Abstract
Experimental evolution affords the opportunity to investigate adaptation to stressful environments. Studies combining experimental evolution with whole-genome resequencing have provided insight into the dynamics of adaptation and a new tool to uncover genes associated with polygenic traits. Here, we selected for starvation resistance in populations of Drosophila melanogaster for over 80 generations. In response, the starvation-selected lines developed an obese condition, storing nearly twice the level of total lipids than their unselected controls. Although these fats provide a ∼3-fold increase in starvation resistance, the imbalance in lipid homeostasis incurs evolutionary cost. Some of these tradeoffs resemble obesity-associated pathologies in mammals including metabolic depression, low activity levels, dilated cardiomyopathy, and disrupted sleeping patterns. To determine the genetic basis of these traits, we resequenced genomic DNA from the selected lines and their controls. We found 1,046,373 polymorphic sites, many of which diverged between selection treatments. In addition, we found a wide range of genetic heterogeneity between the replicates of the selected lines, suggesting multiple mechanisms of adaptation. Genome-wide heterozygosity was low in the selected populations, with many large blocks of SNPs nearing fixation. We found candidate loci under selection by using an algorithm to control for the effects of genetic drift. These loci were mapped to a set of 382 genes, which associated with many processes including nutrient response, catabolic metabolism, and lipid droplet function. The results of our study speak to the evolutionary origins of obesity and provide new targets to understand the polygenic nature of obesity in a unique model system.
Keywords: obesity, evolve and resequence, Drosophila melanogaster, starvation selection, experimental evolution, metabolism
Introduction
Natural selection drives populations to adapt to a wide variety of stressors which are dynamic across evolutionary time. Investigating the mechanistic basis of these adaptations has fundamentally transformed modern science and medicine (Hodgkin and Huxley 1952; Chien et al. 1976; Krebs and Johnson 1980). A complementary approach to studying uniquely adapted organisms in nature is to simulate evolution in the laboratory, where investigators mimic selective pressures an organism may face in the wild with highly controlled, replicated designs (Bennett 2003). Traditionally, experimental evolution has been a powerful tool in testing theories of evolution and understanding the physiological basis of adaptation to novel environments (Garland and Rose 2009; Kawecki et al. 2012).
Recently, experimental evolution has been elevated by the increased accessibility of next-generation sequencing, which has provided insight into the genetic basis of adaptation on a genome-wide level. The “Evolve and Resequence” (E&R, Turner et al. 2011) strategy has been frequently implemented in work with Drosophila, where researchers have analyzed whole-genome data sets for a variety of selected traits including hypoxia tolerance (Zhou et al. 2011), longevity (Remolina et al. 2012), courtship song (via interpulse interval; Turner and Miller 2012), body size (Turner et al. 2011), desiccation resistance (Kang et al. 2016), development time (Burke et al. 2010), temperature tolerance (Orozco-terWengel et al. 2012) and resistance to both the parasitoid Asobara tabida (Jalvingh et al. 2014), and the Drosophila C virus (Martins et al. 2014). While future improvements in experimental design promise to increase the power to detect adaptive loci (Baldwin-Brown et al. 2014; Kofler and Schlotterer 2014; Kessner and Novembre 2015), studies of existing selected Drosophila populations have done a great deal to describe the genomic topography of laboratory-selected phenotypes. They provide evidence that adaptation is driven by standing genetic variation (Burke et al. 2010), and they also find evidence for multiple temporal trajectories for alleles under selection (Orozco-terWengel et al. 2012; but see Burke and Long 2012). Regions of the genome that change in frequency following laboratory selection may harbor adaptive polymorphisms; genes at or near these candidate alleles have been associated with selected traits (Burke et al. 2010; Turner et al. 2011; Zhou et al. 2011; Orozco-terWengel et al. 2012; Remolina et al. 2012; Turner and Miller 2012; Jalvingh et al. 2014; Martins et al. 2014; Kang et al. 2016). Consigning causality to candidate loci remains challenging as linkage disequilibrium may increase in experimentally evolved populations of Drosophila (Teotónio et al. 2009; Franssen et al. 2015). While this may inflate the number of loci detected under selection, Drosophila are a genetically tractable model, which allows for functional analysis to discriminate causal variants. For instance, candidates can be tested using high-throughput screens, with tools such as the GAL4-UAS system, RNA-interference lines and CRISPR, used to modify spatiotemporal gene expression. There have been many E&R studies in other species (reviewed in: Kawecki et al. 2012; Long et al. 2015), however, Drosophila is particularly well suited for evolution experiments due to its reasonably short generation times, low maintenance costs, and genetic tractability. In addition, the relative genetic similarity between flies and mammals provides promise for translational impact (Reiter et al. 2001).
Here, we performed an E&R study on populations of Drosophila melanogaster that were selected for starvation resistance for 83 generations. In response to this selection regime, the starvation-selected lines have evolved an obese condition, storing nearly two times the levels of total lipids as compared with their unselected controls (Reynolds 2013; Masek et al. 2014; Hardy et al. 2015; fig. 1A). They metabolize these fats when starved, allowing them to live ∼11–14 days without food which is 3–4 times longer than the unselected control populations. Obesity in these populations is largely driven by a developmental delay during the third larval instar (Reynolds 2013). This delay provides time for the selected lines to acquire excess nutrients, which are synthesized into lipids as nutrient deprivation during this window has been shown to reduce whole body triglycerides to control levels in adult flies (Masek et al. 2014; Hardy et al. 2015). The selected populations are also adapted to maintain high fat stores as adults, presenting with high whole body triglyceride levels at 4 and 11 days post-eclosion despite consuming fewer overall calories (Masek et al. 2014; Hardy et al. 2015). The starvation-selected populations are genetically programmed to store and retain fat, which comes with many evolutionary costs. The tradeoffs associated with disrupted lipid homeostasis range from depressed metabolic rate accompanied by low activity levels and poor flight performance to low fecundity, dilated cardiomyopathy and excess sleep (Brewer 2013; Reynolds 2013; Masek et al. 2014; Hardy et al. 2015). Many of these phenotypes are reminiscent of other Drosophila models of obesity (Birse et al. 2010; Na et al. 2013) and are also characteristic of metabolic disorder in mammals.
