Abstract
The adenosine-to-inosine (A-to-I) RNA editomes have been systematically characterized in various metazoan species, and many editing sites were found in clusters. However, it remains unclear whether the clustered editing sites tend to be linked in the same RNA molecules or not. By adopting a method originally designed to detect linkage disequilibrium of DNA mutations, we examined the editomes of ten metazoan species and detected extensive linkage of editing in Drosophila and cephalopods. The prevalent linkages of editing in these two clades, many of which are conserved between closely related species and might be associated with the adaptive proteomic recoding, are maintained by natural selection at the cost of genome evolution. Nevertheless, in worms and humans, we only detected modest proportions of linked editing events, the majority of which were not conserved. Furthermore, the linkage of editing in coding regions of worms and humans might be overall deleterious, which drives the evolution of DNA sites to escape promiscuous editing. Altogether, our results suggest that the linkage landscape of A-to-I editing has evolved during metazoan evolution. This present study also suggests that linkage of editing should be considered in elucidating the functional consequences of RNA editing.
Keywords: RNA editing, linkage, adaptive evolution, Drosophila, cephalopods, worms, mice, humans
Introduction
Adenosine-to-inosine (A-to-I) RNA editing, catalyzed by ADAR (adenosine deaminase acting on RNA) enzymes (Sommer et al. 1991; Palladino et al. 2000a; Keegan et al. 2001, 2005; Nishikura 2010; Savva et al. 2012a, 2012b), is an evolutionarily conserved mechanism in metazoans (Paul and Bass 1998; Bass et al. 1997; Bass 2002; Nishikura 2006, 2010, 2016; Jepson and Reenan 2008; Zinshteyn and Nishikura 2009; Jepson et al. 2011; Li and Church 2013). Inosine (I) is recognized as guanosine (G) by various cellular machinery (Wahba et al. 1962; Sommer et al. 1991; Rueter et al. 1999; Flomen et al. 2004; Nishikura 2006, 2010; Jin et al. 2007; Lev-Maor et al. 2007; Liang and Landweber 2007; Borchert et al. 2009; Alon et al. 2012). Therefore an A-to-I RNA editing event often has a similar cellular function as an A-to-G DNA substitution. Compared with the possible pleiotropic effects generated by DNA mutations (He and Zhang 2006; Wagner and Zhang 2011; Qian et al. 2012), RNA editing is hypothesized to facilitate adaptive evolution by increasing proteomic diversities temporally or spatially, in a more flexible manner (Gommans et al. 2009; Nishikura 2010, 2016; Klironomos et al. 2013; Rosenthal 2015).
The A-to-I RNA editing sites have been systematically characterized in various metazoan species during the past decade (Ramaswami and Li 2016; Savva et al. 2016). Despite the deep conservation of this mechanism, the target landscapes of editing have considerably evolved during metazoan evolution. The majority of RNA editing sites are located in clusters in noncoding regions of humans (Blow et al. 2004; Levanon et al. 2004; Li et al. 2009; Peng et al. 2012; Mazin et al. 2013; Ramaswami et al. 2013; Bazak et al. 2014; Sakurai et al. 2014; Picardi et al. 2015, 2017), monkeys (Chen et al. 2014; Yang et al. 2015), mice (Danecek et al. 2012), worms (Zhao et al. 2015; Goldstein et al. 2017), and coral (Porath et al. 2017). Nevertheless, a considerable number of editing sites are clustered in mRNA regions in Drosophila (Graveley et al. 2011; Rodriguez et al. 2012; St Laurent et al. 2013; Mazloomian and Meyer 2015; Yu et al. 2016; Buchumenski et al. 2017; Duan et al. 2017; Zhang et al. 2017), ants (Li et al. 2014) and cephalopod species (Alon et al. 2015; Liscovitch-Brauer et al. 2017). These results suggest that RNA editing modulates the diversity of the transcriptomes and proteomes through different means. Previously, we systematically identified about 2,000A-to-I editing sites in brains of D. melanogaster (Duan et al. 2017). By comparing the observed nonsynonymous (N) to synonymous (S) editing sites versus the expected N/S ratio under randomness (neutrality), we found abundant nonsynonymous editing events in Drosophila brain are adaptive and maintained by natural selection (Duan et al. 2017). The signals of adaptation in nonsynonymous editing sites were also detected by other studies in Drosophila (Yu et al. 2016; Zhang et al. 2017) and cephalopods (Garrett and Rosenthal 2012; Alon et al. 2015; Liscovitch-Brauer et al. 2017). In humans, although the N/S analysis reveals the nonsynonymous editing events in CDSs are overall nonadaptive (Xu and Zhang 2014), the editing events conserved in both humans and mice show the signature of adaptation (Xu and Zhang 2015). Despite the fact that many RNA editing sites are clustered (Alon et al. 2015; Zhao et al. 2015; Yu et al. 2016; Duan et al. 2017; Goldstein et al. 2017; Liscovitch-Brauer et al. 2017; Zhang et al. 2017), most studies, including our own (Duan et al. 2017), focused primarily on the effect of each editing event on amino acid changes and did not consider the possible combinatory effect of the neighboring editing events. Since the effects of multiple editing events in an RNA molecule might interfere with each other, the combined effect of the linked editing events should be taken into account.
Previous studies have demonstrated that many adenosines in a double-stranded RNA (dsRNA) could be edited simultaneously (Nishikura et al. 1991; Polson and Bass 1994; Zhang and Carmichael 2001; Prasanth et al. 2005; St Laurent et al. 2013). With the advent of next-generation sequencing (NGS), it was reported that multiple editing events could be detected in a single NGS read (Carmi et al. 2011; Porath et al. 2014). Moreover, considering such “hyper-edited” reads has helped to identify numerous novel editing sites (Carmi et al. 2011; Porath et al. 2014), such as in human tumors (Han et al. 2015; Paz-Yaacov et al. 2015; Wang et al. 2017), primate Alu sequences (Levanon and Eisenberg 2015) and mammalian transposons (Knisbacher and Levanon 2015), mouse polyomavirus (Garren et al. 2015), Drosophila (Buchumenski et al. 2017), coral (Porath et al. 2017), and cephalopods (Alon et al. 2015; Liscovitch-Brauer et al. 2017). Despite these intriguing discoveries, it remains unclear whether such “hyper-edited” NGS reads are caused by significant linkage of editing events in the same RNA molecules or just by randomness. In principle, if ADAR catalyzes multiple editing sites on an RNA molecule simultaneously, in a cell the edited “I” (read as “G” in the sequencing results) alleles across sites would be linked in the same RNA molecules, and the unedited “A” alleles would be linked in the unedited RNAs. In other words, the linkage of editing events would generate a pattern similar to the DNA haplotype blocks with strong “linkage disequilibrium” (LD) (supplementary fig. S1A, Supplementary Material online). In contrast, if ADAR catalyzes the clustered editing sites randomly, the postedited mRNAs should be a mixture of different haplotypes that do not show any signal of linkage among the edited sites (supplementary fig. S1B, Supplementary Material online). Therefore, observing multiple editing sites clustered within short distances of each other does not necessarily suggest significant linkage of the editing events in these sites. LD describes the nonrandom association between DNA mutations at different loci within a population (Lewontin 1988), and DNA haplotypes with strong LDs often inform the possible epistatic interactions between the mutated sites (Cheng et al. 2017; Slatkin 2008). Here we employed the algorithm (r2) that was originally designed to detect LD of DNA mutations to identify the significantly linked editing sites. Knowing the linkage information is crucial for comprehending the cellular functions of the A-to-I editing events, especially when the editing sites are adjacent to each other.
In this study, we compiled the previously characterized A-to-I editing sites in ten metazoan species (fig. 1A and supplementary table S1, Supplementary Material online). In each species, we investigated whether or not the editing events were significantly linked in the same RNA molecules. Our results suggest that many editing events are linked in flies and cephalopods, presumably associated with the adaptation of the editing events; nevertheless, the patterns are different in worms and humans. We also found the linkage of editing events has different impacts on DNA sequence evolution of these species.
Fig. 1.
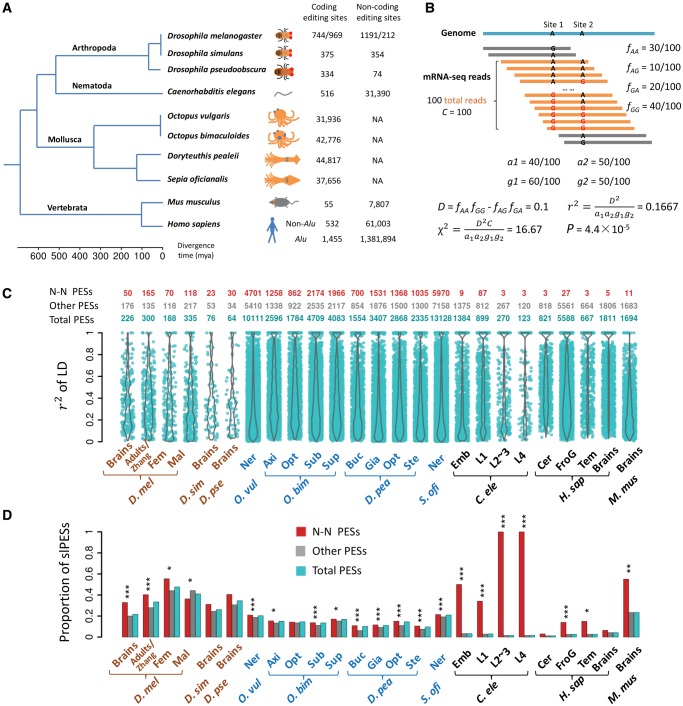
The landscape of linkage between RNA editing events in ten metazoan species. (A) A phylogenetic tree of the ten species we used in this study. The numbers of editing sites in coding and noncoding regions are given next to each species. For Drosophila melanogaster, the editing sites before and after the slash were retrieved from brains (Duan et al. 2017) and adults (Zhang et al. 2017), respectively. (B) A hypothetical example of calculating r2 between two editing sites based on the coverage of the NGS reads. Only reads covering the two editing sites (in orange) were included, and the reads spanning only one editing site (in gray) were discarded. (C) Violin plots of r2 (y-axis) for significantly linked PESs in 25 samples of ten species (x-axis). The numbers of total, N–N and the remaining slPESs in each sample are presented above the plots. (D) The proportions of the PESs that have editing events significantly linked in each sample (adjusted P < 0.05). Besides the total PESs, the fraction for the N–N or the remaining PESs that was significantly linked was also calculated in each sample. The fraction of the N–N PESs that is significantly linked was compared with that of the remaining PESs with the Fisher’s exact test. *P < 0.05; **P < 0.01; ***P < 0.001. For the four species O. bimaculoides, D. pealeii, C. elegans, and H. sapiens, only the top four samples with the largest numbers of slPESs are shown in (C) and (D). Fem, female adults; Mal, male adults; Ner, nerve system; Axi, axial nerve cord; Opt, Optic lobe; Sub, subesophageal brain; Sup, supraesophageal brain; Buc, Buccal ganglia; Gia, giant fiber lobe; Ste, stellate ganglion; Emb, embryos; L1, L1 larvae; L2∼3, L2∼3 larvae; L4, L4 larvae; Cer, cerebellum; FroG, frontal gyrus; Tem, temporal lobe. D. mel, Drosophila melanogaster; D. sim, Drosophila simulans; D. pse, Drosophila pseudoobscura; O. vul, Octopus vulgaris; O. bim, Octopus bimaculoides; D. pea, Doryteuthis pealeii; S. ofi, Sepia oficianalis; C. ele, Caenorhabditis elegans; H. sap, Homo sapiens; M. mus, Mus musculus.
