Abstract
Novel genes arising from random DNA sequences (de novo genes) have been suggested to be widespread in the genomes of different organisms. However, our knowledge about the origin and evolution of de novo genes is still limited. To systematically understand the general features of de novo genes, we established a robust pipeline to analyze >20,000 transcript-supported coding sequences (CDSs) from the budding yeast Saccharomyces cerevisiae. Our analysis pipeline combined phylogeny, synteny, and sequence alignment information to identify possible orthologs across 20 Saccharomycetaceae yeasts and discovered 4,340 S. cerevisiae-specific de novo genes and 8,871 S. sensu stricto-specific de novo genes. We further combine information on CDS positions and transcript structures to show that >65% of de novo genes arose from transcript isoforms of ancient genes, especially in the upstream and internal regions of ancient genes. Fourteen identified de novo genes with high transcript levels were chosen to verify their protein expressions. Ten of them, including eight transcript isoform-associated CDSs, showed translation signals and five proteins exhibited specific cytosolic localizations. Our results suggest that de novo genes frequently arise in the S. sensu stricto complex and have the potential to be quickly integrated into ancient cellular network.
Keywords: de novo gene, novel gene, S. sensu stricto yeast, yeast evolution, transcript isoform, synteny analysis, yeast genomics
Introduction
Recent comparative genomic studies have revealed that most eukaryotic genomes contain sets of genes that are not shared by other related species (Betran et al. 2006; Wang et al. 2006; Sunagawa et al. 2009; Toll-Riera et al. 2009; Bornberg-Bauer et al. 2015). These newly evolved genes have been suggested to play important roles when organisms adapt to different habitats (Khalturin et al. 2009; Arendsee et al. 2014). In some cases, they can even become essential for an organism’s development (Chen et al. 2010). New genes arise through various mechanisms, including gene duplication, retrotransposition, exon shuffling, horizontal gene transfer, gene fission/fusion, and de novo origination (Long et al. 2003). Although most mechanisms for new gene creation have been extensively studied, our understanding of de novo formation remains limited.
De novo origination of a protein-coding gene from a random DNA sequence that exhibits no detectable similarity to existing proteins was considered to be rare. But studies in Drosophila have identified dozens of de novo genes that arose from noncoding DNA (Levine et al. 2006; Begun et al. 2007; Chen et al. 2007, 2010; Zhou et al. 2008). Other than Drosophila, de novo genes have also been discovered in other animals, plants, fungi, bacteria, and viruses (Cai et al. 2008; Heinen et al. 2009; Knowles and McLysaght 2009; Xiao et al. 2009; Li et al. 2010, 2016; Wu et al. 2011; Murphy and McLysaght 2012; Xie et al. 2012; Pavesi et al. 2013; Wissler et al. 2013; Fellner et al. 2015; Ruiz-Orera et al. 2015; Aguilera et al. 2017; Jayasena et al. 2017), suggesting that de novo creation is a common mechanism for generating novel genes among different species. Through a systematic exploration, 11.9% of novel genes in the Drosophila melanogaster species complex were found to be of de novo origins (Zhou et al. 2008). Such a high proportion of de novo genes among the Drosophila novel gene list suggest that the number of de novo genes may be highly underestimated. Indeed, by specifically analyzing the transcriptome of D. melanogaster populations, >200 testis-expressed de novo genes were identified (Zhao et al. 2014).
Even though de novo genes have been observed in diverse species, whether a protein generated from a random sequence has the chance to become functional remains largely uncertain. From studies in Drosophila, de novo genes were found to contribute to organismal fitness and development, indicating that some are indeed functional (Chen et al. 2010; Reinhardt et al. 2013). Another interesting example is the human p19ARF gene, which largely overlaps with an ancient gene, p16INK4a, and is considered a de novo gene because p19ARF is only found in Eutherians (Gil and Peters 2006; Rancurel et al. 2009; Neme and Tautz 2013). The p19ARF protein is known to be involved in the ARF–p53–MDM2 network, suggesting that de novo genes can be integrated into an ancient system (Gil and Peters 2006). Although a few examples of de novo genes have been dissected in detail, the general properties of de novo genes in most species are still unclear. Understanding how these de novo genes impact genome evolution remains a daunting challenge.
Recently, bioinformatics-based methods have been used to identify de novo genes in various mammalian species (Knowles and McLysaght 2009; Wu et al. 2011; Murphy and McLysaght 2012; Xie et al. 2012; Neme and Tautz 2013). Two major approaches have been applied to search for such genes, including phylostratigraphy and synteny-based methods. The phylostratigraphy pattern-based method usually utilizes annotated genome information to find genes that are restricted to certain clades (Domazet-Loso et al. 2007; Neme and Tautz 2013). The synteny- or BLAT-based ortholog detection method searches for similar DNA sequences in closely related species to find clade-specific genes (Knowles and McLysaght 2009; Wu et al. 2011). However, it is relatively challenging to dissect the function of de novo genes and their impact on the evolution of genomes in mammals. To understand the evolutionary trajectories of de novo genes, we analyzed de novo genes of the Saccharomyces sensu stricto complex that includes S. cerevisiae, a species with one of the best-annotated genomes.
Despite several studies having reported de novo gene identification in yeast (Li et al. 2008, 2010; Ekman and Elofsson 2010; Carvunis et al. 2012; Tsai et al. 2012), a few issues related to the origin and evolutionary trajectory of de novo genes remained unanswered from these studies. First, none of these yeast studies performed synteny comparisons and so the origins of reported candidates remain uncertain. Moreover, when gene ages were assigned, they mainly depended on BLAST-based comparisons without further annotating existing coding sequences (CDSs) in other genomes. In many cases, the gene ages were not properly determined because the conserved CDSs were not annotated in other species (OhEigeartaigh et al. 2011). Second, coding sequences overlapping with ancient genes were often excluded from analysis (Carvunis et al. 2012), but several studies have suggested that de novo genes could arise as overprinting genes (Sabath et al. 2012; Neme and Tautz 2013; Fellner et al. 2015). In addition, it is known that removing gene-overlapping regions will shorten candidate sequences and reduce the power of BLAST comparisons (Moyers and Zhang 2016). Obviously, approaches that include overprinting candidates are required if we want to understand the general mechanisms of de novo gene origins. Finally, ribosome profiling presents a difficulty in revealing unannotated de novo genes overprinted with ancient genes in the same direction (e.g., the human p19ARF gene is embedded within the p16INK4a locus on the same strand). Thus, the number of de novo genes is probably underestimated.
In the current study, we developed a de novo gene analysis pipeline to address the abovementioned concerns. In addition, we analyzed CDSs from different sources to obtain a more complete picture about the evolution of de novo genes. We identified 13,211 genes that specifically arose in S. cerevisiae or the S. sensu stricto complex with conserved syntenies in Saccharomycetaceae lineages. This large number of de novo genes reveals that the many de novo genes in yeast originated through transcript isoforms associated with ancient genes.
Results
A Total of 4,340 S. cerevisiae- and 8,871 S. sensu stricto-Specific De Novo Genes Originate from Conserved Syntenic Regions in Saccharomycetaceae Yeasts
The S. sensu stricto complex contains at least five closely related yeast species—S. cerevisiae, S. paradoxus, S. mikatae, S. kudriavzevii, and S. uvarum—whose genomes have all been well sequenced (Scannell et al. 2011). Within the complex, S. cerevisiae has the best annotated genome, so it was chosen as our reference species. Saccharomyces uvarum is the most distantly related of the species, and is estimated to have diverged from the common ancestor of S. cerevisiae ∼20 Ma. To get a comprehensive analysis of de novo genes in S. sensu stricto yeasts, we first collected different sources of S. cerevisiae CDSs including all open reading frames (ORFs) annotated in the Saccharomyces Genome Database (SGD, the most commonly used database in yeast), small translated sequences identified by ribosome profiling (Carvunis et al. 2012), and the first CDSs of the transcripts from transcript isoform sequencing (TIF-seq) data if the CDSs were not previously annotated by SGD (Pelechano et al. 2013). In yeast, protein translation usually starts from the first start codon of the transcript so only the first CDS in each transcript was considered as our candidate. In addition, if the transcripts were overlapping with that of existing genes, only the CDSs that were in different frames from existing genes were selected. Therefore, our candidate CDSs did not consist of truncated or expanded forms of existing genes. Transcript units identified by TIF-seq data provided base-pair resolution transcripts with cap and termination structures, ensuring that they were mature transcript products (Pelechano et al. 2013). These CDSs sourced from TIF-seq data represented an unexplored data source, as they had not been analyzed in previous de novo gene studies. CDSs from these different resources were named sgdCDS, smORF, and txCDS, respectively, for the SGD, ribosome profiling, and TIF-seq data. We filtered out CDSs with possible introns, no obvious transcript evidence, or of short length before further analyses (fig. 1A, left panel, see Materials and Methods for details).
Fig. 1.