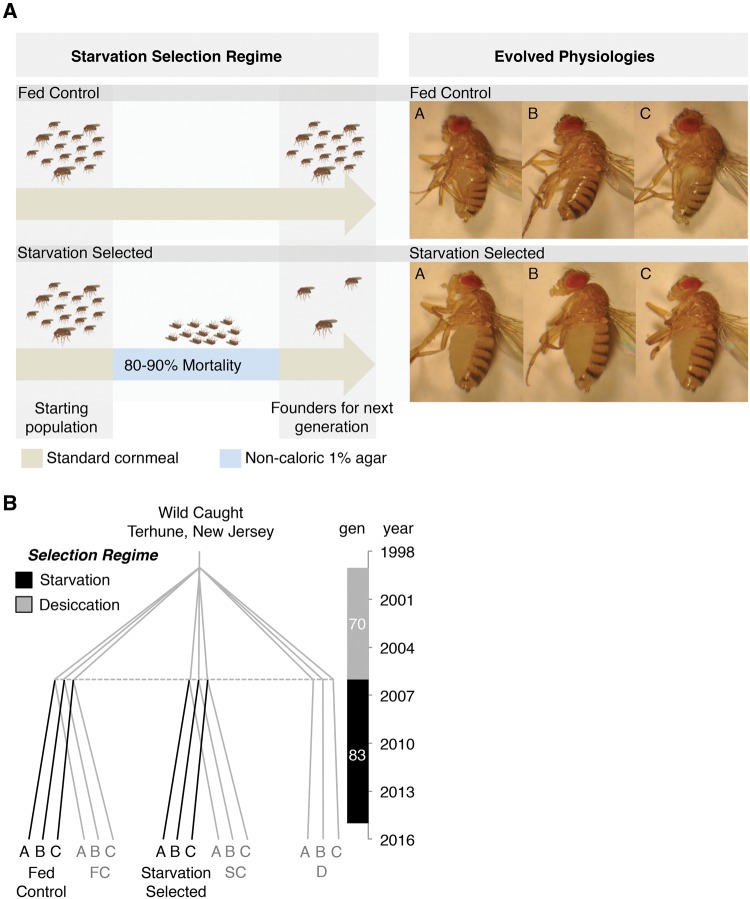
Fig. 1.
Selection regime, phylogeny and evolutionary history of the selected populations. (A) Each generation the starvation-selected populations SA–C were starved until 80–90% had died. The survivors were used as founders for the subsequent generation. At the same time the fed control populations FA–C were handled in parallel and fed ad libitum. The visible differences between FA–C and SA–C as 4–5-day-old adults demonstrate some of the evolved characteristics. (B) Detailed phylogeny and evolutionary history of the starvation-selected lines. Axis to the right displays the number of generations and year. Black lines represent the starvation-selection experiment studied here. To date, the starvation-selection experiment has been running for over 100 generations; however, sequence data were collected at generation 83.
To understand the genetic basis for these complex phenotypes we sequenced pooled genomic DNA from replicate populations of both the starvation-selected and unselected lines. We found many polymorphisms at dissimilar frequencies between the replicate populations of selected lines, which correlated with phenotypic differences in starvation resistance. Genome-wide levels of heterozygosity showed strong signatures of selection, with many large regions of the genome approaching fixation, which were fairly inconsistent across selected replicates. Despite heterogeneity among replicate populations, the differences in allele frequencies between selective treatments were much higher. To look for conserved mechanisms of adaptation in response to selection, we developed an algorithm to filter polymorphisms that were more likely to be divergent due to genetic drift. After filtering, we mapped the remaining loci to a set of 382 genes which were conserved across replicate populations. We found enrichment for genes involved in many biological processes, including nutrient response, catabolic metabolism and lipid droplet function. From an evolutionary perspective, our study is consistent with the thrifty gene hypothesis (TGH), an evolutionary theory of obesity in which alleles that increase nutrient storage and dampen utilization are selected for during periods of famine (Neel 1962). Here, we provide a list of such candidate genes, which can be functionally validated and used as targets for future research.
Results
Genetic Heterogeneity between the Replicates of the Starvation-Selected Populations Correlates with Differences in Starvation Resistance
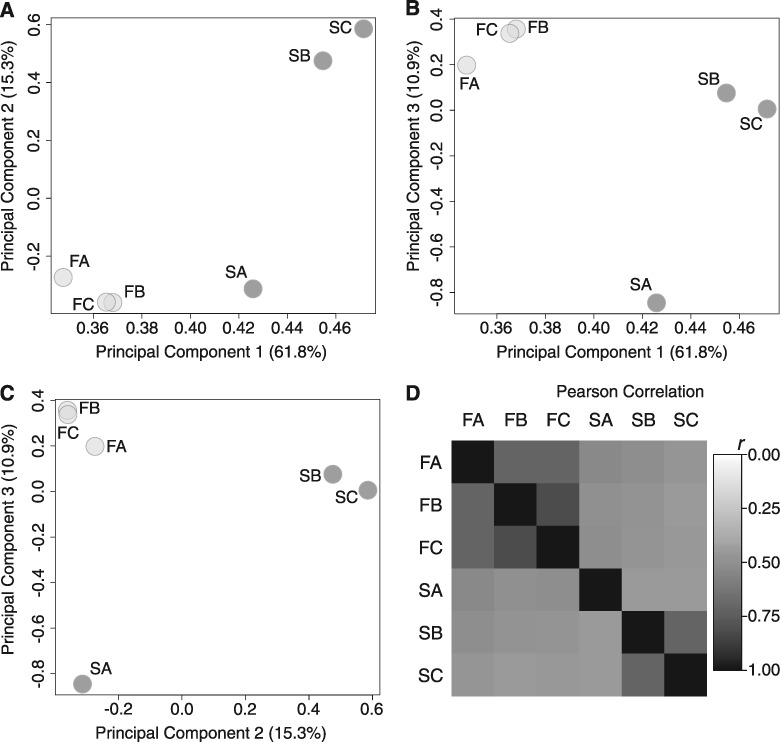
After the 83rd generation of starvation selection we resequenced genomic DNA from pools of 100 females for each of the 3 starvation-selected populations (hereafter “S” populations, subscript “A–C”) and their three unselected fed controls (hereafter “F” populations, subscript “A–C”; see Materials and Methods for detailed evolutionary history; outlined in fig. 1B). The raw sequence reads were mapped and filtered for quality control, resulting in a set of 1,046,373 polymorphisms across all 6 populations (FA–C, SA–C; see Materials and Methods). We performed a principal component analysis (PCA) to visualize the relationships between populations with respect to reference allele frequency. The first three components explained 88.0% of the variation in allele frequency between populations (supplementary table S1, Supplementary Material online). The F populations clustered tightly together, separated from the S populations, suggesting an overall effect of selection on allele frequency (fig. 2A–C). However, the S populations appeared more variable, with the SA replicate clustering independently from the SB and SC populations. We quantified the observed PCA trends by calculating the Pearson product-moment correlation coefficient (r) with respect to reference allele frequency between all six populations in a pairwise manner (fig. 2D, supplementary fig. S1A and B, Supplementary Material online). Correlations in allele frequencies between the F and S populations were relatively low (r = 0.45–0.54, lower left quadrant, fig. 2D) compared with correlations within F replicate populations (r = 0.71–0.82, upper left quadrant, fig. 2D), consistent with a strong effect from the selection treatment. We also found relatively low correlation of allele frequencies between the selected replicate populations (r = 0.45–0.71, lower right quadrant, fig. 2D), compared with intra-population correlations in the F populations (r = 0.71–0.82). This was largely due to poor correlation of the SA population with the other S populations (SA×SB, r = 0.45; SA×SC, r = 0.45) while SB and SC maintained higher relative correlation (r = 0.71). All correlations were statistically significant with Bonferroni adjusted P-values <2.0×10−16 (supplementary fig. S1B, Supplementary Material online).