Results
Data
We collected the previously identified A-to-I RNA editing sites in 45 samples of ten species, including flies, cephalopods, worms, mice, and humans (in each sample, we only considered the editing sites with editing level ≥0.05, supplementary table S1, Supplementary Material online). These ten species were chosen due to the extensive editome characterization in previous studies (fig. 1A). The editing sites of D. melanogaster consist of 1,935 sites in brains, 426 sites in female and 818 sites in male adults of five strains of D. melanogaster we previously characterized (Duan et al. 2017), and 1,181 sites in adults of D. melanogaster identified by Zhang et al. (Zhang et al. 2017). 729 and 408 of the editing sites in brains of D. melanogaster have orthologous sites edited in brains of D. simulans and D. pseudoobscura, respectively (Duan et al. 2017). The editing sites in Caenorhabditis elegans consist of 516 sites in CDSs and 31,390 sites in noncoding regions (Zhao et al. 2015). We also analyzed the previously identified editing sites in CDSs of four cephalopod species (Octopus vulgaris, Octopus bimaculoides, Doryteuthis pealeii, and Sepia oficianalis. The number of sites with editing level ≥0.05 ranges from 31,936 to 44,817 in the four species) (Liscovitch-Brauer et al. 2017). In mammals, we compiled the editing sites in brains of mice (55 in CDSs and 7,807 in noncoding regions) (Danecek et al. 2012; Ramaswami and Li 2014), and about 1.4 million editing sites in brains of humans (Ramaswami and Li 2014; Ramaswami et al. 2013). Based on the phylogenetic tree (Peterson et al. 2004; Telford 2006; Dunn et al. 2014; Liscovitch-Brauer et al. 2017), the divergence time between the species in our analysis spans from a few million (between D. melanogaster and D. simulans, or between O. vulgaris and O. bimaculoides) to ∼600 million years (between vertebrates and the invertebrates). Therefore, our analysis would well reflect the evolutionary dynamics of linkage of RNA editing events at different time scales.
Detecting Linkage of A-to-I RNA Editing Events in Metazoans
For each tissue of a species, we downloaded the raw next-generation sequencing (NGS) data from the original studies (supplementary table S1, Supplementary Material online) and mapped all the sequencing reads to the corresponding reference genome or assembled transcriptome (Materials and Methods). We only considered the NGS reads spanning at least two annotated editing sites (reads spanning only one editing site were discarded, fig. 1B). We also required the editing sites to have sequencing coverage ≥20× and editing levels above 0.05. Then for all the pairwise combinations of editing sites that were covered by the NGS reads, we calculated the frequencies of the four possible “haplotypes” and calculated the D, r2 and P value using the method in calculating LD (fig. 1B). Here we only focused on the linkage of editing events that were mutually associative (D > 0), and did not consider the mutually exclusive editing events (D < 0) that were occasionally (< 1%) observed (Materials and Methods).
At a false discovery rate (FDR) of 0.05, we identified hundreds to thousands of pairs of editing sites (PESs) that had events significantly linked in a sample (see fig. 1C for the numbers and violin plots of r2 for 25 representative samples, and supplementary fig. S2, Supplementary Material online, for the remaining samples; see supplementary table S2, Supplementary Material online, for all the significantly linked PESs [slPESs]). Notably, the fraction of PESs that show significant linkage varies widely among species (fig. 1D). For example, among the 1,087 PESs supported by NGS reads in brains of D. melanogaster, 20.8% (226/1,087) of them show significant linkage after multiple testing corrections (adjusted P < 0.05, fig. 1D). Besides, we also found comparable (or even higher) proportions of the slPESs in adults of D. melanogaster previously characterized by us (Duan et al. 2017) or Zhang et al. (2017), or for the sites that had conserved editing events in brains of D. simulans or D. pseudoobscura (fig. 1D). Moreover, a similar proportion (23.4%) of the PESs was significantly linked in mouse brains (fig. 1D). Intriguingly, we identified thousands of slPESs in neural tissues of cephalopods, which accounts for 9.7–21.0% of all the possible PESs supported by the NGS reads (sequencing coverage C ≥ 20, fig. 1D). In contrast, in various tissues of worms and human brain tissues, only 1.1–4.1% of the PESs supported by the NGS reads were significantly linked (fig. 1D). Despite the difference in the overall patterns of linkage across these clades, we found in each sample, the nonsynonymous–nonsynonymous (N–N) PESs, in general, have considerably higher proportions to be significantly linked compared with the remaining PESs (fig. 1D). These results suggest that the amino acid changes caused by these linked nonsynonymous editing events might have epistatic interactions and be favored by natural selection. In summary, we observed widespread linkage of the editing events in CDSs of flies and cephalopods. Although the N–N PESs tend to be linked in worms and humans, the contribution of these linked N–N PESs to the overall pattern is diluted since the editing sites are predominantly located in noncoding or repetitive regions in these two species (fig. 1D). Therefore, relatively lower fractions of the PESs show evidence of linkage in worms and humans.
The power in detecting linkage of editing events is affected by the sequencing coverage of the editing sites (C), since higher C value would yield a smaller P value even if the r2 and the relative frequencies of the four haplotypes are the same (fig. 1B, Materials and Methods). Indeed, C is generally lower for the human brain samples since most of the editing sites in humans are in noncoding regions that are lowly transcribed (supplementary fig. S3A and B, Supplementary Material online). To exclude the possible detection bias caused by sequencing coverage (C), we used different cutoffs of C in our analysis. By requiring C to be at least 50 or 100, we constantly found a considerably lower fraction of the PESs are significantly linked in humans and worms than in flies, mice, and cephalopods (supplementary fig. S4, Supplementary Material online). The editing sites analyzed in this study were identified based on mRNA-Seq data generated under different Illumina sequencing protocols: the brain editomes of the three Drosophila species were determined by 50-bp single-end sequencing, whereas the remaining libraries were sequenced by 50-bp single-end or ≥70-bp pair-end protocols (supplementary table S1, Supplementary Material online). The median length of the insert size ranged from 150 to 250 bps for most of the pair-end libraries (supplementary fig. S5, Supplementary Material online). Since we collapsed the paired-end RNA-Seq reads whenever applicable, the paired-end reads would in principle recover more distantly located editing sites and yield more power in detecting the linkage of editing events. Thus, it seems unlikely that the difference in NGS read length would cause the fraction of slPESs to be higher in Drosophila brains (50-bp single-end) than in worms or humans (both have pair-end sequences). To further exclude the potential bias caused by the difference in NGS read lengths, we employed a “50 nt mapping” approach by which we extracted the first 50 nt from each NGS reads (the paired-end reads were treated as two independent reads) and redid all the LD analysis (supplementary fig. S6A, Supplementary Material online). As expected, the global trend that higher fractions of PESs were significantly linked in flies and cephalopods than in humans or worms remained intact with this “50 nt mapping” approach (supplementary fig. S6B, Supplementary Material online). In addition, this pattern held true when we only used the ≥ 70-bp pair-end sequencing libraries in the analyses (supplementary fig. S7, Supplementary Material online).
Notably, in both CDSs and noncoding regions, the neighboring editing sites that showed significant linkage had significantly shorter distances compared with those that were not significantly linked in a sample (supplementary fig. S8, Supplementary Material online). To further control for the effects of the distance between editing sites as well as the NGS read lengths on our analyses, we examined the PESs with distances ≤15 nt (this cutoff was chosen because the median distance of slPESs across all the samples was ∼15 nt). At different cutoffs of C (20, 50, or 100), we constantly observed higher proportions of the PESs are significantly linked in flies, mice, and cephalopods than in humans and worms (supplementary fig. S9A, Supplementary Material online). Furthermore, we found that the PESs within 15 nt have remarkably higher proportions to be linked compared with the remaining PESs that have distances >15 nt (supplementary fig. S9A, Supplementary Material online). Analogous results were obtained when we used other cutoffs (10, 20, and 30 nt; see supplementary fig. S9B–D, Supplementary Material online, respectively). These patterns might not be surprising since more distantly located sites are less likely to be simultaneously edited by ADAR (Sommer et al. 1991; Palladino et al. 2000a, 2000b; Keegan et al. 2001, 2005; Nishikura 2006; Nishikura 2010; Savva et al. 2012a, 2012b).
Altogether, our results suggest that abundant editing events are significantly linked in the RNA molecules in metazoans, and this pattern is more pronounced in flies and cephalopods than in worms or humans. Furthermore, the distinct patterns we observed in the former two versus the latter two species are not affected by the difference in sequencing coverage or NGS read length.
Conservation of the Linked Editing Events across Species
Previous studies revealed that the nonsynonymous editing events tend to be conserved between Drosophila (Duan et al. 2017; Zhang et al. 2017) or cephalopod species (Liscovitch-Brauer et al. 2017). Here, we examined the conservation patterns of the slPESs, especially the ones containing nonsynonymous editing events, between closely related species. The conservation of RNA editing events can be manifested at different levels (fig. 2A). Even if the adenosine sites are conserved between two species, a slPES in one species might have three possible patterns in another species (fig. 2A). First, the two orthologous adenosine sites are both edited and significantly linked in the other species (conserved linkage). Second, the orthologous adenosine sites are edited but linked with different editing site in the other species (evolved linkage). Third, neither (or one) of the two orthologous sites is edited, or they do not show linkage with each other even if one both sites are edited in the other species (species-specific linkage). Moreover, the edited adenosine site might evolve at DNA level, which precludes the possibility of editing in the other species (nonconserved adenosines, fig. 2A).
Fig. 2.
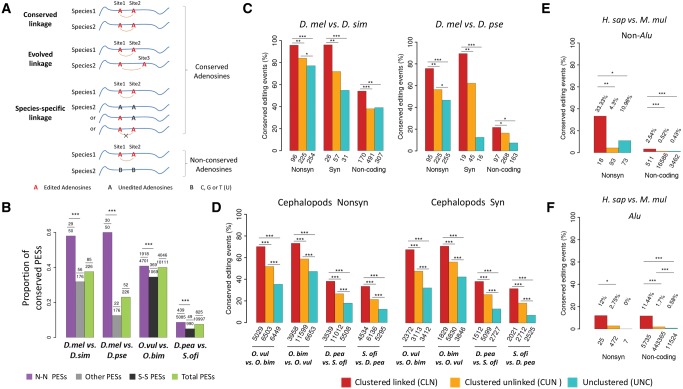
Conservation of linked editing events between species. (A) The possible conservation patterns of a slPES in another species: 1) conserved linkage, for which the two orthologous adenosine sites are both edited and significantly linked in another species; 2) evolved linkage, for which one of the orthologous adenosine sites is edited but linked with a different editing site; 3) species-specific linkage, for which the two orthologous sites might be edited but do not show linkage to each other, and they are not linked with other editing sites as well. It is also possible that the orthologous sites in another species are not adenosines (B, which represents C, T, or G) so that editing would not occur on the orthologous sites. (B) The proportion (y-axis) of the total, the N–N, and the remaining slPESs that are conserved between two Drosophila species or between two cephalopod species (***P < 0.001; Fisher’s exact tests). In each comparison, the numbers of the tested slPESs and the conserved slPESs between species are given above the plot. In flies, all the remaining (Other) slPESs were used to compare with the N–N slPESs to increase the statistical power; and in cephalopods, the S–S (synonymous–synonymous) slPESs were used to compare with the N–N slPESs. (C–F) The proportions of the editing events that are evolutionarily conserved between two species of Drosophila (brains, C), cephalopods (pooled tissues, D), non-Alu (E) and Alu (F) regions between humans and rhesus macaque (prefrontal cortex and cerebellum). Nonsynonymous (Nonsyn), synonymous (Syn) and noncoding adenosine sites are divided into three categories: 1) clustered (within 100 nt) and significantly linked (CLN); 2) clustered but unlinked (CUN); and 3) unclustered (UNC). When comparing editing events in two species, only the sites with editing level ≥ 0.05 and the genomic DNA were adenosines in both species were considered. The numbers of total editing sites in each category used for comparison are presented below the bars. The Fisher’s exact tests were performed to detect statistical significance (*P < 0.05; **P < 0.01; ***P < 0.001).
