Abstract
Murine rodents are excellent models for study of adaptive radiations and speciation. Brown Norway rats (Rattus norvegicus) are successful global colonizers and the contributions of their domesticated laboratory strains to biomedical research are well established. To identify nucleotide-based speciation timing of the rat and genomic information contributing to its colonization capabilities, we analyzed 51 whole-genome sequences of wild-derived Brown Norway rats and their sibling species, R. nitidus, and identified over 20 million genetic variants in the wild Brown Norway rats that were absent in the laboratory strains, which substantially expand the reservoir of rat genetic diversity. We showed that divergence of the rat and its siblings coincided with drastic climatic changes that occurred during the Middle Pleistocene. Further, we revealed that there was a geographically widespread influx of genes between Brown Norway rats and the sibling species following the divergence, resulting in numerous introgressed regions in the genomes of admixed Brown Norway rats. Intriguing, genes related to chemical communications among these introgressed regions appeared to contribute to the population-specific adaptations of the admixed Brown Norway rats. Our data reveals evolutionary history of the Brown Norway rat, and offers new insights into the role of climatic changes in speciation of animals and the effect of interspecies introgression on animal adaptation.
Keywords: Rattus norvegicus, speciation, population genomics, climatic changes, interspecies introgression, chemical communications
Introduction
A fundamental goal in evolutionary biology is to elucidate the interaction between adaptation and speciation in generating biological diversity (Wolf et al. 2010; Fitzpatrick et al. 2015). Although, in the past few years, genetic or genomic variations to explain adaptation and speciation have been reported in a few animal cases (Coyne and Orr 2004; Noor and Feder 2006; Schluter 2009; Li et al. 2015; Li et al. 2016; Marques et al. 2016; Martinez Barrio et al. 2016), we are still far from deciphering the genomic basis underlying these two processes (Henning and Meyer 2014).
Murine rodents, including the genera Rattus and Mus, have genetically adapted to a range of environmental conditions, and they provide an exciting opportunity for study of diverse adaptive radiations and speciation (Verneau et al. 1998; Rowe et al. 2011). The Brown Norway rat (Rattus norvegicus, BN rat), assumed to have evolved on the plains of Northeast China and Mongolia (Silver 1937; Southern 1964; Ness et al. 2012), has spread throughout the world on account of its close association with humans (Robinson 1965). It can colonize any area rapidly with an adequate supply of food and water source (Kurta 1995), and thus it is regarded as the most adaptable mammal on the earth after humans (Fragaszy and Perry 2003). It is notorious for damaging agricultural crops and for transmission of infectious diseases to mankind due to its commensal relationship with humans in both native and introduced regions (Gibbs et al. 2004; Lin et al. 2012). Yet, paradoxically, the rat has made invaluable contributions to human health research. It is the first animal species domesticated for purely scientific reasons and has become a vital model organism with more than 500 laboratory strains for life sciences and biomedical purposes for over 150 years (Jacob 1999; Aitman et al. 2008). Although the phylogeny of the BN rat has been reconstructed using limited mitochondrial gene or LINE-1 retrotransposon sequences (Verneau et al. 1998; Rowe et al. 2011; Chingangbam et al. 2015), the reliable time frame for when BN rats became a separate species (Verneau et al. 1998; Pages et al. 2010), and the genomic information underlying their colonization capabilities still remain unclear. The Himalayan field rat (R. nitidus, HF rat) is a sibling species of the BN rat (Verneau et al. 1998; Pages et al. 2010). In contrast to worldwide distribution of BN rats, HF rats have geographically restricted to Southeast Asia, southern and eastern China, and the Himalayan range (Aplin et al. 2008; Smith et al. 2008), and they are found exclusively in hilly regions especially in various types of forests and cropland (Aplin et al. 2008). Thus far, no genomic information on the HF rat has been reported, and this has hindered a comprehensive understanding of the evolutionary history and speciation of the commonly used BN rat species.
Genetic variation introduced via introgression from closely related species play an increasingly important role in fueling adaptation of some animal species (Abi-Rached et al. 2011; Song et al. 2011; Miller et al. 2012; Liu, Steinberg et al. 2014; Ai et al. 2015; Lamichhaney et al. 2015). For instance, an allele of Vkorc1 shared by old world mice contributes to rodenticide warfarin resistance in the recipient species (Song et al. 2011), and an insecticide resistance mutation has been shared between Anopheles sibling species (Clarkson et al. 2014; Norris et al. 2015). Next-generation sequencing based population genomic approaches can facilitate the understanding of speciation and the genetic interchange between sibling or closely related species (Miller et al. 2012; Martin et al. 2013; Liu, Steinberg et al. 2014; Gante et al. 2016). For instance, recent speciation and rapid evolutionary adaptation in polar bears has been observed in previous population genomic study (Liu, Lorenzen et al. 2014). Similarly, genome-wide introgression among distantly related Heliconius butterfly species has been reported (Zhang et al. 2016). Here, we have generated and analyzed whole-genome sequence data of wild-derived BN rats and their sibling species to: i) estimate when BN rats and their sibling species diverged from the most recent common ancestor; ii) infer the demographic history of both species and their genetic interrelationships following the divergence; and iii) detect selective sweeps that occurred in the populations of the BN rat, so as to provide insight into adaptations to colonization of the rat species.
Results
Genome-Scale Sequencing and Population Relationship of the Two Rat Species
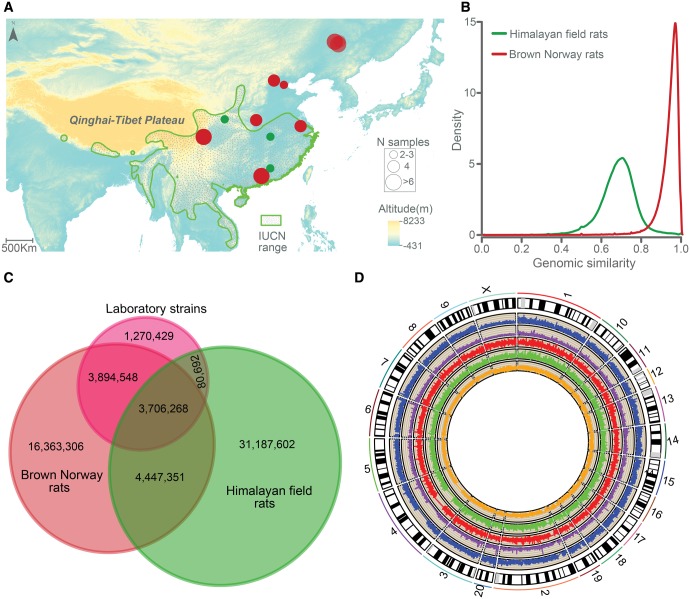
Forty-four BN rat individuals from seven locations across their native regions of China, representing different mitochondrial lineages (Song et al. 2014), were collected (fig. 1A, Supplementary table S1, Supplementary Material online). In addition, we also sequenced their sibling species, the HF rat, from three representative locations in its homeland (Aplin et al. 2008). After aligning short reads to the reference genome (RGSC 5.0) and applying stringent quality control criteria, we identified a total of 28,411,473 single nucleotide polymorphisms (SNPs) within wild BN rats and 39,421,913 within HF rats (fig. 1C), and 8,153,619 SNPs were shared between the two species. Identity score analysis clearly illustrated that HF rats had less genomic similarity to the RGSC 5.0 reference genome than BN rats, especially in X chromosome (fig. 1B, Supplementary fig. S1, Supplementary Material online). The nucleotide diversity in HF rats was higher than that in BN rats in both autosomes (median πHF(Auto)/πBN(Auto)=2.01; P < 2.2 × 10−16, Wilcoxon rank-sum test) and sex chromosome (median πHF(Xchr)/πBN(Xchr)=1.78; P < 2.2 × 10−16, Wilcoxon rank-sum test) (fig. 1D, Supplementary fig. S2, Supplementary Material online). More than 28% of SNPs (11,333,229) in HF rats were fixed, suggesting the divergence between the two species.
Fig. 1.
Genomic variation in Brown Norway and Himalayan field rats. (A) Geographical distribution of Brown Norway (red circle) and Himalayan field (green circle) rats used in this study. The colored areas in green indicate the geographic range of Himalayan field rats based on IUCN distribution (Aplin, et al. 2008). (B) Genomic similarity of Brown Norway and Himalayan field rats to the RGSC 5.0 reference genome. (C) The number of SNPs unique to and shared by Himalayan field, Brown Norway and laboratory rat strains. (D) Genome-wide distribution of nucleotide diversity, Tajima’s D value and recombination rate in the Brown Norway and Himalayan field rats. The two circles adjacent to the karyotypes show lines representing nucleotide diversity in the Brown Norway (purple) and Himalayan field (blue) rat. The innermost circle shows lines representing local recombination rate (cM/Mb) for the Brown Norway rats (orange). The next two circles show lines representing Tajima’s D value in the Brown Norway (green) and Himalayan field rats (red).
We then compared the SNPs that we had identified in wild BN rats with those from the laboratory rat strains (Atanur et al. 2013; Hermsen et al. 2015). The average number of SNPs in the wild BN rats was 6.54 million per individual, which was higher than that in laboratory rat strains (3.46 million SNPs per individual). Approximately 85% (7,600,816) of the total variants in the laboratory strains were observed in our wild SNP data set, whereas more than 73% (20,810,657) of the total variants that we identified in the wild BN rats, including 1,229 nonsense variants and 81,728 missense variants, were absent in laboratory strains. These novel variants substantially expand the reservoir of rat genetic diversity.