Fig. 2.
Genetic heterogeneity in the starvation-selected replicates. (A–C) PCA plots of the first three components, which explain ∼88% of the variation in reference allele frequency between the populations. Dark grey circles represent populations SA–C while the light grey circles represent FA–C. FA–C cluster together and are separated from the S populations. The SA population appears to cluster independently from SB–C. (D) Pairwise Pearson Product Moment Correlation Analysis of all six populations. Darker squares represent a higher degree of correlation in reference allele frequency.
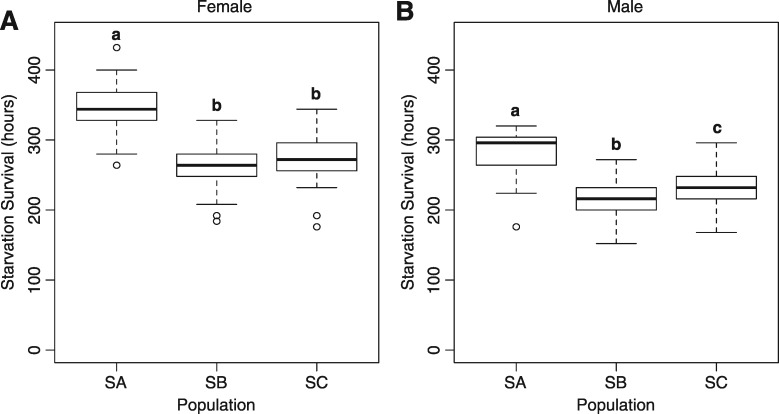
Next we wanted to test if genetic variation between the S populations correlated with differences at the organismal level. To do this we measured starvation resistance in the S populations. On average, SA females survived starvation for 344.6 ± 4.7 h, a 25.8–29.4% increase in duration over SB and SC females (SB: 266.3 ± 4.2 h, P < 2.0×10−16; SC: 274.0 ± 4.7 h, P < 2.0×10−16, fig. 3A). Similarly, SA males survived an average of 283.7 ± 3.9 h of starvation, 20.5–31.6% longer than their sex matched SB and SC controls (SB: 215.5 ± 3.5 h, P < 2.0×10−16; SC: 235.4 ± 4.2 h, P < 2.0×10−16, fig. 3B). We found the SB and SC populations were more similar in starvation survival with no significant difference between SB and SC females (P > 0.46, fig. 3A) and a relatively small 9.3% increase in starvation survival in SC over SB males (P < 1.2×10−3, fig. 3B).
Fig. 3.
Differences in starvation resistance between the starvation-selected populations match patterns of genetic heterogeneity. The SA population is significantly better at surviving starvation than the SB and SC populations for both (A) females and (B) males. Different letters signify statistically different groups where P < 0.05; Tukey Post hoc; ANOVA; N = 47–50.
Genome-Wide Heterozygosity Levels Provide Evidence for Strong Selective Sweeps
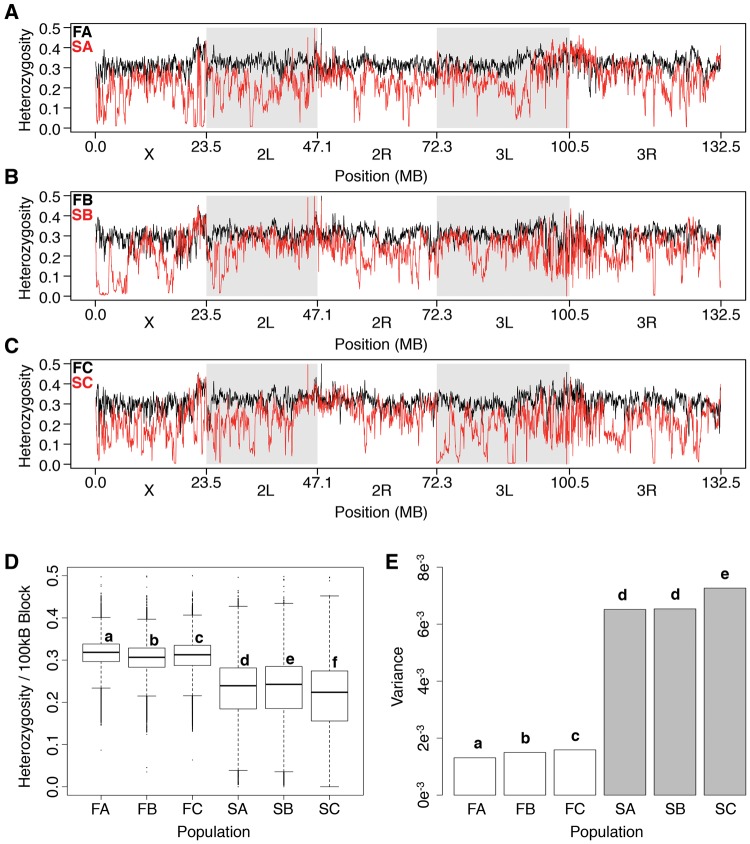
During natural selection, advantageous loci and regions within linkage disequilibrium increase in frequency leading to localized reduction in variance marked by low levels of heterozygosity (Burke 2012). To study the regional effects of selection in the S populations, we calculated average heterozygosity for blocks of SNPs within a 100 kb sliding window with a step size of 2,000 bases across the genome. The F populations demonstrated fairly consistent genome-wide levels of heterozygosity (black lines, fig. 4A–C), as average block heterozygosity was equal to 0.30–0.32 (fig. 4D) with low variance, ranging from 1.3×10−3 to 1.6×10−3 between populations (fig. 4E). The S populations however displayed many regional declines in heterozygosity (red lines, fig. 4A–C), which presented in lower average block heterozygosity (0.21–0.23, P < 2e−16, fig. 4D) and a 4.4–4.9-fold increase in variance over the F populations (P < 2.2e−16, fig. 4E).
Fig. 4.
Genome-wide heterozygosity. (A–C) Genome-wide sliding window heterozygosity plots (100 kb window, with 2 kb steps). SA–C (red) display many regional declines in heterozygosity, while FA–C (black) heterozygosity remains stable. This results in (D) lower mean heterozygosity per 100 kb block and (E) increased levels of variance in the selected lines. Different letters signify statistically different groups where P < 0.05. See Materials and Methods for statistical tests.