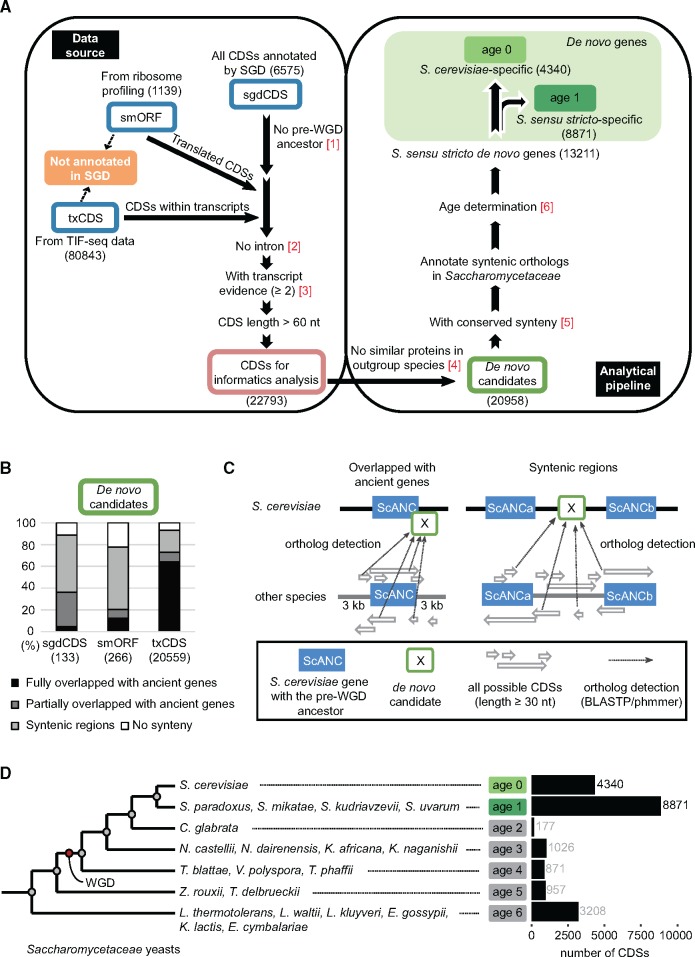
Identification of de novo genes in the Saccharomyces sensu stricto complex. (A) Data sources and analytical pipeline for identification of de novo genes. sgdCDSs, smORFs, and txCDSs (blue boxes) represent CDSs collected from SGD, ribosome profiling, and TIF-seq data, respectively. The pink box represents all collected CDSs after data filtering. The green box indicates genes without protein similarity in non-Saccharomycetaceae species (de novo candidate genes). Filled-green boxes represent S. cerevisiae-specific (age 0) or S. sensu stricto-specific (age 1) de novo genes. Numbers in red represent the analytical steps mentioned in the methods. Numbers in the parentheses are the CDS number in each step or group. (B) The majority of CDSs overlap with ancient genes or localize in syntenic regions. (C) All possible CDSs in the overlapping or syntenic regions are annotated and compared with the de novo candidate genes. For de novo candidates overlapping with ScANCs, we included 3-kb proximal regions of ScANCs to detect possible orthologs. For de novo candidates with conserved synteny, we searched the entire syntenic region between two flanking ancient genes (ScANCa and ScANCb) for possible orthologs. This process was repeated for all 20 Saccharomycetaceae yeasts. (D) Age assignments of de novo candidates. Gray circles indicate phylogenetic nodes of Saccharomycetaceae yeasts determined by a previous study (Marcet-Houben and Gabaldon 2015). The red circle indicates the WGD event. Species belonging to each phylogenetic branch are listed. The numbers of de novo candidate genes with corresponding ages are shown on the right.
Since syntenic information is critical to determine the origins and ages of de novo genes, our analytical pipeline consisted of multiple synteny analysis steps to minimize possible errors in assigning de novo genes (fig. 1A, right panel, see Materials and Methods for details). After excluding CDSs with possible orthologs in Saccharomycetaceae yeasts, the remaining 20,958 CDSs were subjected to synteny comparisons. Previous studies have determined the synteny information from 21 Saccharomycetaceae yeasts and also the S. cerevisiae ancient genes (ScANC), defined as genes derived from a pre-whole genome duplication (WGD) common ancestor (Byrne and Wolfe 2005; Gordon et al. 2009). This information was used to trace the possible origins of our de novo candidates. When the 20,958 candidate CDSs were analyzed, most of them (19,450 out of 20,958 or 93%) were found to be located in syntenic regions or overlapped with ancient genes (fig. 1B). Next, we identified all possible CDSs in the conserved syntenies in other yeast species and then compared these CDSs to our candidate genes to find possible corresponding orthologs (fig. 1C).
After annotating all BLAST hit CDSs in each Saccharomycetaceae yeast genome, the divergence time of the most distantly related species carrying a corresponding homologous CDS was considered as the age of each candidate CDS. We adapted phylogenetic nodes of Saccharomycetaceae yeasts defined by a previous study (Marcet-Houben and Gabaldon 2015), and candidate CDSs with age 0 or age 1 were considered as S. cerevisiae-specific or S. sensu stricto-specific de novo genes, respectively (fig. 1D). In total, 68% of the candidate CDSs (13,211/19,450 = 68%) were confirmed to originate in S. sensu stricto yeasts, including 4,340 age 0 and 8,871 age 1 genes. More than 97% of identified de novo genes were from the txCDS group (supplementary table S1, Supplementary Material online), which had not been considered in previous studies.
Ortholog Detections in Syntenic Regions Provide a Robust Method for Determination of De Novo Genes
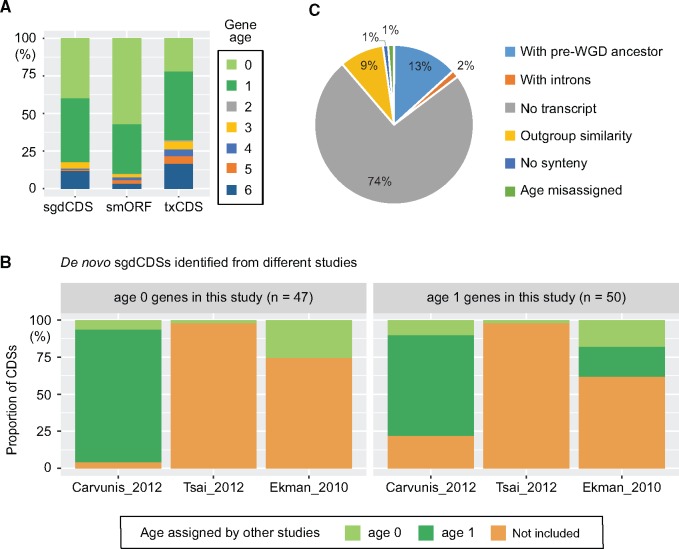
To understand how the identified de novo genes are distributed among the different CDS sources, all candidate genes from the three sources were grouped according to age (fig. 2A). The smORF group had the highest proportion (90%) of age 0 and age 1 genes. To determine whether our pipeline provides a robust tool for identification of de novo genes, we compared our list with three other large-scale de novo yeast gene studies (Ekman and Elofsson 2010; Carvunis et al. 2012; Tsai et al. 2012). Age 0 and age 1 genes from sgdCDSs were first compared since all other large-scale studies have included sgdCDSs in their analyses. Interestingly, only the study by Carvunis et al. (2012) identified a similar cohort of de novo genes, but their age assignments were significantly different from ours, especially for the age 0 group (fig. 2B and supplementary fig. S1A, Supplementary Material online). Many genes classified as S. cerevisiae-specific according to our analysis were considered S. sensu stricto-specific genes by Carvunis et al. (Carvunis et al. 2012). To investigate this discrepancy, we examined each individual case and found that gene ages were often mis-assigned in the previous study; despite similar DNA sequences existing in other S. sensu stricto species, they are either lacking the initiation codon, have early premature stop codons, or contain small deletions that shift most open reading frames (supplementary fig. S1B, Supplementary Material online). Two examples, YCR024C-B and YLR112W, are shown in detail in supplementary figure S2, Supplementary Material online. These results clearly reveal the common problem of age assignments in the study by Carvunis et al. (2012).
Fig. 2.
Comparisons of gene age assignment between different studies. (A) Distribution of gene ages for different CDS sources. (B) Gene age comparisons between different genome-wide studies. In the sgdCDS group, we identified 47 age 0 genes and 50 age 1 genes, many of which were either not identified or assigned different ages in other studies. (C) The factors contributing to the exclusion of 755 de novo sgdCDSs identified in previous genome-wide studies. Also, see supplementary figures S1–S3, Supplementary Material online, for detailed comparisons.
Apart from 97 de novo genes also confirmed by our pipeline, 755 sgdCDSs were considered as de novo genes by previous genome-wide studies but were not included in our list (Ekman and Elofsson 2010; Carvunis et al. 2012; Tsai et al. 2012). We investigated how this inconsistency may have arisen. Our data showed that one major cause of the exclusion of these genes is low transcript levels (fig. 2C). In our pipeline, only candidate genes that had at least two copies of mRNA with complete structures from the TIF-seq data were considered for further analysis. About 74% of the de novo genes identified in earlier studies were excluded by this criterion. However, since the TIF-seq data were obtained from cells growing in glucose- or galactose-containing medium, there is a possibility that those nontranscribed CDSs might be expressed in other more specific conditions. The remaining 24% of the exclusion events were again due to mis-assignment of gene ages in previous studies (fig. 2C and supplementary fig. S1C, Supplementary Material online).
