Abstract
Gene duplication is thought to play a major role in phenotypic evolution. Yet the forces involved in the functional divergence of young duplicate genes remain unclear. Here, we use population-genetic inference to elucidate the role of natural selection in the functional evolution of young duplicate genes in Drosophila melanogaster. We find that negative selection acts on young duplicates with ancestral functions, and positive selection on those with novel functions, suggesting that natural selection may determine whether and how young duplicate genes are retained. Moreover, evidence of natural selection is strongest in protein-coding regions and 3′ UTRs of young duplicates, indicating that selection may primarily target encoded proteins and regulatory sequences specific to 3′ UTRs. Further analysis reveals that natural selection acts immediately after duplication and weakens over time, possibly explaining the observed bias toward the acquisition of new functions by young, rather than old, duplicate gene copies. Last, we find an enrichment of testis-related functions in young duplicates that underwent recent positive selection, but not in young duplicates that did not undergo recent positive selection, or in old duplicates that either did or did not undergo recent positive selection. Thus, our findings reveal that natural selection is a key player in the functional evolution of young duplicate genes, acts rapidly and in a region-specific manner, and may underlie the origin of novel testis-specific phenotypes in Drosophila.
Keywords: duplicate genes, paralogs, neofunctionalization, subfunctionalization, functional evolution
Introduction
Gene duplication is the primary source of new genetic material (Ohno 1970), and has generated large proportions of existing genes in organisms from all three domains of life (Zhang 2003). In the simplest scenario, gene duplication produces an exact copy of an existing gene. Thus, genomes of species that diverged before the duplication event contain only the “ancestral” gene, whereas those that diverged afterward contain an “old” copy that is orthologous to the ancestral gene and a “young” copy that is the product of the duplication event. Theoretical studies predict that redundancy of duplicate genes results in relaxed selective constraint in one copy (Ohno 1970), typically leading to an accumulation of deleterious mutations and its pseudogenization within a few million years (Lynch and Conery 2000). Yet numerous duplicates have surpassed this time window, some by hundreds of millions of years (Ferris and Whitt, 1979; Lundin 1993; Sidow 1996; Brookfield 1997; Nadeau and Sankoff 1997; Postlethwait et al. 1998; Zhang 2003). Moreover, duplicates often have essential biological functions (Holland et al. 1994; Taylor and Raes 2004; Chen et al. 2010) that in many cases are distinct from those of their ancestral genes (Chen et al. 2010; Assis and Bachtrog 2013; Assis and Bachtrog 2015). These observations, along with the sheer abundance of known duplicates, prompt questions about the contribution of natural selection to the functional evolution and long-term retention of duplicate genes.
One hypothesis is that duplicate genes are retained by conservation, whereby both copies maintain the ancestral function after duplication (Ohno 1970). Conservation may occur when increased dosage of the ancestral gene product is beneficial, and thus negative selection acts to preserve the ancestral function in both copies (Ohno 1970; Zhang 2003). Alternatively, the ancestral function may be maintained by gene conversion between duplicates (Zhang 2003), such that conservation is effectively a neutral transient state with a length that is dependent on the rate of nonallelic gene conversion. However, though either strong negative selection or frequent nonallelic gene conversion can potentially result in long-term retention of duplicate genes, conservation results in amplification of the ancestral function, rather than in the acquisition of a new function.
A second hypothesis is that duplicate genes are retained by subfunctionalization, in which the ancestral function is divided between copies (Force et al. 1999; Stoltzfus 1999). There are two popular models of subfunctionalization: escape from adaptive conflict (EAC) and duplication–degeneration–complementation (DDC). Under the EAC model, each duplicate acquires mutations that optimize a different ancestral subfunction, and such mutations are fixed by positive selection (Hittinger and Carroll 2007). In contrast, under the DDC model, degenerative mutations impair different ancestral subfunctions of both copies, but such mutations are selectively neutral due to the functional redundancy of duplicates (Force et al. 1999). This model of subfunctionalization is particularly appealing because it explains how duplicate genes can be retained over millions of years of evolution in the absence of natural selection. Yet like conservation, subfunctionalization via either model cannot explain the acquisition of new duplicate gene functions.
Two hypotheses can explain both the long-term retention and functional novelty of duplicate genes. First, the duplicates can undergo neofunctionalization, in which one copy maintains the ancestral function and the other acquires a new function (Ohno 1970). Under neofunctionalization, beneficial mutations arise in one gene copy either as a product of the duplication event or during the hypothesized period of relaxed constraint after duplication, and such mutations are subsequently fixed by positive selection. The second hypothesis is specialization (i.e., subneofunctionalization), in which rapid subfunctionalization is followed by neofunctionalization (He and Zhang 2005; Rastogi and Liberles 2005). This hypothesis is particularly attractive because the loss of an ancestral subfunction may provide an opportunity for a beneficial function to arise. Thus, subfunctionalization enables fixation of duplicate genes under neutrality, while also uncovering new targets on which natural selection can later act.
Determining the genome-wide roles of these retention mechanisms is the first step in assessing whether natural selection influences the functional evolution of duplicate genes. Though early studies uncovered widespread asymmetric sequence evolution indicative of neofunctionalization (Conant and Wagner 2003; Blanc and Wolfe 2004; Kellis et al. 2004; Li et al. 2005), the recent availability of genome-scale functional data has enabled direct interrogation of duplicate gene functions for the first time. In a landmark study in Drosophila melanogaster, researchers used RNAi knockdown experiments to demonstrate that 30% of young duplicates have new and essential functions that are distinct from those of their ancestral genes (Chen et al. 2010). However, whereas this study highlights the prevalence of neofunctionalization in D. melanogaster, its overall importance is unclear, particularly as such experiments have not been performed in other species. Further, identification and comparison of duplicates retained by different mechanisms is key to disentangling the role of natural selection in their functional evolution.
