taylor JE, Prall SP, Ambu L, Nathan S, Alsisto S, Ramirez D, Escalante AA.. 2015. Accelerated diversification of nonhuman primate malarias in Southeast Asia: adaptive radiation or geographic speciation? Mol Biol Evol. 322:422–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B, Weaver S, Smith MD, Wertheim JO, Murrell S, Aylward A, Eren K, Pollner T, Martin DP, Smith DM, et al. 2015. Gene-wide identification of episodic selection. Mol Biol Evol. 325:1365–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL.. 2012. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 87:e1002764.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson E, Taubert H, Hellgren O, Huang X, Palinauskas V, Markovets MY, Valkiūnas G, Bensch S.. 2016. Multiple cryptic species of sympatric generalists within the avian blood parasite Haemoproteus majoris. J Evol Biol. 299:1812–1826. [DOI] [PubMed] [Google Scholar]
- Njabo KY, Cornel AJ, Bonneaud C, Toffelmier E, Sehgal RN, Valkiūnas G, Russell AF, Smith TB.. 2011. Nonspecific patterns of vector, host and avian malaria parasite associations in a central African rainforest. Mol Ecol. 205:1049–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn CL, Ezenwa VO, Arnold C, Koenig WD.. 2011. Mutualism or parasitism? Using a phylogenetic approach to characterize the oxpecker-ungulate relationship. Evolution 655:1297–1304.http://dx.doi.org/10.1111/j.1558-5646.2010.01212.x [DOI] [PubMed] [Google Scholar]
- Omori S, Sato Y, Hirakawa S, Isobe T, Yukawa M, Murata K.. 2008. Two extra chromosomal genomes of Leucocytozoon caulleryi; complete nucleotide sequences of the mitochondrial genome and existence of the apicoplast genome. Parasitol Res. 1034:953–957. [DOI] [PubMed] [Google Scholar]
- Outlaw DC, Ricklefs RE.. 2010. Comparative gene evolution in haemosporidian (apicomplexa) parasites of birds and mammals. Mol Biol Evol. 273:537–542.http://dx.doi.org/10.1093/molbev/msp283 [DOI] [PubMed] [Google Scholar]
- Outlaw DC, Ricklefs RE.. 2014. Species limits in avian malaria parasites (Haemosporida): how to move forward in the molecular era. Parasitology 14110:1223–1232.http://dx.doi.org/10.1017/S0031182014000560 [DOI] [PubMed] [Google Scholar]
- Outlaw RK, Counterman B, Outlaw DC.. 2015. Differential patterns of molecular evolution among Haemosporidian parasite groups. Parasitology 1424:612–622.http://dx.doi.org/10.1017/S0031182014001668 [DOI] [PubMed] [Google Scholar]
- Pacheco MA, Battistuzzi FU, Junge RE, Cornejo OE, Williams CV, Landau I, Rabetafika L, Snounou G, Jones-Engel L, Escalante AA.. 2011. Timing the origin of human malarias: the lemur puzzle. BMC Evol Biol. 11:299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco MA, Battistuzzi FU, Lentino M, Aguilar RF, Kumar S, Escalante AA.. 2011. Evolution of modern birds revealed by mitogenomics: timing the radiation and origin of major orders. Mol Biol Evol. 28:1927–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco MA, Cranfield M, Cameron K, Escalante AA.. 2013. Malarial parasite diversity in chimpanzees: the value of comparative approaches to ascertain the evolution of Plasmodium falciparum antigens. Malar J. 12:328.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco MA, Reid MJ, Schillaci MA, Lowenberger CA, Galdikas BM, Jones-Engel L, Escalante AA.. 2012. The origin of malarial parasites in orangutans. PLoS One 74:e34990.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelman P, Johnson WE, Roos C, Seuánez HN, Horvath JE, Moreira MA, Kessing B, Pontius J, Roelke M, Rumpler Y, et al. 2011. A molecular phylogeny of living primates. PLoS Genet. 73:e1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins SL. 2008. Molecular systematics of the three mitochondrial protein-coding genes of malaria parasites: corroborative and new evidence for the origins of human malaria. Mitochondrial DNA 196:471–478. [DOI] [PubMed] [Google Scholar]