In our analysis, 76% of the previously identified de novo genes that were found to have pre-WGD ancestors were from Ekman et al.’s study. Ekman and colleagues did not use TBLASTN to find similar coding genes but relied on the original genome annotation in their selected species (Ekman and Elofsson 2010). Therefore, false positive orphan genes might be obtained if orthologous genes in these species were not properly annotated. As an example, the HOR7 gene was classified as a de novo gene in previous studies (Ekman and Elofsson 2010; Carvunis et al. 2012), but was found to be conserved in Saccharomycetaceae yeasts in our pipeline (supplementary fig. S3, Supplementary Material online). Similar protein sequences of HOR7 have also been identified through a previous syntenic ortholog detection approach (Byrne and Wolfe 2005). Only very few de novo genes identified by Tsai et al. (2012) were observed in our list (fig. 2B and supplementary table S1, Supplementary Material online). In their study, a conservative approach was used and only few candidate genes were obtained. However, the majority of them were not supported by the transcript data. Detailed comparisons between our data and other genome-wide studies indicate that only when all possible CDSs are fully annotated in the syntenic regions of each genome and used for the comparison (fig. 1C) can gene age be accurately determined.
De Novo Genes Often Occur in Nonconserved Regions
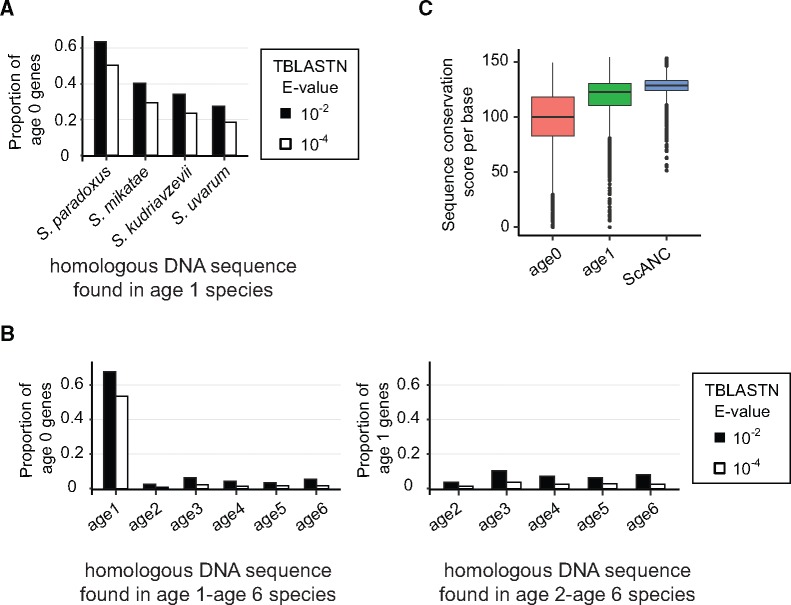
How do de novo genes arise during evolution? We analyzed the corresponding syntenic regions for 21 yeasts to see whether homologous DNA sequences already existed before the emergence of de novo genes. Using TBLASTN to search the syntenic regions, we were able to find homologous sequences of age 0 genes in closely related S. sensu stricto species, supporting the de novo evolution of our candidates. However, the likelihood of finding similar sequences quickly dropped with increasing genetic distance, decreasing from 63% in S. paradoxus to ∼28% in S. uvarum (E value < 10−2, fig. 3A), the most distant species in this group. Beyond the age 1 group of genes, only negligible levels of homologous sequences were found. Similarly, when we searched syntenic regions for homologous sequences of age 1 genes, they were hardly detectable in other age groups (fig. 3B). These results suggest that de novo genes often arose from nonconserved sequences, consistent with a previous observation (Ekman and Elofsson 2010). We further calculated the sequence conservation score (Edgar 2004) and showed that scores of age 0 and age 1 were indeed significantly lower than that of ScANCs (fig. 3C).
Fig. 3.
De novo genes originate from nonconserved sequences. (A) The proportions of age 0 genes with identified homologous DNA sequences drop quickly with respect to species divergence in Saccharomyces sensu stricto species. (B) Only a small proportion of age 0 and age 1 genes have homologous DNA sequences in the syntenic regions of age 1 to age 6 and age 2 to age 6 species, respectively. Two different E value cutoffs (10−2 and 10−4) for TBLASTN were applied, but the results were similar. (C) Sequence conservation is lower in regions containing de novo genes (Mann–Whitney test, P value <2.2e-16, ScANCs vs. age 0, or ScANCs vs. age 1). Conservation scores in different groups of CDSs were calculated using orthologous DNA sequences in S. sensu stricto yeasts.
Most De Novo Genes Are under the Influence of the Structures of Ancient Genes
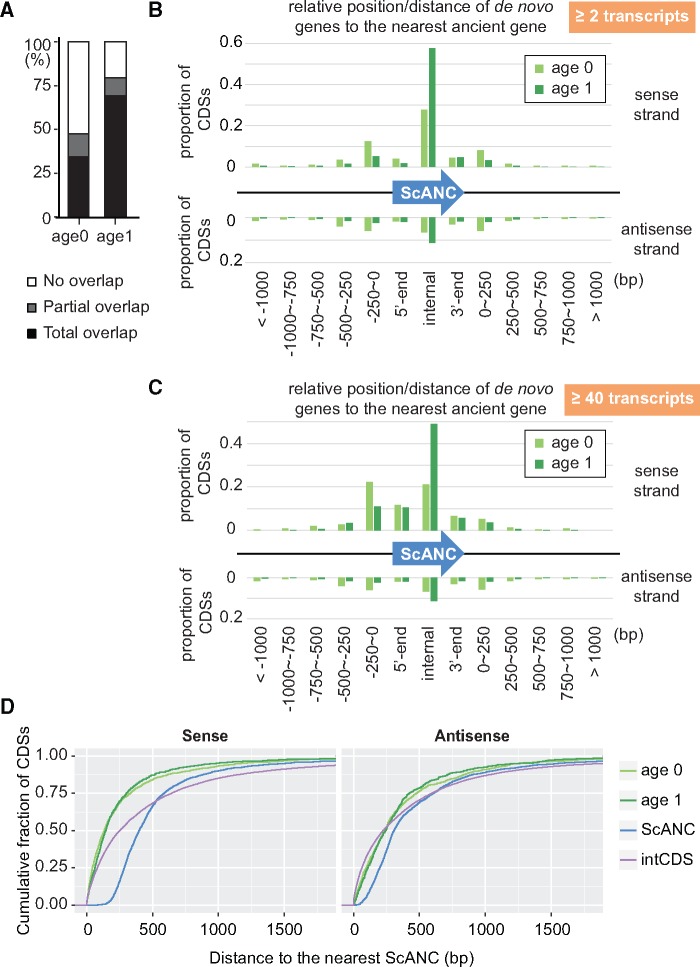
The abundance of de novo genes in the S. sensu stricto complex allows us to analyze their general features. We first investigated how many of our de novo genes overlapped with ScANCs, since some studies indicated that de novo genes tend to overlap with existing ancient genes (Knowles and McLysaght 2009; Carvunis et al. 2012; Murphy and McLysaght 2012; Neme and Tautz 2013). Indeed, a large proportion of our de novo genes overlapped with ScANCs (47% in age 0 genes and 79% in age 1 genes, fig. 4A).
Fig. 4.
Most de novo genes are associated with ancient genes. (A) A large proportion of de novo genes overlap with ancient genes. Ancient genes (ScANC) are defined as the Saccharomyces cerevisiae genes with pre-WGD ancestors. (B, C) Distribution of de novo genes with relative positions or distances to the nearest ScANCs. All CDSs with at least two transcripts were shown in (B) and only CDSs with >40 transcripts were shown in (C). Also, see supplementary figure S4, Supplementary Material online, for the distribution of de novo genes with longer lengths (at least 150 or 300 nt). (D) The association of de novo genes with ancient genes is not simply due to the compact nature of yeast genomes. Intergenic CDSs (intCDS) were used to represent the random origin of CDSs in the yeast genome. Distances to the nearest ScANC of non-ScANC-overlapping CDSs were compared among different group of CDSs. The result showed that both age 0 and age 1 genes were much closer to ScANCs compared with the distances between intCDSs and ScANCs (Mann–Whitney test, P value <2.2e-16) in the sense strand.
When the relative positions and transcription directions of ScANCs and de novo genes were further analyzed, we found that for those de novo genes that did not overlap with ScANCs, most of them were close to ScANCs (i.e., within 250 bp; fig. 4B). In addition, transcription of de novo genes and the overlapping or nearest ScANCs was often in the same direction. We also performed the same analyses using de novo genes expressing at least 40 copies of mRNA in the TIF-seq data or de novo genes with longer CDS lengths (at least 150 or 300 nt). Similar patterns were observed (fig. 4C and supplementary fig. S4, Supplementary Material online), suggesting that the observed features of de novo genes were not biased by different expression levels or gene lengths. It is possible that the association of de novo genes with ScANCs is simply due to the compact nature of yeast genomes. We used the intergenic CDSs (intCDS) to represent the random origin of CDSs in the yeast genome and tested whether they were also closely associated with ScANCs. We identified 34,918 nonoverlapping intCDSs with CDS lengths >60 nt from syntenic regions and measured their distance to the nearest ancient gene. The results showed that age 0 and age 1 genes were much closer to ScANCs compared with intCDSs (Mann–Whitney test, P value < 2.2e-16) in the sense strand (fig. 4D), but not in the antisense strand (P value > 0.5).