We set out to discover the slPESs conserved between species (conserved linkage, fig. 2A). Among the 226 slPESs in brains of D. melanogaster, 85 (37.6%) of them show conserved linkage in brains of D. simulans and 52 (23.0%) of them show conserved linkage in brains of D. pseudoobscura (fig. 2B, see supplementary table S3, Supplementary Material online for details). We found 42 PESs are significantly linked in brains of all the three Drosophila species (supplementary table S4, Supplementary Material online). For the 43 PESs that showed conserved linkage in both D. melanogaster and D. simulans but not in D. pseudoobscura, four PESs had orthologous sites edited in both positions but did not show any linkage and 15 PESs had both orthologous sites conserved (i.e., were also adenosines) but at least one site in a PES was not edited in D. pseudoobscura, and 24 PESs had at least one orthologous site not conserved in D. pseudoobscura at the DNA level (supplementary table S4, Supplementary Material online). We also found 40.0% (4,046 out of 10,111) of the slPESs in O. vulgaris show conserved linkage in O. bimaculoides and 7.5% (825 out of 10,997) of the slPESs in D. pealeii are conserved in Sepia oficianalis (fig. 2B and supplementary table S3, Supplementary Material online). We further found 46 slPESs are conserved in all the four cephalopod species (conserved linkage, supplementary table S5, Supplementary Material online). Consistent with previous observations that only a few editing events are conserved across mammalian species (Pinto et al. 2014), we in total identified 83 sites that had editing events conserved between humans and mice and only two PESs (one in GRIK2 and the other in ZNF397) that showed conserved linkage in both humans and mice (supplementary fig. S10A, Supplementary Material online). Since the majority of the editing sites in human and other primates are located in Alu sequences that are absent in mice (Levanon and Eisenberg 2015), we also examined the editing events conserved between humans and the rhesus macaques that were characterized previously (Chen et al. 2014). We detected 8,434 editing sites (135 in non-Alu and 8,299 in Alu sequences) that had conserved editing events in both humans and macaques (we only focused on the editing sites in the prefrontal cortex and cerebellum since editing information were available in these two tissues for both species). Among the 566 slPESs in humans (47 in non-Alu and 519 in Alu regions, supplementary fig. S10B, Supplementary Material online), only 34 (∼6%) of them (2 in non-Alu and 32 in Alu regions) were also significantly linked in macaques (supplementary table S3, Supplementary Material online), suggesting the fraction of slPESs that is conserved is lower in primates than in Drosophila or cephalopods (fig. 2B). Interestingly, for the two conserved PESs between human and mouse, although the orthologous sites in macaque are conserved adenosine in DNA for the PES in ZNF397, both sites were not edited in prefrontal cortex or cerebellum in macaques. GRIK2 encodes a glutamate receptor for excitatory neurotransmitters and three nonsynonymous editing events (Ile567Val, Tyr571Val, and Gln621Arg) on its mRNA was discovered more than two decades ago (Paschen et al. 1994). We found the editing events of Ile567Val and Tyr571Val are significantly linked in humans, macaques, and mice, which suggests the potential functional importance of this linkage. Strikingly, significantly higher proportions of the N–N PESs showed conserved linkage than the remaining (or S–S) PESs in flies and cephalopods (fig. 2B), and similar results were obtained when we only considered conserved adenosine sites between the closely related species (supplementary fig. S11, Supplementary Material online). Overall, these results further support the hypothesis that the linkage of N–N PESs might result in epistatic amino acid interactions which are preserved by natural selection in flies and cephalopods.
The linkage of editing events can evolve across species (nonconserved or species-specific linkage, fig. 2A). Thus, the conserved linked PESs might only account for a fraction of the conserved editing events across species. For example, among the 348 editing sites in the 226 slPESs in brains of D. melanogaster, 212 sites had editing events conserved in brains of D. simulans; whereas only 140 sites had conserved editing events if we only considered the conserved linked PESs between D. melanogaster and D. simulans. Therefore, 72 sites had editing events conserved in the two Drosophila species, but the linkage among these editing sites might have evolved. Similarly, among the 2,265 sites that had editing events conserved in both D. pealeii and S. oficianalis, only 1,003 of them comprised slPESs that were conserved between these two species, and the remaining 1,262 sites had conserved editing events but the linkage patterns might have evolved between these two species. To broadly test whether linkage is associated with conserved editing events across species, we divided the edited adenosine sites in a species into three categories: 1) clustered (within 100 nt) and significantly linked (CLN), 2) clustered but unlinked (CUN), and 3) unclustered (UNC). To control for the confounding effects caused by DNA evolution, we only focused on the adenosine sites that are conserved between species. We defined an editing site to be linked if it was significantly linked with a neighboring editing site in the LD analysis (adjusted P < 0.05). Since the nonsynonymous, synonymous and noncoding editing sites are under different selective pressures (Xu and Zhang 2015; Duan et al. 2017), we explored the conservation patterns of the editing events in each functional class separately. For the adenosine sites conserved between D. melanogaster and D. simulans and edited in brains of D. melanogaster, the CLN sites have the highest whereas the UNC sites have the lowest proportion of editing events conserved in brains D. simulans (fig. 2C, left panel). This pattern persisted for the nonsynonymous and synonymous editing sites in brains of D. melanogaster. We observed similar patterns when we examined the editing conservation status between D. melanogaster and D. pseudoobscura (fig. 2C, right panel), or between a cephalopod and its sibling species (fig. 2D). Despite the fact that only a small fraction of editing events were conserved between humans and macaques (135/20,798 = 0.65% for non-Alu and 8,299/461,344 = 1.80% for Alu sequences), the CLN sites in both non-Alu and Alu regions tend to have significantly higher proportions of editing events conserved between the two species compared with the CUN or UNC sites (fig. 2E and F).
Taken together, our findings suggest that the linked editing events might be functionally important and thus preserved by natural selection during evolution. These results are well consistent with previous observation in Drosophila that evolutionarily conserved editing events tend to be clustered (Zhang et al. 2017). We also found the linkage of editing events frequently evolve across species. The overall pattern is that on the adenosine sites that are conserved between species at the DNA sequence level, the editing sites that have linked and clustered events (CLN sites) tend to have conserved editing events between two metazoan species.
Opposite Modes of DNA Sequence Evolution Driven by Linkage of Editing Events in Metazoans
The optimized DNA sequence contexts are important for ADAR to exert editing and DNA mutations often affect the levels or status of editing (Ramaswami et al. 2015; Gu et al. 2016; Kurmangaliyev et al. 2016; Liscovitch-Brauer et al. 2017; Park et al. 2017). The majority of the editing events in humans are located in Alu sequences, which are usually not conserved at the DNA sequence level (Pinto et al. 2014; Levanon and Eisenberg 2015). Nevertheless, the adenosine sites where editing occurs are usually conserved across different Drosophila or cephalopod species (Reenan 2005; Yu et al. 2016; Duan et al. 2017; Liscovitch-Brauer et al. 2017; Zhang et al. 2017). Here we questioned whether linkage of the editing sites would affect genome evolution.
First, we examined the phyloP scores (a higher phyloP score means a higher conservation level of a DNA site) for the CLN, CUN, and UNC sites in flies. Notably, for the editing sites identified in brains or whole adults of D. melanogaster, the CLN sites are generally more conserved at the DNA level than the CUN or UNC sites, and this pattern persisted for the nonsynonymous, synonymous, or noncoding editing sites (fig. 3A). Due to the unavailability of phyloP scores for the cephalopods, for each species, we calculated the proportion of adenosines that were conserved in another species to measure the conservation levels (Supplementary Methods). Briefly, we investigated the proportion of the edited adenosine sites in an octopus species (O. vulgaris or O. bimaculoides) whose orthologous sites were also adenosines in squid (D. pealeii). For the squid and cuttlefish (S. oficianalis), we examined whether the edited adenosine sites in one species were also adenosine sites in the other species. For example, in O. vulgaris, 5,784, 7,771, and 7,646 nonsynonymous editing sites belong to CLN, CUN, and UNC category, respectively, and 58.5%, 50.0%, and 40.2% of these sites are also adenosines in D. pealeii, respectively, suggesting the conservation level of the genomic sites decreased in the order of CLN, CUN, and UNC (fig. 3B). Moreover, we observed similar patterns for the nonsynonymous editing sites in O. bimaculoides, D. pealeii, and S. oficianalis (fig. 3B, left panel), and for the synonymous editing sites in each of the four species as well (fig. 3B, right panel). Taken together, these results support that the linkage of editing events exerts further selective constraints on the evolution of the DNA sequences in flies and cephalopods.
Fig. 3.
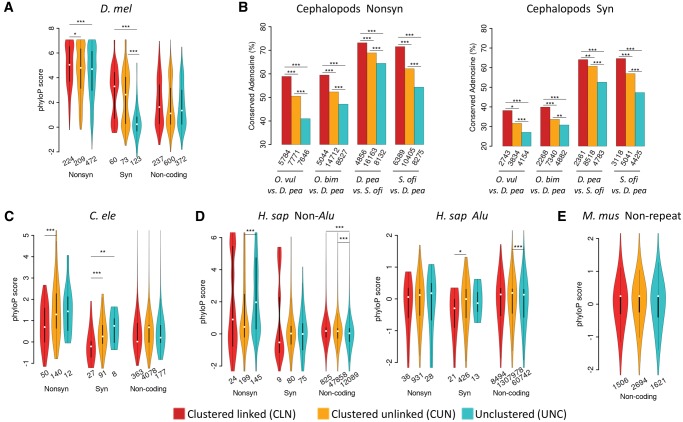
Conservation patterns of the CLN, CUN, and UNC editing sites at DNA level. (A) The violin plots of phyloP scores (y-axis) of the CLN, CUN, and UNC editing sites in the nonsynonymous (Nonsyn), synonymous (Syn) and noncoding functional categories in D. melanogaster. (B) The proportions of adenosine sites that were edited in a cephalopod species (the former in a comparison) and have orthologous sites to be adenosines in another cephalopod species (the latter in a comparison). The Fisher’s exact test was performed to test differences in the proportions of conserved adenosine sites (*P < 0.05; **P < 0.01; ***P < 0.001). (C–E) The violin plots of phyloP scores (y-axis) of the CLN, CUN, and UNC editing sites in the Nonsyn, Syn and noncoding editing functional categories in C. elegans (C), non-Alu and Alu regions of humans (D) and nonrepetitive regions of mice (E). The numbers of total editing sites in each category are given below the violins or bars. For flies, worms, humans, and mice, only editing sites with phyloP scores available are considered and the Wilcoxon rank-sum test was performed to test differences in phyloP scores (*P < 0.05; **P < 0.01; ***P < 0.001).