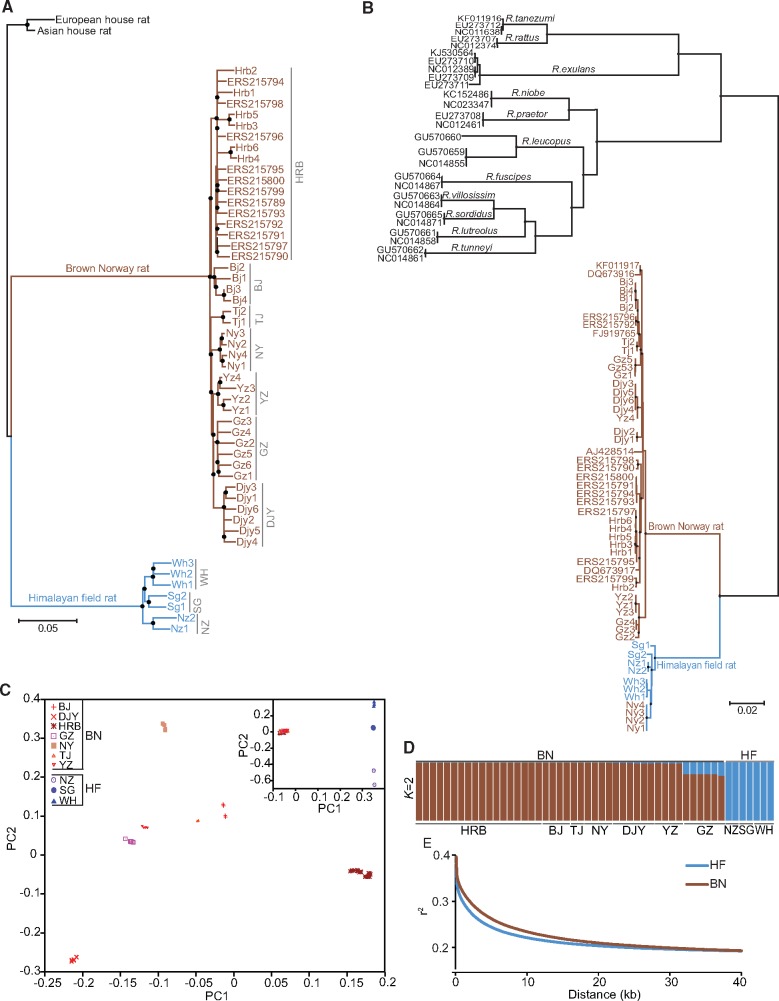
Phylogenetic analysis using assembled mitochondrial genomes of the sequenced wild Rattus individuals and publicly available mitochondrial genomes of Rattus spp. confirmed the sibling relationship between the BN and the HF rat (Verneau et al. 1998; Pages et al. 2010) (fig. 2). Principal component and structure analysis could define the species relationship between these two species (fig. 2). By calculating the pairwise linkage disequilibrium (LD) between polymorphic sites for all autosome regions in each species, we found that LD decreased more rapidly in HF than in BN populations, which was consistent with the higher levels of nucleotide diversity observed in HF rats (fig. 2).
Fig. 2.
Population genetic analyses of Brown Norway (BN) and Himalayan field (HF) rats. (A) Neighbor-joining phylogenetic tree based on whole-genome SNPs, with filled black circles at individual nodes indicating bootstrap support >99%. (B) Bayesian consensus tree reconstructed from mitochondrial genomes, with filled black circles at individual nodes indicating posterior probabilities >0.99 and bootstrap support >99%. Published mitochondrial genome sequences of the Rattus genus were downloaded from GenBank and Accession numbers are indicated. (C) Principal component analysis (PCA) of the first two components. Inset is for all individuals and the large panel is for BN rats only. (D) Population structure plots with K = 2. Individuals are represented as rows partitioned into segments corresponding to the inferred membership as indicated by the colors. The population of each individual is indicated at the bottom of the figure. (E) Decay of linkage disequilibrium of HF and BN populations measured by r2.
Demographic History and Speciation of the Two Rat Species
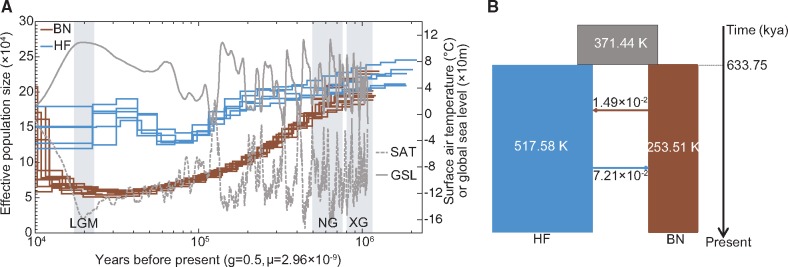
Two models, “migration” and “no migration”, were used in a Bayesian coalescent based approach (Generalized Phylogenetic Coalescent Sampler, G-PhoCS) (Gronau et al. 2011) to infer effective population size (Ne), divergence time, and rates of gene flow between BN and HF rat species. The estimated Ne values for BN rats and HF rats were 2.53 × 105 and 5.18 × 105, respectively, and the inferred ancestral Ne was 3.71 ×105 (fig. 3B). The migration rate from HF rats to BN rats was 7.21 ×10−2 per generation, which was higher than that from BN rats to HF rats (1.49 ×10−2 per generation) (fig. 3B, Supplementary table S2, Supplementary Material online). The estimated time when the two species diverged was 621.62–643.58 thousand years ago (kya), which was compatible with the divergence time inferred informally from pairwise sequentially Markovian coalescent (PSMC) analysis. It should be noted that the divergence coincided with the most extensive glaciation (Naynayxungla Glaciation, 780–500 kya) that occurred in East Asia during the Middle Pleistocene (Zheng et al. 2002). According to data from the National Climatic Data Center (NCDC), both global sea level and surface air temperature fluctuated frequently during the past 1 million years. During the Quaternary Period of climatic changes, a more drastic reduction of Ne occurred in BN rats than in HF rats based on inferences from PSMC analysis, and the most severe bottleneck in the BN rat populations occurred about 20 kya (Deinum et al. 2015), the end of the Last Glacial Maximum (fig. 3A). Our results suggest that the drastic changes in the climate may have contributed to the speciation of these two species.
Fig. 3.
Demographic history of Brown Norway (BN) and Himalayan field (HF) rats. (A) Demographic history inferred using PSMC. Three major glacial events, Xixiabangma Glaciation (XG, 1,170–800 thousand years ago, kya), Naynayxungla Glaciation (NG, 780–500 kya), and the Last Glacial Maximum (LGM, 20 kya), are shaded in grey. SAT: surface air temperature relative to the present; GSL: reverse-transformed global sea level relative to the present. (B) Schematic of demographic scenario modeled using G-PhoCS. The ancestral population is shown in grey, HF is in blue, and BN is in brown. The values beside the arrows indicate the average number of migrants per generation between the HF and BN rat species.
Discordance of Mitochondrial and Nuclear Evolutionary Histories in NY Population of the BN Rat
Although principal component and structure analyses could define the species relationship between these two species, we found that the maternal genetic classification of NY population, which was collected from the Nanyang City in Henan Province of China and morphologically classified as BN rat species, did not follow taxonomic boundaries. In the mitochondrial phylogeny, individuals from this population were more closely related to HF rats than to the rest of BN rats (fig. 2B). However, phylogenetic tree based on autosome-wide SNPs exhibited a pattern different from that revealed from the mitochondrial genome, and the NY group was more closely related to nonNY BN rat individuals than to HF rats, in accordance with their morphological characteristics (fig. 2A). Three evolutionary scenarios, including ancient hybridization, recent hybridization, and incomplete lineage sorting, can be used to explain the discordance. According to the recombination-based test, the probability to retain a 100,000 bp fragment in the NY population due to incomplete lineage sorting was <1.11 × 10−16. More than 100,000 bp introgressed fragments observed in the genomes of NY population indicates that another two scenarios, ancient or recent gene flow, may have occurred in the NY population after the divergence of the two species.
