Abstract
Understanding the structural basis for evolutionary changes in protein function is central to molecular evolutionary biology and can help determine the extent to which functional convergence occurs through similar or different structural mechanisms. Here, we combine ancestral sequence reconstruction with functional characterization and structural modeling to directly examine the evolution of sequence-structure-function across the early differentiation of animal and plant Dicer/DCL proteins, which perform the first molecular step in RNA interference by identifying target RNAs and processing them into short interfering products. We found that ancestral Dicer/DCL proteins evolved similar increases in RNA target affinities as they diverged independently in animal and plant lineages. In both cases, increases in RNA target affinities were associated with sequence changes that anchored the RNA’s 5′phosphate, but the structural bases for 5′phosphate recognition were different in animal versus plant lineages. These results highlight how molecular-functional evolutionary convergence can derive from the evolution of unique protein structures implementing similar biochemical mechanisms.
Keywords: ancestral sequence reconstruction, RNA interference, Dicer evolution
Introduction
Characterizing how protein function evolves over long timespans can inform our understanding of the structural basis for functional differentiation and impact the development of evolutionary theory (Lunzer et al. 2005; Dean and Thornton 2007; Bridgham et al. 2008; Hobbs et al. 2012). Mechanistic studies of functional evolution have demonstrated how historical changes in protein sequence can alter molecular structure, resulting in qualitative and quantitative shifts in ligand preference (Bridgham et al. 2009, 2014; Kratzer et al. 2014; Pugh et al. 2016). However, deriving generalizable information about the evolution of sequence-structure-function requires characterizing a large number of diverse protein families interacting with a variety of ligands. Studies examining functional evolution using modeled—rather than empirically determined—structures may be particularly useful, as they can potentially examine a larger number of functional shifts and could guide future structure-determination efforts by identifying cases in which the structural basis for molecular function may have changed.
Dicer (also called “Dicer-like” or “DCL” in plants) is an enzyme that participates in RNA interference (RNAi) by identifying double-stranded RNA targets and cutting them to specific lengths, producing short RNA molecules that can be loaded onto the RNA-induced silencing complex (RISC) to regulate complementary RNAs (Bernstein et al. 2001; Ketting et al. 2001; Carrington and Ambros 2003; Jaskiewicz and Filipowicz 2008). The Dicer protein family spans Eukarya and exhibits a large degree of diversity in domain architecture and molecular function (Mukherjee et al. 2013). Dicers and DCLs from different model species have been shown to exhibit marked differences in RNA target preference, length of short RNA products and specific interaction partners (Lee et al. 2004; Hiraguri et al. 2005; Lee et al. 2013; Bologna and Voinnet 2014). Although this functional diversity appears important for determining the efficiency and specificity of extant RNAi pathways, we currently know very little about how Dicer/DCL function evolved.
Dicer plays a critical role in the first molecular step of RNA interference, recognizing the RNA target. Dicers from a variety of model organisms can recognize a wide array of double-stranded RNA molecules bearing various structural features on their 5′ and 3′ ends (Meister and Tuschl 2004; Vermeulen et al. 2005; Brodersen and Voinnet 2006). In humans, initial RNA recognition appears primarily facilitated by an extended Dicer PAZ domain, termed the Platform + PAZ + Connector (hereafter, PPC) (Park et al. 2011; Tian et al. 2014). Structural studies have found that the human Dicer PPC domain forms two primary RNA-binding pockets, one anchoring the 5′phosphate of typical microRNAs and the other anchoring the 3′UU overhang (Tian et al. 2014). The Dicer protein from Giardia lamblia lacks much of the Platform and Connector subdomains and appears to only bind the 3′ end of its RNA target (Macrae et al. 2006). Our previous work suggested that the human Dicer 5′phosphate pocket likely originated early in the animal lineage (Mukherjee et al. 2013), but the molecular-evolutionary basis for Dicer’s RNA target recognition has not been investigated mechanistically.
Here, we use a combination of phylogenetic analysis, ancestral sequence resurrection, structural modeling and molecular binding kinetics to characterize how Dicer’s RNA target recognition evolved across the protein family. We find evidence for adaptively driven protein-coding changes in the Dicer Platform + PAZ + Connector domain as the family diversified independently in early animal and plant lineages. Although these changes were not the same, they appear to have led to similar increases in RNA target affinity in animals and plants, suggesting that long-term optimization of RNA-binding function may have occurred convergently via unique structural mechanisms that improved recognition of the RNA target’s 5′phosphate. These findings explicitly link an important aspect of Dicer molecular-functional diversity to specific evolutionary events and suggest that at least some aspects of Dicer function have experienced long-term optimization independently in major eukaryote lineages, possibly due to selection pressure to increase RNAi efficiency.
Results and Discussion
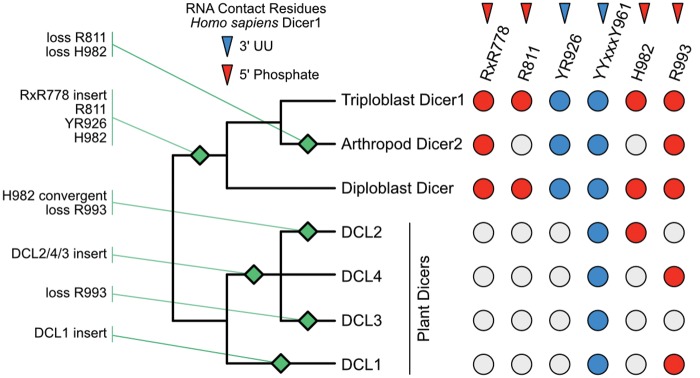
To begin examining the evolution of animal and plant Dicers, we identified full-length Dicer protein sequences from the NCBI nr database, aligned these sequences using a variety of methods and reconstructed a consensus maximum-likelihood phylogeny by combining supermatrix and supertree approaches (see Materials and Methods; all alignments and phylogenies are available at http://purl.org/phylo/treebase/phylows/study/TB2:S20991; last accessed September 22, 2017; NCBI sequence accessions are available in supplementary File IDMAP.txt, Supplementary Material online). We found that Dicer proteins separated into distinct taxonomically defined groups from protozoa, fungi, animals and plants in the consensus tree (fig. 1, supplementary fig. S1, Supplementary Material online). Plant Dicers (aka, “Dicer-like” or DCL1-4) were monophyletic with >0.99 SH-like aLRT support, depending on the alignment and phylogenetic inference strategy. Within the plant Dicer clade, the major DCL groups (DCL1, DCL2, DCL3, and DCL4) were each recovered with maximal support (supplementary fig. S1, Supplementary Material online). All major DCL groups contained monocot and eudicot representatives as well as examples from mosses, suggesting that all the major Dicer duplications occurred prior to the emergence of vascular plants. In our analysis, DCL2 and DCL4 were sisters (support > 0.98), and DCLs 2, 4, and 3 formed a monophyletic group excluding DCL1 (support > 0.97), which was basal to the other plant DCLs.
Fig. 1.
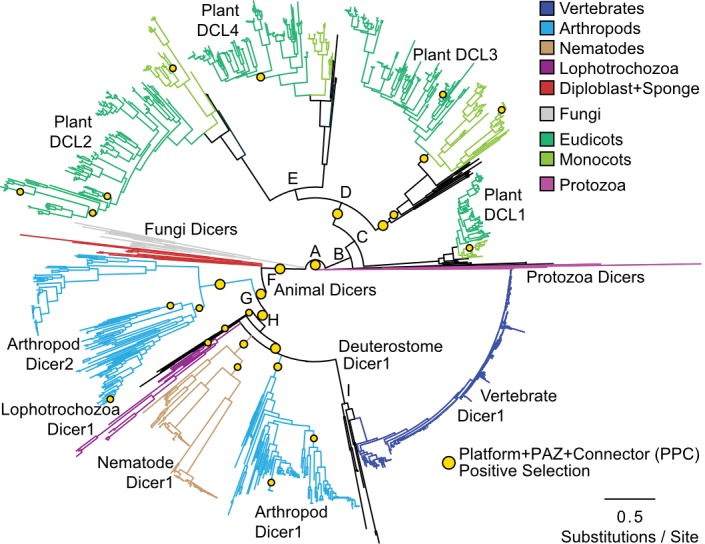
The Dicer protein family diversified independently in early animal and plant lineages, coincident with adaptive protein-coding changes in the Platform + PAZ + Connector (PPC) domain. We show a consensus phylogeny of Dicer and plant Dicer-like (DCL) proteins, inferred using a variety of alignment and tree-inference strategies (see Materials and Methods). Major taxonomic groups are indicated by branch color, and branches are scaled to the average number of substitutions/site across full-length sequence alignments. Support for key nodes (letters A–I) is indicated in supplementary fig. S1, Supplementary Material online. Protein-coding adaptation specific to the Platform + PAZ + Connector (PPC) domain was inferred using a branch-sites test (see Materials and Methods). Yellow circles indicate significant support for adaptive protein-coding changes on the indicated branch (P < 0.05 after correction for multiple tests). Protein-coding adaptation in other domains is shown in supplementary fig. S2, Supplementary Material online.
Animal Dicers were also monophyletic with >0.99 SH-like aLRT (fig. 1, supplementary fig. S1, Supplementary Material online). Within the animal Dicer clade, a group of arthropod Dicers (Dicer2) fell basal to the major Dicer1 clade from triploblasts, including Dicer1 sequences from all major protostome and deuterostome lineages (support > 0.99). Arthropod Dicer2 was distinct from the basal clade of diploblast Dicers, including sequences from cnidarians and ctenophores as well as poriferans (support > 0.89). Although multiple lineage-specific Dicer duplications have been observed among poriferan and diploblast lineages (de Jong et al. 2009), in this study we will use “Dicer2” to refer to the lineage of arthropod-specific Dicer2 sequences. The major animal taxonomic divisions (Lophotrochozoa, Nematoda, Arthropoda, and Deuterostomia) were recovered in the Dicer1 group (support > 0.92), although the branching pattern among these taxa was unresolved.
We calculated statistical clade support using SH-like aLRT scores, which are typically higher than bootstrap proportions but have been shown to exhibit low false-positive rates above ∼0.8 (Anisimova et al. 2011; Simmons and Norton 2014). Our consensus phylogeny is generally congruent with previous investigations of Dicer evolutionary history (de Jong et al. 2009; Mukherjee et al. 2013; Gao et al. 2014). Although researchers have speculated that the two arthropod Dicers may have arisen via an arthropod-specific duplication (de Jong et al. 2009), this parsimonious conclusion has never been supported by phylogenetic analysis. In the current analysis, constraining Dicer1 and Dicer2 as an arthropod-specific duplication resulted in a likelihood that was >213.42 log-units worse than the consensus tree, depending on the alignment. This corresponds to the arthropod-specific duplication tree being >4.87e92 times less likely than the consensus phylogeny (AU test P < 0.0057).