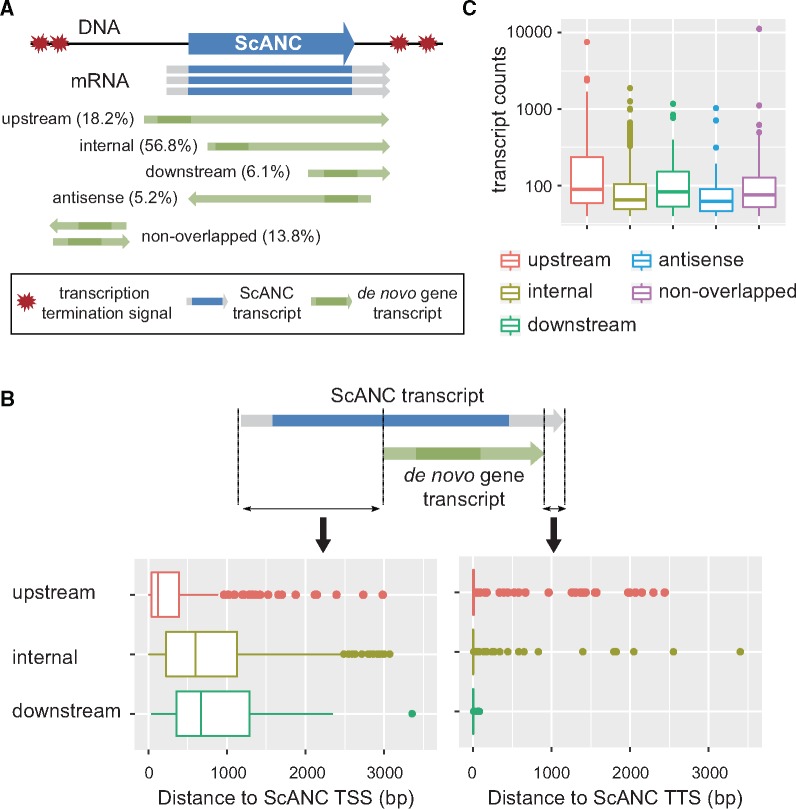
Interestingly, although de novo gene transcripts can start upstream, internally, or downstream of ScANCs, most of them terminate closely to the termination site of their associated ScANCs if they are transcribed in the same direction (supplementary fig. S5, Supplementary Material online). To avoid bias from low-expressing genes which were more susceptible to experimental error, we reanalyzed the de novo genes presenting >40 transcripts in the TIF-seq data. The majority of de novo genes in this group were of the ScANC upstream- and internally initiated types (fig. 5A and supplementary fig. S4C and table S2, Supplementary Material online). Consistent with the general pattern, almost all upstream, internal, and downstream types of de novo genes terminated closely to the termination sites of their associated ScANCs (median distance = 0 bp, fig. 5B). The overlapping nature and cotermination sites suggest that the formation and regulation of de novo genes are likely to be influenced by the structures of ancient genes.
Fig. 5.
Transcript structures of de novo genes are often affected by ScANCs. (A) De novo genes whose transcripts are in the same direction and overlap with that of an associated ScANC were further classified into “upstream,” “internal,” or “downstream” types depending on the relative positions of their initiation and stop codons with respect to that of the associated ScANC. The darker regions of the transcripts correspond to CDSs. Only de novo genes with >40 transcripts in TIF-seq data records are shown here. Also, see supplementary figures S4 and S5, Supplementary Material online, for the distribution of all de novo genes and the example of different types of de novo genes. (B) Proximal de novo genes often terminate at the same sites as associated ScANCs. To avoid bias from low-expressing genes, only genes with >40 transcripts in TIF-seq data were analyzed. TSS, transcription start site. TTS, transcription termination site. (C) The upstream type of de novo genes are expressed at higher levels than other types (Mann–Whitney test, P value <2.2e-16 for all pairwise comparisons).
Since genes with higher expression levels may have a higher likelihood of influencing cell physiology, we also compared the transcript levels of different types of de novo genes. The result of this analysis shows that the upstream type of de novo genes is more likely to be expressed at high levels (fig. 5C).
De Novo Genes Exhibit Different Levels of Structure and Sequence Conservation in S. cerevisiae and S. paradoxus Populations
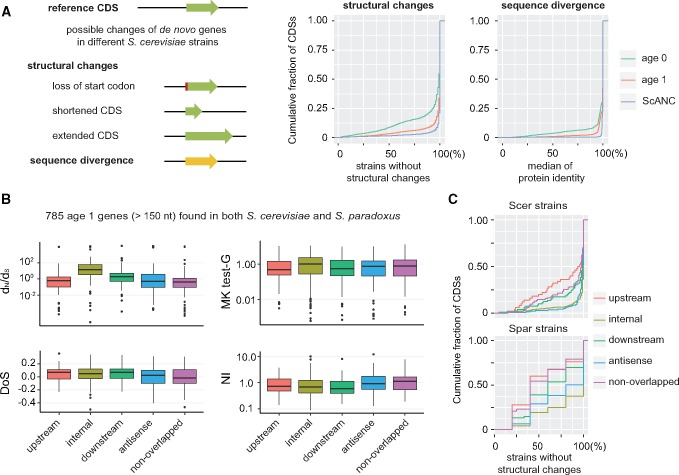
Knowing whether the identified de novo genes are functional remains a challenge. Functional genes usually show signs of positive or negative selection. Population data allow us to assess the conservation level of de novo genes. We analyzed the genome sequences from 93 S. cerevisiae strains collected from various geographic locations and ecological niches (Strope et al. 2015). Since the structures of de novo genes are quite dynamic among populations (Palmieri et al. 2014; Yang et al. 2015), we used multiple criteria to judge their conservation, including the maintenance of start codons, length variation of CDSs, and protein sequence divergence (fig. 6A, see Materials and Methods for details).
Fig. 6.
Population data reveal different protein conservation levels between age 1 genes and age 0 genes. (A) The changes in de novo genes that are commonly observed among 93 different Saccharomyces cerevisiae strains. The structural changes included loss of start codons and CDS-length variation. Only length changes >15 bp were considered true variants. Sequence divergence was determined by protein sequence identity through pairwise alignments. Age 1 genes are more conserved than age 0 genes in terms of both structure and sequence. Also, see supplementary figure S6, Supplementary Material online, for the comparison of de novo genes and intergenic CDSs. (B) Different tests are applied for detecting possible selection forces in different types of age 1 CDSs across species and population. The internal type of de novo genes were significantly different from all other types in dN/dS (Mann–Whitney test, P value ≤3.86e-15). Age 1 genes carrying S. paradoxus orthologs and >150 bp were selected for the analysis. (C) The internal type of age 1 genes were the least variable in structural changes. Different types of age 1 genes were analyzed for CDS structure conservation in S. cerevisiae and S. paradoxus populations.
Despite that both age 0 and age 1 genes have lower scores than ancient genes (fig. 6A), age 0 genes are significantly different from age 1 genes for all three criteria (Mann–Whitney test, maintenance of the start codon, P < 2.2 × 10−16; CDS length variation, P < 2.2 × 10−16; protein sequence identity, P < 2.2 × 10−16) (fig. 6A). The low conservation of age 0 genes in S. cerevisiae populations suggest that age 0 genes are still at an early stage of gene formation. For those that have not evolved functions contributing to cell fitness, they are expected to accumulate random mutations in the populations. Indeed, similar levels of conservation were observed in age 0 genes and intergenic CDSs which are most likely to be nonfunctional (supplementary fig. S6, Supplementary Material online).
Using the population data, we could further dissect whether different types of age 1 genes were subjected to different selection forces. We performed several tests (NI: Neutrality Index test, DoS: Direction of Selection test, MK: McDonald–Kreitman test, and dN/dS) for signs of selection using 785 age 1 genes that are >150 nt and have orthologs shared between 94 S. cerevisiae and 5 S. paradoxus populations (see Materials and Methods for detail). Our data showed that the nonoverlapped type of de novo genes were more likely to be under purifying selection (median of NI = 1.098, median of DoS = −0.017, fig. 6B). In contrast, the internal type of genes were significantly different from other types in dN/dS (Mann–Whitney test, P value ≤ 3.86e-15) and the median of dN/dS is 3.43, suggesting that they were likely under positive selection. Since the CDS structure of the internal type was the least variable (fig. 6C), it suggested that high dN/dS values are not simply due to structural changes. For other types of genes, they were not strongly deviated from neutrality and probably under weak or neutral selection in general.
Finally, we used stringent cutoffs in dN/dS (< 0.5), DoS (< 0), and structural conservation (> 95% most-conserved ScANCs) to collect a group of de novo genes that were more likely to be functional. Fifty-four candidate genes passed these criteria and represented good candidates for further functional assays (supplementary table S3, Supplementary Material online).