- Poinar G., Jr. 2005. Plasmodium dominicana n. sp. (Plasmodiidae: Haemospororida) from Tertiary Dominican amber. Syst Parasitol. 611:47–52.http://dx.doi.org/10.1007/s11230-004-6354-6 [DOI] [PubMed] [Google Scholar]
- Pond SLK, Frost SDW, Muse SV.. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 215:676–679.http://dx.doi.org/10.1093/bioinformatics/bti079 [DOI] [PubMed] [Google Scholar]
- Prum RO, Berv JS, Dornburg A, Field DJ, Townsend JP, Lemmon EM, Lemmon AR.. 2015. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 5267574:569–573. [DOI] [PubMed] [Google Scholar]
- Ramiro RS, Reece SE, Obbard DJ.. 2012. Molecular evolution and phylogenetics of rodent malaria parasites. BMC Evol Biol. 12:219.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rannala B, Yang Z.. 2007. Inferring speciation times under an episodic molecular clock. Syst Biol. 563:453–466.http://dx.doi.org/10.1080/10635150701420643 [DOI] [PubMed] [Google Scholar]
- Rehkopf DH, Gillespie DE, Harrell MI, Feagin JE.. 2000. Transcriptional mapping and RNA processing of the Plasmodium falciparum mitochondrial mRNAs. Mol Biochem Parasitol. 1051:91–103. [DOI] [PubMed] [Google Scholar]
- Ricklefs RE, Outlaw DC.. 2010. A molecular clock for malaria parasites. Science 3295988:226–229.http://dx.doi.org/10.1126/science.1188954 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP.. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 1912:1572–1574.http://dx.doi.org/10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Rutledge GG, Böhme U, Sanders M, Reid AJ, Cotton JA, Maiga-Ascofare O, Djimdé AA, Apinjoh TO, Amenga-Etego L, Manske M, et al. 2017. Plasmodium malariae and P. ovale genomes provide insights into malaria parasite evolution. Nature 5427639:101–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaer J, Perkins SL, Decher J, Leendertz FH, Fahr J, Weber N, Matuschewski K.. 2013. High diversity of West African bat malaria parasites and a tight link with rodent Plasmodium taxa. Proc Natl Acad Sci U S A. 11043:17415–17419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaer J, Reeder DM, Vodzak ME, Olival KJ, Weber N, Mayer F, Matuschewski K, Perkins SL.. 2015. Nycteria parasites of Afrotropical insectivorous bats. Int J Parasitol. 456:375–384. [DOI] [PubMed] [Google Scholar]
- Schlee D. 1990. Das Bernstein-Kabinett. Stuttgarter Beiträge zur Naturkunde (C). 28:1–100. [Google Scholar]
- Seo TK, Kishino H, Thorne JL.. 2004. Estimating absolute rates of synonymous and nonsynonymous nucleotide substitution in order to characterize natural selection and date species divergences. Mol Biol Evol. 217:1201–1213.http://dx.doi.org/10.1093/molbev/msh088 [DOI] [PubMed] [Google Scholar]
- Silva JC, Egan A, Arze C, Spouge JL, Harris DG.. 2015. A new method for estimating species age supports the coexistence of malaria parasites and their Mammalian hosts. Mol Biol Evol. 325:1354–1364.http://dx.doi.org/10.1093/molbev/msv005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Wu Z.. 2014. History of plastid DNA insertions reveals weak deletion and at mutation biases in angiosperm mitochondrial genomes. Genome Biol Evol. 612:3210–3221.http://dx.doi.org/10.1093/gbe/evu253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 309:1312–1313.http://dx.doi.org/10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland CJ, Tanomsing N, Nolder D, Oguike M, Jennison C, Pukrittayakamee S, Dolecek C, Hien TT, do Rosário VE, Arez AP, et al. 2010. Two nonrecombining sympatric forms of the human malaria parasite Plasmodium ovale occur globally. J Infect Dis. 20110:1544–1550. [DOI] [PubMed] [Google Scholar]