In two recent studies, researchers used RNA-seq data to perform the first genome-wide classifications of duplicate gene retention mechanisms in Drosophila (Assis and Bachtrog 2013) and mammals (Assis and Bachtrog 2015). In particular, they quantified and compared spatial expression profiles between pairs of duplicates and their ancestral genes, with the assumption that tissue-specific changes in expression correspond to functional divergence. In Drosophila, they found that 65% of duplicates were retained by neofunctionalization, 19% by conservation, 15% by specialization, and only 1% by subfunctionalization (Assis and Bachtrog 2013). Additionally, new functions often arose within a few million years and primarily (91%) in young copies, particularly those duplicated by RNA-mediated mechanisms and expressed specifically in testis tissue (Assis and Bachtrog 2013; Assis 2014). Thus, in Drosophila, neofunctionalization is the primary duplicate gene retention mechanism, typically occurs rapidly, leads to biased acquisition of new functions in young copies, may often result from beneficial mutations introduced by duplication events, and generates new testis-related functions. In contrast, 58% of mammalian duplicates were retained by conservation, 33% by neofunctionalization, 8% by specialization, and 1% by subfunctionalization (Assis and Bachtrog 2015). Moreover, functional divergence of mammalian duplicates occurred more gradually than in Drosophila, affected young and old copies at equal rates, and resulted in a diversity of novel gene functions (Assis and Bachtrog 2015).
The rapid functional divergence of duplicate genes in Drosophila compared with mammals is reminiscent of their relative rates of protein sequence evolution (Britten 1986; Moriyama 1987; Bustamante et al. 2002; Smith and Eyre-Walker 2002; Sawyer et al. 2003; Bustamante et al. 2005; Chimpanzee Sequencing and Analysis Consortium 2005; Zhang and Li 2005; Charlesworth and Eyre-Walker 2006; Gossmann et al. 2010; Haddrill et al. 2010; Slotte et al. 2010). Faster sequence evolution in Drosophila is thought to be attributed to a greater efficiency of natural selection, which is the product of the effective population size, Ne, and strength of selection (Kimura 1983; Charlesworth 2009). In particular, because the Ne of Drosophila species are at least an order of magnitude larger than those of mammals (Lynch and Conery 2003), they are expected to evolve more quickly even when selection has the same strength. Thus, the difference between rates of functional divergence in Drosophila and mammalian duplicates implicates natural selection in the origin of new duplicate gene functions.
In the present study, we utilize population-genetic analyses to interrogate the role of natural selection in the functional evolution of young Drosophila duplicate genes. In particular, we investigate the types, targets, and timing of natural selection to answer the following questions: Are ancestral functions in young conserved duplicates maintained by negative selection, or by a neutral process such as gene conversion? Is there evidence of positive selection acting on young neofunctionalized or specialized duplicates? Does natural selection target coding or regulatory regions of young duplicates? When does natural selection act on young duplicates? Finally, is positive selection associated with the acquisition of testis-specific functions in young duplicates?
Results and Discussion
Retention Mechanisms of Young Drosophila Duplicate Genes
For our analyses, we required data on the retention mechanisms of young Drosophila duplicate genes. Thus, we utilized the data set of Assis and Bachtrog (2013), which consists of 108 pairs of duplicates that arose in the D. melanogaster lineage after its split from D. pseudoobscura and, for each pair, the identities of young and old copies in D. melanogaster and of their ancestral gene in D. pseudoobscura, the phylogenetic age of the duplication event, and the hypothesized retention mechanism (69 neofunctionalized, 20 specialized, 18 conserved, and 1 subfunctionalized).
It is important to note that Assis and Bachtrog’s (2013) classifications of retention mechanisms were made under the assumption that changes in spatial gene expression profiles correspond to changes in gene function. Though defining biological function is inherently challenging, particularly with any single measure, we chose to use expression profiles as proxies for function for three major reasons. First, RNA-seq data are available for the same six tissues in D. melanogaster and D. pseudoobscura, enabling straightforward comparisons between spatial gene expression profiles in the two species. Second, in contrast to other measures of gene function, expression profiles and differences between them are easily quantifiable. Third, gene expression profiles correlate to other metrics of gene function (e.g., Ge et al. 2001, Zhou et al. 2002, Bhardwaj and Lu 2005, French and Pavlidis 2011; Assis and Kondrashov 2014). Indeed, as expected, differences in protein–protein interactions are consistent with classifications of retention mechanisms in Drosophila (Assis and Bachtrog 2013). Thus, though gene expression profiles are not synonymous with gene function, they are ideal proxies for our study.
Rates of Protein-Coding Sequence Evolution in Young Drosophila Duplicates
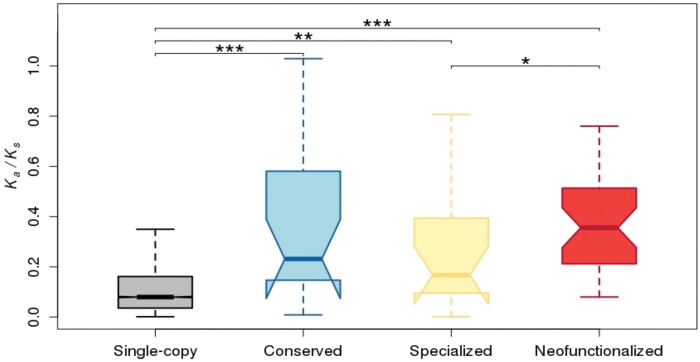
We first examined protein sequence divergence rates of young duplicates by estimating pairwise Ka/Ks ratios between orthologs in D. melanogaster and D. simulans (fig. 1; see Materials and Methods for details). Consistent with previous studies (Zhang 2003; Assis and Bachtrog 2013), all young duplicate genes display elevated Ka/Ks ratios relative to single-copy genes (P < 0.001, permutation test). Further, individual comparisons revealed that neofunctionalized, conserved, and specialized young duplicates each have higher Ka/Ks ratios than single-copy genes (P < 0.01, permutation tests). Thus, regardless of retention mechanism, protein-coding sequence divergence occurs rapidly in young Drosophila duplicates. However, though Ka/Ks ratios of neofunctionalized genes are significantly greater than those of specialized genes (P < 0.05, permutation test), there are no significant differences between Ka/Ks ratios of conserved duplicates and those retained by either neofunctionalization or specialization. Hence, the only conclusion that can be made by comparing Ka/Ks ratios across different retention mechanisms is that neofunctionalized duplicates likely evolve fastest at the protein-coding sequence level.
Fig. 1.