In contrast, we did not find the edited adenosine sites in the CLN or CUN classes are evolutionarily more conserved than the UNC sites in the CDSs of worms (fig. 3C) and humans (fig. 3D), both of which have editing sites predominantly located in noncoding regions. Note that the editing sites in human Alu region are usually nonconserved (fig. 3D, right panel), and for both synonymous and nonsynonymous editing sites, we observed similar patterns in both Alu and non-Alu regions. In mice, the majority of the annotated editing sites are also located in noncoding regions, and we did not observe substantial differences in phyloP scores among the CLN, CUN, and UNC sites as well (fig. 3E).
Altogether, our results suggest that in flies and cephalopods, the linkage of editing events further constrains the evolution of the edited adenosine sites at the DNA level. However, we observed opposite patterns in humans and worms, in particular for the nonsynonymous editing sites. Previous studies suggest the nonsynonymous editing events in humans are generally nonadaptive (Xu and Zhang 2014). It is possible that some linked or clustered nonsynonymous editing events in humans or worms might cause more detrimental effects than the individual editing events, which would drive the adenosine sites to evolve to escape the promiscuous editing. Although some conserved linked PESs or editing events might be maintained by natural selection in worms and humans, such conserved sites only account for a small subset of the editomes. Our hypothesis is further supported by the editing level comparisons. In flies and cephalopods, the editing levels of the CLN sites are higher than the CUN or UNC sites in the nonsynonymous and synonymous functional classes (supplementary fig. S12, Supplementary Material online). However, for the nonsynonymous editing sites in worms and humans, the editing levels of the CLN sites are considerably lower compared with the CUN or UNC sites (supplementary fig. S12, Supplementary Material online). In short, lines of evidence suggest that linkage of nonsynonymous editing events in flies and cephalopods are favored by natural selection so that the adenosine sites are constrained at the DNA level. Nevertheless, the linkage of nonsynonymous editing events in worms and humans is overall selected against which drives those adenosine sites to evolve.
Verifying Linkage of Editing Events with Large-Scale Sanger-Sequencing
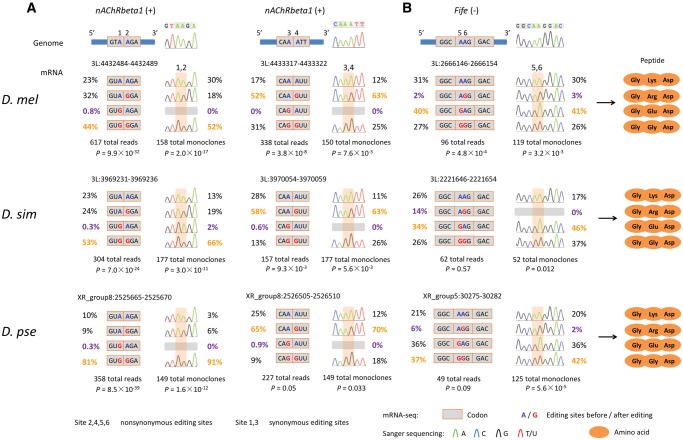
To verify the linkage of editing events revealed by the NGS reads, we carried out extensive Sanger sequencing of the cDNA monoclones (Supplementary Methods). In total, we obtained 2,568 Sanger sequences of single cDNA clones in heads of three Drosophila species (492 sequences for nAChRbeta1, 296 for Fife, 1,002 for qvr, and 778 for NaCP60E). nAChRbeta1 (nicotinic Acetylcholine Receptor β1) is mainly expressed in the nervous system of Drosophila and plays an important role in fast synaptic excitatory transmission (Seeburg 2002; Sattelle et al. 2005; Dupuis et al. 2012). In nAChRbeta1, we observed two PESs that were significantly linked in brains of all the three Drosophila species (fig. 4A). The two linked PESs are located in the N-terminal topological domain of nAChRbeta1, which potentially influence functions of this receptor (Hoopengardner et al. 2003; Sattelle et al. 2005). In Fife we also found one PES that is significantly linked in brains of D. melanogaster (fig. 4B). For each of the three PESs, the relative abundance of each haplotype is similar across three Drosophila species. Notably, the frequencies of the “GG” haplotypes are considerably high in these cases (fig. 4). We further verified the haplotype frequencies and the linkage events of the editing sites by Sanger sequencing of single cDNA clones (fig. 4).
Fig. 4.
Sanger verification of linked editing events in three Drosophila species. (A) In nAChRbeta1, two pairs of adjacent editing sites (sites 1 and 2, and sites 3 and 4) are highly linked in brains of all the three Drosophila species. For each of the two PESs, the first editing site is located at the third base of a codon, and the other linked editing site is located at the first base of next codon. (B) In Fife, a linked pair of editing sites (sites 5 and 6) that are located at the first and second bases of the same codon, is conserved across three Drosophila species. The linkage events revealed by NGS reads were verified by the Sanger sequencing of cDNA monoclones. For each of the three double editing pairs, the genomic location, haplotype frequencies, the resultant amino acids, and the depth of the NGS reads and Sanger sequencing results, as well as examples of Sanger sequencing traces, are shown.
We also identified multiple adjacent editing sites that had editing events linked in the same RNA molecules. For example, qvr is involved in homeostatic regulation of circadian cycle and the voltage-gated potassium channel activity (Koh et al. 2008). There are seven editing sites located in a 24-nt CDS fragment (eight codons) in qvr, and editing on four of these sites causes nonsynonymous changes (site 1, 3, 5, and 7, see supplementary fig. S13, Supplementary Material online). Intriguingly, the most predominant postedited RNA molecules harbor all the four nonsynonymous editing events, and this pattern is consistently observed across all the three Drosophila species (P < 10−5 in all three Drosophila species; supplementary fig. S13A, Supplementary Material online, the yellow haplotype; and supplementary fig. S14, Supplementary Material online, for the remaining haplotypes). Our Sanger sequencing of the single cDNA clones further confirmed the linkage of these editing events in heads of the three Drosophila species (supplementary fig. S13A, Supplementary Material online). The pairwise LD analysis (r2) of these seven sites demonstrated that the four nonsynonymous sites (site 1, 3, 5, and 7) have stronger pairwise linkage than the other pairs in all the three Drosophila species (supplementary fig. S13B, Supplementary Material online). These linked editing events might collectively affect the conformation or activity of QVR, although it is challenging to predict the functional consequences at this moment. Moreover, we detected strong linkage among four nonsynonymous editing events in NaCP60E in all the three Drosophila species (P < 0.001 in each species) and further verified the linked editing events by Sanger sequencing (supplementary fig. S15, Supplementary Material online). Taken together, our Sanger sequencing of single cDNA clones has further confirmed the existence of linked editing events that are also conserved in brains of three Drosophila species.
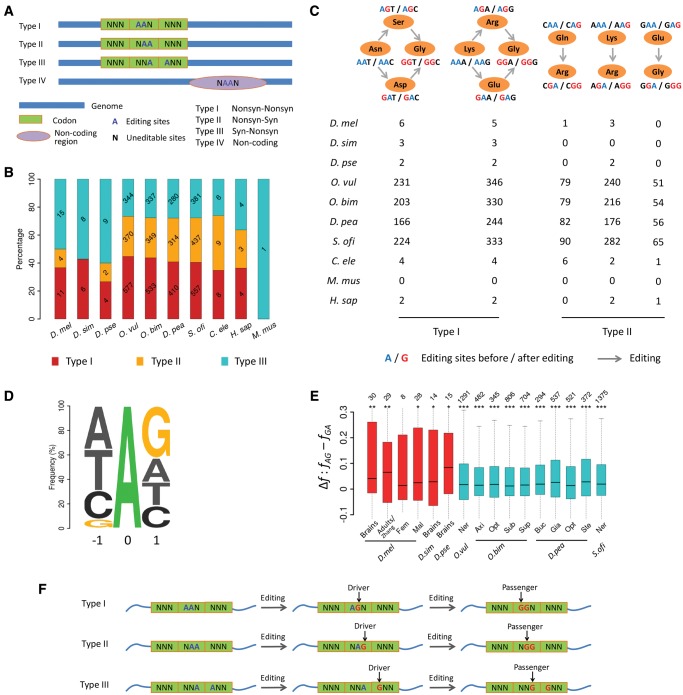
Non-Independent Editing of AA Dinucleotides and the Implication for Functional Prediction
The functional consequence of the “hyper RNA editing” events was hypothesized to compete with RNAi (Scadden and Smith 2001a), facilitate cleavage of double-stranded RNAs (Scadden and Smith 2001b; Scadden 2005), retain RNAs in the nucleus (Zhang and Carmichael 2001; Prasanth et al. 2005), or regulate translation of Alu-containing transcripts (Kim et al. 2004; Osenberg et al. 2009). Although it is challenging to predict the combinatory effects of multiple editing events at this moment, the AA (adenosine–adenosine) double editing sites might shed light on the functional consequences of linkage of editing events.
We classified the significantly linked editing events on the AA double-editing sites into four types: Type I, the AA double-editing sites are located in the first and second bases of the same codon; type II, they are in the second and third bases of the same codon; type III, they are in the third base of an upstream codon and the first base of a downstream codon; and type IV, the AA dinucleotides are located in noncoding regions (fig. 5A and supplementary table S6, Supplementary Material online). Here, we only considered the first three types of AA double editing sites. For each species, we presented the AA pairs that had editing events significantly linked in CDSs in figure 5B. For instance, we found 11 type I, 4 type II, and 15 type III AA double-editing pairs that were significantly linked (adjusted P-value < 0.05) in brains of D. melanogaster. Intriguingly, for the type I AA double-editing sites, the original codon encodes either Asn or Lys, and the resultant amino acid after a single editing event would be Ser/Asp/Arg/Glu. However, the resultant amino acid after double editing would always be Gly (fig. 5C). Although we only observed a few type I AA editing sites in flies, worms, and humans, we found hundreds of such sites in CDSs of cephalopods (fig. 5C), suggesting the combined editing events should be taken into account for amino acid changes in these species (an example was given in supplementary fig. S16, Supplementary Material online). For the type II AA double-editing sites, the resultant amino acids are merely decided by the editing events located in the second base of the codon (fig. 5A and C). The type III AA double-editing sites are located across two consecutive codons, with the “front” editing event causing a synonymous change (except when AUA was edited into AUG), and the “rear” one causes a nonsynonymous change (fig. 5A and C).
Fig. 5.
The nonindependent editing of the AA dinucleotides and the implications for functional prediction. (A) Four types of AA double editing sites that are significantly linked. The first three types are located in CDSs, and the fourth type is in the noncoding region. (B) Numbers (in each bar) and percentages (y-axis) of the three types of AA double editing sites that are significantly linked (adjusted P < 0.05) in each species. (C) Summary of the type I and type II PESs that are significantly linked in the pooled samples of each species. The codons containing type I editing sites originally encode Asn (AAT or AAC) or Lys (AAA or AAG). When both adenosines are edited, the encoded amino acid becomes Gly (GGA, GGC, GGG, or GGT). For codons containing type II editing sites, the amino acids encoded are merely decided by whether the front editing sites are edited or not. (D) The triplet centered with the focal editing sites based on our previous study (Duan et al. 2017). (E) The differences in frequency (Δf) of “AG” (fAG) and “GA” (fGA) haplotypes among all the significantly linked editing events in the AA dinucleotides in CDSs of 16 representative samples. In each sample, the differences in frequency of two haplotypes were compared with the Wilcoxon sign-rank test (*P < 0.05; **P < 0.01; ***P < 0.001). (F) The nonindependent editing of the AA dinucleotides. Editing of the rear adenosine (Driver) facilities editing of the front adenosine (Passenger) in an AA dinucleotide.