Many De Novo Genes with High Transcript Levels Exhibit Evidence of Protein Translation
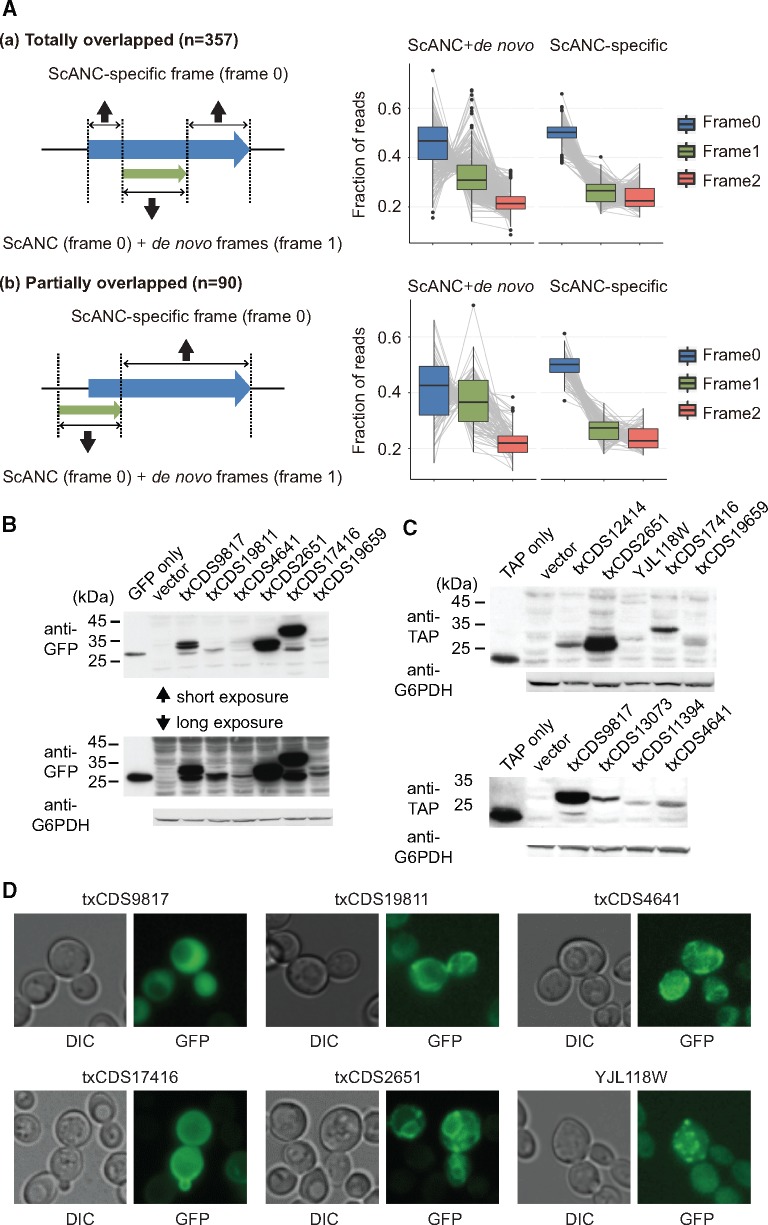
Our analysis indicated that the majority of de novo genes arose from regions overlapping with ancient genes. However, protein evidence for overprinted genes is very limited (supplementary table S1, Supplementary Material online). We searched for evidence of protein translation in the overprinted genes using ribosome profiling data from previous studies (Ingolia et al. 2009; Brar et al. 2012). Since overprinting de novo genes in our list were always in different frames from the overlapped ScANCs, we examined whether ribosome signals were enriched in two different frames in the overprinting regions (examples of totally or partially overlapped genes shown in supplementary fig. S7, Supplementary Material online, and see Materials and Methods for details). Our analysis showed that translation signals were significantly increased in the alternative frame in de novo CDS-containing regions (fig. 7A), suggesting that many overprinting de novo genes might be translated.
Fig. 7.
Many highly expressed de novo genes are translated and exhibit specific protein localization patterns. (A) Ribosome profiling of overprinting CDSs indicates increased translation signals from the alternative frame in de novo CDS-containing regions. The frame with the highest ribosome signals in ScANC-specific region is defined as frame 0. The other frame with enhanced signals in the overprinting region is assigned as frame 1. The remaining frame is frame 2. Frame-specific enrichment was analyzed in CDSs that are (a) totally overlapped (including the internal type of de novo genes) or (b) partially overlapped (including upstream and downstream types) with ScANCs. The results showed that ribosome signals in frame 1 were significant enriched in de novo CDS-containing regions (Mann–Whitney test, ScANC+de novo vs. ScANC-specific, P value <2.2e-16 and =6.8e-16 for totally overlapped and partially overlapped genes, respectively). No enrichment was observed in frame 2 (Mann–Whitney test, ScANC+de novo vs. ScANC-specific, P value=1 and =0.98 for totally overlapped and partially overlapped genes, respectively). In the right panel, the ribosome profile of three different frames in each CDS was represented by a gray line. Also, see supplementary figure S7, Supplementary Material online, for examples of two overprinting genes. (B, C) Detection of full-length proteins of de novo genes by Western blot. To examine whether full-length proteins were translated from de novo genes, we selected different types of candidate genes (table 1) that were expressed for >40 transcripts in the TIF-seq data to perform Western-blot analysis. These candidate genes were fused with GFP (B) or TAP (C) tags and were under the control of their own promoters. Among 14 candidates, 10 exhibited detectable fusion protein signals. Glucose-6-phosphate dehydrogenase (G6PDH) served as a loading control. (D) Some de novo proteins showed special localization patterns. The GFP-fusion protein of txCDS9817 represented a general cytosolic localization pattern. Five other GFP-fusion proteins were observed to localize in mitochondria, cell periphery, ER, or cytosolic punctates. Also, see supplementary figure S8, Supplementary Material online, for the protein features in de novo genes.
To further confirm the translation of de novo genes, we experimentally examined the protein signals of 14 de novo genes that presented at least 40 copies of mRNA in the TIF-seq data set, representing each individual category of transcript structures (table 1). More de novo genes of the internal type (seven genes) were chosen since this type of gene is rarely reported. GFP- or TAP-fusion proteins were constructed and examined using Western blots. Among the chosen genes, 71% of them (10/14), including five internal type de novo genes, exhibited detectable signals in normal growth conditions (table 1; fig. 7B and C).
Table 1.
Summary of De Novo Genes Examined by GFP Localization or Western.
| Type | CDS Name | Gene Age | Gene Length (nt) | Predicted Size of Fusion Protein (kD) | Western | Protein Localization | Protein Feature | Population Conservation |
|---|---|---|---|---|---|---|---|---|
| Internal | txCDS9817 | 1 | 108 | G: 31.04/T: 25.37 | G/T | Cytoplasm | — | Y |
| txCDS13073 | 1 | 135 | G: 31.67/T: 26.00 | T | — | — | Y | |
| txCDS19811 | 1 | 69 | G: 29.43/T: 23.77 | G | Mitochondria | — | Y | |
| txCDS4641 | 1 | 99 | G: 30.67/T: 25.01 | G | Mitochondria | — | Y | |
| txCDS11394 | 1 | 81 | G: 29.93/T: 24.26 | T | — | — | Y | |
| txCDS18399 | 0 | 108 | G: 30.64/T: 24.98 | — | — | — | Y | |
| txCDS15470 | 1 | 138 | G: 31.77/T: 26.10 | — | — | TM, SP | N | |
| Upstream | txCDS17416 | 1 | 225 | G: 35.27/T: 29.61 | G/T | Cell periphery | SP | Y |
| Downstream | txCDS19659 | 1 | 111 | G: 30.83/T: 25.16 | G/T | — | SP | N |
| Antisense | txCDS2651 | 1 | 108 | G: 30.35/T: 24.68 | G/T | ER | TM | N |
| txCDS12414 | 1 | 177 | G: 34.08/T: 28.42 | T | — | — | N | |
| Nonoverlapped | txCDS2240 | 1 | 192 | G: 33.89/T: 28.22 | — | — | TM | Y |
| YLR112W | 0 | 420 | G: 42.63/T: 36.97 | — | — | — | Y | |
| YJL118W | 1 | 660 | G: 52.31/T: 46.65 | T | Punctate composite | — | Y |
Note.—Rows in gray: genes with no protein evidence in this study. Predicted size and Western: G represents GFP size/signal and T represents TAP size/signal. Protein features: TM represents transmembrane domain and SP represents signal peptide. Population conservation: defined by figure 6A.
Potential Integration of De Novo Genes into Current Functional Networks in S. cerevisiae
In addition to translation evidence, we searched for known domain structures in de novo genes. Other than the transmembrane domain that was reported in a previous study (Carvunis et al. 2012), mitochondria-targeting peptides and signal peptides were detected in our de novo proteins (supplementary fig. S8, Supplementary Material online). Interestingly, transmembrane domains and signal peptides were found in proportions about half to those found for ancient genes, suggesting that newly formed proteins can easily generate these two features.
The de novo genes validated in our Western-blot experiments were further examined for their localization patterns. Interestingly, half of these de novo proteins (5/10) exhibited very distinct localization patterns (fig. 7D). Proteins encoded by txCDS19811 and txCDS4641 were both localized to mitochondria. Proteins encoded by txCDS17416 and txCDS2651 were enriched in the cell periphery and the endoplasmic reticulum (ER), respectively. The protein encoded by YJL118W formed punctate structures. These results suggest that de novo proteins are able to interact with the ancient transport system and are stably maintained in different cellular compartments.
Discussion
Previously, only two de novo genes had been experimentally characterized in S. cerevisiae (Cai et al. 2008; Li et al. 2010). In a few recent studies, bioinformatics methods were used to identify de novo genes that have arisen in S. cerevisiae or S. sensu stricto species (Ekman and Elofsson 2010; Carvunis et al. 2012; Tsai et al. 2012). These studies have substantially broadened our understanding of de novo genes in yeast. However, in these studies, synteny information was not included in their analyses so the ages of the de novo genes could not be precisely determined and, as a result, many de novo genes were misidentified or mis-classified. In addition, a large group of CDSs from TIF-seq had not been previously analyzed (Pelechano et al. 2013).