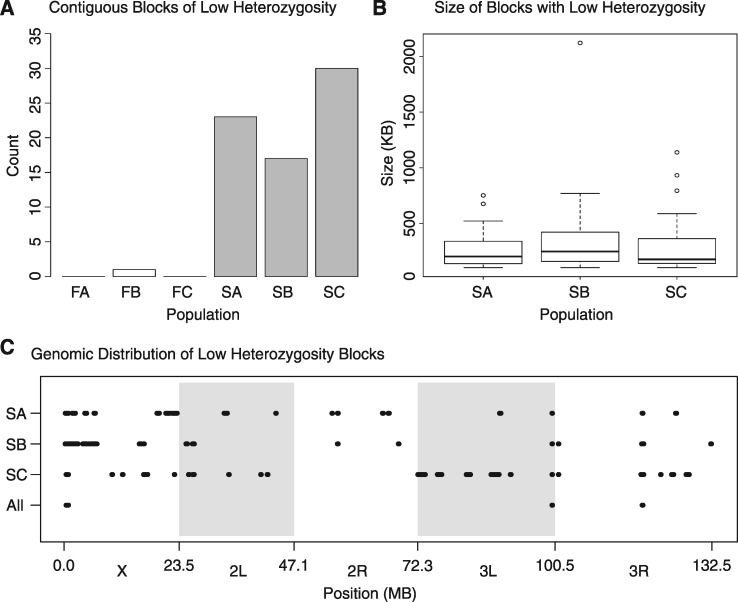
In some instances, we found regions of the genome approaching fixation, as we discovered 1,983–3,184,100 kb blocks in the S populations with average heterozygosity <0.05 and no more than four blocks in any F population (supplementary fig. S2A, Supplementary Material online). Many times these 100 kb blocks overlapped and clustered together into contiguous sequences. For each population (SA–C) we found 17–30 contiguous blocks of low heterozygosity (fig. 5A) which averaged 321 ± 38 kb (S.E.M.) in length and ranged from 104 to 2,120 kb (fig. 5B). We looked for overlapping regions of these contiguous blocks across replicates to look for any repeatable signatures of selection (fig. 5C). While many of the blocks were associated with a specific population, we did find four regions across chromosomes X, 3L and 3R which had low heterozygosity (<0.05) across all replicates which ranged from 72 to 176 kb (bottom row, fig. 5C). We found a set of 12 previously characterized genes within these windows and 36 in total (supplementary table S2, Supplementary Material online). These genes did not cluster into any significant gene ontology (GO) categories. We also visualized genetic differentiation in this data set by plotting FST values for the F and S populations (supplementary fig. S2B, Supplementary Material online). Genome-wide values of FST were higher in the S populations compared to the F controls, consistent with our observation of greater heterogeneity in allele frequencies in the selected populations.
Fig. 5.
Selective sweeps are large and inconsistent in the starvation-selected populations. (A) SA–C have many extended regions of low heterozygosity approaching fixation (2pq<0.05), (B) which average ∼300 kb in size but range from ∼100 to 2,000 kb. (C) The large contiguous blocks of heterozygosity are inconsistent across replicates. On the “All” horizontal axis are the four regions of overlap across the S populations. The regions of low heterozygosity are not to scale; they were made large to visualize the approximate region.
Finding Polymorphisms That Are Consistent with Selection
Localized depressions in heterozygosity are indicative of selection favoring a variant and “sweeping,” or reducing, neutral variation linked to the selected locus. However, alleles associated with complex traits in sexual species may respond to selection through incomplete sweeps and/or dynamic trajectories that do not involve fixation (Burke 2012). Such loci may be difficult to distinguish from those which have changed in frequency due to genetic drift. In order to find loci that were consistent with selection we developed an algorithm to filter SNPs whose difference in allele frequency between selection treatments could be explained by genetic drift. The algorithm models the work of Motoo Kimura, using a diffusion model (eq. 1) to describe genetic drift (Supplemental Code, Kimura 1955). While Kimura’s diffusion approximation has been used to model drift elsewhere (Stemshorn et al. 2011), to our knowledge it has not been adapted for use in E&R studies.
| (1) |
Given the number of generations (t), effective population size (Ne), and initial allele frequency (p), the algorithm generates a posterior probability distribution of allele frequency due to drift. Kimura’s equation requires the Gauss hypergeometric function 2F1, which was calculated using the hyp2f1 function from SciPy (Release 0.14.0). We found that 50 summations, x = 1,000 was sufficient sampling to generate an appropriate distribution with adequate speed.
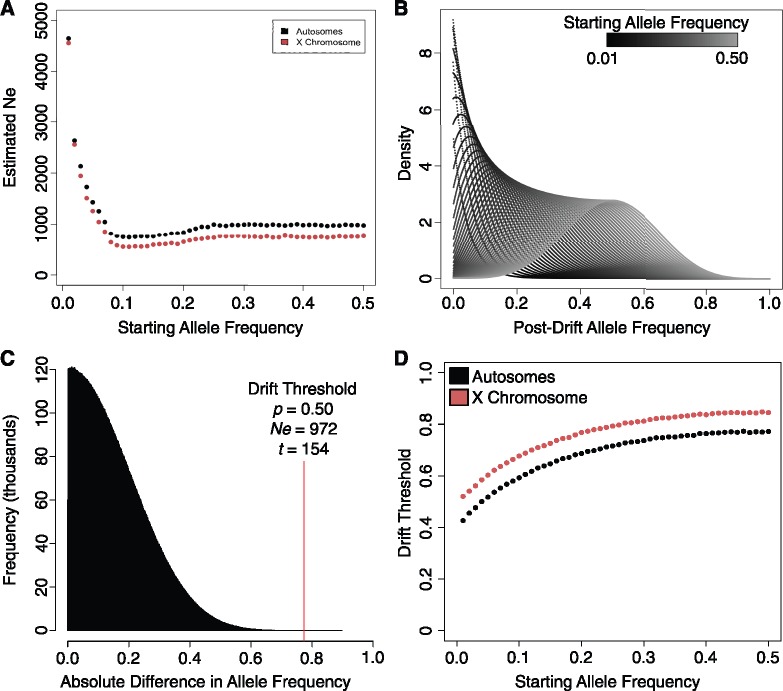
Next we needed to define the parameters (t, Ne, p) for the starvation-selection experiment. Here, we used t = 154 generations as replicates were split for 70 generations prior to the 83 generations of selection for starvation resistance plus the generation removed from selection (see detailed phylogeny in fig. 1B). Effective population size was then estimated by measuring the variance in allele frequency between the unselected (FA–C) populations, using the 1,046,373 quality control filtered SNPs and the theoretical equation for variance in allele frequency after t generations (eq. 2; Templeton 2006).
| (2) |
We grouped SNPs whose average minor allele frequency across populations FA–C was equal when rounded to the nearest hundredth and calculated the variance of each bin. We then calculated Ne for each bin, using the bin’s associated allele frequency P = {0.0, 0.01, 0.02, …, 0.50} and t = 154 generations. Variation calculated from low allele frequency bins (P < 0.07) was much lower than from higher frequencies, which led to suspiciously high Ne values (fig. 6A). However, the bias quickly resolved toward a stable estimate at allele frequencies >0.07. Because of this bias we took the median Ne from all 51 bins. From the data we calculated effective population sizes of 972 for autosomes and 748 for the X chromosome. We believe these estimates are reasonable given the census population size of ∼2,000 individuals (see Materials and Methods). Furthermore, the X to autosome Ne ratio of ∼0.77 is consistent with the theoretical 0.75 ratio, given no evidence of a sex bias in our populations.