It is well recognized that the triplet centered with the focal editing site affect the editing efficiencies in various metazoan species (Kleinberger and Eisenberg 2010; Chen et al. 2014; Porath et al. 2014; Duan et al. 2017; Liscovitch-Brauer et al. 2017; Zhang et al. 2017), with G avoided immediately upstream and favored immediately downstream the focal editing site (fig. 5D). Thus for the AA double-editing sites, the “AG” intermediates would be easier than the “GA” intermediates to be converted into the “GG” molecules. In support of this hypothesis, we found that for the significantly linked AA double editing sites, the frequencies of the AG molecules are significantly higher than the GA molecules in CDSs of flies and cephalopods (fig. 5E). Therefore, our results revealed the nonindependent editing of the AA dinucleotide sites. Although the three types of AA double editing sites in CDSs have different functional consequences, our results suggest the editing events in the rear adenosine might be the driver and the immediately upstream adenosine might be the passenger, since the AG molecules are facilitated to be converted into the GG molecules (fig. 5F). Therefore, the editing trajectories and the linkage information of the AA double-editing sites should be considered in functional annotations of the editing events.
Discussion
By adopting the LD analysis of DNA mutations in population genetics, we detected extensive linkage of editing events in flies and cephalopods, where abundant nonsynonymous A-to-I editing events are adaptive (Garrett and Rosenthal 2012; Alon et al. 2015; Yu et al. 2016; Duan et al. 2017; Liscovitch-Brauer et al. 2017; Zhang et al. 2017). We also carried out large-scale Sanger sequencing of single cDNA clones to verify the linkage of editing events in four genes in three Drosophila species. By contrast, in worms and humans, in which the editing events are mainly in the noncoding regions (Levanon and Eisenberg 2015; Zhao et al. 2015; Goldstein et al. 2017) and no adaptive signals were detected in the nonsynonymous editing sites (Xu and Zhang 2014), we only detected modest proportions of significantly linked editing events. Interestingly, although mice are closely related to humans, the overall linkage patterns are different between mice and humans, presumably because most editing sites in human genomes are in Alu sequences that originated after the divergence between humans and mice (Levanon and Eisenberg 2015).
Since the slPESs tend to have shorter distances than the random neighboring editing sites (supplementary fig. S8, Supplementary Material online), it is possible that editing sites in CDSs have shorter distance to each other than those in noncoding regions, which would cause higher proportions of PESs to be significantly linked in Drosophila and cephalopods since they have higher proportion of editing sites in CDSs than humans or worms. However, we found the distance between two neighboring editing sites in CDSs was overall larger than that in the noncoding regions when we considered all the editing sites (supplementary fig. S17A, Supplementary Material online) or the slPESs (supplementary fig. S17B, Supplementary Material online) in a sample. Therefore, the distance between editing sites is unlikely to cause higher proportions of PESs to be significantly linked in Drosophila and cephalopods than in humans or worms. Moreover, considerably higher proportions of the slPESs were conserved between different Drosophila species or between cephalopods (fig. 2B) than between human and macaques (supplementary fig. S10B, Supplementary Material online). Altogether, our results suggest the linkage of editing events in flies and cephalopods might be associated with the adaptive proteomic changes conferred by RNA editing in these two clades.
We found that linkage constrains the evolution of the adenosine sites at the DNA level in both flies and cephalopods (fig. 3). It is possible that these linked editing sites are located in structurally accessible regions that allow ADAR to edit multiple adenosines simultaneously. These linked editing events, if selectively advantageous, would drive natural selection to maintain the optimized DNA sequence contexts during evolution at the cost of genome conservation, as previously observed in cephalopods (Liscovitch-Brauer et al. 2017). We also found linkage would cause the editing events to be more conserved between different Drosophila or different cephalopod species than the unclustered (UNC) editing sites. In contrast, we found linkage has the opposite effect on the DNA site evolution in CDSs of worms and humans. Although we found a handful of slPESs (supplementary fig. S10B, Supplementary Material online) and thousands of editing events conserved between humans and macaques (fig. 2E and F), such sites only account for a small fraction of the overwhelmingly large number of editing sites identified in humans (Ramaswami and Li 2014; Levanon and Eisenberg 2015). Furthermore, we found the CLN or CUN editing sites are usually less conserved than the UNC sites in the CDSs of worms and humans at the DNA level, in particular for the nonsynonymous editing sites. This result is congruent with previous observations that the editing targets in humans are primarily in the noncoding regions, and the majority of the nonsynonymous editing events in CDSs might be the by-products and nonadaptive (Xu and Zhang 2014). For example, the clustering of five editing events in serotonin 2 C receptor HTR2C is related to human mood, appetite, and behavior (Molineaux et al. 1989; Roth et al. 1998; Pinto et al. 2014). It was demonstrated that the mRNA isoform containing simultaneous editing events of three out of the five sites (site A, C’, and C) is more likely to be associated with suicide or mood disorder than the other isoforms (Gurevich et al. 2002). Notably, we also observed linkage of these editing events based on a limited number of NGS reads (supplementary fig. S18, Supplementary Material online). Therefore, linkage of multiple editing events in the CDSs of humans or worms might be more deleterious, which would drive the adenosine sites to evolve to avoid promiscuous editing.
A previous study suggests that the linkage of RNA editing events might be much weaker than the linkage between DNA mutations (Zhang and Xiao 2015). To compare the extent of linkage between editing sites versus that between SNPs, we retrieved the known SNPs in D. melanogaster (Grenier et al. 2015) and in humans (dbSNP 150) and detected the significantly linked SNPs in the NGS reads with the same procedure used to detect linkage of editing sites (supplementary fig. S19A, Supplementary Material online). In parallel, we also applied the same analytic procedures to detect the linkage between editing events and DNA SNPs supported by the NGS reads (supplementary fig. S19A, Supplementary Material online). Here, we conducted the analyses in human frontal gyrus that has the highest number of slPESs among all the human samples as well as in five different strains of D. melanogaster. At the FDR of 0.05, we detected thousands to tens of thousands of pairs of SNPs that showed significant linkage in a sample, and the majority of the r2 values were above 0.9 (supplementary fig. S19B and C, Supplementary Material online). By contrast, the r2 values for the slPESs were roughly 0.4 in each sample, significantly lower than those of the linked SNPs (P < 0.001 in each sample, Kolmogorov–Smirnov test; supplementary fig. S19B and C, Supplementary Material online). This difference might not be surprising since the linkage between SNPs are coded at DNA level while the linkage of editing events occurs posttranscriptionally. These comparisons also suggest that the slPESs are unlikely to be caused by the erroneous annotations of SNPs as editing sites. Interestingly, we observed prevalent significant linkage between editing events and SNPs with r2 values smaller than those of the SNP–SNP linkage but larger than those of the slPESs (supplementary fig. S19B and C, Supplementary Material online). These results are in good agreement with recent studies showing that some SNPs could influence the editing levels of neighboring sites and generate allele-specific RNA editing by changing the secondary structure or local sequence contexts in mice (Gu et al. 2016), flies (Ramaswami et al. 2015), or humans (Park et al. 2017).
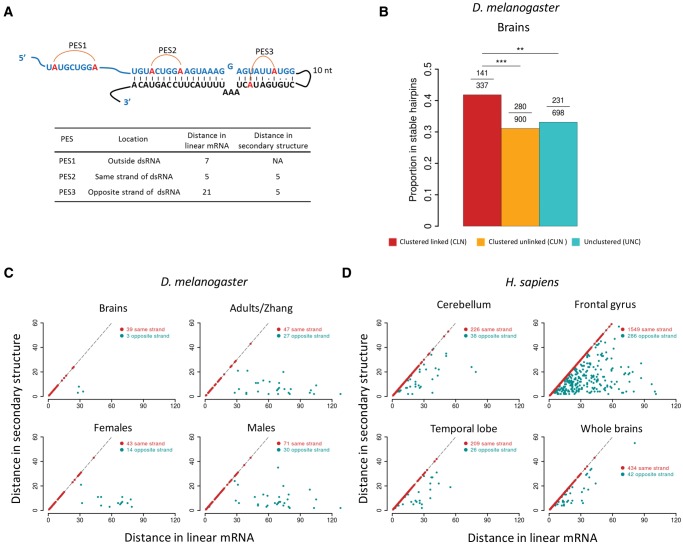
We previously found about one third of the exonic editing sites are located in stable hairpin structures (Duan et al. 2017). Here, we also found the significantly linked editing sites are over-represented in stable hairpin structures compared with the unlinked editing sites in Drosophila brains (Materials and Methods, fig. 6A and B). With the 50-bp single-end sequencing of Drosophila brains, we found ∼90% of the slPESs are located in the same sides of stable hairpin structures. Meanwhile, with the 100-bp pair-end sequencing data in adults of D. melanogaster, we found ∼40% of the significantly linked editing sites in the complementary sides of hairpin structures (fig. 6C). Hence the numbers of slPESs we detected based on the Illumina NGS reads might be conservative since the sites in the complementary sides of the hairpin structures that are edited simultaneously would not be detected if their linear distances exceed the NGS read lengths. This might be particularly true for humans because human editing events mostly happen on Alu regions, which form double strand RNAs with a stem length of ∼300 bp (Athanasiadis et al. 2004). Indeed, we found roughly 10–20% of the slPESs in humans were located on the opposite sides if they were located in the stable hairpin structures (fig. 6D). Therefore, further studies are needed to identify the editing sites that are further apart but significantly linked.
Fig. 6.
The spatial distance of two linked editing sites in a hairpin structure. (A) The scheme illustrating the distance between two linked editing sites in a hairpin structure. Examples of PESs that are outside the hairpin (PES1), in the same (PES2) or opposite sides (PES3) of a stable hairpin structure. The distances between two editing sites in the linear mRNA and the secondary structure are show in the table below. The distance in the secondary structure was calculated as the length of the shortest path between two editing sites by treating the hairpin as a graph. (B) The proportion of editing sites in brains of D. melanogaster that are located in the stable hairpins. The editing sites were divided into CLN, CUN, and UNC categories. In each category, the numbers of total editing sites and those in stable hairpins are given above the bars. Fisher’s exact tests were performed to detect the statistical differences (**P < 0.01; ***P < 0.001). (C) The linear (x-axis) and spatial (y-axis) distance (nt) between the significantly linked editing sites that were located in the same (red) or opposite (cyan) sides of hairpin structures in four samples of D. melanogaster. (D) Similar as (C) but shows the results for four human brain samples.
We have primarily focused on the pairwise linkage of editing events, whereas in qvr and NaCP60E we detected the linkage of editing events on multiple sites that were conserved in different Drosophila species (supplementary figs. S13 and S15, Supplementary Material online). Here we extended our analysis to the linkage of editing events on three editing sites. For any combination of three editing sites in a sample (we required editing level ≥0.05 at each site), we extracted the NGS reads that covered these sites and counted the “GGG” haplotype that was edited at all the three sites. We only focused on the triplets that had coverage ≥20× and compared the observed number of “GGG” haplotype to the expected one obtained with random permutations (Materials and Methods). Among all the 1,455,820 triplets supported by the NGS reads in all the samples, we in total identified 75,432 triplets of editing sites that showed significantly higher “GGG” haplotypes than the expected ones under the assumption of randomness at the FDR of 0.05 (supplementary table S7, Supplementary Material online). Notably, the proportion of significantly linked triplets (the “GGG” haplotype) is higher in flies and cephalopods than in worms or humans (supplementary fig. S20, Supplementary Material online), which is consistent with the trend observed for the pairwise linkage of editing sites (fig. 1D). Hence the linkage of editing events across multiple sites might be a feature of adaptive editing, and further studies are needed to identify more extensively linked editing events across multiple sites and elucidate their functional significance.