Distributions of Ka/Ks ratios between orthologous single-copy, conserved, specialized, and neofunctionalized genes in D. melanogaster and D. simulans. Two-sample permutation tests were used to assess significant differences between each pair of distributions. *P<0.05, **P<0.01, ***P<0.001.
Though informative about sequence evolutionary rates, there are several limitations of using Ka/Ks ratios to draw conclusions about selective forces acting on duplicate genes. First, a Ka/Ks ratios is computed on the entire protein-coding region of a gene. Because positive selection likely acts at specific sites, and negative selection may act at many (and perhaps most) other sites, there must be a strong signal to detect positive selection acting on a gene. Indeed, only three young Drosophila duplicates have Ka/Ks > 1, which is a signature of positive selection. This also means that elevated Ka/Ks ratios can either be due to positive or relaxed selection, both of which have been hypothesized to contribute to the evolution of young duplicate genes. Second, power to detect positive selection may be even lower in our study because the data sets of conserved and specialized duplicates are quite small, as evidenced by overlapping notches in figure 1, and we can only compute a single Ka/Ks ratio for each gene. A last caveat is that Ka/Ks ratios can only detect evolutionary changes occurring in protein-coding regions of genes. Yet natural selection may act on regulatory regions as well, resulting in gene expression divergence, which is thought to play a major role in phenotypic evolution. Thus, Ka/Ks ratios may only provide a glimpse into the evolutionary histories of young duplicate genes.
Role of Natural Selection in the Evolution of Young Drosophila Duplicates
To investigate the types and genic targets of natural selection in young Drosophila duplicates, we implemented a Hudson–Kreitman–Aguadé (HKA) test (Hudson et al. 1987), which uses a X2 test statistic to compare expected and observed counts of substitutions and polymorphisms between putatively neutral and nonneutral genomic regions. However, rather than designating neutral and nonneutral regions, we used a sliding window approach. In particular, we slid over the D. melanogaster genome one nucleotide (nt) at a time, comparing the substitution-to-polymorphism ratio within each 10,000-nt window to the genome-wide ratio, which represents a neutral reference. The benefits of using a sliding window are that it did not require us to decide which genomic regions are neutral or nonneutral, and it also provided us with substantially greater power, in that we obtained a X2 test statistic for every nucleotide of each region of interest, rather than a single test statistic for the entire region (see Materials and Methods for details).
The X2 test statistic from a HKA test is expected to be zero under neutrality, and greater than zero otherwise. However, following the approach of Huber et al. (2016), we assigned each X2 test statistic a positive sign when there is an excess of substitutions, and a negative sign when there is an excess of polymorphisms. The advantage of this modification is that it enabled us to distinguish between evidence of positive and negative selection, which lead to relative excesses of substitutions and polymorphisms, respectively (Charlesworth and Charlesworth 2010). Though an excess of polymorphisms can also be caused by balancing selection (Charlesworth and Charlesworth 2010), we do not believe this to be a viable option in our study for two major reasons. First, our analysis focuses on groups of genes, rather than on individual genes. Because negative selection is widespread and balancing selection is rare, an overall excess of polymorphisms is more likely to be caused by pervasive negative selection than by balancing selection acting on a few genes. Second, a signature of balancing selection is an excess of intermediate-frequency variants. In contrast, site frequency spectra for all classes of genes are skewed toward low-frequency variants (supplementary fig. S1, Supplementary Material online) and are similar to the genome-wide frequency spectrum (Huang et al. 2014), inconsistent with expectations under balancing selection. Using this rationale, we expect the distribution of signed X2 scores for a class of genes to be centered around zero (i.e., equal rates of substitution and polymorphism) under neutrality, a positive value (i.e., excess substitution) under positive selection, and a negative value (i.e., excess polymorphism) under negative selection.
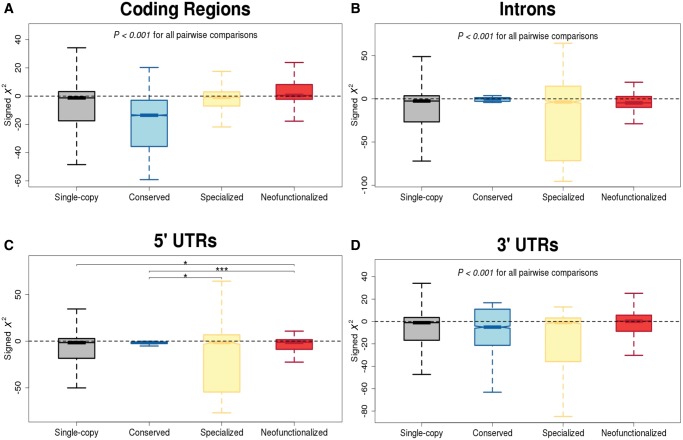
A major advantage of the HKA test is that it can be performed on both coding and noncoding sequences, enabling us to investigate the role of natural selection in different genic regions. Thus, we examined distributions of signed X2 scores in coding regions, introns, 5′ UTRs, and 3′ UTRs of D. melanogaster single-copy, conserved, specialized, and neofunctionalized genes (fig. 2). For single-copy genes, signed X2 scores are negatively biased in all genic regions (P < 0.001 for all regions, sign tests). Thus, consistent with previous findings in Drosophila (Bergman and Kreitman 2001; Halligan et al. 2004; Kohn et al. 2004; Andolfatto 2005; Haddrill et al. 2005; Bachtrog and Andolfatto 2006; Halligan and Keightley 2006; Haddrill et al. 2008), all genic regions appear to be subject to selective constraint, highlighting the important role of negative selection in maintaining both protein function and its various forms of regulation. In contrast, duplicate gene classes display diverse patterns across genic regions.
Fig. 2.
Distributions of signed X2 scores of single-copy, conserved, specialized, and neofunctionalized D. melanogaster genes in their (A) coding regions, (B) introns, (C) 5′ UTRs, and (D) 3′ UTRs. Two-sample permutation tests were used to assess significant differences between each pair of distributions. *P<0.05, **P<0.01, ***P<0.001.