Geographically Wide Introgression between the HF Rat and the BN Rat
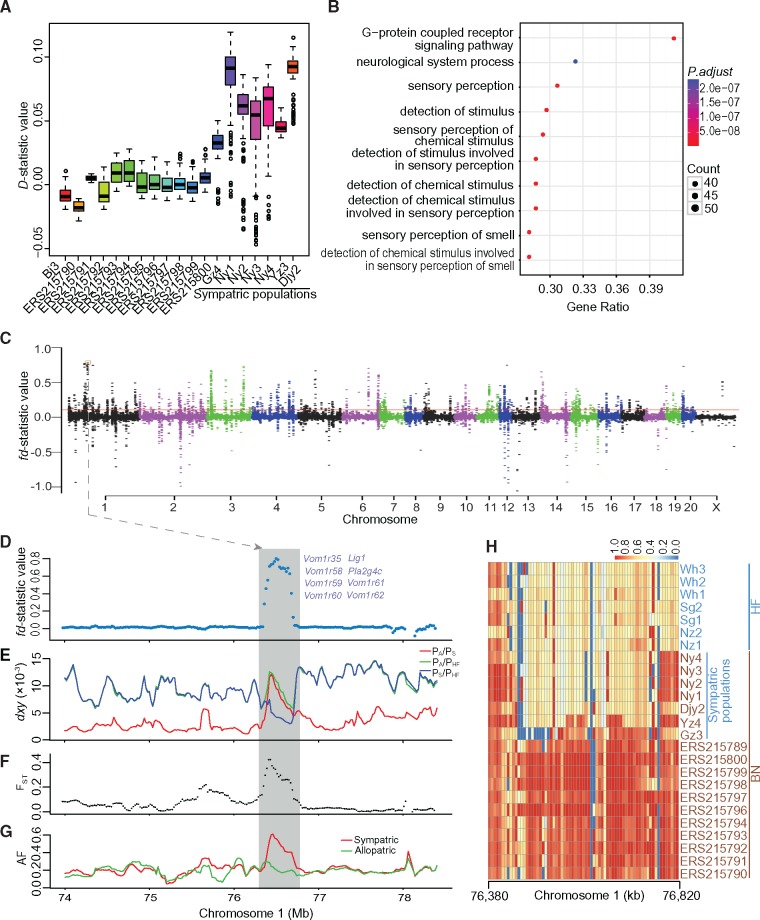
The discrepancy between mitochondrial and nuclear evolutionary histories suggests that, in NY population, gene flow occurred after the divergence of these two species. To determine whether admixture occurred more widely, we used the more sensitive D statistics (Green et al. 2010) to detect the admixture between each of deep-sequenced BN rats and each of HF rats from the genome level. Our D-statistic results provided clear evidence from the genome level that gene flow between the two species had a geographically wide impact, with DNA of HF rat being found within the genomes of sympatrically distributed BN rat individuals (fig. 4A and Supplementary table S3, Supplementary Material online).
Fig. 4.
Identification of introgressed regions in the Brown Norway (BN) rat genome. (A) Patterson’s D-statistic measure of admixture in deep-sequenced BN rats. Box-and-whisker plots showing the range of D-statistic values for a BN individual compared with every other BN individual with every HF rat as a candidate introgressor (Supplementary table S3, Supplementary Material online). Circles indicate data that fall outside of the 25th to 75th percentile range (outliers). Statistically significant D-statistic values indicate that the subject BN individual shares an excess of derived alleles with Himalayan field (HF) rat. (B) Functional enrichment of genes covered in the adaptive introgressed regions. (C) Introgressed regions identified in the BN genome. A modified f-statistic (fd) for 100-kb windows with 20 kb steps is plotted along the chromosomes. Each dot represents a 100-kb window. Red horizontal line corresponds to FDR 5% significance level threshold. The two strongest signals of introgression on chromosome 1 (boxed) are plotted in (D). (E) Mean pairwise sequence divergence (dxy) between different populations. PA and PS represent allopatrically and sympatrically distributed populations of BN rats compared with distributions of HF rats, respectively; PHF, populations of HF rats. (F) Population differentiation (FST) between allopatric and sympatric populations of BN rats. (G) Allele frequency (AF) of allopatric and sympatric populations of BN rats. (H) Identical score of HF and BN individuals of introgressed regions on chromosome 1 as in (D–G) are shown for SNPs within each 50-kb windows.
To further locate the regions of admixture in the genome of sympatrically distributed BN rats, we calculated the modified f-statistic (ƒd) (Martin et al. 2015) value for each 100 kb window with a step size of 20 kb across the genome. In autosomes of sympatrically distributed BN rats that we have sampled, we found 346 admixture blocks with the length from 100,000 bp to 1.42 Mbp (fig. 4C, Supplementary table S4, Supplementary Material online), and the proportion of admixed fragments in the genomes of these rats was about 1.59%. The recombination rates within these admixed regions in allopatric BN rat population were higher than those within nonadmixed regions (P = 1.04 × 10−9, Wilcoxon rank-sum test). If these admixed regions were derived from random assortment of ancestral polymorphisms, they should exist in some populations before the divergence of the two species. As recombination can break up the haplotypes over time due to drift, we estimated the expected length of a shared ancestral sequence over the divergence time of the two rat species. Using 600 thousand years as the divergence time of the two rat species and a recombination rate of 0.0837 cm/Mb, the lower bound of recombination rate in these admixed regions estimated by FastEPRR (Supplementary table S4, Supplementary Material online), we expected that the length of a shared ancestral sequence was about 1,991 bp, and the probability to retain a 40,000 bp fragment was 3.97 × 10−8. Thus, the admixed regions in the genomes of BN rats were unlikely to be retained due to incomplete ancestral lineage sorting. Using the length of each introgressed tracts as the expected length and the inferred recombination rates of local regions, we estimated the ages of each introgressed regions. We found that these admixtures occurred across a broad range of time scales from 300 to 9,500 generations.
Identification and Annotation of Putative Adaptive Introgressed Loci
Statistics based on allelic frequencies of introgressed loci in a specific admixed population related to nonintrogressed populations were particularly efficient in detecting adaptive introgression (Racimo et al. 2015, 2016). Integrative analysis of allele frequencies in sympatric and allopatric populations of BN rats and genetic differentiation between these two populations, we identified 92 putative adaptive introgressed loci from the 346 introgressed regions (Supplementary table S5, Supplementary Material online). Gene ontology analysis of genes covered by the 92 adaptive introgressed regions showed some enrichment of terms such as “sensory perception (GO:0007600)” and “G-protein coupled receptor signaling pathway (GO:0007186)” (fig. 4B and Supplementary table S6, Supplementary Material online). The region with the strongest introgression signal in the chromosome 1 of BN rat contains vomeronasal 1 receptor cluster (fig. 4D–H). In mice, genes related to such chemical communications have been enriched in introgressed haplotypes of its subspecies (Staubach et al. 2012; Janousek et al. 2015), and they are easily subject to population-specific adaptations (Teeter et al. 2008). It is conceivable that the adaptive introgressed regions related to chemical communications may have contributed to the population-specific adaptations of the admixed BN rats.
Another example of adaptive introgressed region is 620,000 bp in chromosome 11, which contains only one gene, Hmgb2. Although Hmgb2 is broadly expressed during mouse embryogenesis, it is restricted in adult lymphoid organs and testes, and male Hmgb2−/− mice have defects in fertility (Ronfani et al. 2001), which indicating the role of Hmgb2 in the differentiation of germ cell (Ronfani et al. 2001) and mesenchymal stem cell (Taniguchi et al. 2011). The introgression of this gene might contribute to an individual’s reproduction ability of the admixed BN rats.
Selective Sweeps in the BN Rat Genome
Based on integrative analysis of multilocus allele frequency differentiation, linkage disequilibrium and genetic polymorphism, we identified 325 candidate selective sweep regions that occurred in the autosomes of the BN rat (fig. 5A and Supplementary table S7, Supplementary Material online). Gene over-representation analysis for protein-coding genes harbored in these regions revealed two major categories: metabolism and immune response (fig. 5D and Supplementary table S8, Supplementary Material online). Genes related to metabolism were particularly interesting, because food source for commensal BN rats provided by humans might have been the driving force for the selective sweep of these kinds of genes. Multiple regions involved in lipid metabolic process (GO:0006629), response to nutrient (GO:0007584), response to drug (GO:0042493), and xenobiotic metabolic process (GO:0006805), showed evidence of positive selection. An example of such region is the one containing Acsl1, liver acyl-CoA levels are reduced when Acsl1 is conditionally knocked out in the mouse liver (Li et al. 2009). Another example is the region harbored Acacb. Mice for a null mutation of Acacb exhibit high levels of fatty acid oxidation, and low levels of fat accumulation in their adipose tissue and liver (Abu-Elheiga et al. 2001). Genes related to immune response might contribute to the defense response of BN rats to their external habitat. Several sweep regions containing genes involved in immune response, including response to interferon-gamma (GO:0034341), complement activation (GO:0006956), innate immune response (GO:0045087), and mast cell mediated immunity (GO:0002448), were observed in our integrative analysis. Three representative genes located in such sweep regions were Trib1, Anxa1 and Serpinb9, and loss of function or mutation of each gene has been associated with defects in immune response (Hannon et al. 2003; Zhang et al. 2006; Yamamoto et al. 2007). For example, mutations in Anxa1 result in increased inflammatory response and decreased macrophage activity (Hannon et al. 2003), and homozygous mice with null mutation of Serpinb9 show defective cytotoxic T lymphocytes immunity (Zhang et al. 2006).
Fig. 5.