Our analyses on the AA double editing sites revealed the nonindependent editing of the AA dinucleotides. Under our interpretation, the rear editing event in the AA double editing sites facilitates editing of the front adenosine site (fig. 5F). Our results suggest that considering the combinatory effect of the AA double-editing events is necessary since the resultant amino acid is different from the one expected with individual editing event (type I, fig. 5A). We also found that synonymous editing events might facilitate the preceding nonsynonymous events (type II, fig. 5A), or facilitated by the downstream nonsynonymous editing events (type III, fig. 5A). Hence, the surrounding editing contexts should also be considered when studying the function of editing sites. Overall, deciphering the linkage of the editing events would help to understand the molecular mechanism underlying editing, and the linkage information would help to better identify the functionally important editing sites.
Materials and Methods
A-to-I Editing Sites in Flies, Cephalopods, Worms, Humans, Macaques, and Mice
In this study, we only considered the editing sites with editing level ≥0.05 in each sample. With this criteria, we obtained 1,935 editing sites in brains of D. melanogaster we previously characterized (eight mRNA-Seq libraries, in total 148.7 million reads), and 729 and 408 of these editing sites had orthologous sites edited (levels ≥ 0.05) in brains of D. simulans (six mRNA-Seq libraries, in total 117.2 million reads) and D. pseudoobscura (six mRNA-Seq libraries, in total 117.5 million reads), respectively (Duan et al. 2017). We also obtained 426 sites in female and 818 sites in male adults we previously characterized (Duan et al. 2017), and 1,181 sites in adults of D. melanogaster identified by Zhang et al. (Zhang et al. 2017). We compiled the RNA editing sites in cephalopods (Liscovitch-Brauer et al. 2017), worms (Zhao et al. 2015), humans (Ramaswami et al. 2013; Ramaswami and Li 2014), rhesus macaques (Chen et al. 2014) and mice (Danecek et al. 2012; Ramaswami and Li 2014) from the original studies (in each sample sites with editing level < 0.05 were filtered throughout this study). For humans, after removing the editing sites overlapping with human SNPs (dbSNP 150), we in total obtained 1,444,884 A-to-G editing sites in seven brain samples (supplementary table S1, Supplementary Material online), including 601,662 sites identified by (Ramaswami et al. 2013) and another 843,222 sites updated in the RADAR database (Ramaswami and Li 2014). We obtained 7,862 editing sites in mouse brains, including 7,065 sites identified by (Danecek et al. 2012) and another 797 sites updated in the RADAR database (Ramaswami and Li 2014). The editing sites of the four cephalopod species were based on the transcriptomes of Octopus vulgaris, Octopus bimaculoides, Doryteuthis pealeii, or Sepia oficianalis that were assembled by a previous study (Liscovitch-Brauer et al. 2017) and downloaded from www.tau.ac.il/∼elieis/squid; last accessed June 9, 2017. The functional annotations of the editing sites were parsed from the original studies or inferred with the software SnpEff version 4.3 (Cingolani et al. 2012) whenever necessary.
Processing of NGS Reads
For each of the samples above, we downloaded the raw RNA sequencing data of each study (supplementary table S1, Supplementary Material online) and employed STAR (2.4.2a) (Dobin et al. 2013) with default parameters to map all the sequencing reads to the corresponding reference genome or transcriptome. The genomic coordinates of editing sites, sequences and annotations of the genomes were based on the following assemblies: D. melanogaster (r6.04), D. simulans (r1.4), and D. pseudoobscura (r3.2) from FlyBase (www.flybase.org; last accessed October 4, 2017); human (hg19) and mouse (mm10) from UCSC Genome Browser (genome.ucsc.edu); worm (Ensembl v63) and rhesus macaque (Ensembl v89) from Ensembl Genome Browser (www.ensembl.org; last accessed October 4, 2017). In case multiple libraries of mRNA-Seq for a sample of a particular species are available, we pooled the sequencing reads together to increase the statistical power in detecting linkage of the editing events.
For each BAM sequence alignment file, we extracted all the uniquely mapped reads spanning at least two editing sites with SAMtools 1.3.1 (Li 2011) and Sam2Tsv (Pierre 2015). Soft clipping bases in the alignments were not considered, and reads spanning only one editing sites were discarded. In each sample, we required the editing sites to have sequencing coverage ≥20× and calculated the editing levels for each site with the NGS reads. For the pair-end sequencing, we assembled the reads and treated the pair-end sequences as one single read. Then for all the pairwise combinations of editing sites that were covered by the NGS reads, we calculated the frequencies of the four possible “haplotypes” and calculated the r2 and P value using the method in calculating LD (see below for details). For each pair-end library, the RNA insert size was estimated based on the paired-end read pairs that were uniquely aligned against the reference genome.
Linkage of Editing Events between Editing Sites
The calculation follows the algorithm depicted by Lewontin (Lewontin 1988). Briefly, let us suppose:
the total number of reads covering the two editing sites in a sample is N;
the four combinatory molecules for the two sites are AA, AG, GA, and GG; and the frequency is fAA, fAG, fGA, and fGG, respectively;
the frequency for the unedited (A) and edited (G) molecules for site 1 is a1 and g1 respectively, and the frequency for the unedited (A) and edited (G) molecules for site 2 is a2 and g2 respectively.
Then the LD coefficient D is calculated as D = fAA fGG − fAG fGA.
The correlation coefficient for the two editing sites is , with the χ2 statistics for testing LD written as . The significance of LD (P value) is determined by the χ2 value and df of 1 (Hill and Robertson 1968). We corrected the P values with the Benjamini and Hochberg method (Benjamini and Hochberg 1995). At the adjusted P < 0.05, we in total identified 50,800 slPESs in all the species, and the majority of them (99.15%) were mutually associative (with D > 0), and only a small number of them (433 PESs) were mutually exclusive (D < 0). We only considered the slPESs that were mutually associative throughout this study. The analyses were performed under the R environment (www.r-project.org; last accessed October 4, 2017). The haplotypes of editing events were displayed with Haploview (Barrett et al. 2005) whenever necessary.
Conservation of Editing Events and Genomic DNA Sites between Species
The orthologous sites between D. melanogaster and D. simulans, or between D. melanogaster and D. pseudoobscura were obtained from our previous study (Duan et al. 2017). The orthologous sites between H. sapiens and M. musculus or between H. sapiens and M. mulatta were determined with liftOver (Hinrichs et al. 2006) based on the pairwise genomic alignments downloaded from the UCSC Genome Browser. The procedures in aligning the CDS alignments between pairwise cephalopod species were described in Supplementary Methods.
When defining conserved editing events between species, we required the editing sites in one species (e.g., O. vulgaris) to have editing level ≥0.05 in at least one sample, and the orthologous site in the other species (e.g., O. bimaculoides) should also be edited and have editing level ≥0.05 in at least one sample. When defining the slPESs that were conserved between species, we required the PESs to have q value (adjusted P value) < 0.05 in at least one sample of one species (e.g., D. melanogaster) and a P value < 0.05 in the other species (e.g., D. simulans).
The phyloP scores for each site of D. melanogaster, C. elegans, H. sapiens, and M. musculus were obtained from UCSC Genome Browser.
The “50-nt Mapping” Approach
We performed the 50 nt mapping procedure in which the pair-end libraries were used as single-end libraries by treating the two mates of a pair-end read as two independent single-end reads. In each library, the first 50 nt of each NGS read was mapped to the reference genome or transcriptome with STAR (2.4.2a), and the obtained BAM sequence alignments were subjected to the same analytic procedures as the brain samples of Drosophila which were sequenced with the 50-bp single-end method.
The Distance of Two Linked Editing Sites in a Hairpin Structure
The stable hairpin structures harboring the editing sites in D. melanogaster were identified previously (Duan et al. 2017). Specifically, we folded the flanking sequences of each editing sites with RNALfold (Lorenz et al. 2011) to search for the stable hairpin structures (z score < −1.5, ΔG < −15 kcal/mol, and the stem length > 50 nt) that harbored the editing sites. For each slPES in humans, the sequence between the two editing sites in the slPES and 200 bp flanking sequences at each side were extracted and subjected to RNALfold to search for the stable hairpin structures with the same criteria as in Drosophila. If two editing sites are located in the same side of a hairpin structure, we assumed the spatial distance to be identical as the linear distance in the mRNA. For the editing sites located on the opposite sides of a hairpin structure, a path graph was generated for the sequence of the hairpin and base-pairings between nucleotides in the hairpin were added as new edges. Then the spatial distance of the PES was calculated as the length of the shortest path between the two sites, which was derived with NetworkX library in python (https://networkx.github.io; last accessed October 4, 2017). Therefore, the two editing sites in the opposite sides of a hairpin structure would have a shorter spatial distance than the linear distance (e.g., see fig. 6A).
Identifying Significantly Linked Events on Three Editing Sites
In each sequence alignment BAM file, we extracted the uniquely mapped NGS reads that covered any combination of three editing sites (we required editing level ≥0.05 for each site) and counted the “GGG” haplotype (edited at all of the three sites). We only considered the triplets that had a coverage ≥20× and at least one NGS read covering the “GGG” haplotype.
For a triplet, the edited and unedited alleles for each of the three sites were randomly shuffled, and the number of GGG haplotype was counted after shuffling. This procedure was repeated for 100,000 times, and the number of replicates (n) in which the observed number of GGG haplotype was no larger than the simulated ones was counted. The P value for significant linkage of a triplet was calculated as n/100,000, and the adjusted P value was calculated for all the triplets in a sample to control for false discovery rate. Significantly linked triplets were determined at adjusted P value < 0.05. We also employed similar permutation tests to estimate the significance of the linkage of editing events across multiple sites as exemplified in supplementary figures S13 and S15, Supplementary Material online.
Linkage between SNPs or between SNP-Editing Site
The editing sites in female and male adults of five strains (B12, I17, N10, T07, and ZW155) of D. melanogaster were identified previously (Duan et al. 2017), and the genome-wide SNPs of these five strains were retrieved from (Grenier et al. 2015). The SNPs in humans were downloaded from dbSNP Build 150 (http://www.ncbi.nlm.nih.gov/SNP/; last accessed September 13, 2017). We chose human frontal gyrus as a representative example since this sample has the highest number of slPESs among all the human samples. We examined the linkage between SNPs or between a SNP and an editing site with the same procedure used to detect linkage of editing sites as illustrated in supplementary figure S19A, Supplementary Material online.
Data Accession of Sanger Sequencing
The Sanger sequencing data of the four genes (nAChRbeta1, Fife, qvr, and NaCP60E) are submitted to NCBI with the accession numbers MF504162–MF506729.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by grants from the Ministry of Science and Technology of the People’s Republic of China (2016YFA0500800), National Natural Science Foundation of China (No. 31771411, 91431101 and 31571333), and the Peking-Tsinghua Center for Life Science to J.L.