Fig. 6.
Filtering for genetic drift. (A) Effective population size was estimated from the data using equation (2), plotted here as a function of the starting allele frequency. (B) Posterior density distribution of post-drift allele frequency for each starting allele frequency in {0.01, 0.02, …, 0.50}. These distributions were obtained using Kimura’s diffusion equation. (C) Histogram, showing the 1.0×107 values of absolute difference in allele frequency obtained from simulating two populations under drift. The 99.999% quantile of each distribution became the Drift Threshold. (D) Relationship between the starting allele frequency and the Drift Threshold for autosomes (black) and the X chromosome (red).
We used Kimura’s equation described above using parameters t = 154 and Ne = {972|748} for each starting allele frequency p = {0.01, 0.02, 0.03, …, 0.50}. For each value of P we obtained a probability distribution of allele frequencies, which described the likelihood of posterior allele frequencies due to genetic drift from 0 to 1 (fig. 6B). We randomly sampled from these probability distributions to perform simulations of genetic drift. Specifically, for each value of p we simulated two populations under drift by generating a set of 3.0×107 values for each population by randomly sampling from its associated probability distribution. We then calculated the absolute difference between the indices of both sets, creating a distribution of absolute difference in allele frequency between two populations under drift (fig. 6C). We calculated the 99.999% quantile of each distribution to observe extreme absolute changes in allele frequency (fig. 6C). The 99.999% value became the “Drift Threshold,” which was calculated for each value of p, to generate a curve which described the relationship between starting allele frequency and the maximum allele frequency difference expected between two populations by drift (fig. 6D). To corroborate our approach, we adjusted the number of simulations in our algorithm to match the first 13,000 runs of a forward simulation of genetic drift (supplementary fig. S3A, Supplementary Material online). We found a high degree of similarity between our algorithm and the forward simulation (supplementary fig. S3B and C, Supplementary Material online, autosomes: r2 = 0.96, P < 5.9×10−36; X: r2 = 0.93, P < 4.5×10−29).
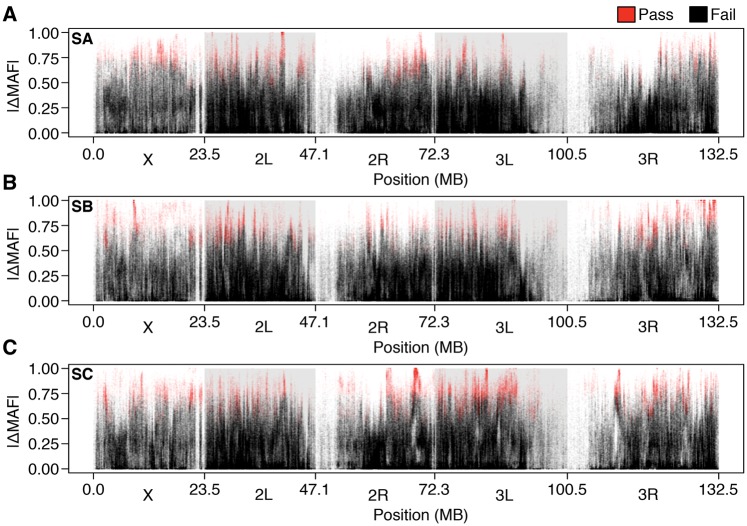
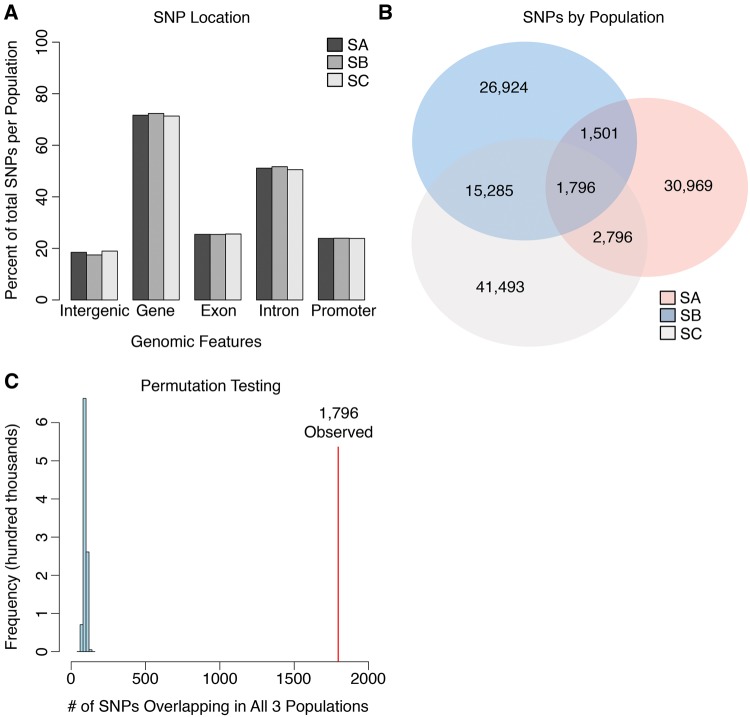
Starting allele frequency for each SNP was estimated as the average minor allele frequency across F replicates. This value was rounded to the nearest hundredth and used as the starting allele frequency to find the Drift Threshold for that SNP. If the absolute difference in allele frequency between the independent replicates (e.g. |MAFFA–MAFSA|) was less than the Drift Threshold, the SNP was discarded. After applying the algorithm to our data set we were left with 37,062, 45,506 and 61,370 high-quality SNPs for replicates A, B, and C whose differences between selection treatments were consistent with selection. We plotted the absolute difference in minor allele frequency between the F and S populations for all SNPs across the genome and found many peaks across all three replicates (red points, fig. 7A–C). We mapped SNPs from each population (SA–C) to genomic features, including total gene region, introns, exons, promoter regions (1 kb up or down stream) and intergenic regions. We found a nearly identical distribution of SNPs by genomic feature across populations (fig. 8A). In total, 120,764 unique SNPs passed our filter in at least one replicate population. Most of these SNPs (82.3%) were unique to a single replicate, further demonstrating genetic dissimilarity among starvation-selected replicates. There were however 19,582 SNPs, that were detected in at least two replicate populations. The bulk of these SNPs (15,285) were shared between the SB and SC populations. The SA population had fewer SNPs in common with the SB and SC populations, sharing just 1,501 and 2,796 SNPs, respectively, consistent with our previous measurements of low allelic correlation (fig. 2). In total only 1,796 SNPs (1.5%) were consistent with selection across all populations (fig. 8B).