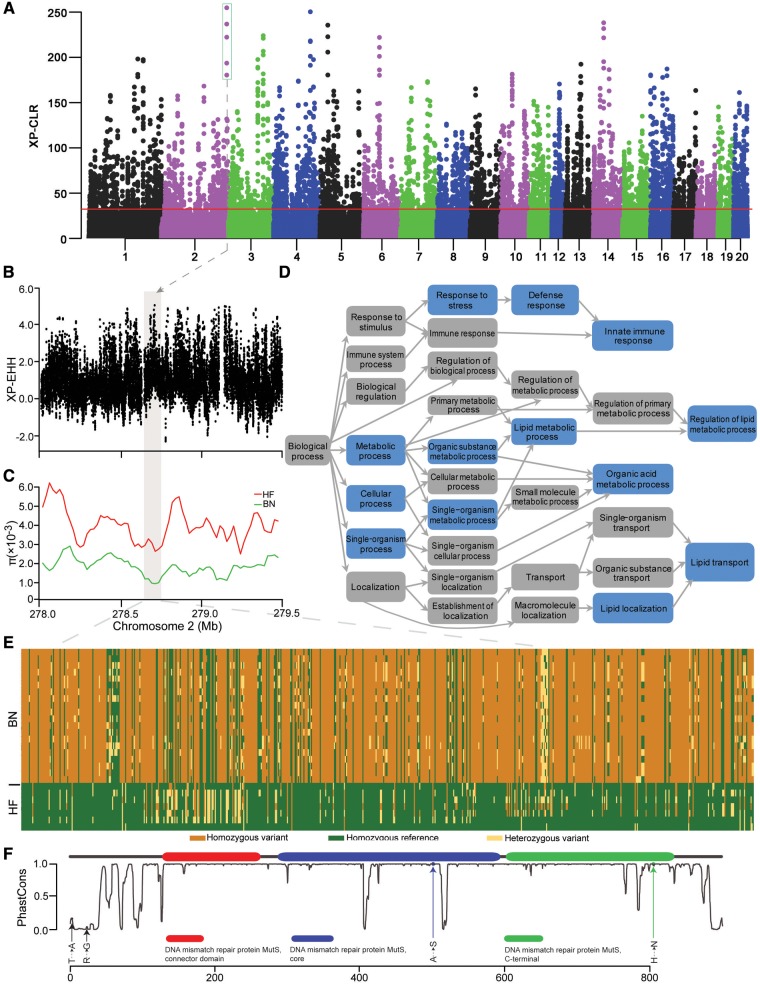
Genomic regions with selective sweep signals in the Brown Norway (BN) rat. (A) Distribution of cross-population composite likelihood ratio (XP-CLR) for 100 kb windows with 20 kb steps. Red horizontal line corresponds to FDR 5% significance level threshold. (B) Score of standardized cross-population extended haplotype homozygosity (XP-EHH) for each SNP along chromosome 2. (C) Nucleotide diversity (π) values of Himalayan field (HF) and BN rat are plotted using a 100-kb sliding window. The box shaded in grey in (B, C) represents the strongest signals of XP-CLR on chromosome 2. (D) Functional enrichment of genes covered in the sweep regions. Blue shading indicates significantly enriched categories in the BN rat lineage. (E) Genotypes of SNPs in putative selective sweeps containing the Msh4 gene in the BN and HF rats. (F) The distribution of fixed nonsynonymous BN rat mutations (arrows) compared with the HF rat. The gray curve shows the amino acid conservation scores; higher scores indicate higher conservation across 13 vertebrate species. The x axis shows the amino acid position from the N-terminal.
The most prominent signature of allele frequency differentiation was located on chromosome 2 (fig. 5A), which was further supported by the strong evidence of cross-population extended haplotype homozygosity (fig. 5B) and reduced nucleotide diversity (fig. 5C). The gene harbored in this region was Msh4, which has been implicated in reproduction of mice. As a member of the DNA mismatch repair mutS family, Msh4 plays a role in directing the recombinational repair of DNA double-strand breaks during meiosis (Novak et al. 2001; Nishant et al. 2010). Both male and female mice for a Msh4 null mutation show sterility and defects in pairing during meiosis (Kneitz et al. 2000). Multiple SNPs within this regions were found in the wild BN rat genome (fig. 5E), and two fixed missense mutations were present at conserved sites within the Msh4 gene (fig. 5F). Although their functional effect need further characterized, these missense mutations provided an intriguing resource for understanding the adaptation of the species.
Neural activities related to alertness, anxiety, and aggression are essential for survival of animals (Steimer 2002). Three anxiety- and aggression-related regions were observed in our research. One such region is denoted by the Gad2 gene in chromosome 17, which is responsible for GABA synthesis for synaptic release, and mutations of this gene have been implicated in anxiety-like behavior and intermale aggression (Stork et al. 2000). A further two regions harbored protein-coding genes, Vamp7 and Park2. Mice for Vamp7 knock-out exhibit increased anxiety (Danglot et al. 2012), and transmission of dopamine and glutamate is impaired in Park2−/− mice, leading to decreased exploratory behavior (Cremer et al. 2015). Additionally, several regions involved in sensing local environmental stimuli (hearing or smell) were also identified to be under positive selection in the BN rat genome. The results suggest that functional categories involved in metabolism, immune, and defense response might contribute to the adaptation of wild BN rats to their habitats.
Discussion
In the present study, using whole-genome sequencing, we generated a comprehensive resource on rat genetic variation and identified more than 20 million genetic variants that were absent in laboratory strains. These novel SNPs provide a valuable resource for identifying functionally important variants that contribute to the complex phenotypes of this model organism. Further, we performed de novo genome assembly for three Rattus species (R. nitidus, R. tanezumi, and R. rattus), and generated 2.5–2.7 Gb genome survey sequences of each species, which provided an ideal data set for comparative genomic analyses of the Rattus genus. Two important parameters in population genetics, Ne and recombination rate, were estimated in our study. Based on 4-fold degenerate sites of 12 wild-caught BN rats within one population, Deinum et al. estimated the recent Ne of BN rat was 1.24 × 105 (Deinum et al. 2015). Using a Bayesian coalescent analysis based on 4,542 autosomal, unlinked, and neutrally evolving 1-kb loci from multiple populations, we inferred Ne value for BN rats was 2.53 × 105. It was about 2-fold but the same order-of-magnitude as that in Deinum et al.’s study. Using machine learning approach based on the intraspecific DNA polymorphism data of autosome (Lin et al. 2013; Gao et al. 2016), we estimated that the recombination rates for 100 kb windows in wild BN rats ranged from 0.0073 to 0.5832 cM/Mb with an average of 0.3873 cM/Mb when assuming Ne of 2.53 × 105. This average rate was about 62% of that (0.62 cM/Mb for autosome) reported in Jensen-Seaman et al.’s study (Jensen-Seaman et al. 2004). If a smaller Ne of 1.24 × 105 was used as suggested by Deinum et al. (2015), this rate would increase by a factor of two. In Jensen-Seaman et al.’s study, it was estimated based on intercrosses between specific inbred rat strains without considering variation within the species (Jensen-Seaman et al. 2004). Given that there is variation in the rate of recombination between individuals of other species (Koehler et al. 2002; Kong et al. 2002), Jensen-Seaman et al.’s estimation might not hold for the species generally (Jensen-Seaman et al. 2004). The recombination rate we provided herein for BN rats were based on multiple wild populations, and it may be a more reliable estimation for the species, which may facilitate future studies on evolutionary genomics and dissection of complex traits of the rat species.
Compared with their geographically restricted sibling species, the globally distributed species of BN rats exhibited the lower levels of nucleotide diversity in accordance with their more drastic declining of Ne during period of Quaternary climatic changes. Based on the extant distribution of the HF rat (Aplin et al. 2008; Smith et al. 2008) (fig. 1A), we inferred that the less intensive response of ancestral Ne to Quaternary climatic changes in HF rats might have resulted from their distribution around the Qinghai-Tibet Plateau. Similarly, multiple glacial refuges around the Plateau during Pleistocene climate fluctuations have been reported in some animals (Qu et al. 2010; Zhan et al. 2011; Hofmann 2012). The impact of Plio-Pleistocene climatic oscillations on the current genetic structure of many animals is widely recognized (Roy et al. 1996; Liu et al. 2011; Zhan et al. 2011; Hofmann 2012; Miller et al. 2012; Stewart and Stringer 2012; Zhao et al. 2013), but their importance in driving speciation remains elusive (Lovette 2005; Hoskin et al. 2011). Through a Bayesian coalescent-based approach, we inferred that the estimated time of divergence between the two species coincided with the period of most extensive glaciation in east Asia during the Middle Pleistocene (Zheng et al. 2002). During that time, the global sea level and the surface air temperature fluctuated frequently. Climate may drive speciation through at least two mechanisms: firstly, the extreme climatic conditions impose divergent selection on populations that occupy different habitats, and secondly, differences in climate over geographical space serve as a barrier to dispersal between allopatric populations (Hua and Wiens 2013). Although we cannot identify the specific mechanism of the speciation, we here provide evidence to support the climatically induced speciation model.
It is surprising that little is known regarding the genetic background of the adaptations of the BN rat, the most common and successful mammalian species (Fragaszy and Perry 2003). In the present study, we found that multiple genes involved in metabolism, immune and defense response had undergone positive selection in its genome, which suggests the essential role of these kinds of genes in the adaptation of wild BN rats to their habitats. Introgression from closely related species is another important source of genetic variation in natural populations, and adaptive introgression may lead to rapid adaptation of some mammalian species than the process with mutation and selection alone (Abi-Rached et al. 2011; Miller et al. 2012; Ai et al. 2015; Racimo et al. 2016). For example, introgression of one Vkorc1 allele contributes to rodenticide resistance in the recipient mouse species (Song et al. 2011), and an mutation related to insecticide resistance has been introgressed between Anopheles sibling species (Clarkson et al. 2014; Norris et al. 2015). Even for humans, some research found modern humans had inherited alleles critical to immune function from an ancient admixture with archaic humans (Abi-Rached et al. 2011). Through our population genomic analysis, we found that admixture had occurred between the BN rat and its sibling species, and the recombination rates within the admixed regions were significantly higher than those within nonadmixed regions. Considering the divergence time of the two species, the length and local recombination rate of the admixed regions, we inferred that the admixed regions resulted not from ancestral polymorphism, but from the gene flow after the divergence of these two sibling species. Increased allelic frequencies of introgressed loci in a specific admixed population due to positive selection, were particularly useful for detecting adaptive introgression (Racimo et al. 2015, 2016). Using this strategy, we identified 92 putative adaptive introgressed loci in the sympatric BN rat population. Functional enrichment analysis of candidate genes included in the adaptive introgressed regions revealed “chemosensory perception” terms. Similarly, these gene terms have been enriched in the introgressed haplotypes between house mouse subspecies (Staubach et al. 2012; Janousek et al. 2015). Rodent olfactory receptors acting at the cell periphery often undergo rapid molecular evolution and extensive segmental duplications (Godfrey et al. 2004; Park et al. 2011), and in mice they are easily subject to population-specific adaptations (Teeter et al. 2008; Staubach et al. 2012; Janousek et al. 2015). The adaptive introgressed regions related to chemical communications might contribute to the population-specific adaptations of the admixed BN rats. Our study provided the first example that the effect of interspecific introgression on the adaptation of the rat species. In addition to our case, other interspecies hybridization or introgression events might exist in the species. Similarly, multiple introgression events have been reported between house mouse subspecies (M. m. domesticus and M. m. musculus) (Staubach et al. 2012; Janousek et al. 2015), and between M. m. domesticus and M. spretus (Song et al. 2011; Liu, Steinberg et al. 2014). However, consequence of natural selection on other kinds of genetic variations, such as copy number variation and structural variation, might also contribute to the adaptation of the species, and their roles in adaptation and colonization of the widespread species need to be further investigated.