Although the animal phylogeny is not known with certainty, the inferred consensus Dicer tree implies that arthropod Dicer2 must have been lost independently from deuterostomes, lophotrochozoans, and nematodes, while being maintained in the stem lineage that gave rise to arthropods. Alternatively, the placement of arthropod Dicer2 basal to the main Dicer1 clade could be a case of long-branch attraction, as Dicer2 branch lengths are generally longer than those of Dicer1 and more similar to the long branches of diploblast and fungi Dicers. Although it is impossible to completely rule out phylogenetic artifacts, our previous analysis of Dicer evolutionary history examined a number of factors contributing to potential long-branch attraction and could find no evidence for such artifacts (Mukherjee et al. 2013). The addition of new sequence data, alignments and tree inference strategies in the current analysis further reinforced the major features of our previously inferred phylogeny, arguing in favor of its general robustness. Our previous studies suggest that Argonaute (AGO) and double-stranded RNA-binding protein (DRB) phylogenies exhibit similar patterns of duplications and losses as our Dicer tree (Mukherjee et al. 2013; Dias et al. 2017), suggesting a general model in which major components of the RNA interference pathway may have duplicated in early animals, with the duplicate pathway being retained only in the arthropod lineage. Of course, new sequence data or major advances in phylogenetic methods could alter our current view of Dicer evolutionary history.
Protein-Coding Adaptation May Have Repeatedly Affected Dicer Functional Domains Anchoring the RNA Target
The core catalytic domain architecture of the Dicer protein consists of a PAZ domain that anchors the end of a double-stranded RNA target molecule, followed by twin RIBOc (aka, “RNase III”) domains that cut the RNA backbone. This “catalytic core” is sufficient for basic Dicer function and constitutes the full-length protein in some species (Macrae et al. 2006). Human Dicer encodes an extended PAZ domain, termed the Platform + PAZ + Connector (PPC), which anchors both the 5′ and 3′ ends of the dsRNA target and is essential for proper RNA processing in humans (Park et al. 2011; Tian et al. 2014).
We identified the extended Platform + PAZ + Connector (PPC) domain in our multiple sequence alignments, using structural information from human Dicer1 PPC as a guide (Tian et al. 2014). Although the PPC is too short to reconstruct a completely reliable phylogeny, we did find that trees reconstructed using only PPC domain alignments were congruent with the consensus Dicer protein family tree (fig. 1), arguing that domain-shuffling or other complex evolutionary events are unlikely to have played a major role in PPC domain evolution (AU test P > 0.38). Other functional domains were identified by sequence search of the NCBI Conserved Domain Database (see Materials and Methods, supplementary File DOMANNOT.csv.txt, Supplementary Material online)
Our previous work suggested that the Dicer PAZ domain was a strong target for adaptive protein-coding evolution (Mukherjee et al. 2013). Consistent with this previous finding, we found significant support for adaptive protein-coding changes in the PPC and PAZ domains along early branches in animal and plant Dicer lineages (fig. 1, supplementary fig. S2, Supplementary Material online; see Materials and Methods for details). The HelicC domain was often identified as exhibiting adaptive protein-coding changes across the animal lineage but was less frequently a target for protein-coding adaptation in plants, which tended to exhibit adaptive changes in the RIBOc and C-terminal dsrm domains (supplementary fig. S2, Supplementary Material online). Although the Dicer protein binding domain (PBD) was a target for recurrent protein-coding adaptation across animals (supplementary fig. S2, Supplementary Material online), this domain is not present in plant or other Dicer sequences (see supplementary File DOMANNOT.csv.txt, Supplementary Material online). Together, these results suggest that the Dicer PPC domain likely experienced adaptive evolutionary pressures driving changes in protein sequence early in animal and plant evolutionary history, with adaptive changes in other domains being largely confined to specific Dicer/DCL lineages.
The branch-sites test we used to identify protein-coding adaptation is generally considered robust (Zhang et al. 2005; Yang and dos Reis 2011; Gharib and Robinson-Rechavi 2013; Lu and Guindon 2014). We further employed a conservative approach that controlled the overall false-discovery rate (FDR) and identified branches exhibiting protein-coding adaptation only when significance exceeded a specified threshold (P < 0.05 after FDR correction) using three separate sequence alignments (see Materials and Methods for details). However, concerns have been raised that the branch-sites test may be unreliable under some circumstances (Suzuki 2008; Nozawa et al. 2009). To examine the potential robustness of the branch-sites test in this case, we simulated codon sequence data along the Dicer consensus phylogeny (fig. 1), with branch lengths and other model parameters estimated from the empirical coding-sequence data. We found that the rate of false-positive inferences of protein-coding adaptation was below 0.05 when codon sequences were simulated under a neutral model and even lower when stabilizing selection was imposed at the protein level along the branches not being tested (supplementary fig. S3, Supplementary Material online). Although these simulations cannot capture the complexity of the actual evolutionary process, the do suggest that the basic evolutionary dynamics of the Dicer protein family do not appear to interfere with the reliability of the branch-sites test.
That we identified different patterns of adaptive protein-coding changes in different Dicer functional domains (see supplementary fig. S2, Supplementary Material online) further supports the general reliability of our results, as biases inherent to the statistical approach are expected to exert a similar impact across all functional domains. Nonetheless, we remain cautious in our interpretation of the branch-sites test results, as other changes in evolutionary dynamics could be misinterpreted as protein-coding adaptation by the statistical test. We do feel our results suggest that the PPC domain, in particular, appears to have experienced unique long-term evolutionary pressures across animal and plant lineages, in contrast to other Dicer functional domains, although these evolutionary pressures may not have been adaptive and may not have always occurred on the specific branches identified in our analysis.
Residues Anchoring the Target RNA’s 5′phosphate Are Animal-Specific, While 3′-Anchoring Residues Are Conserved across Animals and Plants
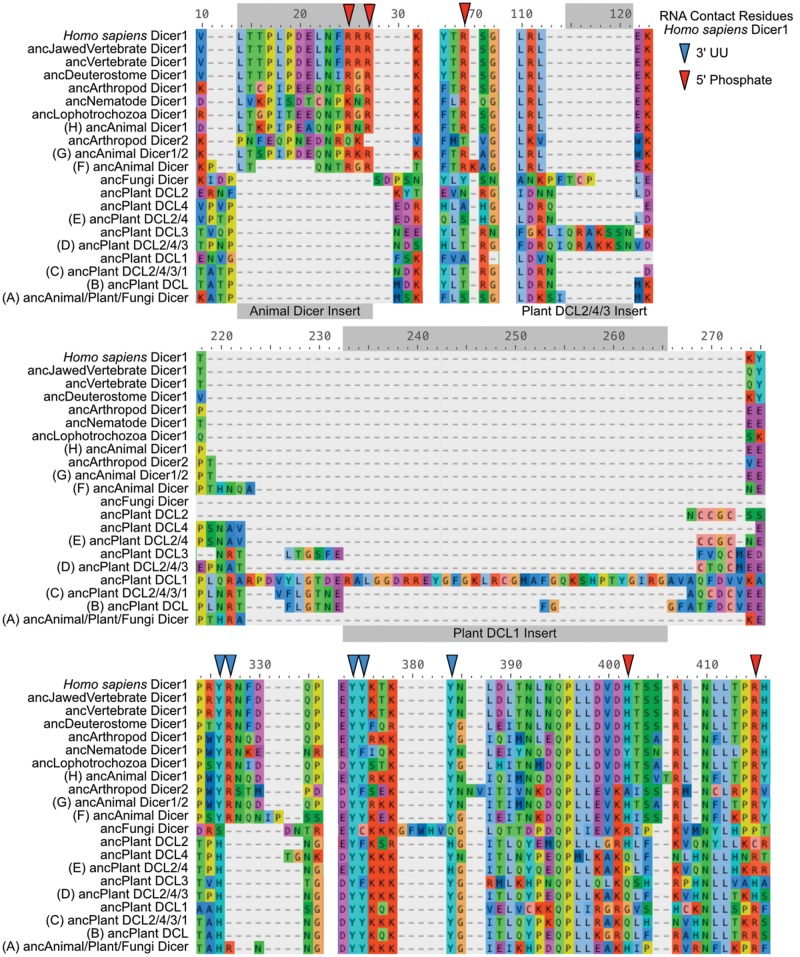
We found that much of the PPC domain was conserved across animal, plant and fungi Dicers, although many of the 5′-anchoring residues identified in studies of human Dicer1 were not conserved outside the animal Dicer1 clade (fig. 2, supplementary fig. S4, Supplementary Material online). For example, an RxR778 motif in the N-terminal Platform domain thought to contribute to anchoring the RNA’s 5′phosphate (Tian et al. 2014) is conserved across vertebrate, arthropod and mollusc/annelid Dicer1 PPCs but is not found outside this group, including a change to KxR in nematode Dicer1 (supplementary fig. S4, Supplementary Material online, alignment positions 21–23). Although some animal Dicer2 and diploblast Dicers have sequence that aligns to the RxR778 motif, this motif appears to be part of a ∼10-residue insertion that is not present in any plant or fungi Dicer sequences. Downstream of this animal Dicer1 RxR778 motif, another key 5′phosphate contact residue (R811, position 57 in supplementary fig. S4, Supplementary Material online) is also only conserved across animal Dicer1, although it is present in the diploblast, Hydra vulgaris. We additionally note the presence of two plant-specific inserts in the Dicer Platform domain, a short one in DCL3 (spanning positions ∼106–117 in supplementary fig. S4, Supplementary Material online), and a longer one in DCL1 (positions ∼187–231).
Fig. 2.
Dicer Platform + PAZ + Connector (PPC) domain evolved through gains and losses of RNA contact residues in animals and plants. We plot a schematic of the key events in Dicer Platform + PAZ + Connector (PPC) domain evolution, inferred by examination of protein sequence alignment (supplementary fig. S4, Supplementary Material online) and ancestral reconstruction (fig. 3, supplementary figs. S7–S9, Supplementary Material online). A schematic of the Dicer protein family phylogeny is shown (see fig. 1). Sequence motifs thought to anchor the RNA’s 3′UU overhang (blue) and 5′ phosphate (red) in human Dicer1 are indicated (Tian et al. 2014); colored circles indicate the presence of the motif in extant sequences from each Dicer/DCL group, whereas gray circles indicate absence of the motif. Inferred gains and losses of specified sequence features are indicated by annotated green diamonds.
Within the Dicer PAZ domain, a 5′phosphate-binding H982 (position 336 in supplementary fig. S4, Supplementary Material online) is only found in animal Dicer1 sequences—although it is not strictly conserved in all animal Dicer1s—and in some plant DCL2 sequences, suggesting a potential case of molecular-evolutionary convergence (fig. 2). The nearby R993 5′-contact residue (position 347 in supplementary fig. S4, Supplementary Material online) is found across many animal and plant (particularly DCL1,4) sequences, suggesting it may have arisen early and was lost in some Dicer lineages.