In coding regions (fig. 2A), signed X2 scores of conserved duplicate genes are also negatively biased (median =−13.56; P = 4.94 ×10−324, sign test), but are significantly more negative than those of single-copy genes (P< 0.001, permutation test). Thus, coding regions of conserved duplicates appear to be under increased selective constraint, which is expected if amplification of the ancestral function is beneficial, but not if conservation is simply a product of a neutral process such as nonallelic gene conversion. Though signed X2 scores in coding regions of specialized duplicates are negatively biased as well (median =−0.88; P = 1.43 ×10−277, sign test), they are significantly less negative than those of conserved (P< 0.001, permutation test) and single-copy (P< 0.001, permutation test) genes. This pattern is consistent with the combined action of relaxed constraint and positive selection hypothesized to underlie specialization. Moreover, in contrast to all other classes, signed X2 scores in coding regions of neofunctionalized duplicates are positively biased (median =0.53; P = 9.88 ×10−324, sign test), and are also significantly greater than those of all other classes of genes (P< 0.001, permutation tests). Hence, this result strongly supports the hypothesis that positive selection drives neofunctionalization.
In introns (fig. 2B), signed X2 scores are negatively biased for all classes of duplicates, with an ordering of distributions opposite to that observed in coding regions. In particular, signed X2 scores are most negatively biased in introns of neofunctionalized genes (median =−4.71; P = 4.94 ×10−324, sign test), and are significantly more negative than those of single-copy genes (P< 0.001, permutation test). Signed X2 scores in introns of specialized duplicates (median =−3.67; P = 2.85 ×10−5, sign test) are also significantly more negative than those of single-copy genes (P< 0.001, permutation test), though less so than those of neofunctionalized duplicates (P< 0.001, permutation test). Finally, signed X2 scores in introns of conserved duplicates are least negatively biased (median = −0.02; P = 2.48 ×10−5, sign test), and also significantly less negative than those of any other class of genes (P< 0.001, permutation tests). Thus, selective constraint appears to be increased in introns of duplicates with new functions, and reduced in those with ancestral functions. However, the interpretation of this result is complicated by the strong association between intron presence and the retention mechanism of young Drosophila duplicates (Assis and Bachtrog 2013). In particular, conserved and specialized duplicates typically arose by DNA-mediated duplication, which produces complete gene copies, whereas neofunctionalized duplicates often originated via RNA-mediated duplication, which creates copies lacking introns and cis-regulatory sequences (Assis and Bachtrog 2013). Hence, many young neofunctionalized duplicates lost their introns as an immediate consequence of RNA-mediated duplication, which may have contributed to their acquisition of new functions. As a result, 66% of conserved, 85% of specialized, and 45% of neofunctionalized duplicates in our contain introns. Thus, comparing levels of selective constraint among intron sequences of different classes may be unfair, in that one must consider that selection likely also acted on the presence of the introns themselves, rather than solely on their sequence content.
In 5′ UTRs (fig. 2C), signed X2 scores are negatively biased for all classes of duplicates as well. However, though differences between pairs of distributions are not all significant, the ordering of median signed X2 scores is consistent with that of coding regions. In particular, signed X2 scores in 5′ UTRs of conserved duplicates are most negatively biased (median =−1.62; P < 10−325, sign test), though they are not significantly more negative than those of single-copy genes (P> 0.05, permutation test). Moreover, signed X2 scores in 5′ UTRs of specialized duplicates are also negatively biased (median =−1.47; P = 1.52 ×10−40, sign test), and are significantly less negative than those of conserved (P< 0.05, permutation test), but not single-copy (P> 0.05, permutation test), genes. Signed X2 scores in 5′ UTRs of neofunctionalized duplicates are least negatively biased (median =−1.28; P = 1.44 ×10−77, sign test), and are also significantly less negative than both conserved (P< 0.001, permutation test) and single-copy (P< 0.05, permutation test), but not specialized (P> 0.05, permutation test), genes. Thus, the patterns are weak, but generally supportive of the ordering observed in coding regions. Yet the only group that is significantly different from single-copy genes is neofunctionalized duplicates, which also have negatively biased X2 scores. Therefore, this difference suggests that divergence of 5′ UTRs of duplicate genes may occur via either relaxed constraint or weak positive selection.
In 3′ UTRs (fig. 2D), both the biases and ordering of distributions of signed X2 scores are consistent with those of coding regions. Specifically, signed X2 scores in 3′ UTRs of conserved duplicates are most negatively biased (median = −5.02; P = 3.41 ×10−74, sign test), and are significantly more negative than those of single-copy genes (P< 0.001, permutation test). Signed X2 scores in 3′ UTRs of specialized duplicates are also negatively biased (median = −0.99; P = 1.05 ×10−43, sign test), though significantly less negative than those of both conserved (P< 0.001, permutation test) and single-copy (P< 0.001, permutation test) genes. Finally, signed X2 scores in 3′ UTRs of neofunctionalized duplicates are positively biased (median = 0.26; P = 9.57 ×10−17, sign test), and significantly greater than those of all other classes of genes (P< 0.001, permutation tests). Thus, these findings support the hypotheses that conservation is driven by negative selection, specialization by a period of relaxed selection followed by positive selection, and neofunctionalization by positive selection. Further, these trends suggest that 3′ UTRs play an integral role in the functional divergence of young duplicate genes, perhaps implicating miRNAs and regulatory binding proteins in their evolution.
Timing of Natural Selection after Duplication
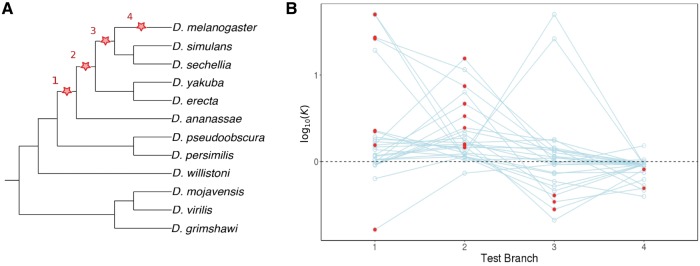
Next, we wanted to investigate the timing of natural selection on Drosophila duplicate genes. In particular, we were interested in whether duplicates generally experience a period of relaxed constraint after duplication, and when and how strongly natural selection acts. To address these questions, we applied the tree-based method RELAX, which tests whether selection is relaxed or intensified on focal branches relative to reference branches in a predefined tree (Wertheim et al. 2015). RELAX groups sites into two categories based on their Ka/Ks ratios, and then computes the selection intensity parameter K, which is based on a comparison of the distributions of Ka/Ks ratios between focal and reference branches. In particular, K = 1 under the null model and is allowed to differ under the alternative model, and a likelihood ratio test is used to compare models. K < 1 indicates relaxed selection, whereas K > 1 indicates directional (either positive or negative) selection, with larger K implying stronger selection.