In summary, we reveal the speciation between the BN rat and its sibling species, and present evidence that interspecies introgression has contributed to the population-specific adaptations of BN rats. Our data offers new insights into the role of climatic changes in speciation of animals and the effect of interspecies introgression on animal adaptation.
Materials and Methods
Samples, DNA Extraction, and Library Preparation
Ninety-two BN rat and HF rat individuals from the original homeland of these two species (Silver 1937; Southern 1964; Aplin et al. 2008; Ness et al. 2012; Song et al. 2014), were trapped and identified using mitochondrial cytochrome oxidase subunit I (COI) as barcode in 2014, and relatedness of samples from the same locality were assessed using sequences of mitochondrial COI, cytochrome B, and D-loop regions before whole genome sequencing. And then, 32 BN rat individuals from seven locations and seven HF rat individuals from three populations were selected for whole genome sequencing (fig. 1A). An individual of Asian house rat (R. tanezumi) was also used in this study as an outgroup (Teng et al. 2016). Genomic DNA samples were extracted from small pieces of tail using a TailGen DNA extraction Kit (CWBIO, Beijing, China). The quality and integrity of the extracted DNA were checked by measuring the A260/A280 ratio, using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA), and by agarose gel electrophoresis. Libraries with an insert size of ∼300 bp were prepared, and sequenced at 2 × 100 bp on an Illumina HiSeq 2000 instrument in Beijing Institutes of Life Science, Chinese Academy of Sciences (Beijing, China), or 2 × 150 bp on HiSeq X Ten instrument in Novogene Bioinformatics Institute (Beijing, China), respectively. The raw sequencing data sets are deposited in SRA (http://www.ncbi.nlm.nih.gov/sra/) under accession number: SRP078989. Our previously generated 12 whole genome sequences of wild-caught BN rat individuals from their potential ancestral ranges and the whole-genome sequence of an individual of the European house rat (R. rattus), also known as the black rat, were reanalyzed in this study (Deinum et al. 2015). After filtering out raw sequencing reads containing adapters, and reads of low quality, the remaining clean reads were aligned to the RGSC5.0 reference genome using BWA version 0.7.5a-r405 with default parameters (Li and Durbin 2009).
Small-Variant Calling
SNPs were detected using the Genome Analysis Toolkit (GATK version 3.2-2-gec30cee) HaplotypeCaller (McKenna et al. 2010) according to the recommendation of GATK Best Practice (https://www.broadinstitute.org/gatk/guide/best-practices.php). Briefly, before variants calling, the aligned data were processed with the following steps: i) duplicate reads were masked independently for each library using Picard v1.119 (http://sourceforge.net/projects/picard/); ii) reads were realigned around the InDels; iii) 1,726,154 SNPs shared between the top 30% of high-quality SNPs in the data set of laboratory rat strains (Atanur et al. 2013) and dbSNP138 were used as the training set for the variant quality score recalibration function of GATK to exclude sequencer, alignment, or data-processing artifacts (DePristo et al. 2011). Then, multi-sample instead of single-sample calling was used by GATK-HaplotypeCaller to improve the accuracy of variant calling. Variant calling data sets in the VCF format are available at http://159.226.67.237:6080/sun/Rnor/Rnorvegicus.Rnitidus.SNP.Chrall.vcf.
Genome-Wide Identity Scores
Similarities of the deep-sequenced genomes to the RGSC5.0 reference genome were calculated and visualized as the average of the identity scores of individual SNPs within 50 kb windows along the genome, according to the formula described by Ai et al. (2015).
De Novo Assembly of Nuclear and Mitochondrial Genome
The sequence reads of a representative individual from each of the three nonreference species, R. nitidus, R. tanezumi, and R. rattus, were assembled separately using SOAPdenovo V 2.04 (Luo et al. 2012) with the following parameters: -K 41 -R -M 2 -m 63 -E -F -w -k 41. In addition, we used BWA version 0.7.5a-r405 with default parameters to align the reads from each sample to the RGSC5.0 mitochondrial reference sequence. These aligned reads were used as input to assemble the mtDNA genome of each sample using the reference-based assembler mitoMaker v1.14 (http://sourceforge.net/projects/mitomaker/).
Population Structure Analyses
To investigate the population relationship within HF and BN rat species, a neighbor-joining phylogenetic tree was constructed by means of MEGA-CC (Kumar et al. 2012), with P-distance model and bootstrap analysis using all identified SNPs. Published mitochondrial genome sequences of Rattus genus (accession numbers listed in fig. 2B) were downloaded from GenBank. Phylogenetic analyses based on the sequences of 13 mitochondrial protein-coding genes were conducted using MrBayes v3.22 (Ronquist et al. 2012). Genetic structure was assessed using principal component analysis (PCA) with all bi-allelic SNPs (Patterson et al. 2006; Price et al. 2006). ADMIMXTURE was also employed to analyze the population structure (Alexander et al. 2009). To mitigate the effects of linkage disequilibrium (LD) on genetic structure, we pruned the markers using the “–indep-pairwise 50 5 0.05” option of PLINK (Purcell et al. 2007).
Genetic Diversity and LD Analyses
Pairwise nucleotide diversity and Tajima’s D value within BN or HF rat species were calculated using VCFtools version 0.1.13 (Danecek et al. 2011) by means of the π (-site-pi) and the Tajima’s D (-TajimaD) with 100 kb sliding window, respectively. All individuals of HF rats were used to estimate LD. To mitigate the effect of sample size on LD, seven randomly selected deep-sequenced individuals of BN rats were used to calculate the correlation coefficient (r2) between any two loci on each autosome by means of Plink v1.90 (Purcell et al. 2007) with the option of “–r2 -ld-window 3000 -ld-window-kb 200 -ld-window-r2 0 -maf 0.05 -hwe 0.05.” The average r2 was calculated for pairwise SNPs in a 20-bp window with an in-house Perl script.
Analysis of Local Recombination Rate
To obtain local recombination rate of different regions across the BN rat genome, we performed recombination rate analysis using a machine learning approach implemented in FastEPRR with the nonoverlapped sliding window length of 100 kb (Lin et al. 2013; Gao et al. 2016). The local recombination rate was estimated based on the intraspecific DNA polymorphism data of whole BN rat population according to the formula: ρ = 4Ne × r. To exclude the effect of introgression on the estimation, we also estimated the local recombination rate based on the DNA polymorphism data of allopatric BN rat population. Our previously reported Ne (Ness et al. 2012; Deinum et al. 2015) and the size inferred by the following G-PhoCS was both used in the estimation.
Inference of Demographic History and Population Divergence Time
In order to obtain a detailed population history and changes in ancestral population sizes in response to Quaternary climatic change, we used PSMC (Li and Durbin 2011) to analyze our deep-sequenced genomes, using 0.5 years for the generation time, and 2.96 × 10−9 per site per generation for the mutation rate according to our previous report (Ness et al. 2012; Deinum et al. 2015). We downloaded data (ID: noaa-recon-11932) of atmospheric surface air temperature relative to present (°C) and global sea level relative to present (10 m) for the past 1 million years from the NCDC (http://www.ncdc.noaa.gov/). We also used G-PhoCS (Gronau et al. 2011) with a model of no gene flow and bi-directional gene flow to infer Ne, migration rate, and divergence time between the BN and HF rats. A total of 4,542 autosomal neutral loci of 1 kb in length were selected along the genome according to the following criteria: i) these loci contained no coding DNA, no EST, and no LincRNA (NONCODEv4) (Xie et al. 2014); ii) no RepeatMasked and simple repeat elements and no segmental duplications in the reference genome; iii) no N’s in the reference genome; iv) mean score of PhastCons within 1kb of <0.5; v) at least 100 kb away from any known gene; and vi) interlocus distance of these loci was more than 50 kb. One million iterations were run with the first 10% iterations discarded as burn-in, and the convergence of Markov chain Monte Carlo (MCMC) algorithms was evaluated by Tracer v1.6 (Rambaut et al. 2014). Ne for each species was calculated according to the formula: Ne = θ/(4 × µ), where θ refers to the mutation rate of the species. The divergence time (T) between the two species was inferred using the same generation time, and the same mutation rate as described above according to our previous report (Ness et al. 2012; Deinum et al. 2015). G-PhoCS infers the migration rate from the source species (S) to the target species (T) as mS−T, which can be converted into migrants from S to T per generation (MS−T) with MS−T = mS−T × θT, where θT is the population mutation rate of the target population.