Using a refined analytical pipeline and multiple CDS sources, we have identified 4,340 S. cerevisiae-specific (age 0) genes and 8,871 S. sensu stricto-specific (age 1) genes in S. cerevisiae. Most of these genes were not identified in previous studies (supplementary table S1, Supplementary Material online). Since the transcript structures and profiles from the TIF-seq data were integrated into our analyses, all our identified de novo genes are associated with full-length experimentally confirmed transcripts. In addition, we compared our de novo gene list with those from previous genome-wide studies and revealed that gene age mis-assignment and lack of transcript evidence are two major reasons for inconsistencies between different studies.
The large number of de novo genes in S. cerevisiae allowed us to investigate their general features and evolutionary trajectories. One striking observation is that many de novo genes arose from transcript isoforms of ancient genes (figs. 4B and C and5A). In yeast, over 26 transcript isoforms per protein-coding gene were detected in half of the yeast genome. Moreover, the levels of different transcript isoforms could vary in response to growth conditions (Pelechano et al. 2013). It has been speculated that the abundance of transcript isoforms may increase gene diversity and accelerate genome evolution. Our results provide further evidence that transcript isoforms are one major origin of de novo genes. Interestingly, several recent studies using ribosome profiling also identified various unannotated ORFs associated with ancient genes in mouse and human cells, suggesting that novel proteins can be generated through similar patterns in both unicellular and multicellular organisms (Fields et al. 2015; Ji et al. 2015; Ingolia 2016).
Despite that de novo genes could be found in various types of transcript isoforms, our analysis showed that de novo genes upstream of ancient genes were more likely to be expressed at high levels (fig. 5C). Studies in different organisms have revealed that many small open reading frames (ORFs) exist upstream of annotated genes and that these ORFs are often translated (Ingolia et al. 2009; Brar et al. 2012; Fields et al. 2015; Ji et al. 2015; Ingolia 2016). Furthermore, some upstream ORFs have been shown to regulate the protein translation or mRNA stability of downstream genes under specific physiological conditions (Hood et al. 2009). It is possible that many of the upstream type of de novo genes have evolved similar functions and, therefore, increased expression levels have been selected for.
Currently, the cellular functions of identified de novo genes are largely uncharacterized. However, a few lines of indirect evidence suggest that some of them have evolved functions. First, our population data showed that age 1 genes exhibit a different level of conservation from age 0 genes. In addition, some of them are strongly deviated from neutrality and likely under positive or negative selection. Second, many de novo genes contain signal peptides, mitochondria-targeting peptides, or transmembrane domains that may provide them with the basic structure to evolve further functions. Third, our experimental data validate that some de novo gene products can indeed be transported into mitochondria, localize to the ER or cell periphery, or form punctate structures. These results confirm that de novo genes have the ability to be integrated into existing transport systems or functional networks.
Conclusions
Only a few of the de novo genes presented here have been previously studied. Since many of these de novo genes probably have not yet evolved functions, it will be tedious to characterize all of them. However, our analysis provides the age, transcript abundance, and conservation levels of age 1 and age 0 genes, helping us to judge which de novo genes may have become functional. By deleting these candidate genes in the original species or expressing them in a closely related species in which the genes are absent, we can better understand how these genes influence or are integrated into existing functional networks.
Materials and Methods
Collecting CDSs for Informatics Analysis
Three different sources of CDSs were employed for our de novo gene analyses, including sgdCDSs (CDSs annotated by SGD), smORFs (translated CDSs identified by ribosome profiling), and txCDSs (first unannotated CDS within transcripts from TIF-seq data) (Carvunis et al. 2012; Pelechano et al. 2013). To ensure that each CDS represented as a unique record to simplify our analysis pipeline, each CDS was classified to only one category: whenever a CDS was already annotated by SGD, it was defined as sgdCDS. If a CDS was found in both smORFs and txCDSs, it was defined as smORF. txCDSs only contained CDSs not found in other two groups (supplementary table S1, Supplementary Material online). All collected CDSs were processed through several filtering steps before synteny and age analyses, and the numbers of candidate CDSs in each step could be found in supplementary figure S9, Supplementary Material online. For sgdCDSs, only genes lacking corresponding orthologs in pre-WGD ancestors were included (step 1 in fig. 1A) (Gordon et al. 2009). CDSs overlapping with intron-containing genes were removed due to insufficient information on intron structures from the TIF-seq data (step 2 in fig. 1A). The majority of yeast genes lack introns, so this removal step should not radically influence our later analyses. CDSs with no solid transcript evidence from TIF-seq (< 2 transcript counts) or with lengths shorter than 60 nucleotides were excluded (step 3 in fig. 1A). All remaining CDSs were subjected to further informatics analysis.
Searching for Possible Homologs in Non-Saccharomycetaceae Species
To make sure that the de novo genes were not derived from distant species, we searched for all possible orthologs in ten distant outgroup yeast species and the NCBI nr protein database using BLASTP/TBLASN with a relaxed E value threshold (10−2), which was also used in a previous study (Carvunis et al. 2012) (step 4 in fig. 1A). The ten distant outgroup yeasts were: Candida albicans, Cryptococcus neoformans, Debaryomyces hansenii, Fusarium graminearum, Malassezia globose, Neurospora crassa, Ogataea parapolymorpha, Pichia sorbitophila, Schizosaccharomyces pombe, and Yarrowia lipolytica.
Annotating Syntenic Orthologs in Saccharomycetaceae Yeasts
Genome sequence and ortholog assignment data of 21 Saccharomycetaceae species were obtained from the YGOB data set (Byrne and Wolfe 2005). Synteny information was obtained from a previous study (step 5 in fig. 1A) (Gordon et al. 2009). For candidate genes overlapping with ScANCs, 3 kb-proximal regions of ScANCs were considered as syntenic sequences in each Saccharomycetaceae species (fig. 1C).
To search for possible syntenic orthologs, we reannotated all possible CDSs in syntenic regions across 20 Saccharomycetaceae yeasts. In each species, all possible CDSs (from start to stop codons) >30 nucleotides in syntenic regions were first identified by GetORF (a program in the EMBOSS suite) (Rice et al. 2000). All syntenic CDS sequences were then compared with the corresponding S. cerevisiae protein by BLASTP or phmmer (a hidden Markov model-based method in HMMER3) (Eddy 2011) (fig. 1C). An E value cutoff (10−4) generally used for ortholog determination was applied in our analysis (Knowles and McLysaght 2009; Guerzoni and McLysaght 2016). Once protein homology passed the E value threshold, the protein lengths of candidates were further compared with the lengths of corresponding S. cerevisiae proteins. The length threshold is critical for assigning the gene age. In a study of human de novo genes, at least 80% of the reference protein size was used as a filtering criterion for true orthologs in nonhuman primates (Wu et al. 2011). Using the yeast ancient genes as a reference set, we found that such a length threshold allowed us to recover >90% of ScANCs in all species, suggesting that it is a reasonable threshold. Therefore, we applied this ortholog length criterion in our ortholog detection pipeline. However, if the BLASTP/phmmer-positive candidates encoded proteins longer than 200 amino acids, the length filter was not applied.
Age Determination and DNA Sequence Analysis
We assigned gene age by determining the most distantly related group of species carrying corresponding orthologs (step 6 in fig. 1A), which is similar to the approach used by a previous study (Carvunis et al. 2012). However, since we used the ortholog information instead of BLAST information, gene ages were often reassigned compared with earlier studies (fig. 2). Gene ages were described according to relative divergence of S. cerevisiae to other Saccharomycetaceae yeasts. Nodes of phylogenetic trees in figure 1D were adapted from a previous study (Marcet-Houben and Gabaldon 2015). Genes only found in S. cerevisiae itself were assigned as age 0.
To understand the origin and evolution of de novo genes, TBLASTN was applied to search for homologous DNA sequences in syntenic regions of closely related species with two different E value cutoffs (10−2 and 10−4). For age 0 genes, species of age 1 to age 6 were considered as outgroup species. For age 1 genes, species from age 2 to age 6 were considered as outgroup species. For conservations of DNA sequences in S. sensu stricto yeasts (fig. 3C), synteny DNAs from five species were aligned by MUSCLE (e.g., fig. S2) and regions covering S. cerevisiae CDS were assayed for the DNA conservation (Edgar 2004). Conservation scores for each base were reported by MUSCLE program by using the “-scorefile” parameter. Gene-specific score was represented by the average of scores in S. cerevisiae CDS region.
Classification of Different Types of De Novo Genes
Transcript profiles from S. cerevisiae cells growing in YPD (glucose-containing) and YPGal (galactose-containing) conditions were downloaded from the TIF-seq data (Pelechano et al. 2013). Transcript data were displayed by Integrative Genomics Viewer (Robinson et al. 2011).
Identified de novo genes were first divided into distal and proximal groups, depending on whether most of their transcripts overlapped with that of the associated ScANC. In the proximal group, genes were classified as “antisense” type if the transcripts were from the opposite strand of the associated ScANC. For the remaining proximal genes, they were classified into “upstream,” “internal,” or “downstream” types depending on the relative positions of their initiation and stop codons with respect to the nearest ScANC (see examples in supplementary fig. S5, Supplementary Material online). The relative distance of de novo genes to the nearest ancient gene was the shortest distance between the boundaries (start or stop codons) of de novo genes and nearby ancient genes.