References
- Alon S, Garrett SC, Levanon EY, Olson S, Graveley BR, Rosenthal JJC, Eisenberg E.. 2015. The majority of transcripts in the squid nervous system are extensively recoded by A-to-I RNA editing. eLife 4:e05198.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon S, Mor E, Vigneault F, Church GM, Locatelli F, Galeano F, Gallo A, Shomron N, Eisenberg E.. 2012. Systematic identification of edited microRNAs in the human brain. Genome Res. 22(8): 1533–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiadis A, Rich A, Maas S.. 2004. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2(12): e391.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ.. 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21(2): 263–265.http://dx.doi.org/10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- Bass BL. 2002. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 71:817–846.http://dx.doi.org/10.1146/annurev.biochem.71.110601.135501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass BL, Nishikura K, Keller W, Seeburg PH, Emeson RB, O’Connell MA, Samuel CE, Herbert A.. 1997. A standardized nomenclature for adenosine deaminases that act on RNA. RNA 3:947–949. [PMC free article] [PubMed] [Google Scholar]
- Bazak L, Haviv A, Barak M, Jacob-Hirsch J, Deng P, Zhang R, Isaacs FJ, Rechavi G, Li JB, Eisenberg E, Levanon EY.. 2014. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 24(3): 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 57:289–300. [Google Scholar]
- Blow M, Futreal PA, Wooster R, Stratton MR.. 2004. A survey of RNA editing in human brain. Genome Res. 14(12): 2379–2387.http://dx.doi.org/10.1101/gr.2951204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchert GM, Gilmore BL, Spengler RM, Xing Y, Lanier W, Bhattacharya D, Davidson BL.. 2009. Adenosine deamination in human transcripts generates novel microRNA binding sites. Hum Mol Genet. 18(24): 4801–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchumenski I, Bartok O, Ashwal-Fluss R, Pandey V, Porath HT, Levanon EY, Kadener S.. 2017. Dynamic hyper-editing underlies temperature adaptation in Drosophila. PLoS Genet. 13(7): e1006931.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmi S, Borukhov I, Levanon EY.. 2011. Identification of widespread ultra-edited human RNAs. PLoS Genet. 7(10): e1002317.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-Y, Peng Z, Zhang R, Yang X-Z, Tan BC-M, Fang H, Liu C-J, Shi M, Ye Z-Q, Zhang YE, et al. 2014. RNA editome in Rhesus Macaque shaped by purifying selection. PLoS Genet. 10:e1004274.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S-J, Shi F-Y, Liu H, Ding Y, Jiang S, Liang N, Gao G.. 2017. Accurately annotate compound effects of genetic variants using a context-sensitive framework. Nucleic Acids Res. 45(10): e82–e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM.. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly 6(2): 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Nellaker C, McIntyre RE, Buendia-Buendia JE, Bumpstead S, Ponting CP, Flint J, Durbin R, Keane TM, Adams DJ.. 2012. High levels of RNA-editing site conservation amongst 15 laboratory mouse strains. Genome Biol. 13(4): 26.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y, Dou S, Luo S, Zhang H, Lu J.. 2017. Adaptation of A-to-I RNA editing in Drosophila. PLoS Genet. 13(3): e1006648.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn CW, Giribet G, Edgecombe GD, Hejnol A.. 2014. Animal Phylogeny and Its Evolutionary Implications. Annu Rev Ecol Evol Syst. 45(1): 371..http://dx.doi.org/10.1146/annurev-ecolsys-120213-091627 [Google Scholar]
- Dupuis J, Louis T, Gauthier M, Raymond V.. 2012. Insights from honeybee (Apis mellifera) and fly (Drosophila melanogaster) nicotinic acetylcholine receptors: from genes to behavioral functions. Neurosci Biobehav Rev. 36(6): 1553–1564.http://dx.doi.org/10.1016/j.neubiorev.2012.04.003 [DOI] [PubMed] [Google Scholar]
- Flomen R, Knight J, Sham P, Kerwin R, Makoff A.. 2004. Evidence that RNA editing modulates splice site selection in the 5‐HT2C receptor gene. Nucleic Acids Res. 32(7): 2113–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garren SB, Kondaveeti Y, Duff MO, Carmichael GG.. 2015. Global analysis of mouse polyomavirus infection reveals dynamic regulation of viral and host gene expression and promiscuous viral RNA editing. PLoS Pathog. 11(9): e1005166.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett S, Rosenthal JJ.. 2012. RNA editing underlies temperature adaptation in K+ channels from polar octopuses. Science 335(6070): 848–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B, Agranat-Tamir L, Light D, Ben-Naim Zgayer O, Fishman A, Lamm AT.. 2017. A-to-I RNA editing promotes developmental stage-specific gene and lncRNA expression. Genome Res. 27(3): 462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommans WM, Mullen SP, Maas S.. 2009. RNA editing: a driving force for adaptive evolution?. BioEssays 31(10): 1137–1145.http://dx.doi.org/10.1002/bies.200900045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471(7339): 473–479.http://dx.doi.org/10.1038/nature09715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier JK, Arguello JR, Cardoso Moreira M, Gottipati S, Mohammed J, Hackett SR, Boughton R, Greenberg AJ, Clark AG.. 2015. Global diversity lines: a five-continent reference panel of sequenced Drosophila melanogaster strains. G3 5(4):593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu T, Gatti DM, Srivastava A, Snyder EM, Raghupathy N, Simecek P, Svenson KL, Dotu I, Chuang JH, Keller MP, et al. 2016. Genetic architectures of quantitative variation in RNA editing pathways. Genetics 202(2): 787.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich I, Tamir H, Arango V, Dwork AJ, Mann JJ, Schmauss C.. 2002. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron 34(3): 349–356. [DOI] [PubMed] [Google Scholar]
- Han L, Diao L, Yu S, Xu X, Li J, Zhang R, Yang Y, Werner Henrica MJ, Eterovic AK, Yuan Y, et al. 2015. The genomic landscape and clinical relevance of A-to-I RNA editing in human cancers. Cancer Cell 28(4): 515–528.http://dx.doi.org/10.1016/j.ccell.2015.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zhang J.. 2006. Toward a molecular understanding of pleiotropy. Genetics 173(4): 1885–1891.http://dx.doi.org/10.1534/genetics.106.060269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Robertson A.. 1968. Linkage disequilibrium in finite populations. Theor Appl Genet. 38(6): 226–231.http://dx.doi.org/10.1007/BF01245622 [DOI] [PubMed] [Google Scholar]
- Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, Furey TS, Harte RA, Hsu F, et al. 2006. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 34:D590–D598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopengardner B, Bhalla T, Staber C, Reenan R.. 2003. Nervous system targets of RNA editing identified by comparative genomics. Science 301(5634): 832..http://dx.doi.org/10.1126/science.1086763 [DOI] [PubMed] [Google Scholar]
- Jepson JE, Savva YA, Yokose C, Sugden AU, Sahin A, Reenan RA.. 2011. Engineered alterations in RNA editing modulate complex behavior in Drosophila: regulatory diversity of adenosine deaminase acting on RNA (ADAR) targets. J Biol Chem. 286(10): 8325–8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepson JEC, Reenan RA.. 2008. RNA editing in regulating gene expression in the brain. Biochim Biophys Acta 1779(8): 459–470.http://dx.doi.org/10.1016/j.bbagrm.2007.11.009 [DOI] [PubMed] [Google Scholar]
- Jin Y, Tian N, Cao J, Liang J, Yang Z, Lv J.. 2007. RNA editing and alternative splicing of the insect nAChR subunit alpha6 transcript: evolutionary conservation, divergence and regulation. BMC Evol Biol. 7:98.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan LP, Brindle J, Gallo A, Leroy A, Reenan RA, O’Connell MA.. 2005. Tuning of RNA editing by ADAR is required in Drosophila. EMBO J. 24(12): 2183–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan LP, Gallo A, O’Connell MA.. 2001. The many roles of an RNA editor. Nat Rev Genet. 2(11): 869–878. [DOI] [PubMed] [Google Scholar]
- Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A.. 2004. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 14(9): 1719–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinberger Y, Eisenberg E.. 2010. Large-scale analysis of structural, sequence and thermodynamic characteristics of A-to-I RNA editing sites in human Alu repeats. BMC Genomics 11:453.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klironomos FD, Berg J, Collins S.. 2013. How epigenetic mutations can affect genetic evolution: model and mechanism. BioEssays 35(6): 571–578.http://dx.doi.org/10.1002/bies.201200169 [DOI] [PubMed] [Google Scholar]
- Knisbacher BA, Levanon EY.. 2015. DNA and RNA editing of retrotransposons accelerate mammalian genome evolution. Ann N Y Acad Sci. 1341(1): 115–125. [DOI] [PubMed] [Google Scholar]
- Koh K, Joiner WJ, Wu MN, Yue Z, Smith CJ, Sehgal A.. 2008. Identification of SLEEPLESS, a sleep-promoting factor. Science 321(5887): 372–376.http://dx.doi.org/10.1126/science.1155942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurmangaliyev YZ, Ali S, Nuzhdin SV.. 2016. Genetic determinants of RNA editing levels of ADAR targets in Drosophila melanogaster. G3 6:391–396.http://dx.doi.org/10.1534/g3.115.024471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev-Maor G, Sorek R, Levanon EY, Paz N, Eisenberg E, Ast G.. 2007. RNA-editing-mediated exon evolution. Genome Biol. 8(2): R29.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon EY, Eisenberg E.. 2015. Does RNA editing compensate for Alu invasion of the primate genome?. Bioessays 37(2): 175–181.http://dx.doi.org/10.1002/bies.201400163 [DOI] [PubMed] [Google Scholar]
- Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. 2004. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol. 22(8): 1001–1005. [DOI] [PubMed] [Google Scholar]
- Lewontin RC. 1988. On measures of gametic disequilibrium. Genetics 120(3): 849–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27(21): 2987–2993.http://dx.doi.org/10.1093/bioinformatics/btr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Church GM.. 2013. Deciphering the functions and regulation of brain-enriched A-to-I RNA editing. Nat Neurosci. 16(11): 1518–1522.http://dx.doi.org/10.1038/nn.3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Levanon EY, Yoon JK, Aach J, Xie B, Leproust E, Zhang K, Gao Y, Church GM.. 2009. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science 324(5931): 1210–1213. [DOI] [PubMed] [Google Scholar]
- Li Q, Wang Z, Lian J, Schiøtt M, Jin L, Zhang P, Zhang Y, Nygaard S, Peng Z, Zhou Y, et al. 2014. Caste-specific RNA editomes in the leaf-cutting ant Acromyrmex echinatior. Nat Commun. 5:4943.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Landweber LF.. 2007. Hypothesis: RNA editing of microRNA target sites in humans? RNA 13:463–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscovitch-Brauer N, Alon S, Porath HT, Elstein B, Unger R, Ziv T, Admon A, Levanon EY, Rosenthal JJ, Eisenberg E.. 2017. Trade-off between transcriptome plasticity and genome evolution in cephalopods. Cell 169(2): 191–202.e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Honer Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL.. 2011. ViennaRNA Package 2.0. Algorithms Mol Biol. 6:26.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin P, Xiong J, Liu X, Yan Z, Zhang X, Li M, He L, Somel M, Yuan Y, Phoebe Chen YP, Li N, Hu Y, Fu N, Ning Z, Zeng R, Yang H, Chen W, Gelfand M, Khaitovich P.. 2013. Widespread splicing changes in human brain development and aging. Mol Syst Biol 9:633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazloomian A, Meyer IM.. 2015. Genome-wide identification and characterization of tissue-specific RNA editing events in D. melanogaster and their potential role in regulating alternative splicing. RNA Bio. 12(12): 1391–1401.http://dx.doi.org/10.1080/15476286.2015.1107703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molineaux SM, Jessell TM, Axel R, Julius D.. 1989. 5-HT1c receptor is a prominent serotonin receptor subtype in the central nervous system. Proc Natl Acad Sci U S A. 86(17): 6793–6797.http://dx.doi.org/10.1073/pnas.86.17.6793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K. 2006. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat Rev Mol Cell Biol. 7(12): 919–931.http://dx.doi.org/10.1038/nrm2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K. 2010. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 79:321–349.http://dx.doi.org/10.1146/annurev-biochem-060208-105251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K. 2016. A-to-I editing of coding and non-coding RNAs by ADARs. Nat Rev Mol Cell Biol. 17(2): 83–96.http://dx.doi.org/10.1038/nrm.2015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K, Yoo C, Kim U, Murray JM, Estes PA, Cash FE, Liebhaber SA.. 1991. Substrate specificity of the dsRNA unwinding/modifying activity. EMBO J. 10(11): 3523–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osenberg S, Dominissini D, Rechavi G, Eisenberg E.. 2009. Widespread cleavage of A-to-I hyperediting substrates. RNA 15(9): 1632–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino MJ, Keegan LP, O’Connell MA, Reenan RA.. 2000a. dADAR, a Drosophila double-stranded RNA-specific adenosine deaminase is highly developmentally regulated and is itself a target for RNA editing. RNA 6(7):1004–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino MJ, Keegan LP, O’Connell MA, Reenan RA.. 2000b. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell 102:437–449. [DOI] [PubMed] [Google Scholar]
- Park E, Guo J, Shen S, Demirdjian L, Wu YN, Lin L, Xing Y.. 2017. Population and allelic variation of A-to-I RNA editing in human transcriptomes. Genome Biol. 18(1): 143.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschen W, Hedreen JC, Ross CA.. 1994. RNA editing of the glutamate receptor subunits GluR2 and GluR6 in human brain tissue. J Neurochem. 63(5): 1596–1602. [DOI] [PubMed] [Google Scholar]
- Paul MS, Bass BL.. 1998. Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. EMBO J 17(4): 1120–1127.http://dx.doi.org/10.1093/emboj/17.4.1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Yaacov N, Bazak L, Buchumenski L, Porath HT, Danan-Gotthold M, Knisbacher BA, Eisenberg E, Levanon EY.. 2015. Elevated RNA editing activity is a major contributor to transcriptomic diversity in tumors. Cell Rep. 13(2): 267–276. [DOI] [PubMed] [Google Scholar]
- Peng Z, Cheng Y, Tan BC, Kang L, Tian Z, Zhu Y, Zhang W, Liang Y, Hu X, Tan X, et al. 2012. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat Biotechnol. 30(3): 253–260. [DOI] [PubMed] [Google Scholar]
- Peterson KJ, Lyons JB, Nowak KS, Takacs CM, Wargo MJ, McPeek MA.. 2004. Estimating metazoan divergence times with a molecular clock. Proc Natl Acad Sci U S A. 101(17): 6536–6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picardi E, D’Erchia AM, Lo Giudice C, Pesole G.. 2017. REDIportal: a comprehensive database of A-to-I RNA editing events in humans. Nucleic Acids Res. 45(D1): D750–D757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picardi E, Manzari C, Mastropasqua F, Aiello I, D’Erchia AM, Pesole G.. 2015. Profiling RNA editing in human tissues: towards the inosinome Atlas. Sci Rep. 5:14941.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre L. 2015. JVarkit: java-based utilities for Bioinformatics. doi: 10.6084/m9.figshare.1425030.