Detection of Admixture
Patterson’s D statistic (Green et al. 2010; Durand et al. 2011) was used to examine whether some individuals of BN rat shared more alleles with HF rats than with other individuals of BN rats. A tree topology from sites covered by the four individuals as [[[H1, H2], H3], O], using European house rat as an outgroup (O), was used to test whether two conspecific individuals, H1 and H2, shared more alleles with a candidate introgressor, H3. For sites where European house rat possessed the ancestral allele “A” and H3 possessed the derived allele “B,” we calculated the presence of the “ABBA” pattern, versus the “BABA” pattern in the topology. A 5-Mb block size was used to calculate the “ABBA” and “BABA” pattern, and the standard error for both estimates was calculated using a weighted block jack-knife (Green et al. 2010). Analyses for all combinations of pairs of deep-sequenced individuals of BN rats and candidate introgressors of HF rats were performed. Following the detection of gene flow among individuals at the genome level, we used a modified ƒ(d) to locate introgressed loci (Martin et al. 2015) at the population level, using a 100-kb sliding window with 20-kb stepping. The frequency of the derived “ABBA” and “BABA” allele at each site (Martin et al. 2015) in each PA, PS, PHF, and outgroup population was used, where PA and PS represent allopatrically and sympatrically distributed populations of BN rats compared with distributions of HF rat population (PHF), respectively. Then, P values were estimated based on Z-transformed ƒ(d) values using the standard normal distribution, and were further corrected by multiple testing using the Benjamini–Hochberg false discovery rate (FDR) method (Benjamini and Hochberg 1995). Windows with positive ƒ(d) values and FDR values <0.05 were selected, and then the selected windows that were ≤100 kb apart were merged into a single region and regarded as introgressed regions. Mean pairwise sequence divergence (dxy) between the two sibling species was also calculated using a 100-kb sliding window with 20-kb stepping (Martin et al. 2015).
Probability and Expected Length of Introgressed Regions from Shared Ancestral Lineage
The distribution of local recombination rate of admixed blocks in the BN rat genome was examined. A recombination-based test (Huerta-Sanchez et al. 2014) was then used to assess the incomplete lineage sorting model under the hypothesis of the putative introgressive haplotypes existed in some BN rat genomes before the divergence of the HF and BN rat species. The expected length of a shared ancestral sequence (L) can be calculated as 1/(r × t) (Huerta-Sanchez et al. 2014), where r is the recombination rate per generation per base pair of local region, t is the generation since admixture. Assuming an exponential distribution of the length of admixed tracts, the expected length of observing the sibling species’ nucleotide at a specific position of BN rats is the sum of two exponential random variables with expected lengths L, and it follows a Gamma distribution with shape parameter 2 and rate parameter 1/L (Huerta-Sanchez et al. 2014). Therefore, the probability of a length of at least m bp is 1-Gamma (m, shape = 2, rate = 1/L). We estimated such probability for each putative introgressive region.
Differences in Allele Frequency of Introgressed Regions between Populations of BN Rats
Allele frequencies in selected regions can be increased over time due to positive selection. Among the approaches used for detecting selection on introgressed DNA, the approach based on significantly high frequencies of an introgressed fragment in a specific population, may be less likely to be misled by introgression than other methods based on pattern of polymorphism and haplotype structure (Racimo et al. 2015). Since the pattern within introgressed genomic region may be different than expected under a neutral model with no introgression, which may be misinterpreted as evidence of selection by other methods. For each SNP, population differentiation (Weir and Cockerham estimator of FST) between allopatrically and sympatrically distributed populations of BN rats and allele frequencies in each population were calculated using vcflib (https://github.com/vcflib/vcflib, September 2014). Then, the mean allele frequency and FST in 100 kb sliding windows with a step size of 20 kb across the genome were calculated. Introgressed regions containing high-FST outliers (corresponded to the upper 5% of the empirical genome-wide distribution of FST) and higher allele frequency in sympatric than allopatric populations of BN rats were identified as adaptive introgressed regions. To characterize the molecular functions of genes contained in adaptive introgressed regions, we performed functional enrichment analysis using the clusterProfiler toolkit (Yu et al. 2012).
Detection of Selective Sweep in the BN Rat Genome
Three different approaches that based on decay of haplotype homozygosity, multilocus allele frequency differentiation or reduced nucleotide diversity were used in our study. First, we performed a multilocus allele frequency differentiation scan using an updated cross-population composite likelihood approach XP-CLR, which statistic is particularly robust to ascertainment bias and population demography (Chen et al. 2010). A 0.039 cM sliding window with 2,000 bp steps across the whole genome was used for scanning, and the number of SNPs assayed in each window was set to 100. Then, the mean likelihood scores in 100 kb sliding windows with a step size of 20 kb across the genome were calculated. To determine cutoffs for sweep statistics under a neutral scenario, we employed whole-genome simulation of the discrete time Wright–Fisher process using ARGON (Palamara 2016). We simulated 500 regions of 10 Mb size using ARGON (Palamara 2016) with the parameter of each species as inferred by G-PhoCS and PSMC. Based on these simulated data, we performed a multilocus allele frequency differentiation scan using XP-CLR with the same parameters that were used to analyze the real data. The P value of each observed data was assigned as the proportion of the simulated XP-CLR values that exceed the value of the statistic actually observed(Bramanti et al. 2009; Vatsiou et al. 2016), and were further corrected by multiple testing using the Benjamini–Hochberg false discovery rate (FDR) method (Benjamini and Hochberg 1995). Windows with FDR values <0.05 were selected, and adjacent windows with a distance of <100 kb were grouped to represent a single selective sweep.
Cross-population extended haplotype homozygosity (XP-EHH) test was also used in our study to scan putative regions of positive selection that occurred in BN rat genomes. Standardized XP-EHH statistics for each SNPs with cross-population comparison of BN rats with HF rats were estimated using XP-EHH (Sabeti et al. 2007). P value of each SNPs was estimated using the normal distribution of standardized XP-EHH value (Sabeti et al. 2007). The significant SNPs (P value < 0.05) with a distance of <100 kb were clustered as candidate regions for further analysis.
A reduced level of genetic polymorphism in one subpopulation may be indicative of a recent selective sweep (Malinsky et al. 2015). Genome regions show reduced nucleotide diversity in the BN rat species were identified if they corresponded to the upper 5% quantile of the ratio of πHF/πBN from the genome-wide distribution. The overlapping regions detected by all the three approaches were determined as candidate regions under positive selection. To characterize the molecular functions of the genes contained in selective sweep regions, we performed functional enrichment analysis using the clusterProfiler toolkit (Yu et al. 2012).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
J. Z., Z. S., and F. Z. conceived and designed the study. H. T., Y. Z., J. Z., Z. S., and F. Z. wrote the manuscript. H. T., Y. Z., C. S., F. M., and F. Z. analyzed the data. H. T., L. L., W. C., Y. Z. J. Z., and C. S. prepared the samples, collected data and performed genotyping experiments and sequencing.
Supplementary Material
Acknowledgments
This work was supported by the grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (No. XDB11010400), the National Natural Science Foundation of China (No. 91231107 and 31572277). We thank Lin Cong, Quansheng Liu, Bo Zou, Yingjuan Liu, Zhishu Xiao, Wanhong Wei, and Hongchuan Liang for their helps in wild sample collection.
References
- Abi-Rached L, Jobin MJ, Kulkarni S, McWhinnie A, Dalva K, Gragert L, Babrzadeh F, Gharizadeh B, Luo M, Plummer FA, et al. 2011. The shaping of modern human immune systems by multiregional admixture with archaic humans. Science 334:89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Elheiga L, Matzuk MM, Abo-Hashema KA, Wakil SJ.. 2001. Continuous fatty acid oxidation and reduced fat storage in mice lacking acetyl-CoA carboxylase 2. Science 291:2613–2616. [DOI] [PubMed] [Google Scholar]
- Ai H, Fang X, Yang B, Huang Z, Chen H, Mao L, Zhang F, Zhang L, Cui L, He W, et al. 2015. Adaptation and possible ancient interspecies introgression in pigs identified by whole-genome sequencing. Nat Genet. 47:217–225. [DOI] [PubMed] [Google Scholar]
- Aitman TJ, Critser JK, Cuppen E, Dominiczak A, Fernandez-Suarez XM, Flint J, Gauguier D, Geurts AM, Gould M, Harris PC, et al. 2008. Progress and prospects in rat genetics: a community view. Nat Genet. 40:516–522. [DOI] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19:1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aplin K, Lunde D, Molur S.. 2008. Rattus nitidus The IUCN Red List of Threatened Species:e.T19352A8866576.