Conservation of De Novo Candidate Genes in 93 S. cerevisiae Strains
Genomic sequences of 93 S. cerevisiae strains were downloaded from a previous study (Strope et al. 2015). Coding regions plus 1 kb of upstream and downstream sequences of de novo and ancient genes were used to find similar regions in these 93 S. cerevisiae strains by BLASTN. BLASTN-positive sequences (E value ≤ 10−10) were aligned to the reference strain by MUSCLE with the default parameters. In addition to protein sequence divergence, two other types of variation were often observed, that is, loss of start codons and CDS-length variation. We decided to examine these three types of changes in individual strains. First, by aligning to the reference genome, corresponding positions of the start codon were determined and confirmed. If corresponding start codons in query strains could be found, complete CDS sequences were extracted and further analyzed for CDS length fluctuations and sequence divergence. For CDS-length fluctuations, CDS lengths that changed by >15 nucleotides were considered true variants. Sequence divergence was determined by protein sequence identity through pairwise alignments.
The rate of nonsynonymous to synonymous substitutions (dN/dS) was used in a previous study to predict purifying selection of de novo genes among eight S. cerevisiae strains (Carvunis et al. 2012). We applied similar calculations for our candidate genes in the 93 S. cerevisiae strains. For testing dN/dS ratios, protein sequences aligned by MUSCLE were further analyzed by codeml module from PAML (Yang 2007). However, our dN and dS values of de novo and ancient genes exhibited a strange pattern (supplementary fig. S10A, Supplementary Material online). To understand why a large proportion of de novo genes have lower dN and dS values than ancient genes, we examined the distributions of dN and dS values and found that ∼75% of the dN or dS values of de novo genes were equal to zero. The zero mutation rate was most likely due to the short lengths of de novo genes (supplementary fig. S10C and D, Supplementary Material online). Similarly, when compared with ancient genes, more strains encoded identical protein sequences of de novo genes (supplementary fig. S10B, Supplementary Material online). These results suggest that without setting a gene length threshold, the dN/dS ratio may not be an ideal indicator for detecting purifying selection in S. cerevisiae populations. Consistent with our observation, a recent study also indicated that statistical tests of natural selection might be powerless for analyses of de novo genes (Moyers and Zhang 2016). In our later analysis, only CDSs longer than 150 nt were used.
Molecular Evolution of Age 1 Genes in S. cerevisiae and S. paradoxus
In total, 785 age 1 genes with S. paradoxus orthologs and long CDS (> 150 nt) were chosen for the analysis. Genome sequences of five resequenced S. paradoxus strains were downloaded from the previous study (Yue et al. 2017). CDS structure and sequence conservation of S. paradoxus orthologs were determined by the same procedure mentioned in the previous method section. Coding sequences from all S. cerevisiae and S. paradoxus strains were aligned by MUSCLE with the default parameters. Macdonald–Kreitman tests for examining population evolution were performed following the setting used in previous studies (McDonald and Kreitman 1991; Egea et al. 2008). G- and P values were obtained by an R package, RVAideMemoire (https://cran.r-project.org/web/packages/RVAideMemoire/index.html). Selection directions of chosen age 1 candidates were estimated by the Neutrality Index (NI) and the Direction of Selection (DoS) indicated in previous studies (Scannell et al. 2011).
Protein Translation of De Novo Genes
Protein translation data were obtained from several previous studies in which various methods, including Western-blot detection, GFP fusion protein detection, peptide identification, and ribosome profiling (Ghaemmaghami et al. 2003; Huh et al. 2003; Brar et al. 2012; Carvunis et al. 2012). We also collected peptide evidence from one summary study (Cai et al. 2008), BioGRID (Stark et al. 2006), Global Proteome Machine Database (gpmDB), PRoteomics IDEntifications (PRIDE) database, and PeptideAtlas database. Data from BioGRID included “Affinity Capture-MS,” “Co-purification,” and “Affinity Capture-Western” data. For S. cerevisiae peptides identified from gpmDB, PRIDE, and PeptideAtlas, we searched matched and nonidentical peptides from other coding genes to support protein translation.
Ribosome Profiling of Overprinting Candidates
For ribosome profiling analysis, we downloaded sequencing read data from previous studies and used the same parameters for our analysis procedures (Ingolia et al. 2009; Brar et al. 2012). The first 25 nucleotides of each profiling read were collected by Trimmomatic (Bolger et al. 2014) and further mapped to the genome of S. cerevisiae SK1 strain by Bowtie2 (Langmead and Salzberg 2012). Translation signals for each CDS were collected from 16 nucleotides before the start codon to 14 nucleotides before the last base of the CDS by SAMtools (Li et al. 2009). For overprinting genes, we first defined ScANC-specific and overprinting regions (supplementary fig. S7, Supplementary Material online). In ScANC-specific region, all profiling reads were divided into three different frames and the frame with the highest signal was determined as frame 0. Next, we considered signals enriched in the frame that encode the de novo gene in the overprinting region as translation signals of de novo genes and defined this frame as frame 1. The remained frame was defined as frame 2.
Expression of GFP- or TAP-Fused De Novo Candidate Genes
We selected 14 de novo genes that were expressed for >40 transcripts in the TIF-seq data (table 1). These genes were fused with GFP or TAP tags in C-terminal regions. In the TIF-seq data, 80% of ScANCs have >40 transcripts and the median transcript number of ScANCs is 244. The expression levels of de novo genes are relatively low compared with ScANCs. In order to increase the chance of detection, we decided to put the fusion genes with the endogenous promoters in a 2-micron plasmid (pRS426). The plasmids were transformed into a S. cerevisiae W303 strain (MATa/α ura3-1 his3-11, 15 leu2-3, 112 trp1-1 can1-100) and protein expression was examined. Protein localizations of de novo candidate genes were revealed by GFP localization. Western signals of de novo candidate genes were detected by anti-TAP or anti-GFP antibodies.
Detecting Protein Features of De Novo Proteins
Three tools were used to detect different features of de novo candidates: TMHMM2.0, SignalP4.1, and TargetP1.1 (Sonnhammer et al. 1998; Emanuelsson et al. 2007; Petersen et al. 2011). TMHMM2.0 was used to detect transmembrane regions of proteins (TM). TargetP1.1 was used to detect mitochondria-targeting peptides (mTP), with candidate genes exhibiting >0.9 specificity being considered as positive. SignalP4.1 was used to detect the presence and location of signal peptide cleavage sites.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank members of the Leu and Lin labs for helpful discussions and comments on the manuscript. We also thank Hsin-Chou Yang for statistical analysis consultation and John O’Brien for manuscript editing. J.Y.L. was supported by Academia Sinica of Taiwan (grant no. 2316-1050-300) and the Taiwan Ministry of Science and Technology (MOST105-2321-B-001-030). W.C.L. was supported by the Taiwan Ministry of Science and Technology (MOST104-2311-B-001-010).
Author Contributions
J.Y.L. conceived the study. T.C.L., W.C.L., and J.Y.L. designed analyses and interpreted results. T.C.L. performed the experiments. T.C.L. and W.C.L. performed computational analyses. T.C.L., W.C.L., and J.Y.L. wrote the paper. All authors read and approved the final manuscript.