To examine when selection acts post-duplication, we applied RELAX to D. melanogaster duplicates that arose after its divergence from D. pseudoobscura and D. persimilis, but before its divergence from D. ananassae (see Materials and Methods for details). Using this age group enabled us to assess selection on four branches representing distinct evolutionary time points after duplication (fig. 3A). For each analysis, we set one of these as the test branch and the remaining as reference branches. Thus, we obtained K-values and their associated P-values for each of the four test branches (fig. 3B). For most duplicates, K > 1 on branches 1 and 2, supporting the hypothesis that natural selection generally acts immediately or soon after duplication. Thus, the hypothesized period of relaxed constraint after duplication appears to be either short or absent in Drosophila, which is consistent with efficient natural selection (Lynch and Conery 2003), the significance of duplication-induced mutations in the functional divergence of duplicate genes (Assis and Bachtrog 2013; Assis 2014), and the bias toward origin of new functions in young duplicates (Assis and Bachtrog 2013; Assis 2014). Moreover, K varies on branch 3, but tends to be smaller than on branches 1 and 2, and K < 1 on branch 4 for nearly all duplicates. Thus, K decreases with distance from the branch on which the duplication event occurred, suggesting that the strength of directional selection on duplicate genes weakens over time.
Fig. 3.
Assessment of natural selection on branches representing four post-duplication time points for duplicates that arose in the D. melanogaster lineage after its divergence from D. pseudoobscura and D. persimilis. (A) Phylogenetic tree of 12 sequenced Drosophila species, with red stars indicating test branches used for each of four RELAX runs. (B) Intensity of selection for RELAX analyses with test branches 1–4 from (A). Circles indicate log10 (K) for each duplicate gene pair in a RELAX run with the indicated test branch, and lines connect circles from the same pair of duplicates. The horizontal dashed line depicts neutrality (K = 1), with points below the dashed line (K < 1) indicating relaxed selection, and points above the dashed line (K > 1) indicating intensified directional selection. Red circles indicate P < 0.05 from the likelihood ratio test implemented by RELAX.
Association between Natural Selection and Functions of Young Drosophila Duplicates
Finally, we were interested in determining whether natural selection is associated with the acquisition of testis-specific functions in young Drosophila duplicate genes. However, we could not answer this question by assessing functions of duplicates that underwent selection in the past because we do not have knowledge of their past functions. In contrast, we do have insight about the current functions of genes. Thus, to directly address the association between natural selection and the current biological functions of duplicate genes, we examined functional enrichment in duplicates that underwent recent positive selection.
We identified genes that underwent recent positive selection with the haplotype-based statistic nSL, which can detect genomic regions that recently underwent either hard or soft selective sweeps, has higher power than other haplotype-based statistics under most selection scenarios and parameter values, and is robust to recombination rate estimation error and a variety of demographic factors (Ferrer-Admetlla et al. 2014). We applied nSL to phased haplotypes from D. melanogaster (see Materials and Methods for details) and ranked normalized |nSL| scores for each chromosome. Of the 108 pairs of duplicates in our data set, there are 25 young and 33 old copies with at least one normalized |nSL| score within the top 5% of all scores. Accordingly, we split duplicates into those with (in top 5%) and without (not in top 5%) evidence of recent positive selection.
To assess the functional enrichment of these duplicate genes, we used GOrilla (Eden et al. 2007, 2009), which compares gene ontology (GO) terms between target and background gene lists (see Materials and Methods for details). We used single-copy genes as our background and considered four targets: the 25 young duplicates with evidence of recent positive selection, the 83 young duplicates without evidence of recent positive selection, the 33 old duplicates with evidence of recent positive selection, and the 75 old duplicates without evidence of recent positive selection. Interestingly, the only target with significant GO enrichment is the one containing young duplicates with evidence of recent positive selection. Thus, we did not observe any functional enrichment in young duplicates without evidence of recent positive selection, or in old duplicates with or without evidence of recent positive selection. Moreover, functional enrichment was only found for GO terms related to reproduction (GO: 0032504, P < 5.46 ×10−5; GO: 0000003, P < 8.05 ×10−5; GO: 0044703; P < 6.71 ×10−5), and four of the enriched genes (Acp36DE, Obp56i, CG31413, CG6690) encode seminal fluid proteins. This specific enrichment in young duplicates with evidence of recent positive selection suggests that positive selection may be associated with the acquisition of testis-related functions in young duplicates.
Conclusions
The role of natural selection in the evolution of duplicate genes has been a topic of extensive debate during the past few decades (e.g., Lynch and Conery 2000, 2003; Shiu et al. 2006). On one hand, the theoretical basis for the fixation and long-term retention of duplicate genes under neutrality is strong (Lynch and Conery 2000, 2003). However, empirical findings contrast such predictions, with numerous examples of widespread asymmetric sequence divergence between duplicates (Conant and Wagner 2003; Blanc and Wolfe 2004; Kellis et al. 2004; Li et al. 2005), natural selection acting on young duplicates (Malik and Henikoff 2001; Llopart et al. 2002; Zhang et al. 2002; Betrán and Long 2003; Long et al. 2003; Han et al. 2009), and gene expression divergence consistent with neofunctionalization (Chen et al. 2010; Assis and Bachtrog 2013, 2015). Yet whereas such findings suggest that natural selection underlies the long-term retention of many duplicate genes, they do not provide insight into its types, targets, timing, or association with functional outcomes.