- Pinto Y, Cohen HY, Levanon EY.. 2014. Mammalian conserved ADAR targets comprise only a small fragment of the human editosome. Genome Biol. 15(1): R5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson AG, Bass BL.. 1994. Preferential selection of adenosines for modification by double-stranded RNA adenosine deaminase. EMBO J. 13(23): 5701–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porath HT, Carmi S, Levanon EY.. 2014. A genome-wide map of hyper-edited RNA reveals numerous new sites. Nat Commun. 5:4726.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porath HT, Schaffer AA, Kaniewska P, Alon S, Eisenberg E, Rosenthal J, Levanon EY, Levy O.. 2017. A-to-I RNA editing in the earliest-diverging eumetazoan phyla. Mol Biol Evol. 34(8): 1890–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL.. 2005. Regulating gene expression through RNA nuclear retention. Cell 123(2): 249–263. [DOI] [PubMed] [Google Scholar]
- Qian W, Ma D, Xiao C, Wang Z, Zhang J.. 2012. The genomic landscape and evolutionary resolution of antagonistic pleiotropy in yeast. Cell Rep. 2(5): 1399–1410.http://dx.doi.org/10.1016/j.celrep.2012.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami G, Deng P, Zhang R, Anna Carbone M, Mackay TFC, Billy Li J.. 2015. Genetic mapping uncovers cis-regulatory landscape of RNA editing. Nat Commun. 6:8194.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami G, Li JB.. 2016. Identification of human RNA editing sites: A historical perspective. Methods 107:42–47.http://dx.doi.org/10.1016/j.ymeth.2016.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami G, Li JB.. 2014. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 42(Database issue): D109–D113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami G, Zhang R, Piskol R, Keegan LP, Deng P, O’Connell MA, Li JB.. 2013. Identifying RNA editing sites using RNA sequencing data alone. Nat Methods 10(2): 128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reenan RA. 2005. Molecular determinants and guided evolution of species-specific RNA editing. Nature 434(7031): 409–413.http://dx.doi.org/10.1038/nature03364 [DOI] [PubMed] [Google Scholar]
- Rodriguez J, Menet JS, Rosbash M.. 2012. Nascent-seq indicates widespread cotranscriptional RNA editing in Drosophila. Mol Cell 47(1): 27–37.http://dx.doi.org/10.1016/j.molcel.2012.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal JJC. 2015. The emerging role of RNA editing in plasticity. J Exp Biol. 218(Pt 12): 1812.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth BL, Willins DL, Kristiansen K, Kroeze WK.. 1998. 5-Hydroxytryptamine2-family receptors (5-hydroxytryptamine2A, 5-hydroxytryptamine2B, 5-hydroxytryptamine2C): where structure meets function. Pharmacol Ther. 79(3): 231–257.http://dx.doi.org/10.1016/S0163-7258(98)00019-9 [DOI] [PubMed] [Google Scholar]
- Rueter SM, Dawson TR, Emeson RB.. 1999. Regulation of alternative splicing by RNA editing. Nature 399(6731): 75–80.http://dx.doi.org/10.1038/19992 [DOI] [PubMed] [Google Scholar]
- Sakurai M, Ueda H, Yano T, Okada S, Terajima H, Mitsuyama T, Toyoda A, Fujiyama A, Kawabata H, Suzuki T.. 2014. A biochemical landscape of A-to-I RNA editing in the human brain transcriptome. Genome Res. 24(3): 522–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattelle DB, Jones AK, Sattelle BM, Matsuda K, Reenan R, Biggin PC.. 2005. Edit, cut and paste in the nicotinic acetylcholine receptor gene family of Drosophila melanogaster. Bioessays 27(4): 366–376. [DOI] [PubMed] [Google Scholar]
- Savva YA, Jepson JE, Sahin A, Sugden AU, Dorsky JS, Alpert L, Lawrence C, Reenan RA.. 2012. Auto-regulatory RNA editing fine-tunes mRNA re-coding and complex behaviour in Drosophila. Nat Commun. 3:790.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savva YA, Laurent GS, Reenan RA.. 2016. Genome-wide analysis of A-to-I RNA editing In: Dassi E, editor. Post-transcriptional gene regulation. New York: Springer; p. 255–268. [DOI] [PubMed] [Google Scholar]
- Savva YA, Rieder LE, Reenan RA.. 2012b. The ADAR protein family. Genome Biol. 13:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scadden ADJ. 2005. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat Struct Mol Biol. 12(6): 489–496.http://dx.doi.org/10.1038/nsmb936 [DOI] [PubMed] [Google Scholar]
- Scadden ADJ, Smith CWJ.. 2001. RNAi is antagonized by A → I hyper-editing. EMBO Rep. 2(12): 1107–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scadden ADJ, Smith CWJ.. 2001. Specific cleavage of hyper-edited dsRNAs. EMBO J. 20(15): 4243–4252.http://dx.doi.org/10.1093/emboj/20.15.4243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeburg PH. 2002. A-to-I editing: new and old sites, functions and speculations. Neuron 35(1): 17–20.http://dx.doi.org/10.1016/S0896-6273(02)00760-2 [DOI] [PubMed] [Google Scholar]
- Slatkin M. 2008. Linkage disequilibrium—understanding the evolutionary past and mapping the medical future. Nat Rev Genet. 9(6): 477–485.http://dx.doi.org/10.1038/nrg2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B, Köhler M, Sprengel R, Seeburg PH.. 1991. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67(1): 11–19. [DOI] [PubMed] [Google Scholar]
- St Laurent G, Tackett MR, Nechkin S, Shtokalo D, Antonets D, Savva YA, Maloney R, Kapranov P, Lawrence CE, Reenan RA.. 2013. Genome-wide analysis of A-to-I RNA editing by single-molecule sequencing in Drosophila. Nat Struct Mol Biol. 20(11): 1333–1339. [DOI] [PubMed] [Google Scholar]
- Telford MJ. 2006. Animal phylogeny. Curr Biol. 16(23): R981–R985. [DOI] [PubMed] [Google Scholar]
- Wagner GP, Zhang J.. 2011. The pleiotropic structure of the genotype-phenotype map: the evolvability of complex organisms. Nat Rev Genet. 12(3): 204–213.http://dx.doi.org/10.1038/nrg2949 [DOI] [PubMed] [Google Scholar]
- Wahba AJ, Basilio C, Speyer JF, Lengyel P, Miller RS, Ochoa AS.. 1962. Synthetic polynucleotides and the amino acid code. VI. Proc Natl Acad Sci U S A. 48:1683–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xu X, Yu S, Kang KJ, Zhou Z, Han L, Tsang YH, Li J, Chen H, Mangala LS, et al. 2017. Systematic characterization of A-to-I RNA editing hotspots in microRNAs across human cancers. Genome Res. 27(7):1112–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Zhang J.. 2014. Human coding RNA editing is generally nonadaptive. Proc Natl Acad Sci U S A. 111(10): 3769–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Zhang J.. 2015. In search of beneficial coding RNA editing. Mol Biol Evol. 32(2): 536–541.http://dx.doi.org/10.1093/molbev/msu314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X-Z, Chen J-Y, Liu C-J, Peng J, Wee YR, Han X, Wang C, Zhong X, Shen QS, Liu H, et al. 2015. Selectively constrained RNA editing regulation crosstalks with piRNA biogenesis in primates. Mol Biol Evol. 32:3143–3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Zhou H, Kong Y, Pan B, Chen L, Wang H, Hao P, Li X.. 2016. The landscape of A-to-I RNA editome is shaped by both positive and purifying selection. PLoS Genet. 12(7): e1006191.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Xiao X.. 2015. Genome sequence-independent identification of RNA editing sites. Nat Methods 12(4): 347–350.http://dx.doi.org/10.1038/nmeth.3314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Deng P, Jacobson D, Li JB.. 2017. Evolutionary analysis reveals regulatory and functional landscape of coding and non-coding RNA editing. PLoS Genet. 13(2): e1006563.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Carmichael GG.. 2001. The fate of dsRNA in the nucleus: A p54(nrb)-containing complex retention of promiscuously mediates the nuclear A-to-I edited RNAs. Cell 106(4): 465–475.http://dx.doi.org/10.1016/S0092-8674(01)00466-4 [DOI] [PubMed] [Google Scholar]
- Zhao HQ, Zhang P, Gao H, He X, Dou Y, Huang AY, Liu XM, Ye AY, Dong MQ, Wei L.. 2015. Profiling the RNA editomes of wild-type C. elegans and ADAR mutants. Genome Res. 25(1): 66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinshteyn B, Nishikura K.. 2009. Adenosine-to-inosine RNA editing. Wiley Interdiscip. Rev. 1(2): 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.