- Atanur SS, Diaz AG, Maratou K, Sarkis A, Rotival M, Game L, Tschannen MR, Kaisaki PJ, Otto GW, Ma MC, et al. 2013. Genome sequencing reveals loci under artificial selection that underlie disease phenotypes in the laboratory rat. Cell 154:691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57:289–300. [Google Scholar]
- Bramanti B, Thomas MG, Haak W, Unterlaender M, Jores P, Tambets K, Antanaitis-Jacobs I, Haidle MN, Jankauskas R, Kind CJ, et al. 2009. Genetic discontinuity between local hunter-gatherers and central Europe’s first farmers. Science 326:137–140. [DOI] [PubMed] [Google Scholar]
- Chen H, Patterson N, Reich D.. 2010. Population differentiation as a test for selective sweeps. Genome Res. 20:393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chingangbam DS, Laishram JM, Suzuki H.. 2015. Molecular phylogenetic characterization of common murine rodents from Manipur, Northeast India. Genes Genet Syst. 90:21–30. [DOI] [PubMed] [Google Scholar]
- Clarkson CS, Weetman D, Essandoh J, Yawson AE, Maslen G, Manske M, Field SG, Webster M, Antao T, MacInnis B, et al. 2014. Adaptive introgression between Anopheles sibling species eliminates a major genomic island but not reproductive isolation. Nat Commun. 5:4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA.. 2004. Speciation. Sunderland, MA; [Great Britain: ]: Sinauer Associates. [Google Scholar]
- Cremer JN, Amunts K, Schleicher A, Palomero-Gallagher N, Piel M, Rosch F, Zilles K.. 2015. Changes in the expression of neurotransmitter receptors in Parkin and DJ-1 knockout mice: a quantitative multireceptor study. Neuroscience 311:539–551. [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danglot L, Zylbersztejn K, Petkovic M, Gauberti M, Meziane H, Combe R, Champy MF, Birling MC, Pavlovic G, Bizot JC, et al. 2012. Absence of TI-VAMP/Vamp7 leads to increased anxiety in mice. J Neurosci. 32:1962–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deinum EE, Halligan DL, Ness RW, Zhang YH, Cong L, Zhang JX, Keightley PD.. 2015. Recent evolution in Rattus norvegicus is shaped by declining effective population size. Mol Biol Evol 32:2547–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand EY, Patterson N, Reich D, Slatkin M.. 2011. Testing for ancient admixture between closely related populations. Mol Biol Evol. 28:2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick B, Ryan M, Johnson J, Corush J, Carter E.. 2015. Hybridization and the species problem in conservation. Curr Zool. 61:206–216. [Google Scholar]
- Fragaszy DM, Perry S.. 2003. The biology of traditions: models and evidence. Cambridge: Cambridge University Press. [Google Scholar]
- Gante HF, Matschiner M, Malmstrom M, Jakobsen KS, Jentoft S, Salzburger W.. 2016. Genomics of speciation and introgression in Princess cichlid fishes from Lake Tanganyika. Mol Ecol. 25:6143–6161. [DOI] [PubMed] [Google Scholar]
- Gao F, Ming C, Hu W, Li H.. 2016. New Software for the Fast Estimation of Population Recombination Rates (FastEPRR) in the Genomic Era. G3 (Bethesda) 6:1563–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, et al. 2004. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428:493–521. [DOI] [PubMed] [Google Scholar]
- Godfrey PA, Malnic B, Buck LB.. 2004. The mouse olfactory receptor gene family. Proc Natl Acad Sci U S A. 101:2156–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, et al. 2010. A draft sequence of the Neandertal genome. Science 328:710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronau I, Hubisz MJ, Gulko B, Danko CG, Siepel A.. 2011. Bayesian inference of ancient human demography from individual genome sequences. Nat Genet. 43:1031–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon R, Croxtall JD, Getting SJ, Roviezzo F, Yona S, Paul-Clark MJ, Gavins FN, Perretti M, Morris JF, Buckingham JC, et al. 2003. Aberrant inflammation and resistance to glucocorticoids in annexin 1−/− mouse. Faseb J 17:253–255. [DOI] [PubMed] [Google Scholar]
- Henning F, Meyer A.. 2014. The evolutionary genomics of cichlid fishes: explosive speciation and adaptation in the postgenomic era. Annu Rev Genomics Hum Genet. 15:417–441. [DOI] [PubMed] [Google Scholar]
- Hermsen R, de Ligt J, Spee W, Blokzijl F, Schafer S, Adami E, Boymans S, Flink S, van Boxtel R, van der Weide RH, et al. 2015. Genomic landscape of rat strain and substrain variation. BMC Genomics 16:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann S. 2012. Population genetic structure and geographic differentiation in the hot spring snake Thermophis baileyi (Serpentes, Colubridae): indications for glacial refuges in southern-central Tibet. Mol Phylogenet Evol. 63:396–406. [DOI] [PubMed] [Google Scholar]
- Hoskin CJ, Tonione M, Higgie M, Mackenzie JB, Williams SE, Vanderwal J, Moritz C.. 2011. Persistence in peripheral refugia promotes phenotypic divergence and speciation in a rainforest frog. Am Nat. 178:561–578. [DOI] [PubMed] [Google Scholar]
- Hua X, Wiens JJ.. 2013. How does climate influence speciation? Am Nat. 182:1–12. [DOI] [PubMed] [Google Scholar]
- Huerta-Sanchez E, Jin X, Asan Bianba Z, Peter BM, Vinckenbosch N, Liang Y, Yi X, He M, Somel M, et al. 2014. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 512:194–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob HJ. 1999. Functional genomics and rat models. Genome Res. 9:1013–1016. [DOI] [PubMed] [Google Scholar]
- Janousek V, Munclinger P, Wang L, Teeter KC, Tucker PK.. 2015. Functional organization of the genome may shape the species boundary in the house mouse. Mol Biol Evol. 32:1208–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen-Seaman MI, Furey TS, Payseur BA, Lu Y, Roskin KM, Chen CF, Thomas MA, Haussler D, Jacob HJ.. 2004. Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 14:528–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneitz B, Cohen PE, Avdievich E, Zhu L, Kane MF, Hou H Jr, Kolodner RD, Kucherlapati R, Pollard JW, Edelmann W.. 2000. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev. 14:1085–1097. [PMC free article] [PubMed] [Google Scholar]
- Koehler KE, Cherry JP, Lynn A, Hunt PA, Hassold TJ.. 2002. Genetic control of mammalian meiotic recombination. I. Variation in exchange frequencies among males from inbred mouse strains. Genetics 162:297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, et al. 2002. A high-resolution recombination map of the human genome. Nat Genet. 31:241–247. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Peterson D, Tamura K.. 2012. MEGA-CC: computing core of molecular evolutionary genetics analysis program for automated and iterative data analysis. Bioinformatics 28:2685–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurta A. 1995. Mammals of the Great Lakes region. Ann Arbor, MI: The University of Michigan Press. [Google Scholar]
- Lamichhaney S, Berglund J, Almen MS, Maqbool K, Grabherr M, Martinez-Barrio A, Promerova M, Rubin CJ, Wang C, Zamani N, et al. 2015. Evolution of Darwin’s finches and their beaks revealed by genome sequencing. Nature 518:371–375. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2011. Inference of human population history from individual whole-genome sequences. Nature 475:493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Hong W, Jiao H, Wang GD, Rodriguez KA, Buffenstein R, Zhao Y, Nevo E, Zhao H.. 2015. Sympatric speciation revealed by genome-wide divergence in the blind mole rat Spalax. Proc Natl Acad Sci U S A. 112:11905–11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Wang H, Cai Z, Wang L, Xu Q, Lovy M, Wang Z, Nevo E.. 2016. Sympatric speciation of spiny mice, Acomys, unfolded transcriptomically at Evolution Canyon, Israel. Proc Natl Acad Sci U S A. 11329:8254–8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LO, Ellis JM, Paich HA, Wang S, Gong N, Altshuller G, Thresher RJ, Koves TR, Watkins SM, Muoio DM, et al. 2009. Liver-specific loss of long chain acyl-CoA synthetase-1 decreases triacylglycerol synthesis and beta-oxidation and alters phospholipid fatty acid composition. J Biol Chem. 284:27816–27826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Futschik A, Li H.. 2013. A fast estimate for the population recombination rate based on regression. Genetics 194:473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin XD, Guo WP, Wang W, Zou Y, Hao ZY, Zhou DJ, Dong X, Qu YG, Li MH, Tian HF, et al. 2012. Migration of Norway rats resulted in the worldwide distribution of Seoul hantavirus today. J Virol 86:972–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Tatarenkov A, Beacham TD, Gorbachev V, Wildes S, Avise JC.. 2011. Effects of Pleistocene climatic fluctuations on the phylogeographic and demographic histories of Pacific herring (Clupea pallasii). Mol Ecol. 20:3879–3893. [DOI] [PubMed] [Google Scholar]
- Liu KJ, Steinberg E, Yozzo A, Song Y, Kohn MH, Nakhleh L.. 2014. Interspecific introgressive origin of genomic diversity in the house mouse. Proc Natl Acad Sci U S A. 112:196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Lorenzen ED, Fumagalli M, Li B, Harris K, Xiong Z, Zhou L, Korneliussen TS, Somel M, Babbitt C, et al. 2014. Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears. Cell 157:785–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovette IJ. 2005. Glacial cycles and the tempo of avian speciation. Trends Ecol Evol. 20:57–59. [DOI] [PubMed] [Google Scholar]