In our study, we address these questions by directly linking natural selection with the evolutionary retention mechanisms of young Drosophila duplicate genes. Together, our findings implicate natural selection as a key player in the functional evolution of young Drosophila duplicates. In particular, HKA tests are generally consistent with the hypotheses that conservation is driven by negative selection, specialization by both relaxed constraint and positive selection, and neofunctionalization by positive selection. Moreover, support for these hypotheses is strongest in protein-coding regions and 3′ UTRs of duplicates. Thus, selection may act primarily on encoded proteins and regulatory sequences found in 3′ UTRs, which most notably include miRNA binding sites. Further, our RELAX results suggest that natural selection acts immediately after duplication and weakens over time. This finding is consistent with efficient natural selection in Drosophila (Lynch and Conery 2003), an important role of duplication-induced mutations in the functional evolution of duplicate genes (Assis and Bachtrog 2013; Assis 2014), and the bias toward origin of new functions in young duplicates (Assis and Bachtrog 2013; Assis 2014). Finally, young duplicate genes predicted to have undergone recent positive selection by the haplotype-based nSL statistic are enriched in reproduction and spermatogenesis-related functions. Thus, this analysis supports an association between positive selection and testis-related functions in Drosophila. In summary, our findings indicate that natural selection likely determines whether and how young duplicates will be retained, primarily targets their protein-coding regions and 3′ UTRs, acts immediately after duplication and weakens over time, and may be involved in the origin of novel testis-specific phenotypes in Drosophila.
Materials and Methods
Genomic Data Sets
Reference genome annotation and sequence data from D. melanogaster (version 5.49), D. simulans (version 2.01), D. sechellia (version 1.3), D. yakuba (version 1.05), D. erecta (version 1.05), D. ananassae (version 1.05), D. pseudoobscura (version 3.03), D. persimilis (version 1.3), D. willistoni (version 1.3), D. mojavensis (version 1.3), D. virilis (version 1.2), and D. grimshawi (version 1.3) were downloaded from FlyBase at http://www.flybase.org, last accessed September 5, 2017. Polymorphism data for 205 D. melanogaster inbred lines were downloaded from the Drosophila Genetic Reference Panel (DGRP) website at http://dgrp2.gnets.ncsu.edu, last accessed September 5, 2017 (Mackay et al. 2012; Huang et al. 2014). A list of 7,131 single-copy orthologs in D. melanogaster and D. pseudoobscura, and a data set of 108 pairs of D. melanogaster duplicate genes that arose after its split from D. pseudoobscura (including identities of young and old copies and of ancestral D. pseudoobscura genes, inferred phylogenetic ages, and classifications of evolutionary retention mechanisms), were obtained from Assis and Bachtrog (2013). For each pair of duplicates in this data set, Assis and Bachtrog (2013) distinguished the young and old copy by the presence and absence, respectively, of orthologs in the 12 sequenced Drosophila species, which were assigned based on sequence conservation and synteny (Drosophila 12 Genomes Consortium 2007).
Sequence Analyses
One-to-one orthologs between all protein-coding genes in D. melanogaster and each of the other 11 sequenced Drosophila species were obtained from a recent table of FlyBase orthologs (2016, version 1), which were assigned based on sequence conservation and synteny (Drosophila 12 Genomes Consortium 2007). We used MACSE (Ranwez et al. 2011) to perform pairwise sequence alignments of all D. melanogaster and D. simulans orthologs, as well as multiple alignments of D. melanogaster duplicate genes and all of their available Drosophila orthologs.
Evolutionary Analyses
Of the 108 young duplicates in our data set, 18 arose after the divergence of D. melanogaster from D. simulans and D. sechellia (Assis and Bachtrog 2013), and therefore do not have orthologs in D. simulans and could not be used in Ka/Ks or HKA analyses. Pairwise Ka/Ks ratios between protein-coding sequences of all D. melanogaster and D. simulans orthologs were estimated using PAML (Yang 2007).
We used reference genome sequences of D. melanogaster and D. simulans, as well as D. melanogaster DGRP polymorphism data, to perform the described sliding window HKA test. To minimize the impact of small sample size on variance, we used a step size of 1 nt and a sliding window of 10,000 nt, though window sizes of 1,000, 50,000, and 100,000 nt all produced similar patterns. Reference annotation data were used to map signed X2 scores to coding regions, introns, 5′ UTRs, and 3′ UTRs of D. melanogaster genes. Introns containing “nested” protein-coding genes, which represent ∼10% of D. melanogaster genes (Assis et al. 2008), were removed. To maximize power, we included all X2 scores obtained from windows for which the middle position fell within a particular region of interest, resulting in 10,551,784 X2 scores in protein-coding sequences, 2,346,748 X2 scores in 5′ UTRs, 4,088,184 X2 scores in 3′ UTRs, and 29,345,299 X2 scores in introns.
For RELAX analyses, we reconstructed the most complete species phylogeny for each pair of D. melanogaster duplicate genes. Specifically, we required the presence of both members of a pair of duplicates on all four test branches of the tree (see fig. 3A). We also required the presence of an ortholog of the ancestral D. pseudoobscura gene in at least one outgroup species of D. melanogaster and D. pseudoobscura (D. willistoni, D. mojavensis, D. virilis, or D. grimshawi), and included all available orthologs to ensure maximum power. Using this approach, we were able to construct trees for 28 of the 29 duplicates in this age group.
We used selscan (Szpiech and Hernandez 2014) to calculate the haplotype-based statistic nSL (Ferrer-Admetlla et al. 2014) and scan the D. melanogaster genome for recent soft and hard selective sweeps. Haplotypes were obtained from DGRP polymorphism data, and Beagle (Browning and Browning 2009) was used to impute missing data. All scans were run with default parameters, and unstandardized nSL scores were frequency-normalized over all chromosomes. For individual chromosome arms, the standard nSL was calculated as described by Ferrer-Admetlla et al. (2014).
GO Analyses
We performed all GO analyses with the GOrilla tool found at http://cbl-gorilla.cs.technion.ac.il/, last accessed September 5, 2017 (Eden et al. 2007, 2009). We ran GOrilla four times on two unranked lists of genes, with single-copy genes as the background list for all runs. As our target sets, we used young duplicates with evidence of recent positive selection, young duplicates without evidence of recent positive selection, old duplicates with evidence of recent positive selection, and old duplicates without evidence of recent positive selection. For each run, we output results for all enriched GO categories (process, function, and component) and set the P-value threshold to P = 10−3.