In contrast to the general lack of conservation in 5′phosphate-binding residues, anchoring of the RNA’s 3′UU overhang appears more strongly conserved across animal and plant Dicer PAZ domains (Tian et al. 2014). A critical 3′UU-anchoring motif (YYxxxY961) is broadly conserved across animal and plant Dicers, although the C-terminal Tyrosine is not present in fungi Dicers and is Histidine in some sequences (fig. 2, supplementary fig. S4, Supplementary Material online, positions 313–320). The other described 3′UU anchoring motif (YR926, position 271 in supplementary fig. S4, Supplementary Material online) is strongly conserved across animal Dicer1, weakly conserved in Dicer2 and diploblast Dicers and is not found in plant Dicers, although H926 is conserved across plants, suggesting it may play a role in 3′ anchoring.
Ancestral Sequence Reconstructions Suggest 5′phosphate Binding Arose in Early Animals
To explicitly investigate the evolutionary origins of RNA-contact residues within the Dicer PPC domain, we reconstructed ancestral PPC sequences at key nodes on the Dicer phylogeny, using an approach that incorporates uncertainty in both the phylogeny and the alignment (see Materials and Methods). We found that all ancestral sequences were reconstructed with high confidence, despite phylogenetic and alignment uncertainty, consistent with previous reports that phylogenetic ambiguity is unlikely to strongly impact ancestral sequence reconstruction (Hanson-Smith et al. 2010). Averaged across all reconstructed sequences, 86.5% of sites were reconstructed with posterior probability >0.95, and no sequence had <80% of sites reconstructed with posterior probability >0.95 (supplementary fig. S5, Supplementary Material online). Across all sequences, only 1.3% of sites had a plausible alternative reconstruction with posterior probability >0.3, and the vast majority of these (80.5%) were biochemically conservative, suggesting little potential impact on protein function (supplementary table S1, Supplementary Material online). Only one sequence (ancestral Plant DCL1) had >2% of sites with a plausible alternative reconstruction. When we plotted positions with plausible alternative reconstructions on the structure of the human PPC domain, we found that only two of the potentially ambiguous positions were at proposed RNA-contact residues (supplementary fig. S6, Supplementary Material online). Each potentially ambiguous RNA-contact residue was present in only one ancestral sequence, and both were conservative amino-acid changes, suggesting that ancestral sequence ambiguity is unlikely to strongly impact inferences about the PPC-RNA interface.
Ancestral sequence reconstruction suggested that many of the key 5′phosphate anchoring residues identified from human Dicer1 originated in the earliest ancestral animal Dicer (figs. 2 and 3, supplementary fig. S7, Supplementary Material online), implying that losses of these contact residues in Dicer2 and fast-evolving diploblast Dicers may be primarily responsible for the observed pattern of conservation among extant Dicer sequences (supplementary fig. S4, Supplementary Material online). All three contact residues in the N-terminal Platform domain (RxR778 and R811) are present in the ancestral animal Dicer and the last common ancestor before the Dicer 1/2 split, although both motifs are altered in the ancestral arthropod Dicer2 sequence (fig. 3, alignment positions 25–27 and 70, respectively; see also fig. 2). The animal-specific Platform insert (positions 14–27 in fig. 3) appears to have occurred in two steps, one in the ancestral animal Dicer—which introduced the RxR778 motif—and the other in ancestral Dicer1/2, which introduced a six-residue proline-rich region upstream of RxR778. Within the PAZ domain, the 5′-anchoring H982 and R993 residues (positions 402 and 415, respectively, in fig. 3) and all of the 3′-anchoring contacts are conserved across most of the ancestral-reconstructed animal Dicers (see figs. 2 and 3).
Fig. 3.
Residues anchoring the RNA 5′ phosphate in human Dicer1 originated in the ancestral animal Dicer. We show sections of aligned ancestral-reconstructed Dicer Platform + PAZ + Connector (PPC) domains from key nodes on the Dicer/DCL protein family phylogeny. Letters in parentheses correspond to node identifiers in figure 1. Residues thought to anchor the RNA’s 3′UU overhang and 5′phosphate in human Dicer1 are indicated by blue and red arrows, respectively (Tian et al. 2014). Identified animal- and plant-specific insertions are also indicated. Complete alignment is shown in supplementary fig. S7, Supplementary Material online. Alternative alignments are shown in supplementary figs. S8 and S9, Supplementary Material online.
A recent study has suggested that arthropod Dicer2 may have evolved a unique 5′phosphate binding pocket, perhaps independently from the 5′binding pocket of human Dicer1 (Kandasamy and Fukunaga 2016). Interestingly, only one of the 5′-anchoring residues identified in that study of Drosophila melanogaster Dicer2 was reconstructed in the ancestral arthropod Dicer2 sequence (H743, R752, R759, R943, and R956, corresponding to alignment positions 32, 45, 52, 401 and 415 in supplementary fig. S7, Supplementary Material online). Only the D. melanogaster R956 residue—which we aligned with the highly conserved R993 5′-contact—was reconstructed as Arginine in the ancestral arthropod Dicer2 (position 415 in supplementary fig. S7, Supplementary Material online). These results suggest that the 5′phosphate binding pocket identified in D. melanogaster Dicer2 likely evolved later in the Dicer2 lineage, after Dicer2 diverged from Dicer1.
Other than the 3′-anchoring YYxxxY961 motif (alignment positions 374–384 in fig. 3), most of the RNA-contact residues identified in human Dicer1 were not found in ancestral-reconstructed Dicers from plants, suggesting that plant Dicers may anchor their RNA targets using different structural mechanisms (see fig. 2). Although plant sequences appear to encode a Platform domain, they lack the 14-residue animal-specific insertion harboring the 5′-anchoring RxR778 motif, and R811 is not present in plants (see fig. 3, positions 14–28 and 70, respectively). However, plant Dicers do have an Arginine close to R811 in animals, which originated in the ancestral plant DCL and is conserved in ancestral-reconstructed DCLs 1, 3, and 2 (position 72 in fig. 3). The ancestral plant DCL2/3/4 sequence also appears to have evolved a highly basic seven-residue insertion within the Platform domain—conserved in plant DCL3—which could contribute to anchoring the RNA target (positions 115–121 in fig. 3; see also fig. 2). The ancestral plant DCL1 also has a unique insertion containing a number of basic residues (positions 233–265 in fig. 3; see also fig. 2). Although these plant-specific insertions might provide the capacity to anchor the RNA target’s 5′phosphate, their functions are currently unknown.
We reconstructed ancestral insertions and deletions (indels) using a simple binary likelihood model applied to the presence–absence protein sequence alignment (see Materials and Methods). This model is unrealistic, in that it treats individual columns in the presence–absence alignment as statistically independent, whereas biological indels potentially involve multiple contiguous residues. However, reconstructed indels were equivalent when we used a model that incorporates multisite insertions and deletions (Ashkenazy et al. 2012), suggesting that using a simplified indel model did not impact our results.
Indel reconstruction may be particularly sensitive to sequence alignment. Although our ancestral reconstructions incorporated uncertainty in the sequence alignment (see Materials and Methods), we further examined the robustness of reconstructed indels to sequence alignment by reconstructing ancestral PPC domains using an alternative alignment strategy designed to align divergent sequences (Notredame et al. 2000; Chang et al. 2012). Although we did observe some differences in ancestral sequences reconstructed from this alternative alignment (supplementary fig. S8, Supplementary Material online), these differences did not impact our conclusions about the major events in Dicer PPC’s evolutionary history. The presence or absence of 5′- and 3′-contacting residues was equivalent across ancestral sequences reconstructed using our original approach (supplementary fig. S7, Supplementary Material online) and our alternative alignment (supplementary fig. S8, Supplementary Material online), and the major animal- and plant-specific insertion events were reconstructed similarly. Reconstructing ancestral PPC sequences using a Bayesian approach that integrates over alignments and tree topologies (Suchard and Redelings 2006) also resulted in slight sequence differences but no major changes in inferred RNA-contact residues or large insertion/deletion events (supplementary fig. S9, Supplementary Material online). Although the specific alignment positions of RNA-contact motifs differed across approaches, ancestral reconstructions of the motifs, themselves, were equivalent, and the major lineage-specific insertions identified in figure 3 were found across alignments and reconstruction methods (see supplementary figs. S7–S9, Supplementary Material online). Together, these results suggest that our findings are generally robust to alignment uncertainty and different ancestral reconstruction methodologies.
Dicer Platform + PAZ + Connector Increased Affinity for RNA Targets in Early Animal and Plant Lineages
The Dicer Platform + PAZ + Connector (PPC) domain anchors the end of the target RNA molecule and appears to play important roles in target selection, processing efficiency and determining the length of the processed RNA product (Park et al. 2011; Tian et al. 2014; Kandasamy and Fukunaga 2016). To begin examining the evolution of RNA recognition by Dicer, we resurrected ancestral PPC domains and measured their affinity for two types of dsRNA molecules with end structures modeling known Dicer targets (Zhang et al. 2002; Vermeulen et al. 2005; Lee and Collins 2007; Cenik et al. 2011; Nagano et al. 2014). We used “microRNA”-like RNAs having a 5′monophosphate and 3′UU overhang and “viralRNA”-like molecules exhibiting a 5′triphosphate moiety (see supplementary fig. S10, Supplementary Material online for RNA sequences). Although the 5′triphosphate (5′ppp) ligand is not commonly considered a major Dicer target, previous studies suggest that animal Dicers can efficiently process 5′ppp dsRNA in vitro (Zhang et al. 2002; Cenik et al. 2011), and Dicer targeting of 5′ppp dsRNAs may be important for protozoan RNAi in vivo (Lee and Collins 2007). Viral-derived RNAs bearing 5′ppp moieties have been shown to activate cellular immune responses (Hornung et al. 2006; Pichlmair et al. 2006; Plumet et al. 2007) and could be important for initial antiviral RNA recognition by Dicer (Kandasamy and Fukunaga 2016).