Fig. 7.
Candidate loci (A–C) Absolute difference in allele frequency between the replicate F–S pairs. Red points indicate SNPs whose absolute difference in allele frequency is greater than the Drift Threshold, making them consistent with selection. The loci in black could not be distinguished from drift.
Fig. 8.
Characterization of SNPs under selection. (A) The S populations had nearly identical distribution of SNPs in different genomic elements. (B) Selection targeted largely dissimilar loci across S replicates, but converged on a core set of 1,796 SNPs. (C) We created a null distribution of overlapping SNPs, by randomly sampling SNPs for each population from our total pool. We found that the core set of 1,796 overlapping SNPs (indicated in red), were much higher than would be expected by chance (P < 1×10−6, light blue distribution).
The Central Mechanisms of Adaptation Are Related to a Wide Variety of Cellular Processes Including Nutrient Response, Catabolic Metabolism, and Lipid Droplet Function
While the 1,796 SNPs were a low overall fraction of the entire pool, we found the number to be highly significant when compared with a distribution of randomly overlapping SNP counts given our experimental parameters (P < 1.0×10−6, fig. 8C). In brief, the values in the null distribution were calculated by first randomly sampling SNPs for three groups of 37,062, 45,506, and 61,370 SNPs from our total pool of 1,046,373 SNPs. We then calculated the number of SNPs that overlapped across all groups, performing 106 such iterations (see Permutation Testing in Materials and Methods). We decided to focus our analyses on these candidates to explore the central mechanisms of adaptation. First the 1,796 SNPs were mapped to a set of 382 genes (supplementary table S3, Supplementary Material online). We then used the set of 1,796 candidate SNPs as our input for Gowinda, a GO tool that corrects gene-length bias (Kofler and Schlotterer 2012). In total, this resulted in 279 GO terms with P < 0.05. We corrected this set for hierarchical clustering using GO-Module (Yang et al. 2011) and limited the analysis to GO categories that contained five or more genes. Ultimately, this resulted in a set of 77 enriched GO terms from our 1,796 SNPs (supplementary table S4, Supplementary Material online). These terms encompassed a broad range of biological processes, with the top categories largely involved in oxireductase/glutathione transferase activity, nutrient response/catabolic metabolism, immune function, ion transport, COP-II vesicle function, small RNA gene silencing, development, and lipid droplet function (table 1).
Table 1.
Top GO Enrichments.
| GO terms | GO accession | P |
|---|---|---|
| Oxidoreductase activity, acting on a sulfur group of donors | GO: 0016667 | 2.60E−04 |
| Response to nutrient | GO: 0007584 | 5.70E−04 |
| Immune response-regulating signaling pathway | GO: 0002764 | 1.40 E − 03 |
| Glutathione transferase activity | GO: 0004364 | 2.40 E − 03 |
| Cellular catabolic process | GO: 0044248 | 2.40 E − 03 |
| Organic substance catabolic process | GO: 1901575 | 3.10 E − 03 |
| Solute: cation antiporter activity | GO: 0015298 | 3.50 E − 03 |
| COPII vesicle coat | GO: 0030127 | 3.60 E − 03 |
| Regulation of production of small RNA involved in gene silencing by RNA | GO: 0070920 | 4.70 E − 03 |
| Positive regulation of biosynthetic process of antibacterial peptides active against Gram-negative bacteria | GO: 0006964 | 5.00 E − 03 |
| Post-embryonic organ development | GO: 0048569 | 5.60 E − 03 |
| Positive regulation of Toll signaling pathway | GO: 0045752 | 5.80 E − 03 |
| Threonine-type endopeptidase activity | GO: 0004298 | 6.00 E − 03 |
| Lipid particle | GO: 0005811 | 6.20 E − 03 |
| Localization within membrane | GO: 0051668 | 6.20 E − 03 |
| Cytoskeletal anchoring at plasma membrane | GO: 0007016 | 6.70 E − 03 |
| Steroid dehydrogenase activity | GO: 0016229 | 6.80 E − 03 |
| Glutathione metabolic process | GO: 0006749 | 7.90 E − 03 |
| Structural constituent of cytoskeleton | GO: 0005200 | 8.10 E − 03 |
| Purine ribonucleoside metabolic process | GO: 0046128 | 8.50 E − 03 |
Discussion
Here, we report the first E&R study to look at the genome-wide effects of experimental selection for starvation resistance in D. melanogaster. We found 1,046,373 polymorphic sites across populations FA–C and SA–C with 120,764 unique loci whose frequency diverged greater than expected by drift between selection treatments. Allelic frequencies at these loci varied between SA–C, although we found a significantly enriched core set of 1,796 SNPs that associated with 382 genes across replicate populations. Here, we discuss the evolutionary forces driving genetic adaptation to starvation resistance, highlight the allelic responses of genes under selection and consider how these data speak to the evolutionary origins of obesity.
Evolutionary Dynamics of Adaptation to Starvation Selection
Laboratory adaptation in Drosophila generally occurs through selection on standing genetic variation, typically resulting in apparent convergent evolution across experimental replicates (Burke 2012; Long et al. 2015; but see Kang et al. 2016). In asexual species, adaptation occurs via a different mechanism, sometimes called “hard” selective sweeps, where beneficial de novo mutations arise then are driven toward fixation (Burke 2012). Hard sweeps in asexual E&R studies tend to be heterogeneous across replicate populations and implicate candidate alleles private to a single replicate (e.g. Tenaillon et al. 2012; reviewed in Long et al. 2015). Here, we observe heterogeneous sweep patterns among our evolutionary replicates (SA–C) with large (100–2,000 kb) footprints of low heterozygosity that approach fixation. While this may be consistent with a hard sweep model, it is extremely unlikely that de novo mutations are driving adaptation due to the small population sizes (∼103) and relatively low number of generations in our experiment.
Near-fixation of large portions of the genome may also be caused by genetic drift, especially in a population with low Ne. While we developed an algorithm to control for the effects of genetic drift, one potential limitation of our approach is that Ne was estimated from the unselected control populations. If the starvation-selection treatment resulted in lower Ne, potentially due to more genetic similarity between survivors, our method could under-estimate the effects of drift. In order to help mitigate such an effect, each S populations was maintained at ∼10,000 individuals at the start of each generation so that after selection there were still ∼2,000 individuals left to breed (see Materials and Methods). While this is a relatively small population size when considering the possibility of a de novo mutation, it is one of the largest population sizes in an E&R study on sexual organisms to date (see populations sizes of: 320, Turner et al. 2011; 331, Turner and Miller 2012; ∼242, Jalvingh et al. 2014; < ∼600, Martins et al. 2014; ∼150–300, Kang et al. 2016). These methods left the S and F populations with roughly the same number of breeding individuals each generation. While the F populations were under drift for the same number of generations with roughly equal population sizes, we found little evidence of large-scale fixation. We therefore suspect that much of the genetic differentiation is due to selection. While we cannot completely eliminate the effects of genetic drift within our data set, we consciously reduced these effects by using an extremely stringent drift threshold (99.999%) and limiting our downstream analysis to SNPs that were only found in all three S replicate populations.