References
- Aguilera F, McDougall C, Degnan BM.. 2017. Co-option and de novo gene evolution underlie Molluscan shell diversity. Mol Biol Evol. 34:779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendsee ZW, Li L, Wurtele ES.. 2014. Coming of age: orphan genes in plants. Trends Plant Sci. 19:698–708. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Lindfors HA, Kern AD, Jones CD.. 2007. Evidence for de novo evolution of testis-expressed genes in the Drosophila yakuba/Drosophila erecta clade. Genetics 176:1131–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betran E, Bai Y, Motiwale M.. 2006. Fast protein evolution and germ line expression of a Drosophila parental gene and its young retroposed paralog. Mol Biol Evol. 23:2191–2202. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornberg-Bauer E, Schmitz J, Heberlein M.. 2015. Emergence of de novo proteins from ‘dark genomic matter’ by ‘grow slow and moult’. Biochem Soc Trans. 43:867–873. [DOI] [PubMed] [Google Scholar]
- Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, Weissman JS.. 2012. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335:552–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne KP, Wolfe KH.. 2005. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 15:1456–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Zhao R, Jiang H, Wang W.. 2008. De novo origination of a new protein-coding gene in Saccharomyces cerevisiae. Genetics 179:487–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvunis AR, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N, Charloteaux B, Hidalgo CA, Barbette J, Santhanam B, et al. 2012. Proto-genes and de novo gene birth. Nature 487:370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhang YE, Long M.. 2010. New genes in Drosophila quickly become essential. Science 330:1682–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ST, Cheng HC, Barbash DA, Yang HP.. 2007. Evolution of hydra, a recently evolved testis-expressed gene with nine alternative first exons in Drosophila melanogaster. PLoS Genet. 3:e107.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Loso T, Brajkovic J, Tautz D.. 2007. A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet. 23:533–539. [DOI] [PubMed] [Google Scholar]
- Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol. 7:e1002195.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea R, Casillas S, Barbadilla A.. 2008. Standard and generalized McDonald-Kreitman test: a website to detect selection by comparing different classes of DNA sites. Nucleic Acids Res. 36:W157–W162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekman D, Elofsson A.. 2010. Identifying and quantifying orphan protein sequences in fungi. J Mol Biol. 396:396–405. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H.. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2:953–971. [DOI] [PubMed] [Google Scholar]
- Fellner L, Simon S, Scherling C, Witting M, Schober S, Polte C, Schmitt-Kopplin P, Keim DA, Scherer S, Neuhaus K.. 2015. Evidence for the recent origin of a bacterial protein-coding, overlapping orphan gene by evolutionary overprinting. BMC Evol Biol. 15:283.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields AP, Rodriguez EH, Jovanovic M, Stern-Ginossar N, Haas BJ, Mertins P, Raychowdhury R, Hacohen N, Carr SA, Ingolia NT, et al. 2015. A regression-based analysis of ribosome-profiling data reveals a conserved complexity to mammalian translation. Mol Cell 60:816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS.. 2003. Global analysis of protein expression in yeast. Nature 425:737–741. [DOI] [PubMed] [Google Scholar]
- Gil J, Peters G.. 2006. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 7:667–677. [DOI] [PubMed] [Google Scholar]
- Gordon JL, Byrne KP, Wolfe KH.. 2009. Additions, losses, and rearrangements on the evolutionary route from a reconstructed ancestor to the modern Saccharomyces cerevisiae genome. PLoS Genet. 5:e1000485.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerzoni D, McLysaght A.. 2016. De novo genes arise at a slow but steady rate along the primate lineage and have been subject to incomplete lineage sorting. Genome Biol Evol. 8:1222–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinen TJAJ, Staubach F, Haming D, Tautz D.. 2009. Emergence of a new gene from an intergenic region. Curr Biol. 19:1527–1531. [DOI] [PubMed] [Google Scholar]
- Hood HM, Neafsey DE, Galagan J, Sachs MS.. 2009. Evolutionary roles of upstream open reading frames in mediating gene regulation in fungi. Annu Rev Microbiol. 63:385–409. [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK.. 2003. Global analysis of protein localization in budding yeast. Nature 425:686–691. [DOI] [PubMed] [Google Scholar]
- Ingolia NT. 2016. Ribosome footprint profiling of translation throughout the genome. Cell 165:22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS.. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasena AS, Fisher MF, Panero JL, Secco D, Bernath-Levin K, Berkowitz O, Taylor NL, Schilling EE, Whelan J, Mylne JS.. 2017. Stepwise evolution of a buried inhibitor peptide over 45 My. Mol Biol Evol. 34:1505–1516. [DOI] [PubMed] [Google Scholar]
- Ji Z, Song RS, Regev A, Struhl K.. 2015. Many lncRNAs, 5′ UTRs, and pseudogenes are translated and some are likely to express functional proteins. Elife 4:e08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TCG.. 2009. More than just orphans: are taxonomically-restricted genes important in evolution? Trends Genet. 25:404–413. [DOI] [PubMed] [Google Scholar]
- Knowles DG, McLysaght A.. 2009. Recent de novo origin of human protein-coding genes. Genome Res. 19:1752–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine MT, Jones CD, Kern AD, Lindfors HA, Begun DJ.. 2006. Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis-biased expression. Proc Natl Acad Sci U S A. 103:9935–9939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Dong Y, Jiang Y, Jiang HF, Cai J, Wang W.. 2010. A de novo originated gene depresses budding yeast mating pathway and is repressed by the protein encoded by its antisense strand. Cell Res. 20:408–420. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R.. Genome Project Data Processing S. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QR, Carvunis AR, Yu HY, Han JDJ, Zhong Q, Simonis N, Tam S, Hao T, Klitgord NJ, Dupuy D, et al. 2008. Revisiting the Saccharomyces cerevisiae predicted ORFeome. Genome Res. 18:1294–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZW, Chen X, Wu Q, Hagmann J, Han TS, Zou YP, Ge S, Guo YL.. 2016. On the origin of de novo genes in Arabidopsis thaliana populations. Genome Biol Evol. 8:2190–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long MY, Deutsch M, Wang W, Betran E, Brunet FG, Zhang JM.. 2003. Origin of new genes: evidence from experimental and computational analyses. Genetica 118:171–182. [PubMed] [Google Scholar]
- Marcet-Houben M, Gabaldon T.. 2015. Beyond the whole-genome duplication: phylogenetic evidence for an ancient interspecies hybridization in the Baker’s yeast lineage. PLoS Biol. 13:e1002220.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M.. 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351:652–654. [DOI] [PubMed] [Google Scholar]
- Moyers BA, Zhang JZ.. 2016. Evaluating phylostratigraphic evidence for widespread de novo gene birth in genome evolution. Mol Biol Evol. 33:1245–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DN, McLysaght A.. 2012. De novo origin of protein-coding genes in murine rodents. PLoS One 7:e48650.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neme R, Tautz D.. 2013. Phylogenetic patterns of emergence of new genes support a model of frequent de novo evolution. BMC Genomics 14:377.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OhEigeartaigh SS, Armisen D, Byrne KP, Wolfe KH.. 2011. Systematic discovery of unannotated genes in 11 yeast species using a database of orthologous genomic segments. BMC Genomics 12:377.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri N, Kosiol C, Schlotterer C.. 2014. The life cycle of Drosophila orphan genes. Elife 3:e01311.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi A, Magiorkinis G, Karlin DG.. 2013. Viral proteins originated de novo by overprinting can be identified by codon usage: application to the “Gene Nursery” of deltaretroviruses. Plos Comput Biol. 9:e1003162.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, Wei W, Steinmetz LM.. 2013. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature 497:127–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H.. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. [DOI] [PubMed] [Google Scholar]
- Rancurel C, Khosravi M, Dunker AK, Romero PR, Karlin D.. 2009. Overlapping genes produce proteins with unusual sequence properties and offer insight into de novo protein creation. J Virol. 83:10719–10736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt JA, Wanjiru BM, Brant AT, Saelao P, Begun DJ, Jones CD.. 2013. De novo ORFs in Drosophila are important to organismal fitness and evolved rapidly from previously non-coding sequences. Plos Genet. 9:e1003860.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A.. 2000. EMBOSS: the European molecular biology open software suite. Trends Genet. 16:276–277. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP.. 2011. Integrative genomics viewer. Nat Biotechnol. 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Orera J, Hernandez-Rodriguez J, Chiva C, Sabido E, Kondova I, Bontrop R, Marques-Bonet T, Alba MM.. 2015. Origins of de novo genes in human and chimpanzee. PLoS Genet. 11:e1005721.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabath N, Wagner A, Karlin D.. 2012. Evolution of viral proteins originated de novo by overprinting. Mol Biol Evol. 29:3767–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell DR, Zill OA, Rokas A, Payen C, Dunham MJ, Eisen MB, Rine J, Johnston M, Hittinger CT.. 2011. The awesome power of yeast evolutionary genetics: new genome sequences and strain resources for the Saccharomyces sensu stricto genus. G3 Genes Genomes Genet. 1:11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer EL, von Heijne G, Krogh A.. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 6:175–182. [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M.. 2006. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 34:D535–D539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strope PK, Skelly DA, Kozmin SG, Mahadevan G, Stone EA, Magwene PM, Dietrich FS, McCusker JH.. 2015. The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Res. 25:762–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa S, DeSalvo MK, Voolstra CR, Reyes-Bermudez A, Medina M.. 2009. Identification and gene expression analysis of a taxonomically restricted cysteine-rich protein family in reef-building corals. PLoS One 4:e4865.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toll-Riera M, Bosch N, Bellora N, Castelo R, Armengol L, Estivill X, Alba MM.. 2009. Origin of primate orphan genes: a comparative genomics approach. Mol Biol Evol. 26:603–612. [DOI] [PubMed] [Google Scholar]
- Tsai ZTY, Tsai HK, Cheng JH, Lin CH, Tsai YF, Wang DY.. 2012. Evolution of cis-regulatory elements in yeast de novo and duplicated new genes. BMC Genomics 13:717.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zheng HK, Fan CZ, Li J, Shi JJ, Cai ZQ, Zhang GJ, Liu DY, Zhang JG, Vang S, et al. 2006. High rate of chimeric gene origination by retroposition in plant genomes. Plant Cell 18:1791–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissler L, Gadau J, Simola DF, Helmkampf M, Bornberg-Bauer E.. 2013. Mechanisms and dynamics of orphan gene emergence in insect genomes. Genome Biol Evol. 5:439–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu DD, Irwin DM, Zhang YP.. 2011. De novo origin of human protein-coding genes. Plos Genet. 711: e1002379.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao WF, Liu HB, Li Y, Li XH, Xu CG, Long MY, Wang SP.. 2009. A rice gene of de novo origin negatively regulates pathogen-induced defense response. PLoS One 42: e4603.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Zhang YE, Chen JY, Liu CJ, Zhou WZ, Li Y, Zhang M, Zhang RL, Wei LP, Li CY.. 2012. Hominoid-specific de novo protein-coding genes originating from long non-coding RNAs. PLoS Genet. 89: e1002942.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HW, He BZ, Ma HJ, Tsaur SC, Ma CY, Wu Y, Ting CT, Zhang YE.. 2015. Expression profile and gene age jointly shaped the genome-wide distribution of premature termination codons in a Drosophila melanogaster population. Mol Biol Evol. 32:216–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yue JX, Li J, Aigrain L, Hallin J, Persson K, Oliver K, Bergstrom A, Coupland P, Warringer J, Lagomarsino MC, et al. 2017. Contrasting evolutionary genome dynamics between domesticated and wild yeasts. Nat Genet. 49:913–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Saelao P, Jones CD, Begun DJ.. 2014. Origin and spread of de novo genes in Drosophila melanogaster populations. Science 343:769–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Zhang GJ, Zhang Y, Xu SY, Zhao RP, Zhan ZB, Li X, Ding Y, Yang SA, Wang W.. 2008. On the origin of new genes in Drosophila. Genome Res. 18:1446–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.