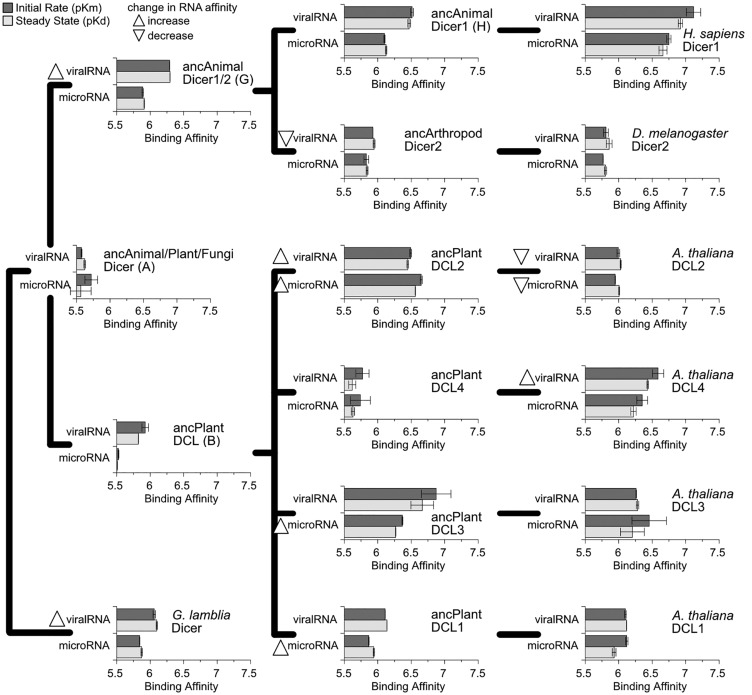
We observed an overall pattern of PPC-RNA affinity evolution in which an ancestral lower-affinity PPC domain evolved increased affinity for RNA targets in early animal and plant lineages, followed by later possible “tuning” of PPC-RNA affinities within major Dicer/DCL groups (fig. 4, supplementary fig. S11, Supplementary Material online). The resurrected ancestral animal/plant/fungi Dicer PPC domain (node A in fig. 1) had relatively low affinity for both RNA types, although in vitro PPC-RNA affinity was strong enough to suggest plausible biological activity (Kd = 2.91 μM, Km = 1.95 μM for microRNA; Kd = 2.39 μM, Km = 2.67 μM for viralRNA). The protozoan, Giardia lamblia, had similar affinity for microRNA (Kd = 1.34 μM, Km = 1.44 μM; P > 0.29), although its affinity for viralRNA-like molecules was greater by ∼3-fold (Kd = 0.80 μM, Km = 0.87 μM; P < 0.0082). We observed a ∼5-fold increase in affinity for viralRNA in the ancestral animal Dicer1/2 PPC domain (node G in fig. 1), compared with ancAnimal/Plant/Fungi Dicer (Kd = 0.51 μM, Km = 0.51 μM; P < 0.012), and no change in its affinity for microRNA (P > 0.26). Although the ancestral plant DCL (node B) did appear to exhibit a slight increase in affinity for viralRNA, this was not statistically significant (P > 0.08). Following the diversification of plant DCLs, we observed increases in microRNA affinities in ancestral DCL2, DCL3, and DCL1 (fig. 4; >2.4-fold increase; P < 0.0029) as well as a ∼4-fold increase in viralRNA affinity in ancDCL2 (P < 0.043). No change in RNA affinity was observed along the branch leading to ancDCL4 (P > 0.11). Within the animal lineage, ancDicer1 did not exhibit a major change in RNA affinity, compared with ancDicer1/2 (<1.64-fold change; P > 0.07), and ancDicer2 slightly reduced its affinity for viralRNA (∼2.3-fold change; P < 0.025).
Fig. 4.
The affinity of the Dicer Platform + PAZ + Connector (PPC) domain for RNA targets increased independently in animal and plant lineages. We measured the binding kinetics of extant and ancestral-reconstructed Platform + PAZ + Connector domains to synthetic RNAs modeling endogenous microRNAs or viral-like RNA products (see Materials and Methods; RNA sequences are shown in supplementary fig. S10, Supplementary Material online). We plot steady-state binding (pKd, light bars) and initial binding rates (pKm, dark bars) to each RNA type, with longer bars indicating tighter binding. Standard errors are shown. Significant increases or decreases in RNA affinity along each branch on the simplified phylogeny are indicated by upward- or downward-pointing triangles, respectively (P < 0.05). Kinetics curves are shown in supplementary fig. S11, Supplementary Material online.
Together, these results are consistent with a model in which an ancestral low-affinity Dicer PPC domain evolved elevated affinities for RNA targets early in the evolutionary histories of animals and plants. Previous ancestral-resurrection studies examining other protein families have observed the same pattern of an ancestral “low-affinity, promiscuous” receptor evolving increased affinity and ligand specificity in descendant lineages (Carroll et al. 2008; Levin et al. 2009; Khersonsky and Tawfik 2010; Risso et al. 2013). Although this pattern of “ligand optimization” could be a real evolutionary phenomenon, errors in ancestral sequence reconstruction—which may correlate with the molecular “age” of the node being reconstructed—could generate the same pattern by artifactually introducing residues that reduce ligand affinity and specificity.
To examine the impact of ancestral sequence ambiguity on our results, we introduced plausible alternative residues (see supplementary table S1, Supplementary Material online) into ancestral PPC protein sequences and characterized the impact on PPC-RNA affinities. We found that simultaneously introducing all plausible alternative residues into each ancestral PPC domain had very little impact on PPC-RNA affinity (supplementary fig. S12, Supplementary Material online). Only two of the alternative reconstructions had significantly different PPC-RNA affinities, compared with their respective maximum-likelihood ancestral sequences (ancPlant DCL3, P < 0.032; ancPlant DCL1, P < 0.019). Both differences were small in absolute value (<1.16-fold), and alternative reconstructions generally had weaker RNA affinities, compared with maximum-likelihood sequences, consistent with previous reports that alternative reconstructions may introduce sequence errors that degrade protein function (Hobbs et al. 2012). The remaining alternative reconstructions had <1.56-fold differences in RNA affinities, compared with maximum-likelihood reconstructions (P > 0.11), suggesting ancestral reconstruction ambiguity did not strongly impact our results. Ancestral proteins reconstructed using an alternative alignment (supplementary fig. S8, Supplementary Material online) also had very similar RNA affinities to those of their original respective ancestral proteins (supplementary fig. S13, Supplementary Material online; <1.5-fold change; P > 0.062). Although it is impossible to completely rule out errors in ancestral sequence reconstruction, potential errors do not appear to have greatly impacted our findings.
Constraining the animal Dicer1-Dicer2 duplication to occur in arthropods was strongly rejected by statistical topology tests (see results above; AU test P < 0.0057). However, this alternative evolutionary history is more parsimonious in terms of gene loss events. To examine the robustness of our functional inferences to the position of the Dicer1-Dicer2 duplication, we reconstructed ancestral animal/plant/fungi Dicer, animal Dicer1/2, Dicer1, and Dicer2 PPC domains, constraining the Dicer1-Dicer2 duplication to occur early in the arthropod lineage. We observed a similar pattern of increased affinity for viralRNA in the ancestral Dicer1/2, followed by reduced viralRNA binding in ancDicer2 (supplementary fig. S14, Supplementary Material online), suggesting our primary findings concerning the patterns of RNA affinity evolution in animal Dicers are largely robust to plausible alternative evolutionary histories.
Comparison of the ancestral progenitors of the major animal and plant Dicer/DCL lineages (ancDicer1, ancDicer2 in animals; ancDCL1-4 in plants) to extant examples from model organisms suggests that further “tuning” of PPC-RNA affinities likely occurred within major Dicer/DCL groups, particularly in the plant lineage (fig. 4, supplementary fig. S11, Supplementary Material online). Specifically, we observed a ∼3–5-fold loss of general RNA affinity in Arabidopsis thaliana DCL2, compared with the ancestral DCL2 (P < 0.011) and a ∼6.6-fold increase in viralRNA affinity along the lineage leading to A. thaliana DCL4 (P < 0.035). The RNA affinities of A. thaliana DCL3 and DCL1 did not change, relative to their respective ancestral progenitors (P > 0.22). Within the animal lineage, we did not observe a change in the RNA affinity of Drosophila melanogaster Dicer2 PPC, compared with the ancestral Dicer2 (P > 0.18), and the >3-fold increases in RNA affinity in Homo sapiens Dicer1 (compared with the ancestral Dicer1) were not statistically significant (P > 0.06).
Overall, these results demonstrate extensive quantitative changes in PPC-RNA affinities across animal and plant Dicer/DCL lineages. Some of these changes are large enough to suggest potentially important changes in Dicer molecular function (i.e., a > 12-fold increase in miRNA affinity arising in ancDCL2; see fig. 4). However, the potential biological significance of these changes in molecular function is unclear. Although the Dicer PPC domain has been shown to play important roles in Dicer’s RNA targeting and processing (Park et al. 2011; Tian et al. 2014; Kandasamy and Fukunaga 2016), the RNAi process can be strongly affected by partner proteins, which can modulate the activities of Dicer (Lee et al. 2004; Hiraguri et al. 2005; Curtin et al. 2008; Dong et al. 2008; Marques et al. 2010; Fukudome et al. 2011; Hartig and Forstemann 2011; Fukunaga et al. 2012; Lee et al. 2013). Subcellular localization can also play a role in determining Dicer’s functional role; for example, plant DCL1 is typically localized to the nucleus, where it appears to function primarily in microRNA biogenesis (Papp et al. 2003; Song et al. 2007). Binding to target RNAs by the PPC domain is the first necessary step in Dicer molecular function; although examining this function in isolation cannot directly assess biological significance, our results do suggest that sequence changes in the PPC domain have impacted PPC-RNA affinity across the evolutionary history of animal and plant Dicers.
Platform Domain Insert Introduced 5′ RNA Binding Pocket in Early Animal Dicer
We determined which sequence changes were primarily responsible for observed changes in Dicer PPC-RNA affinities (see fig. 4) by introducing historical substitutions into ancestral PPC domains via site-directed mutagenesis and observing the effects on PPC-RNA affinity. Potential structural mechanisms through which sequence changes affected Dicer PPC function were assessed using structural homology modeling (see Materials and Methods).
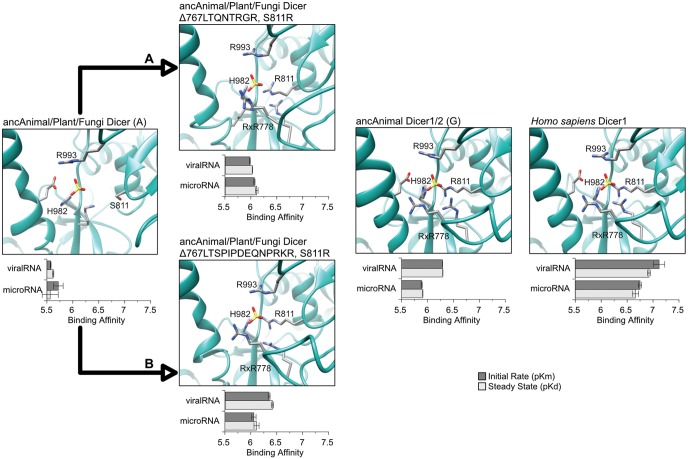
We observed a marked increase in the affinity of the ancestral animal Dicer PPC domain (ancAnimal Dicer1/2) for viral-like RNA, after it diverged from the ancestral animal/plant/fungi Dicer (see fig. 4). All of the 3′ and 5′ RNA-contact residues identified within the PAZ domain of human Dicer are strongly conserved across animal Dicer1 sequences and are present in the ancestral animal/plant/fungi Dicer (figs. 2 and 3). However, the three 5′-contact residues within the Platform subdomain are not present in ancAnimal/Plant/Fungi Dicer, although they are conserved across animal Dicer1s (figs. 2 and 3). We hypothesized that the introduction of the 5′-binding RxR778 motif (part of the animal-specific Platform insert) and R811—both of which occurred in ancAnimal Dicer following its divergence from ancAnimal/Plant/Fungi Dicer—were primarily responsible for the observed increase in viralRNA affinity in ancAnimal Dicer1/2.