Statistical Analyses
All statistical tests were performed in the R software environment (R Core Team 2013). Two-sample permutation tests were used to assess pairwise differences between all distributions compared in figures 1 and 2. For each test, the median was used as the test statistic, and 1,000 permutations were performed. This procedure was repeated at least three times to ensure that P-values were consistent across tests. Sign tests were used to assess whether distributions of signed X2 scores deviate significantly from zero.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We would like to thank two anonymous referees and the journal editor for their valuable feedback. This work was supported by the National Science Foundation (DEB-1555981). Portions of this research were conducted with Advanced CyberInfrastructure resources provided by the Institute for CyberScience at Pennsylvania State University.
References
- Andolfatto P. 2005. Adaptive evolution of non-coding DNA in Drosophila. Nature 4377062:1149–1152. [DOI] [PubMed] [Google Scholar]
- Assis R. 2014. Drosophila duplicate genes evolve new functions on the fly. Fly 82:91–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis R, Bachtrog D.. 2013. Neofunctionalization of young duplicate genes in Drosophila. Proc Natl Acad Sci U S A. 11043:17409–17414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis R, Bachtrog D.. 2015. Rapid divergence and diversification of mammalian duplicate gene functions. BMC Evol Biol. 15:138.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis R, Kondrashov AS.. 2014. Conserved proteins are fragile. Mol Biol Evol. 312:419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis R, Kondrashov AS, Koonin EV, Kondrashov FA.. 2008. Nested genes and increasing organizational complexity of metazoan genomes. Trends Genet. 2410:475–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, Andolfatto P.. 2006. Selection, recombination and demographic history in Drosophila miranda. Genetics 1744:2045–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman CM, Kreitman M.. 2001. Analysis of conserved noncoding DNA in Drosophila reveals similar constraints in intergenic and intronic sequences. Genome Res. 118:1335–1345. [DOI] [PubMed] [Google Scholar]
- Betrán E, Long M.. 2003. Dntf-2r, a young Drosophila retroposed gene with specific male expression under positive Darwinian selection. Genetics 164:977–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj N, Lu H.. 2005. Correlation between gene expression profiles and protein–protein interactions within and across genomes. Bioinformatics 21:2730–2738. [DOI] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH.. 2004. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 167:1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ. 1986. Rates of DNA sequence evolution differ between taxonomic groups. Science 2314744:1393–1399. [DOI] [PubMed] [Google Scholar]
- Brookfield JF. 1997. Genetic redundancy. Adv Genet. 36:137–155. [DOI] [PubMed] [Google Scholar]
- Browning BL, Browning SR.. 2009. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 842:210–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante CD, Fledel-Alon A, Williamson S, Nielsen R, Hubisz MT, Glanowski S, Tanenbaum DM, White TJ, Sninsky JJ, Hernandez RD, et al. 2005. Natural selection on protein-coding genes in the human genome. Nature 4377062:1153–1157. [DOI] [PubMed] [Google Scholar]
- Bustamante CD, Nielsen R, Sawyer SA, Olsen KM, Purugganan MD, Hartl DL.. 2002. The cost of inbreeding in Arabidopsis. Nature 4166880:531–534. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. 2009. Effective population size and patterns of molecular evolution and variation. Nat Rev Genet. 103:195–205. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D.. 2010. Elements of evolutionary genetics. Greenwood Village (CO: ): Roberts and Company Publishers. [Google Scholar]
- Charlesworth J, Eyre-Walker A.. 2006. The rate of adaptive evolution in enteric bacteria. Mol Biol Evol. 237:1348–1356. [DOI] [PubMed] [Google Scholar]
- Chen S, Zhang YE, Long M.. 2010. New genes in Drosophila quickly become essential. Science 3306011:1682–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium. 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437:69–87. [DOI] [PubMed] [Google Scholar]
- Conant GC, Wagner A.. 2003. Asymmetric sequence divergence of duplicate genes. Genome Res. 139:2052–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium. 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450:203–218. [DOI] [PubMed] [Google Scholar]
- Eden E, Lipson D, Yogev S, Yakhini Z.. 2007. Discovering motifs in ranked lists of DNA sequences. PLoS Comput Biol. 33:e39.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z.. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 10:48.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Admetlla A, Liang M, Korneliussen T, Nielsen R.. 2014. On detecting incomplete soft or hard selective sweeps using haplotype structure. Mol Biol Evol. 315:1275–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris SD, Whitt GS.. 1979. Evolution of the differential regulation of duplicate genes after polyploidization. J Mol Evol. 124:267–317. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan Y, Postlethwait J.. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 1514:1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French L, Pavlidis P.. 2011. Relationships between gene expression and brain wiring in the adult rodent brain. PLoS Comput Biol. 7:e1001049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Liu Z, Church GM, Vidal M.. 2001. Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nature Genet. 294:482–486. [DOI] [PubMed] [Google Scholar]
- Gossmann TI, Song BH, Windsor AJ, Mitchell-Olds T, Dixon CJ, Kapralov MV, Filatov DA, Eyre-Walker A.. 2010. Genome wide analyses reveal little evidence for adaptive evolution in many plant species. Mol Biol Evol. 278:1822–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill PR, Bachtrog D, Andolfatto P.. 2008. Positive and negative selection on noncoding DNA in Drosophila simulans. Mol Biol Evol. 259:1825–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill PR, Charlesworth B, Halligan DL, Andolfatto P.. 2005. Patterns of intron sequence evolution in Drosophila are dependent upon length and GC content. Genome Biol. 68:R67.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill PR, Loewe L, Charlesworth B.. 2010. Estimating the parameters of selection on nonsynonymous mutations in Drosophila pseudoobscura and D. miranda. Genetics 1854:1381–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halligan DL, Eyre-Walker A, Andolfatto P, Keightley PD.. 2004. Patterns of evolutionary constraints in intronic and intergenic DNA of Drosophila. Genome Res. 142:273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MV, Demuth JP, McGrath CL, Casola C, Hahn MW.. 2009. Adaptive evolution of young gene duplicates in mammals. Genome Res. 19:859–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halligan DL, Keightley PD.. 2006. Ubiquitous selective constraints in the Drosophila genome revealed by a genome-wide interspecies comparison. Genome Res. 167:875–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zhang J.. 2005. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics 1692:1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hittinger CT, Carroll SB.. 2007. Gene duplication and the adaptive evolution of a classic genetic switch. Nature 4497163:677–681. [DOI] [PubMed] [Google Scholar]