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinsky M, Challis RJ, Tyers AM, Schiffels S, Terai Y, Ngatunga BP, Miska EA, Durbin R, Genner MJ, Turner GF.. 2015. Genomic islands of speciation separate cichlid ecomorphs in an East African crater lake. Science 350:1493–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques DA, Lucek K, Meier JI, Mwaiko S, Wagner CE, Excoffier L, Seehausen O.. 2016. Genomics of rapid incipient speciation in sympatric threespine stickleback. PLoS Genet. 12:e1005887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SH, Dasmahapatra KK, Nadeau NJ, Salazar C, Walters JR, Simpson F, Blaxter M, Manica A, Mallet J, Jiggins CD.. 2013. Genome-wide evidence for speciation with gene flow in Heliconius butterflies. Genome Res. 23:1817–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SH, Davey JW, Jiggins CD.. 2015. Evaluating the use of ABBA-BABA statistics to locate introgressed loci. Mol Biol Evol. 32:244–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Barrio A, Lamichhaney S, Fan G, Rafati N, Pettersson M, Zhang H, Dainat J, Ekman D, Hoppner M, Jern P, et al. 2016. The genetic basis for ecological adaptation of the Atlantic herring revealed by genome sequencing. Elife 5: pii: e12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W, Schuster SC, Welch AJ, Ratan A, Bedoya-Reina OC, Zhao F, Kim HL, Burhans RC, Drautz DI, Wittekindt NE, et al. 2012. Polar and brown bear genomes reveal ancient admixture and demographic footprints of past climate change. Proc Natl Acad Sci U S A. 109:E2382–E2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ness RW, Zhang YH, Cong L, Wang Y, Zhang JX, Keightley PD.. 2012. Nuclear gene variation in wild brown rats. G3 (Bethesda) 2:1661–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishant KT, Chen C, Shinohara M, Shinohara A, Alani E.. 2010. Genetic analysis of baker’s yeast Msh4-Msh5 reveals a threshold crossover level for meiotic viability. PLoS Genet. 6: pii: e1001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MA, Feder JL.. 2006. Speciation genetics: evolving approaches. Nat Rev Genet. 7:851–861. [DOI] [PubMed] [Google Scholar]
- Norris LC, Main BJ, Lee Y, Collier TC, Fofana A, Cornel AJ, Lanzaro GC.. 2015. Adaptive introgression in an African malaria mosquito coincident with the increased usage of insecticide-treated bed nets. Proc Natl Acad Sci U S A. 112:815–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak JE, Ross-Macdonald PB, Roeder GS.. 2001. The budding yeast Msh4 protein functions in chromosome synapsis and the regulation of crossover distribution. Genetics 158:1013–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pages M, Chaval Y, Herbreteau V, Waengsothorn S, Cosson JF, Hugot JP, Morand S, Michaux J.. 2010. Revisiting the taxonomy of the Rattini tribe: a phylogeny-based delimitation of species boundaries. BMC Evol Biol. 10:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palamara PF. 2016. ARGON: fast, whole-genome simulation of the discrete time Wright-fisher process. Bioinformatics 32:3032–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Podlaha O, Grus WE, Zhang J.. 2011. The microevolution of V1r vomeronasal receptor genes in mice. Genome Biol Evol. 3:401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D.. 2006. Population structure and eigenanalysis. PLoS Genet. 2:e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D.. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 38:904–909. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Lei F, Zhang R, Lu X.. 2010. Comparative phylogeography of five avian species: implications for Pleistocene evolutionary history in the Qinghai-Tibetan plateau. Mol Ecol. 19:338–351. [DOI] [PubMed] [Google Scholar]
- Racimo F, Marnetto D, Huerta-Sanchez E.. 2016. Signatures of Archaic Adaptive Introgression in Present-Day Human Populations. Mol Biol Evol. 34:509–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racimo F, Sankararaman S, Nielsen R, Huerta-Sanchez E.. 2015. Evidence for archaic adaptive introgression in humans. Nat Rev Genet. 16:359–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Suchard MA, Xie D, Drummond AJ.. 2014. Tracer v1.6. Available from: http://beast.bio.ed.ac.uk/Tracer.
- Robinson R. 1965. Genetics of the Norway rat. Oxford: Peragon. [Google Scholar]
- Ronfani L, Ferraguti M, Croci L, Ovitt CE, Scholer HR, Consalez GG, Bianchi ME.. 2001. Reduced fertility and spermatogenesis defects in mice lacking chromosomal protein Hmgb2. Development 128:1265–1273. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP.. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe KC, Aplin KP, Baverstock PR, Moritz C.. 2011. Recent and rapid speciation with limited morphological disparity in the genus Rattus. Syst Biol. 60:188–203. [DOI] [PubMed] [Google Scholar]
- Roy K, Valentine JW, Jablonski D, Kidwell SM.. 1996. Scales of climatic variability and time averaging in Pleistocene biotas: implications for ecology and evolution. Trends Ecol Evol. 11:458–463. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R, et al. 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 449:913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter D. 2009. Evidence for ecological speciation and its alternative. Science 323:737–741. [DOI] [PubMed] [Google Scholar]
- Silver J. 1937. The House Rat. Washington: United States Department of Agriculture.
- Smith AT, Xie Y, Hoffmann RS.. 2008. A guide to the mammals of China. Princeton; Oxford: Princeton University Press. [Google Scholar]
- Song Y, Endepols S, Klemann N, Richter D, Matuschka FR, Shih CH, Nachman MW, Kohn MH.. 2011. Adaptive introgression of anticoagulant rodent poison resistance by hybridization between old world mice. Curr Biol. 21:1296–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Lan Z, Kohn MH.. 2014. Mitochondrial DNA phylogeography of the Norway rat. PLoS One 9:e88425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern HNE. 1964. The handbook of British mammals. [S.l.]: Mammal Society of the British Isles.
- Staubach F, Lorenc A, Messer PW, Tang K, Petrov DA, Tautz D.. 2012. Genome patterns of selection and introgression of haplotypes in natural populations of the house mouse (Mus musculus). PLoS Genet. 8:e1002891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steimer T. 2002. The biology of fear- and anxiety-related behaviors. Dialogues Clin Neurosci. 4:231–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JR, Stringer CB.. 2012. Human evolution out of Africa: the role of refugia and climate change. Science 335:1317–1321. [DOI] [PubMed] [Google Scholar]
- Stork O, Ji FY, Kaneko K, Stork S, Yoshinobu Y, Moriya T, Shibata S, Obata K.. 2000. Postnatal development of a GABA deficit and disturbance of neural functions in mice lacking GAD65. Brain Res. 865:45–58. [DOI] [PubMed] [Google Scholar]
- Taniguchi N, Carames B, Hsu E, Cherqui S, Kawakami Y, Lotz M.. 2011. Expression patterns and function of chromatin protein HMGB2 during mesenchymal stem cell differentiation. J Biol Chem. 286:41489–41498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeter KC, Payseur BA, Harris LW, Bakewell MA, Thibodeau LM, O’Brien JE, Krenz JG, Sans-Fuentes MA, Nachman MW, Tucker PK.. 2008. Genome-wide patterns of gene flow across a house mouse hybrid zone. Genome Res. 18:67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng H, Zhang Y, Shi C, Mao F, Hou L, Guo H, Sun Z, Zhang J.. 2016. Whole-Genome Sequencing Reveals Genetic Variation in the Asian House Rat. G3 (Bethesda) 6:1969–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatsiou AI, Bazin E, Gaggiotti OE.. 2016. Detection of selective sweeps in structured populations: a comparison of recent methods. Mol Ecol 25:89–103. [DOI] [PubMed] [Google Scholar]
- Verneau O, Catzeflis F, Furano AV.. 1998. Determining and dating recent rodent speciation events by using L1 (LINE-1) retrotransposons. Proc Natl Acad Sci U S A. 95:11284–11289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JB, Lindell J, Backstrom N.. 2010. Speciation genetics: current status and evolving approaches. Philos Trans R Soc Lond B Biol Sci. 365:1717–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Yuan J, Li H, Li M, Zhao G, Bu D, Zhu W, Wu W, Chen R, Zhao Y.. 2014. NONCODEv4: exploring the world of long non-coding RNA genes. Nucleic Acids Res. 42:D98–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Uematsu S, Okamoto T, Matsuura Y, Sato S, Kumar H, Satoh T, Saitoh T, Takeda K, Ishii KJ, et al. 2007. Enhanced TLR-mediated NF-IL6 dependent gene expression by Trib1 deficiency. J Exp Med. 204:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY.. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Zheng Y, Wei F, Bruford MW, Jia C.. 2011. Molecular evidence for Pleistocene refugia at the eastern edge of the Tibetan Plateau. Mol Ecol. 20:3014–3026. [DOI] [PubMed] [Google Scholar]
- Zhang M, Park SM, Wang Y, Shah R, Liu N, Murmann AE, Wang CR, Peter ME, Ashton-Rickardt PG.. 2006. Serine protease inhibitor 6 protects cytotoxic T cells from self-inflicted injury by ensuring the integrity of cytotoxic granules. Immunity 24:451–461. [DOI] [PubMed] [Google Scholar]
- Zhao S, Zheng P, Dong S, Zhan X, Wu Q, Guo X, Hu Y, He W, Zhang S, Fan W, et al. 2013. Whole-genome sequencing of giant pandas provides insights into demographic history and local adaptation. Nat Genet. 45:67–71. [DOI] [PubMed] [Google Scholar]
- Zhang W, Dasmahapatra KK, Mallet J, Moreira GR, Kronforst MR.. 2016. Genome-wide introgression among distantly related Heliconius butterfly species. Genome Biol. 17:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng BX, Xu QQ, Shen YP.. 2002. The relationship between climate change and Quaternary glacial cycles on the Qinghai-Tibetan Plateau: review and speculation. Quat Int. 97–8:93–101. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.