To test this hypothesis, we first introduced the “shortened” animal-specific Dicer insert bearing an RGR778 motif (originating in ancAnimal Dicer, positions 14–27 in fig. 3) and the S811R substitution (position 70 in fig. 3) into the ancAnimal/Plant/Fungi Dicer background and measured the affinity of this “mutant” PPC domain for microRNA and viralRNA (fig. 5A;supplementary fig. S15, Supplementary Material online). We found that these substitutions occurring along the branch leading from ancAnimal/Plant/Fungi Dicer to the ancestral animal Dicer were sufficient to increase affinity for viralRNA-like molecules (>2.52-fold increase, P < 0.012). A similar apparent >2.25-fold increase in microRNA affinity was not statistically significant (P > 0.16). Similar results were found when replacing the short animal-specific Dicer Platform insert with the longer insert found in ancAnimal Dicer1/2 (fig. 5B;supplementary fig. S15, Supplementary Material online); affinity for viralRNA increased >3.1-fold (P < 0.0098), whereas microRNA affinity remained unchanged (P > 0.12). Introduction of the combined ancAnimal Dicer1/2 Platform insert and the S811R substitution into the ancAnimal/Plant/Fungi Dicer background was sufficient to recapitulate ancAnimal Dicer1/2’s RNA affinity profile (P > 0.085), suggesting that these substitutions were primarily responsible for the observed affinity shift between the animal/plant/fungi ancestor and the derived ancAnimal Dicer1/2 protein.
Fig. 5.
Substitutions within the Dicer Platform subdomain increased affinity for viral-like RNA early in animal evolutionary history by forming a conserved 5′ phosphate binding pocket. We measured the binding kinetics of ancestral Platform + PAZ + Connector (PPC) domains to synthetic RNAs modeling microRNAs or viralRNA products (see Materials and Methods; RNA sequences are shown in supplementary fig. S10, Supplementary Material online). We plot steady-state binding (pKd, light bars) and initial binding rates (pKm, dark bars) to each RNA, with longer bars indicating tighter binding. Standard errors are shown, with kinetics curves shown in supplementary fig. S15, Supplementary Material online. We introduced the historical S811R substitution and either short (A) or long (B) animal-specific insertions into the ancAnimal/Plant/Fungi Dicer background (see figs. 2 and 3) and plot the orientations of 5′ phosphate RNA-contact residues by structural homology modeling. Letters in parentheses next to protein names indicate ancestral node identifiers (see fig. 1). Human Dicer1 is shown for comparison.
Structural modeling of ancestral and mutant PPC domains revealed that broadly conserved 5′-contact residues within the PAZ domain (H982, R993; see figs. 2 and 3) are present in the proposed 5′-phosphate binding pocket in ancAnimal/Plant/Fungi Dicer (fig. 5). However, S811 does not contact the potential ligand, and the missing RxR778 motif is not replaced with other potential ligand-binding residues. Introduction of the S811R substitution created a potentially favorable 5′-phosphate contact, and there was little difference in the general orientation of the RxR778 ligand-binding motif in the short versus long Platform inserts (fig. 5A and B). The increased flexibility of the long Platform insert does appear to orient R778 and R780 residues toward the ligand in a manner more similar to what is observed in the modeled ancAnimal Dicer1/2 and crystalized human Dicer (fig. 5B), which may explain the slightly higher viralRNA affinities of PPC domains containing this longer insert, compared with the shorter insert (P < 0.021).
Together, these results suggest that the introduction of an insert in the Platform domain bearing RxR778 and an accompanying S811R substitution in early animal Dicer generated a conserved 5′-phosphate RNA-binding pocket that increased Dicer’s affinity for dsRNA targets and may have impacted Dicer processing (Park et al. 2011; Tian et al. 2014). The RxR778 insert and S811R substitutions were also reconstructed using alternative sequence alignments, suggesting that these results are robust to ancestral reconstruction and alignment uncertainty (supplementary figs. S8 and S9, Supplementary Material online). Further supporting this conclusion, introduction of a single R778A mutation was sufficient to decrease microRNA affinity in the ancAnimal Dicer1/2, ancAnimal Dicer1 and Homo sapiens Dicer1 backgrounds (>2.0-fold reduction, P < 0.03; supplementary fig. S16, Supplementary Material online). Similar mutations in human Dicer1 have been previously shown to affect the lengths of short RNA products (Park et al. 2011). Our analysis suggests that this “5′-counting rule” observed in human Dicer may have arisen very early in the animal lineage and is likely conserved across animal Dicer1 sequences.
Structurally Unique Insertions Impacted Plant DCL-RNA Affinities
We observed marked increases in PPC-RNA affinities along the branches leading to ancestral plant DCLs 2,3 and 1, compared with the ancestral animal/plant/fungi Dicer (see fig. 4). Although plant sequences lack the animal-specific 5′phosphate contacts within the Platform subdomain, we did observe a short plant-specific insertion within the α3–β5 loop occurring early in the DCL2/4/3 lineage and a longer plant-specific insertion between α4 and α5, which is particularly extended in DCL1 (see figs. 2 and 3, supplementary figs. S4, S7, and S9, Supplementary Material online). Although the structural features of these plant-specific insertions are unknown, sequence conservation is generally high across a broad taxonomic range, suggesting they are likely functional (supplementary fig. S4, Supplementary Material online). These plant-specific insertions were also reconstructed using alternative alignments, suggesting they are generally robust to alignment and reconstruction uncertainty (supplementary figs. S8 and S9, Supplementary Material online). A number of conserved basic residues within these insertions further suggests they might play a role in PPC-RNA interactions, leading us to hypothesize that the plant-specific α3–β5 and α4–α5 insertions were primarily responsible for observed changes in PPC-RNA affinities arising in early DCL2, DCL3 and DCL1 lineages.
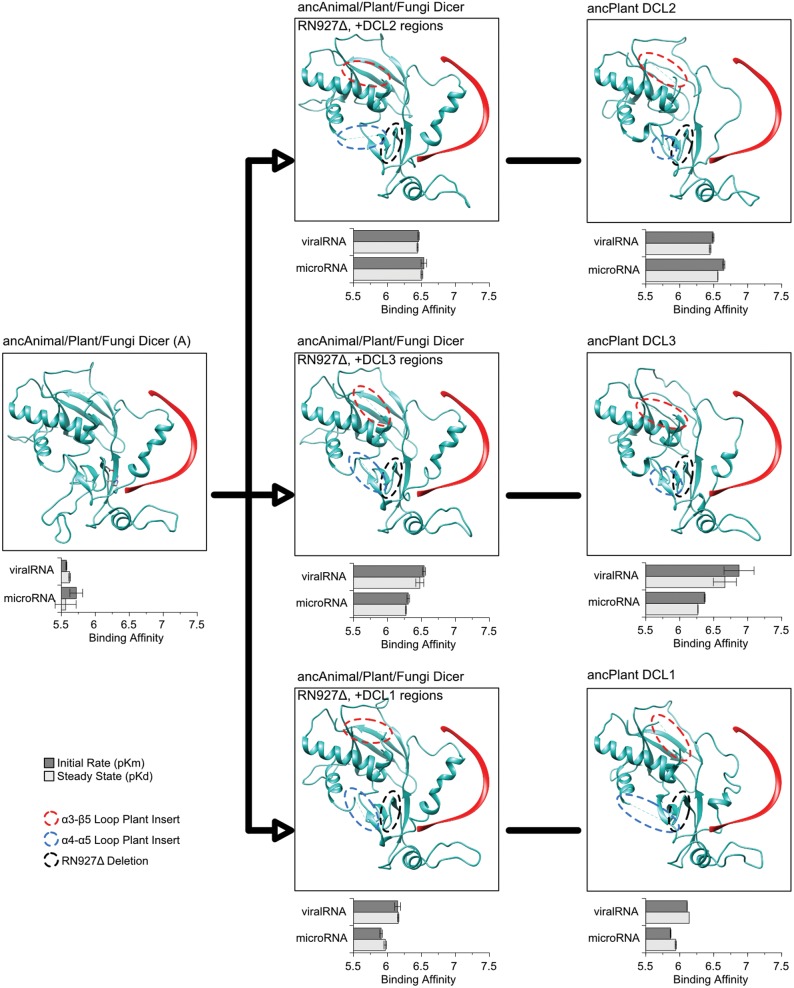
To test this hypothesis, we replaced the α3–β5 and α4–α5 loops in ancAnimal/Plant/Fungi Dicer with the corresponding regions from ancPlant DCL2, ancPlant DCL3 and ancPlant DCL1 (alignment positions 109–149 and 190–305 in supplementary fig. S7, Supplementary Material online for the α3–β5 and α4–α5 loops, respectively) and characterized the resulting impact on PPC-RNA affinities. In addition to introducing these plant-specific insertions, we also deleted RN927, which is present in ancAnimal/Plant/Fungi Dicer and all animal Dicer sequences but absent from all plant sequences (position 327 in fig. 3).
We found that, although these plant-specific changes were on the opposite side of the PPC structure from the animal-specific 5′-binding pocket, they were sufficient to shift PPC-RNA affinity from low-affinity in ancAnimal/Plant/Fungi Dicer to the higher affinities observed in ancPlant DCL2, ancPlant DCL3, and ancPlant DCL1 (fig. 6, supplementary figs. S17 and S18, Supplementary Material online). In the ancAnimal/Plant/Fungi DicerRN927Δ background, introducing ancPlant DCL2’s α3–β5 and α4–α5 loop regions increased affinity for microRNA-like molecules ∼6.6-fold (P < 0.041), and viralRNA affinity increased ∼7.1-fold (P < 7.9e−4). Similarly, introduction of the ancDCL3 α3-β5 and α4-α5 regions increased affinity for viralRNA ∼8.1-fold (P < 0.037), and the ancDCL1 inserts increased viralRNA affinity ∼3.6-fold (P < 0.042). In all cases, incorporation of the derived α3–β5 and α4–α5 loop sequences into the ancAnimal/Plant/Fungi DicerRN927Δ background resulted in RNA affinity profiles that were statistically indistinguishable from those of the corresponding derived ancDCL proteins (<1.91-fold difference in RNA affinity; P > 0.14; fig. 6, supplementary fig. S18, Supplementary Material online). These results suggest that historical changes in the α3–β5 and α4–α5 regions of plant DCL PPC domains—in coordination with the RN927Δ deletion—were primarily responsible for observed changes in PPC-RNA affinities during the early differentiation of plant DCLs.
Fig. 6.