- Holland PW, Garcia-Fernández J, Williams NA, Sidow A.. 1994. Gene duplications and the origins of vertebrate development. Development 1994:125–133. [PubMed] [Google Scholar]
- Huang W, Massouras A, Inoue Y, Peiffer J, Rámia M, Tarone AM, Turlapati L, Zichner T, Zhu D, Lyman RF, et al. 2014. Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 247:1193–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber CD, DeGiorgio M, Hellmann I, Nielsen R.. 2016. Detecting recent selective sweeps while controlling for mutation rate and background selection. Mol Ecol. 251:142–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguadé M.. 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 1161:153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES.. 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 4286983:617–624. [DOI] [PubMed] [Google Scholar]
- Kimura M. 1983. The neutral theory of molecular evolution. Cambridge: Cambridge University Press. [Google Scholar]
- Kohn MH, Fang S, Wu CI.. 2004. Inference of positive and negative selection on the 5′ regulatory regions of Drosophila genes. Mol Biol Evol. 212:374–383. [DOI] [PubMed] [Google Scholar]
- Li WH, Yang J, Gu X.. 2005. Expression divergence between duplicate genes. Trends Genet. 2111:602–607. [DOI] [PubMed] [Google Scholar]
- Llopart A, Comeron JM, Brunet FG, Lachaise D, Long M.. 2002. Intron presence–absence polymorphism in Drosophila driven by positive Darwinian selection. Proc Natl Acad Sci USA. 99:8121–8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Betrán E, Thornton K, Wang W.. 2003. The origin of new genes: glimpses from the young and old. Nat Rev Genet. 4:865–875. [DOI] [PubMed] [Google Scholar]
- Lundin LG. 1993. Evolution of the vertebrate genome as reflected in paralogous chromosomal regions in man and the house mouse. Genomics 161:1–19. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS.. 2000. The evolutionary fate and consequences of duplicate genes. Science 2905494:1151–1155. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS.. 2003. The origins of genome complexity. Science 3025649:1401–1404. [DOI] [PubMed] [Google Scholar]
- Mackay TF, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han Y, Magwire MM, Cridland JM, et al. 2012. The Drosophila melanogaster genetic reference panel. Nature 4827384:173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik HS, Henikoff S.. 2001. Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 1573:1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama EN. 1987. Higher rates of nucleotide substitution in Drosophila than in mammals. Jpn J Genet. 622:139–147. [Google Scholar]
- Nadeau JH, Sankoff D.. 1997. Comparable rates of gene loss and functional divergence after genome duplications early in vertebrate evolution. Genetics 1473:1259–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. Berlin: Springer Science & Business Media. [Google Scholar]
- Postlethwait JH, Yan YL, Gates MA, Horne S, Amores A, Brownlie A, Donovan A, Egan ES, Force A, Gong Z, et al. 1998. Vertebrate genome evolution and the zebrafish gene map. Nat Genet. 184:345–349. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2013.
- Ranwez V, Harispe S, Delsuc F, Douzery EJ.. 2011. MACSE: Multiple Alignment of Coding SEquences accounting for frameshifts and stop codons. PLoS ONE. 69:e22594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi S, Liberles DA.. 2005. Subfunctionalization of duplicated genes as a transition state to neofunctionalization. BMC Evol Biol. 5:28.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer S, Kulathinal R, Bustamante C, Hartl D.. 2003. Bayesian analysis suggests that most amino acid replacements in Drosophila are driven by positive selection. J Mol Evol. 57:S154–S164. [DOI] [PubMed] [Google Scholar]
- Shiu SH, Byrnes JK, Pan R, Zhang P, Li WH.. 2006. Role of positive selection in the retention of duplicate genes in mammalian genomes. Proc Natl Acad Sci U S A. 1037:2232–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidow A. 1996. Gen(om)e duplications in the evolution of early vertebrates. Curr Opin Genet Dev. 66:715–722. [DOI] [PubMed] [Google Scholar]
- Slotte T, Foxe JP, Hazzouri KM, Wright SI.. 2010. Genome-wide evidence for efficient positive and purifying selection in Capsella grandiflora, a plant species with a large effective population size. Mol Biol Evol. 278:1813–1821. [DOI] [PubMed] [Google Scholar]
- Smith NG, Eyre-Walker A.. 2002. Adaptive protein evolution in Drosophila. Nature 4156875:1022–1024. [DOI] [PubMed] [Google Scholar]
- Stoltzfus A. 1999. On the possibility of constructive neutral evolution. J Mol Evol. 492:169–181. [DOI] [PubMed] [Google Scholar]
- Szpiech ZA, Hernandez RD.. 2014. Selscan: an efficient multi-threaded program to perform EHH-based scans for positive selection. Mol Biol Evol. 3110:2824–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JS, Raes J.. 2004. Duplication and divergence: the evolution of new genes and old ideas. Annu Rev Genet. 38:615–643. [DOI] [PubMed] [Google Scholar]
- Wertheim JO, Murrell B, Smith MD, Pond SLK, Scheffler K.. 2015. RELAX: detecting relaxed selection in a phylogenetic framework. Mol Biol Evol. 323:820–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: a program package for phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1568–1591. [DOI] [PubMed] [Google Scholar]
- Zhang J. 2003. Evolution by gene duplication: an update. Trends Ecol Evol. 186:292–298. [Google Scholar]
- Zhang L, Li WH.. 2005. Human SNPs reveal no evidence of frequent positive selection. Mol Biol Evol. 2212:2504–2507. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang YP, Rosenberg HF.. 2002. Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nat Genet. 30:411–415. [DOI] [PubMed] [Google Scholar]
- Zhou X, Kao MCJ, Wong WH.. 2002. Transitive functional annotation by shortest-path analysis of gene expression data. Proc Natl Acad Sci U S A. 99:12783–12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.