We suspect that differences in experimental design play a large role in the differences we observe from other E&R experiments in Drosophila. For instance the starvation-selection regime imposes extreme selection pressures (∼85% mortality/generation) which are lethal, analogous to experimental culling or domestication (Rose et al. 1990; Bennett 2003). While a few artificial selection studies in Drosophila have also reported fixation, the degree of fixation in these studies is not as pervasive as reported here and generally localized to small numbers of ∼10–50 kb windows, which is consistent with complete “soft” sweeps on standing genetic variation (Turner et al. 2011; Zhou et al. 2011). A recent analysis of desiccation-selected Drosophila also found evidence for hard selective sweeps, but at a lower frequency than reported here (Kang et al. 2016). A more appropriate comparison may then be made with studies on domestication, such as genomic data from domesticated rabbits which similarly demonstrate large troughs in heterozygosity that are inconsistent across populations/strains (Carneiro et al. 2014). The large sweeps (∼300 kb) in our data set are consistent with high selection pressure, as studies in domesticated rice have used the size of selective sweeps to estimate the strength of selection—with large selective sweeps associated with high selection strength (Olsen et al. 2006).
Selection can lead to rapid changes in allele frequency especially as selection coefficients approach 1, and rapid fixation would allow less time for recombination events to break apart linked loci, leading to larger sweep sizes. While we do not have genetic data to track allele frequencies over the course of selection, experimental selection for starvation resistance in the S populations and in similar studies have revealed rapid adaptation with evolved responses within as few as five generations despite differences in selective strengths ranging from ∼85% mortality/generation in the S populations (data unpublished) to ∼50% mortality/generation elsewhere (Harshman and Schmid 1998). In nature, Drosophila must adapt rapidly to seasonal changes in climate and polymorphisms with large effects on starvation resistance phenotypes have been shown to vary in their frequency by nearly 20% per season (Bergland et al. 2014; Paaby et al. 2014). It is therefore possible that starvation selection in Drosophila is unique in that it selects on loci with an evolutionary history of rapid adaptation. Because selection pressures in the laboratory are likely much higher than experienced in the wild, these alleles may be pulled toward fixation rapidly. Rapid adaptation is also consistent with epigenetic inheritance as parental nutritional status may alter chromatin structure in both humans and Drosophila (Stöger 2008; Buescher et al. 2013; Öst et al. 2014). However, we have found no evidence for parental effects on starvation resistance or lipid storage in our lines, suggesting a largely genetic response (data not published). The high strength of starvation selection was also observed in studies where reverse evolution of starvation-selected Drosophila failed to fully recover ancestral states, suggesting the possibility of fixation and loss of genetic variation during forward evolution (Teotonio and Rose 2000).
The inconsistency in sweeps across SA–C is also different from most E&R experiments in Drosophila and may be a consequence of selection producing multiple adaptive solutions to the same evolutionary problem. This has been observed in laboratory selection experiments for high levels of physical activity in mice, where half of the replicate selected populations have evolved a “mini-muscle” phenotype that confers an increase in oxidative capacity (Garland et al. 2002; Garland 2003). Differential selection may be influenced by founder effects as subtle variations in allele frequency in the starting population may promote disparate evolutionary trajectories (Garland 2003). Here, small allelic differences in the founding populations may have become further differentiated during the 70-generation “lead in” to starvation selection when the S populations served as controls for a desiccation selection experiment with mild selection pressures (∼20% mortality/generation, fig. 1B; see Materials and Methods). Adaptation by drift or selection during this period may have amplified ancestral allelic differences in the S populations, leading to divergent evolution when severe starvation selection was imposed.
The physiological mechanisms to survive starvation are diverse and may offer many independent targets for selection. For instance, Drosophila may adapt to starvation selection by increasing energy reserves, reducing energy consumption rates or increasing the tolerance of organ systems to starvation stress (Rion and Kawecki 2007; Gibbs and Reynolds 2012; Schwasinger-Schmidt et al. 2012). These physiological mechanisms are likely built upon complex genetic networks that may be convergent, but may also function independently. This is supported by studies in iso-female populations of Drosophila where these genetic networks may be isolated. Such studies have found that genes underlying energy storage and utilization are independent, as metabolic rate and stored TG correlate poorly across strains (Jumbo-Lucioni et al. 2010). However, the interaction between these networks is important for starvation survival as neither metabolic rate nor stored TG correlated well with starvation resistance in iso-female lines (Hoffmann et al. 2001; Jumbo-Lucioni et al. 2010). In natural populations and in large outbred evolution experiments, these traits are highly correlated with starvation resistance, suggesting the ability to properly utilize energy stores during starvation is dependent on genetic background and epistasis (Ballard et al. 2008; Schwasinger-Schmidt et al. 2012; Reynolds 2013). The genes controlling the broad physiological mechanisms such as reduction of energy expenditure may also be diverse as multiple adaptations such as lower metabolic rate, reduced physical activity and disrupted sleep cycles have been reported in starvation-selected Drosophila (Schwasinger-Schmidt et al. 2012; Reynolds 2013; Masek et al. 2014). This potentially offers more selective targets as some of these phenotypes may be genetically independent (e.g. an allele that disrupts flight muscle may reduce physical activity but have no effect on sleep duration). While increased lipid stores and decreased energy expenditure are consistent adaptations in the S populations (Reynolds 2013; Masek et al. 2014), the genetic basis of these phenotypes may be driven by different mechanisms, which could help explain the variation in allele frequency among S populations.
While highly unlikely, our data are consistent with hard selective sweeps and could suggest beneficial de novo mutations arising within each S population. This hypothesis is difficult to address because we lack sequence information from the founding populations. Even when founding populations are sequenced, it is hard to determine if a low frequency variant existed in the starting population due to sampling or sequencing error. The likelihood of a de novo mutation could be higher if any part of the starvation-selection regime inadvertently increased mutagenesis/genomic re-arrangements such as starvation-induced mutagenesis in prokaryotes and yeast (Coyle and Kroll 2008) or diet-induced genomic rearrangements in Drosophila (Aldrich and Maggert 2015). One way to test these hypotheses is to restart evolution with the intended purpose of resequencing, using recently developed guidelines for increased replication and time-series sampling (Baldwin-Brown et al. 2014; Kofler and Schlotterer 2014).