Plant-specific insertions within the Dicer Platform subdomain increased RNA affinity in DCL2, DCL3, and DCL1 lineages. We measured the binding kinetics of ancestral Platform + PAZ + Connector (PPC) domains to synthetic RNAs modeling microRNAs or viralRNAs (see Materials and Methods; RNA sequences are shown in supplementary fig. S10, Supplementary Material online). We plot steady-state binding (pKd, light bars) and initial binding rates (pKm, dark bars) to each RNA, with longer bars indicating tighter binding. Standard errors are shown, with kinetics curves shown in supplementary figs. S11 and S18, Supplementary Material online. We introduced the historical RN927Δ deletion (black dotted outline) in concert with DCL3-, DCL2-, and DCL1-specific α3–β4 (red dotted outline) and α4–α5 (blue dotted outline) loop regions into the ancAnimal/Plant/Fungi Dicer background (see figs. 2 and 3) and plot the structural orientation of each Dicer protein (green ribbon) bound to RNA (red ribbon), inferred by structural homology modeling (see Materials and Methods). Protein regions too diverged from the structural template to ensure reliable modeling are indicated by dotted lines.
The structural features of these plant-specific DCL regions are currently unknown, but the lack of proline-rich sequences expected to disrupt secondary structure suggests that these regions could form relatively stable structural elements (see supplementary fig. S7 and S9, Supplementary Material online). All plant DCLs appear to have α4–α5 loop sequences that are very different from the homologous region in human Dicer1, which forms a short disordered loop in crystal structures (Tian et al. 2014). The human-like loop appears similar to that in ancAnimal/Plant/Fungi Dicer (see fig. 6, supplementary fig. S17, Supplementary Material online), suggesting that plant DCLs may have evolved unique structural features in this region of the PPC domain. All plant DCLs additionally lost the nearby RN927 3′-contact residues, which are present in ancAnimal/Plant/Fungi Dicer and conserved in animal Dicers (see figs. 3 and 6, supplementary fig. S17, Supplementary Material online). Plant DCLs also lack the short animal-specific insertion in this same region; lack of this insertion appears to create additional space in the plant PPC structure (see fig. 6, supplementary fig. S17, Supplementary Material online), which could alter the orientation of the RNA’s 3′ end. Although speculative at this point, these results suggest that 3′ RNA anchoring may function differently in plants, compared with animals and the ancestral animal/plant/fungi Dicer, and that the RNA may orient differently when bound to plant versus animal PPC domains. Further supporting this conclusion, introduction of a K1055A mutation into A. thaliana DCL4 was sufficient to reduce its affinity for microRNA by ∼2.6-fold (P < 0.049) and its affinity for viralRNA by ∼4.7-fold (P < 0.048; supplementary fig. S19, Supplementary Material online). This mutation is on the opposite side of the PPC structure as the animal 5′phosphate pocket, suggesting that plant DCL4 likely binds RNA in a different orientation than animal Dicer1. Future structural and functional studies will be required to characterize potential plant-specific structural elements in DCL PPC domains and determine their effects on RNA processing.
Conclusions
Ancestral protein resurrection is one of the few methods available that can directly investigate how evolution of a molecule’s sequence impacts its function through changes in tertiary structure (Dean and Thornton 2007; Harms and Thornton 2013). Here we have used this general approach to characterize how the first step in RNA interference—recognition of target RNAs by the Dicer/DCL Platform + Paz + Connector (PPC) domain—diversified during the divergence of animal and plant lineages. Our results suggest that 5′phosphate recognition—essential for RNA processing by human Dicer (Park et al. 2011; Tian et al. 2014)—evolved in the earliest animal Dicers and was lost in arthropod Dicer2 before being regained through a novel structural mechanism in D. melanogaster (Kandasamy and Fukunaga 2016). Similarly high affinity for dsRNA targets bearing 5′phosphates or triphosphates evolved early in the plant DCL lineage via lineage-specific insertions in the α3–β5 and α4–α5 loops. Although the structural features of these plant-specific DCL regions are currently unknown, our results do suggest that plant DCLs may bind target RNAs in an orientation that is different from that found in human Dicer, and that the structural bases for Dicer/DCL–RNA interactions are different in animal versus plant lineages.
Although broad in evolutionary scope, our study is a highly reductionist “first step” toward understanding the molecular-functional evolution of the Dicer protein family. We focused on specifically characterizing the functional evolution of the Platform + PAZ + Connector domain, which has been shown to play key roles in RNA target recognition through specific structural interfaces contacting moieties at the end of the dsRNA target (Park et al. 2011; Tian et al. 2014; Kandasamy and Fukunaga 2016). Although little evidence suggests the Dicer helicase domain could participate in RNA target recognition, other functional domains may contribute to RNA preference (Wu et al. 2007; Liu et al. 2013; Suarez et al. 2015). Examining the potential modulating effects of other functional domains on Dicer target recognition will be important for more fully characterizing the evolution of Dicer function.
Although we used microRNA-like and viralRNA-like ligands to probe the molecular-functional evolution of Dicer/DCL proteins, our study was not explicitly designed to examine functional differentiation into miRNA- and siRNA-specific Dicers. The number of Dicer paralogs is too small to draw strong conclusions about how ligand affinity might correlate with functional specificity. Nonetheless, we did not observe strong differences in affinities across RNA types, even for Dicer paralogs thought to exhibit functional specificity. For example, plant DCL4 and DCL1 are thought to have specialized in antiviral immunity and microRNA processing, respectively. However, neither ancestral nor extant proteins exhibited strong RNA preferences among our model ligands (see fig. 4). Similarly, arthropod Dicer2 is commonly considered an siRNA specialist, but neither ancestral nor D. melanogaster Dicer2 exhibited a strong preference for viralRNA-like ligands. Dicer/DCL paralogs do appear to have overlapping functional repertoires, suggesting that functional differentiation among Dicers may not be very strict, at least in some cases (Gasciolli et al. 2005; Patrick et al. 2009). Nonetheless, our results suggest Dicer functional specificity may not be primarily controlled by PPC-RNA binding but could be modulated by higher-order processes like subcellular localization or interactions with partner proteins (Zhou et al. 2009; Zhang et al. 2017).
Dicer molecular function is complex and involves at least three broad conceptual steps: 1) initial recognition of target RNAs, 2) processing of target RNAs through length-specific cleavage and 3) transfer of processed RNAs to the RNA-induced silencing complex (Siomi and Siomi 2009; Axtell et al. 2011; Castel and Martienssen 2013). Each of these steps may be facilitated by interactions with specific partner proteins. For example, double-stranded RNA binding proteins (DRBs) have been shown to interact with Dicers to impact RNAi, although the mechanisms by which DRBs modulate Dicer function are poorly understood and are likely to differ across species (Hiraguri et al. 2005; Curtin et al. 2008; Marques et al. 2010; Lee et al. 2013).
Proteins never function in isolation, and a thorough characterization of the functional evolution of Dicer would have to be done within the context of the larger RNAi pathway in which it functions. Ancestral protein resurrection (ASR) is technically challenging and resource-intensive, requiring examination of a potentially large number of alternative protein sequences to insure robustness. For this reason, existing studies have focused exclusively on characterizing the molecular-functional evolution of proteins whose ligands or interaction partners are not thought to have changed significantly over evolutionary timescales. Examining the functional evolution of groups of evolutionarily labile protein families whose collective function is determined by changes in specific interaction partners will likely require advances in ASR methodology and/or high-throughput methods for protein functional characterization. These advances will be necessary to begin dissecting the functional evolution of complex molecular systems like RNAi.
Materials and Methods
Sequence Identification and Alignments
Protein sequences were identified by rpsblast search of the nr database (Marchler-Bauer and Bryant 2004; Marchler-Bauer et al. 2015; Coordinators 2016). Dicers and Dicer-like proteins were identified as full-length protein sequences containing a single PAZ domain (CD02843, CD02844, or CD00949) followed by two Ribonuclease-C domains (RIBOc, CD00593), each with e-value <0.01. Each sequence’s functional domains were annotated by sequence search of the Conserved Domain Database using an e-value cutoff of 0.01 (Marchler-Bauer et al. 2015).
Full-length protein sequences were aligned using Clustal Omega v1.2.3 (Sievers et al. 2011), MUSCLE v3.8.31 (Edgar 2004), and mafft-einsi v7.215 (Katoh and Standley 2013), with default parameters. Alignments of only the conserved PAZ + RIBOc + RIBOc regions were also produced using the same methods. Alignments were left unprocessed or processed by Gblocks v0.91 to remove potentially ambiguous regions (Talavera and Castresana 2007). We set the minimum number of sequences for a flank position (−b2) equal to 3/5 the total number of sequences in the alignment. The maximum number of contiguous nonconserved positions (−b3) was set to 10. The minimum block length (−b4) was 5, and gap positions were allowed (−b5 = a). Other Gblocks parameters were left at default values.
Phylogenetic Analyses and Ancestral Sequence Reconstruction
Initial maximum likelihood phylogenies were constructed from each alignment using FastTree v2.1.7 with default parameters (Price et al. 2010). Initial trees were used as starting trees for full maximum-likelihood reconstruction using RAxML v8.2.8 (Stamatakis 2014), with the best-fit evolutionary model selected from each alignment using AIC in ProtTest v3 (Darriba et al. 2011). Clade support was evaluated by SH-like aLRT scores (Anisimova and Gascuel 2006). Maximum-likelihood phylogenies produced from each alignment were converted to a clade presence-absence matrix using the Super Tree Toolkit v0.1.2 (Hill and Davis 2014), and a supertree was inferred from this matrix using the BINCAT model in RAxML (Nguyen et al. 2012). We also concatenated all individual alignments into a single supermatrix and reconstructed the maximum-likelihood protein family phylogeny using RAxML, with the best-fit evolutionary model selected by AIC (Wheeler et al. 1995). We present a consensus of “supertree” and “supermatrix” results. The significance of variation in tree topology support was evaluated using the AU test (Shimodaira 2002).
Protein-coding adaptation was assessed using the branch-sites model in PAML v4.9a, which uses a mixture distribution to model a combination of negatively selected, neutral, and positively selected positions in the protein sequence (Zhang et al. 2005; Yang 2007). Coding nucleotide sequences were mapped to each protein sequence alignment (Clustal, Muscle and mafft-einsi; see above). For each branch on the consensus phylogeny, we tested the hypothesis that some codons experienced adaptive protein-coding substitutions against the null hypothesis of neutral evolution using a likelihood ratio test. P values were calculated using the χ2 distribution (Zhang et al. 2005), and multiple testing was corrected for using a false-discovery rate (FDR) correction (Benjamini and Hochberg 1995). Tests for protein-coding adaptation were conducted separately using each alignment, and branches were determined to be under positive selection if all three alignments had P < 0.05 after FDR correction. We tested each protein functional domain for adaptation separately.