- Tamura K, Battistuzzi FU, Billing-Ross P, Murillo O, Filipski A, Kumar S.. 2012. Estimating divergence times in large molecular phylogenies. Proc. Natl Acad Sci U S A. 10947:19333–19338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JE, Pacheco MA, Bacon DJ, Beg MA, Machado RL, Fairhurst RM, Herrera S, Kim JY, Menard D, Póvoa MM, et al. 2013. The evolutionary history of Plasmodium vivax as inferred from mitochondrial genomes: parasite genetic diversity in the Americas. Mol Biol Evol. 30:2050–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford SR., Jr 2009. Hemoparasites of the reptilia. Boca Raton (FL): CRC Press, Taylor & Francis Group. [Google Scholar]
- Templeton TJ, Asada M, Jiratanh M, Ishikawa SA, Tiawsirisup S, Sivakumar T, Namangala B, Takeda M, Mohkaew K, Ngamjituea S, et al. 2016. Ungulate malaria parasites. Sci Rep. 6:23230.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton TJ, Martinsen E, Kaewthamasorn M, Kaneko O.. 2016. The rediscovery of malaria parasites of ungulates. Parasitology 14312:1501–1508. [DOI] [PubMed] [Google Scholar]
- Vaidya AB, Mather MW.. 2009. Mitochondrial evolution and functions in malaria parasites. Annu Rev Microbiol. 63:249–267.http://dx.doi.org/10.1146/annurev.micro.091208.073424 [DOI] [PubMed] [Google Scholar]
- Vaishya S, Kumar V, Gupta A, Siddiqi MI, Habib S.. 2016. Polypeptide release factors and stop codon recognition in the apicoplast and mitochondrion of Plasmodium falciparum. Mol Microbiol. 1006:1080–1095. [DOI] [PubMed] [Google Scholar]
- Valkiūnas G. 2005. Avian malaria parasites and other haemosporidia. Boca Raton (FL: ): CRC Press. [Google Scholar]
- Waldenström J, Bensch S, Hasselquist D, Ostman O.. 2004. A new nested polymerase chain reaction method very efficient in detecting Plasmodium and Haemoproteus infections from avian blood. J Parasitol. 901:191–194. [DOI] [PubMed] [Google Scholar]
- Weber MG, Wagner CE, Best RJ, Harmon LJ, Matthews B.. 2017. Evolution in a community context: on integrating ecological interactions and macroevolution. Trends Ecol Evol. 324:291–304. [DOI] [PubMed] [Google Scholar]
- Wertheim JO, Murrell B, Smith MD, Kosakovsky Pond SL, Scheffler K.. 2015. RELAX: detecting relaxed selection in a phylogenetic framework. Mol Biol Evol. 323:820–832.http://dx.doi.org/10.1093/molbev/msu400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong WSW, Yang Z, Goldman N, Nielsen R.. 2004. Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics 1682:1041–1051.http://dx.doi.org/10.1534/genetics.104.031153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X. 2017. DAMBE6: new tools for microbial genomics, phylogenetics and molecular evolution. J Hered. 1084:431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Xie Z, Salemi M, Chen L, Wang Y.. 2003. An index of substitution saturation and its application. Mol Phylogenet Evol. 261:1–7.http://dx.doi.org/10.1016/S1055-7903(02)00326-3 [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 248:1586–1591.http://dx.doi.org/10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z.. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2212:2472–2479.http://dx.doi.org/10.1093/molbev/msi237 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.