Ancestral protein sequences were reconstructed using an empirical Bayesian method to integrate over plausible tree topologies (Hanson-Smith et al. 2010). We collected all maximum-likelihood trees inferred from any sequence alignment and estimated the posterior probability of each topology—assuming a given alignment—using Bayes’ rule, assuming a flat prior over topologies. Given a topology and alignment, we inferred the marginal posterior-probability distribution over ancestral sequences at each node using RAxML, which implements an empirical Bayesian ancestral reconstruction algorithm (Yang et al. 1995). Next, we integrated over topologies by weighting each ancestral reconstruction by the posterior probability of that tree, given the alignment. Clustal, MUSCLE and mafft-einsi alignments were mapped to one another using the mafft –merge option. Alignment uncertainty was incorporated by combining ancestral sequence reconstructions from each alignment using a flat prior over alignments.
For each ancestral node, n, we calculated the probability of residue r at position i in the combined alignment using:
where Σa is over all three alignments, Σt is over all tree topologies, and is the probability of residue r at position i, node n, given alignment a and tree t. 1/3 is the probability of alignment a, assuming equal priors across alignments, and P(t | a) is the probability of tree t, given alignment a.
Ancestral insertions and deletions (indels) were reconstructed by converting each sequence alignment to a presence–absence matrix and reconstructing ancestral presence/absence states using the BINCAT model in RAxML, which calculates the posterior probability of a “gap” at each position in the alignment, for each ancestral node on the phylogenetic tree. Presence–absence reconstructions generated using each tree and alignment were combined using the same approach used to combine sequence reconstructions. We additionally reconstructed ancestral indels using FastML v3.0, which uses a similar approach to indel reconstruction but models contiguous gaps as a single insertion/deletion event, rather than considering each alignment position as statistically independent (Ashkenazy et al. 2012).
Alternative ancestral reconstructions were generated using T-Coffee v10.0 to align protein sequences (Notredame et al. 2000). We used the psicoffee alignment mode, with ancestral sequences reconstructed directly from this alignment, assuming the consensus phylogeny. Finally, we reconstructed alternative ancestral sequences using BAli-Phy v3.0 beta1, which uses a Bayesian approach to sample sequence alignments and trees from the combined posterior probability distribution (Redelings and Suchard 2005; Suchard and Redelings 2006). BAli-Phy analysis was run using the LG + F substitution model and the RS07 indel model, with among-site rate variation modeled using an 8-category discrete gamma approximation. Other parameters were left at default values. We discarded samples taken during the burnin period and terminated the sampling run after the effective independent sample size was >100 for all model parameters.
Structural Modeling
We used MODELLER v9.14 (Eswar et al. 2008) to infer structural models of the Platform + PAZ + Connector domain bound to RNA, using human Dicer (PDB IDs: 4NGb, 4NGC, 4NGD, 4NGF, 4NGG, 4NH3, 4NH5, 4NH6, 4NHA) as a template (Tian et al. 2014). Using the combined templates, we constructed 100 potential structural models and selected the best one using the modeller objective function (molpdf), DOPE and DOPEHR scores (Shen and Sali 2006; Larsson et al. 2008). Each score was rescaled to units of standard-deviation across the 100 models, and we selected the best model as that with the best average of rescaled scores.
Each initial protein-RNA structural model was used as a starting point for a short molecular dynamics simulation using GROMACS v5.1.2 (Pronk et al. 2013). We used the amber99sb-ildn force field and the tip3p water model. Initial dynamics topologies were generated using the GROMACS pdb2gmx algorithm with default parameters. Topologies were relaxed into simulated solvent at pH = 7 using a 50,000-step steepest-descent energy minimization. The system was then brought to 300K using a 50-picosecond dynamics simulation under positional restraints, followed by pressure stabilization for an additional 50 picoseconds. Simulations were run using Particle-Mesh Ewald electrostatics with cubic interpolation and grid spacing of 0.12 nanometers. Van der Waals forces were calculated using a cutoff of 1.0 nanometer. We used Nose-Hoover temperature coupling, with protein, RNA and solvent systems coupled separately and the period of temperature fluctuations set to 0.1 picoseconds. Pressure coupling was applied using the Parrinello–Rahman approach, with a fluctuation period of 2.0 picoseconds. Nonbonded cutoffs were treated using buffered Verlet lists. We selected the lowest-energy complex sampled during the last 20 picoseconds of each pressure stabilization simulation.
Experimental Measurement of Protein-RNA Affinity
We generated double-stranded viral-like and microRNA-like RNA molecules (see supplementary fig. S7, Supplementary Material online). The top strand of the viralRNA and both strands of the microRNA were synthesized by Integrated DNA Technologies, Inc. (Coralville, Iowa). The bottom strand of viralRNA was generated from synthetic DNA template using the TranscriptAid T7 High Yield Transcription Kit (Thermo Scientific, Catolog # K0441) and purified following the kit’s instructions. Single-stranded RNAs were annealed to produce double-stranded RNA by combining at 1:1 ratio in nuclease-free duplex buffer (30 mM HEPES, pH 7.5, 100 mM Potassium Acetate), heating to 95 °C for 5 min and then cooling to 25 °C.
Ancestral and extant Platform + PAZ + Connector domains with an N-terminal Flag tag were expressed in E. coli BL21 (DE3) cells using pET-22b(+) and verified by Sanger sequencing. Proteins were purified by HisPur Cobalt Resin (Thermo Scientific, Catalog # 89964), visualized by SDS-page stained with 1% coomassie and confirmed by western blot using antiflag antibody. Protein concentrations were measured using a linear-transformed Bradford assay (Zor and Selinger 1996).
We measured protein-RNA binding using a label-free in vitro kinetics assay (Abdiche et al. 2008; Frenzel and Willbold 2014). Biotinylated RNA molecules were bound to a series of eight streptavidin probes for 5 min, until saturation was observed. Probes were exposed to 25 µg/ml biocytin to bind any remaining free streptavidin and then washed. Each probe was then exposed to active protein at increasing concentrations in HBS-EP buffer (10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% Tween 20) for 6 min, followed by dissociation in HBS-EP Buffer for an additional 4 min before exposure to the next concentration of protein (Frenzel and Willbold 2014). Molecular binding at each concentration over time was measured as the change in laser wavelength when reflected through the probe in solution, sampled every 3 ms. Two probes were not exposed to protein as controls to evaluate system fluctuation across the time of the experiment; measurements from these control probes were averaged and subtracted from each analysis probe.
For each replicate experiment, we estimated the protein concentration at which ½-maximal steady-state RNA binding was achieved (Kd) by fitting a one-site binding curve to the steady-state laser wavelengths measured across protein concentrations at saturation, using nonlinear regression. We additionally fit 1-site association/dissociation curves to the full time-course data in order to estimate the initial rates of RNA binding across protein concentrations and used these rates to calculate the protein concentration at which the ½-maximal RNA-binding rate was achieved (Km). Kds and Kms were –log10 transformed to facilitate visualization, and standard errors across three experimental replicates were calculated. We calculated the statistical significance of differences between Kds and Kms using the two-tailed unpaired t test, assuming unequal variances.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation (Molecular and Cellular Biology, grant number 1412442). We are grateful to Dr Jennifer Doudna at University of California, Berkley for providing the Giardia dicer construct.
References
- Abdiche Y, Malashock D, Pinkerton A, Pons J.. 2008. Determining kinetics and affinities of protein interactions using a parallel real-time label-free biosensor, the Octet. Anal Biochem. 377:209–217. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Gascuel O.. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol. 55:539–552. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Gil M, Dufayard JF, Dessimoz C, Gascuel O.. 2011. Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst Biol. 60:685–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H, Penn O, Doron-Faigenboim A, Cohen O, Cannarozzi G, Zomer O, Pupko T.. 2012. FastML: a web server for probabilistic reconstruction of ancestral sequences. Nucleic Acids Res. 40:W580–W584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Westholm JO, Lai EC.. 2011. Vive la difference: biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 12:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B. 57:289–300. [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ.. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363–366. [DOI] [PubMed] [Google Scholar]
- Bologna NG, Voinnet O.. 2014. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu Rev Plant Biol. 65 65:473–503. Vol [DOI] [PubMed] [Google Scholar]
- Bridgham JT, Brown JE, Rodriguez-Mari A, Catchen JM, Thornton JW.. 2008. Evolution of a new function by degenerative mutation in cephalochordate steroid receptors. PLoS Genet. 4:e1000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgham JT, Keay J, Ortlund EA, Thornton JW.. 2014. Vestigialization of an allosteric switch: genetic and structural mechanisms for the evolution of constitutive activity in a steroid hormone receptor. PLoS Genet. 10:e1004058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgham JT, Ortlund EA, Thornton JW.. 2009. An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature 461:515–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Voinnet O.. 2006. The diversity of RNA silencing pathways in plants. Trends Genet. 22:268–280. [DOI] [PubMed] [Google Scholar]
- Carrington JC, Ambros V.. 2003. Role of microRNAs in plant and animal development. Science 301:336–338. [DOI] [PubMed] [Google Scholar]
- Carroll SM, Bridgham JT, Thornton JW.. 2008. Evolution of hormone signaling in elasmobranchs by exploitation of promiscuous receptors. Mol Biol Evol. 25:2643–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel SE, Martienssen RA.. 2013. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 14:100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenik ES, Fukunaga R, Lu G, Dutcher R, Wang Y, Tanaka Hall TM, Zamore PD.. 2011. Phosphate and R2D2 restrict the substrate specificity of Dicer-2, an ATP-driven ribonuclease. Mol Cell 42:172–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JM, Di Tommaso P, Taly JF, Notredame C.. 2012. Accurate multiple sequence alignment of transmembrane proteins with PSI-Coffee. BMC Bioinformatics 13 (Suppl 4): S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coordinators NR. 2016. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 44:D7–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin SJ, Watson JM, Smith NA, Eamens AL, Blanchard CL, Waterhouse PM.. 2008. The roles of plant dsRNA-binding proteins in RNAi-like pathways. FEBS Lett. 582:2753–2760. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong D, Eitel M, Jakob W, Osigus HJ, Hadrys H, Desalle R, Schierwater B.. 2009. Multiple dicer genes in the early-diverging metazoa. Mol Biol Evol. 26:1333–1340. [DOI] [PubMed] [Google Scholar]
- Dean AM, Thornton JW.. 2007. Mechanistic approaches to the study of evolution: the functional synthesis. Nat Rev Genet. 8:675–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias R, Manny A, Kolaczkowski O, Kolaczkowski B.. 2017. Convergence of domain architecture, structure, and ligand affinity in animal and plant RNA-binding proteins. Mol Biol Evol. 34:1429–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z, Han MH, Fedoroff N.. 2008. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc Natl Acad Sci U S A. 105:9970–9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswar N, Eramian D, Webb B, Shen MY, Sali A.. 2008. Protein structure modeling with MODELLER. Methods Mol Biol. 426:145–159. [DOI] [PubMed] [Google Scholar]
- Frenzel D, Willbold D.. 2014. Kinetic titration series with biolayer interferometry. PLoS One 9:e106882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukudome A, Kanaya A, Egami M, Nakazawa Y, Hiraguri A, Moriyama H, Fukuhara T.. 2011. Specific requirement of DRB4, a dsRNA-binding protein, for the in vitro dsRNA-cleaving activity of Arabidopsis Dicer-like 4. RNA 17:750–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga R, Han BW, Hung JH, Xu J, Weng Z, Zamore PD.. 2012. Dicer partner proteins tune the length of mature miRNAs in flies and mammals. Cell 151:533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Wang M, Blair D, Zheng Y, Dou Y.. 2014. Phylogenetic analysis of the endoribonuclease Dicer family. PLoS One 9:e95350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H.. 2005. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol. 15:1494–1500. [DOI] [PubMed] [Google Scholar]
- Gharib WH, Robinson-Rechavi M.. 2013. The branch-site test of positive selection is surprisingly robust but lacks power under synonymous substitution saturation and variation in GC. Mol Biol Evol. 30:1675–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson-Smith V, Kolaczkowski B, Thornton JW.. 2010. Robustness of ancestral sequence reconstruction to phylogenetic uncertainty. Mol Biol Evol. 27:1988–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms MJ, Thornton JW.. 2013. Evolutionary biochemistry: revealing the historical and physical causes of protein properties. Nat Rev Genet. 14:559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig JV, Forstemann K.. 2011. Loqs-PD and R2D2 define independent pathways for RISC generation in Drosophila. Nucleic Acids Res. 39:3836–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J, Davis KE.. 2014. The Supertree Toolkit 2: a new and improved software package with a Graphical User Interface for supertree construction. Biodivers Data J. e1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraguri A, Itoh R, Kondo N, Nomura Y, Aizawa D, Murai Y, Koiwa H, Seki M, Shinozaki K, Fukuhara T.. 2005. Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol. Biol. 57:173–188. [DOI] [PubMed] [Google Scholar]
- Hobbs JK, Shepherd C, Saul DJ, Demetras NJ, Haaning S, Monk CR, Daniel RM, Arcus VL.. 2012. On the origin and evolution of thermophily: reconstruction of functional precambrian enzymes from ancestors of Bacillus. Mol Biol Evol. 29:825–835. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314:994–997. [DOI] [PubMed] [Google Scholar]
- Jaskiewicz L, Filipowicz W.. 2008. Role of Dicer in posttranscriptional RNA silencing. Curr Top Microbiol Immunol. 320:77–97. [DOI] [PubMed] [Google Scholar]
- Kandasamy SK, Fukunaga R.. 2016. Phosphate-binding pocket in Dicer-2 PAZ domain for high-fidelity siRNA production. Proc Natl Acad Sci U S A. 113:14031–14036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH.. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15:2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khersonsky O, Tawfik DS.. 2010. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu Rev Biochem. 79:471–505. [DOI] [PubMed] [Google Scholar]
- Kratzer JT, Lanaspa MA, Murphy MN, Cicerchi C, Graves CL, Tipton PA, Ortlund EA, Johnson RJ, Gaucher EA.. 2014. Evolutionary history and metabolic insights of ancient mammalian uricases. Proc Natl Acad Sci U S A. 111:3763–3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson P, Wallner B, Lindahl E, Elofsson A.. 2008. Using multiple templates to improve quality of homology models in automated homology modeling. Protein Sci. 17:990–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Zhou K, Smith AM, Noland CL, Doudna JA.. 2013. Differential roles of human Dicer-binding proteins TRBP and PACT in small RNA processing. Nucleic Acids Res. 41:6568–6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Collins K.. 2007. Physical and functional coupling of RNA-dependent RNA polymerase and Dicer in the biogenesis of endogenous siRNAs. Nat Struct Mol Biol. 14:604–610. [DOI] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW.. 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117:69–81. [DOI] [PubMed] [Google Scholar]
- Levin KB, Dym O, Albeck S, Magdassi S, Keeble AH, Kleanthous C, Tawfik DS.. 2009. Following evolutionary paths to protein-protein interactions with high affinity and selectivity. Nat Struct Mol Biol. 16:1049–U1067. [DOI] [PubMed] [Google Scholar]
- Liu Q, Yan Q, Liu Y, Hong F, Sun Z, Shi L, Huang Y, Fang Y.. 2013. Complementation of HYPONASTIC LEAVES1 by double-strand RNA-binding domains of DICER-LIKE1 in nuclear dicing bodies. Plant Physiol. 163:108–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Guindon S.. 2014. Performance of standard and stochastic branch-site models for detecting positive selection among coding sequences. Mol Biol Evol. 31:484–495. [DOI] [PubMed] [Google Scholar]
- Lunzer M, Miller SP, Felsheim R, Dean AM.. 2005. The biochemical architecture of an ancient adaptive landscape. Science 310:499–501. [DOI] [PubMed] [Google Scholar]
- Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA.. 2006. Structural basis for double-stranded RNA processing by Dicer. Science 311:195–198. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH.. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327–W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, et al. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res. 43:D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques JT, Kim K, Wu PH, Alleyne TM, Jafari N, Carthew RW.. 2010. Loqs and R2D2 act sequentially in the siRNA pathway in Drosophila. Nat Struct Mol Biol. 17:24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Tuschl T.. 2004. Mechanisms of gene silencing by double-stranded RNA. Nature 431:343–349. [DOI] [PubMed] [Google Scholar]
- Mukherjee K, Campos H, Kolaczkowski B.. 2013. Evolution of animal and plant dicers: early parallel duplications and recurrent adaptation of antiviral RNA binding in plants. Mol Biol Evol. 30:627–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano H, Fukudome A, Hiraguri A, Moriyama H, Fukuhara T.. 2014. Distinct substrate specificities of Arabidopsis DCL3 and DCL4. Nucleic Acids Res. 42:1845–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen N, Mirarab S, Warnow T.. 2012. MRL and SuperFine+MRL: new supertree methods. Algorithms Mol Biol. 7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J.. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 302:205–217. [DOI] [PubMed] [Google Scholar]
- Nozawa M, Suzuki Y, Nei M.. 2009. Reliabilities of identifying positive selection by the branch-site and the site-prediction methods. Proc Natl Acad Sci U S A. 106:6700–6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp I, Mette MF, Aufsatz W, Daxinger L, Schauer SE, Ray A, van der Winden J, Matzke M, Matzke AJ.. 2003. Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol. 132:1382–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN.. 2011. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 475:201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick KL, Shi H, Kolev NG, Ersfeld K, Tschudi C, Ullu E.. 2009. Distinct and overlapping roles for two Dicer-like proteins in the RNA interference pathways of the ancient eukaryote Trypanosoma brucei. Proc Natl Acad Sci U S A. 106:17933–17938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, Weber F, Sousa CRe.. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997–1001. [DOI] [PubMed] [Google Scholar]
- Plumet S, Herschke F, Bourhis JM, Valentin H, Longhi S, Gerlier D.. 2007. Cytosolic 5′-triphosphate ended viral leader transcript of measles virus as activator of the RIG I-mediated interferon response. PLoS One 2:e279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP.. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk S, Pall S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, Shirts MR, Smith JC, Kasson PM, van der Spoel D, et al. 2013. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29:845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh C, Kolaczkowski O, Manny A, Korithoski B, Kolaczkowski B.. 2016. Resurrecting ancestral structural dynamics of an antiviral immune receptor: adaptive binding pocket reorganization repeatedly shifts RNA preference. BMC Evol Biol. 16:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redelings BD, Suchard MA.. 2005. Joint Bayesian estimation of alignment and phylogeny. Syst Biol. 54:401–418. [DOI] [PubMed] [Google Scholar]
- Risso VA, Gavira JA, Mejia-Carmona DF, Gaucher EA, Sanchez-Ruiz JM.. 2013. Hyperstability and substrate promiscuity in laboratory resurrections of Precambrian beta-lactamases. J Am Chem Soc. 135:2899–2902. [DOI] [PubMed] [Google Scholar]
- Shen MY, Sali A.. 2006. Statistical potential for assessment and prediction of protein structures. Protein Sci. 15:2507–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H. 2002. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 51:492–508. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons MP, Norton AP.. 2014. Divergent maximum-likelihood-branch-support values for polytomies. Mol Phylogenet Evol. 73:87–96. [DOI] [PubMed] [Google Scholar]
- Siomi H, Siomi MC.. 2009. On the road to reading the RNA-interference code. Nature 457:396–404. [DOI] [PubMed] [Google Scholar]
- Song L, Han MH, Lesicka J, Fedoroff N.. 2007. Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc Natl Acad Sci U S A. 104:5437–5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez IP, Burdisso P, Benoit MP, Boisbouvier J, Rasia RM.. 2015. Induced folding in RNA recognition by Arabidopsis thaliana DCL1. Nucleic Acids Res. 43:6607–6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard MA, Redelings BD.. 2006. BAli-Phy: simultaneous Bayesian inference of alignment and phylogeny. Bioinformatics 22:2047–2048. [DOI] [PubMed] [Google Scholar]
- Suzuki Y. 2008. False-positive results obtained from the branch-site test of positive selection. Genes Genet Syst. 83:331–338. [DOI] [PubMed] [Google Scholar]
- Talavera G, Castresana J.. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56:564–577. [DOI] [PubMed] [Google Scholar]
- Tian Y, Simanshu DK, Ma JB, Park JE, Heo I, Kim VN, Patel DJ.. 2014. A phosphate-binding pocket within the platform-PAZ-connector helix cassette of human Dicer. Mol Cell 53:606–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen A, Behlen L, Reynolds A, Wolfson A, Marshall WS, Karpilow J, Khvorova A.. 2005. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA 11:674–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler WC, Gatesy J, DeSalle R.. 1995. Elision: a method for accommodating multiple molecular sequence alignments with alignment-ambiguous sites. Mol Phylogenet Evol. 4:1–9. [DOI] [PubMed] [Google Scholar]
- Wu F, Yu L, Cao W, Mao Y, Liu Z, He Y.. 2007. The N-terminal double-stranded RNA binding domains of Arabidopsis HYPONASTIC LEAVES1 are sufficient for pre-microRNA processing. Plant Cell 19:914–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z, dos Reis M.. 2011. Statistical properties of the branch-site test of positive selection. Mol Biol Evol. 28:1217–1228. [DOI] [PubMed] [Google Scholar]
- Yang Z, Kumar S, Nei M.. 1995. A new method of inference of ancestral nucleotide and amino acid sequences. Genetics 141:1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W.. 2002. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 21:5875–5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z.. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 22:2472–2479. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Guo X, Ge C, Ma Z, Jiang M, Li T, Koiwa H, Yang SW, Zhang X.. 2017. KETCH1 imports HYL1 to nucleus for miRNA biogenesis in Arabidopsis. Proc Natl Acad Sci U S A. 114:4011–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Czech B, Brennecke J, Sachidanandam R, Wohlschlegel JA, Perrimon N, Hannon GJ.. 2009. Processing of Drosophila endo-siRNAs depends on a specific Loquacious isoform. RNA 15:1886–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zor T, Selinger Z.. 1996. Linearization of the Bradford protein assay increases its sensitivity: theoretical and experimental studies. Anal Biochem. 236:302–308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.