Experimental Selection for Starvation Resistance Alters the Frequency of Alleles Associated with Metabolism and Fat Storage
Analysis of the candidate genes revealed the potential for many adaptive mechanisms which will require further study. For instance, evolved changes in locomotion and behavior could be affected by candidates such as couch potato (cpo, supplementary tables S2 and S3, Supplementary Material online), which is a well-known regulator of these phenotypes (Bellen et al. 1992; Schmidt et al. 2008). Selection on natural variation at the cpo locus has been previously observed in nature and associates with latitudinal cines in diapause (Schmidt et al. 2008; Cogni et al. 2014). Because diapause is associated with changes in fecundity, starvation resistance and lipid content, further analysis of cpo in the S populations could provide insight into these associations. In addition, the S flies are developmentally delayed and have disrupted 20-hydroxyecdysone (20E) hormone signaling (Reynolds 2013). These evolved differences in development could be influenced by the set of genes associated with post-embryonic organ development (P < 5.6×10−3, table 1, supplementary table S4, Supplementary Material online). This gene set contains many developmentally important transcription factors, including Sin 3 A, which has been shown to bind and co-repress 20E inducible genes during development (Sharma et al. 2008).
Selection also targeted candidates directly related to metabolism and fat storage as we found enrichment for genes associated with nutrient response (P < 5.7×10−4), catabolism (P < 2.4 ×10−4) and the lipid droplet (P < 6.2×10−3, table 1). One of the most prominent candidates is the insulin receptor (InR). Disruptive mutations in InR result in profound metabolic consequences, with pleiotropic effects in varying tissues, as ecological factors such as nutrition and stress are decoupled from the intracellular metabolic machinery (Brogiolo et al. 2001; Tatar et al. 2001; Kayashima et al. 2013). While receptors in general are highly conserved across Drosophila species, they have evolved under relatively low levels of selective constraint compared to other protein families in Drosophila (Clark et al. 2007). These relaxed constraints may have allowed intra-species variation to persist despite evidence of positive selection driving adaptive changes in receptor sequences across species (Schmidt et al. 2000; Guirao-Rico and Aguade 2009). This seems to be the case for InR, as polymorphisms in natural populations of D. melanogaster show clinal patterns of positive selection and associate with variation in fecundity, development time, lipid weight and starvation resistance (Paaby et al. 2010, 2014). In the S populations there were 71 total loci within InR that were consistent with selection across replicates. Many of these SNPs were found in non-coding regions which have the potential to alter regulation of the receptors themselves, but could also affect remote targets. Two of these polymorphisms were predicted to induce missense substitutions His1150Tyr and Asp1192Gly. These regions are found in the second fibronectin type-III domain, an extracellular region which has been associated with Ilp ligand binding (Garza-Garcia et al. 2007; Sajid et al. 2011). Another candidate of interest is Sirtuin 1 (Sirt1), an evolutionarily conserved NAD+-dependent deacetylase that has been shown to regulate metabolic homeostasis through transcriptional regulation of the insulin signaling pathway (Banerjee et al. 2012; Palu and Thummel 2016). Mutations that disrupt Sirt1 function lead to changes in lipid storage, lipid droplet size, starvation resistance, and insulin sensitivity (Banerjee et al. 2012; Palu and Thummel 2016). We also found candidates which could directly impact the structure and function of lipid droplets, such as lipid storage droplet 1 (Lsd-1). Lsd-1 is an evolutionarily conserved perilipin—a lipid droplet membrane-bound protein responsible for lipid mobilization under stressful conditions (Grönke et al. 2007; Beller et al. 2010). Mutations to Lsd-1 can cause an obese phenotype with large lipid droplets and increased starvation resistance, reminiscent of phenotypes observed in the S populations (Beller et al. 2010; Hardy et al. 2015).
Evolutionary Implications
Theories on the evolutionary origins of obesity began with the proposition of the TGH, which postulates that genes that are efficient in storing and retaining energy are selected for during periods of famine (Neel 1962). Critics have argued that famines severe enough to drive evolution are relatively rare events in human history and cite the lack of candidate obesity genes under selection as weaknesses of the theory (Speakman 2008; Stöger 2008; Genné-Bacon 2014). Clearly our experimental design supports the TGH as a generalized model given the extensive evolutionary history of severe famine which has led to adaptive “thrifty” physiologies (i.e. low metabolism, higher energy storage) in the S populations. Here, we provide further support for the TGH as we report a wide range of genetic loci with widely divergent allelic frequencies due to the selective treatment. Whether the TGH contributes to the current obesity epidemic in humans is still up for debate, but its effects here are pervasive and may speak more generally to the evolutionary forces controlling obesity.
Materials and Methods
Evolutionary History of the Starvation-Selected Lines
The starvation-selected lines were derived from the controls of a desiccation-selection experiment which began in 1999 as previously described (Gefen and Gibbs 2009) and diagramed in figure 1B. In brief, the founders for the desiccation-selection experiment were derived from ∼400 adult female D. melanogaster that were collected from the wild in Terhune New Jersey in 1998. These animals were acclimated to lab conditions for 1 year, then split into three large cohorts (“D,” “SC,” and “FC”), each with three replicates (“A,” “B,” and “C”) for a total of nine populations (fig. 1B, gray phylogeny). DA–C were selected for desiccation tolerance, with 4-day-old adults from each generation being deprived of food and water and stressed with artificial desiccant until 80–90% of the population had died. Survivors were re-fed, bred and used as founders for the next generation. Because the desiccation treatment also removes food, DA–C were moderately selected for starvation resistance. To control for the effects of starvation, SCA–C were starved during the selection phase of each generation, given non-caloric agar to provide water, but otherwise handled in parallel with DA–C. FCA–C was ostensibly unselected, a handling control given ad libitum food and water each generation.
The starvation-selection experiment studied here started in 2006, 70 generations after the start of the desiccation-selection experiment. The starvation-selected populations (hereafter “SA–C”) were derived from SCA–C as previously described (Reynolds 2013; Masek et al. 2014; Hardy et al. 2015). Each generation ∼10,000 S adults were raised for 4 days, then food was replaced with non-caloric agar until 80–90% of the population had died. Survivors were allowed to breed and served as founders for the next generation. At the same time, subpopulations were derived from FCA–C of the desiccation-selection experiment and maintained in parallel with SA–C. These lines were ostensibly unselected, with ∼2,000 flies given unrestricted access to food and water each generation (Fed-controls, hereafter “F”; fig. 1B, black phylogeny).
Sample Preparation
Flies were removed from selection for one generation following the 83rd generation of starvation selection to control for parental effects. Pooled samples of 100, 4-day-old adult females were then collected from all 6 populations (FA–C, SA–C). Pooled samples were homogenized using glass tissue homogenizers in a proprietary buffer (ATL) in accordance with the DNeasy Blood and Tissue DNA Extraction Kit (Qiagen, Valencia, CA). We followed the manufacturer’s protocol for purification of total DNA from animal tissues including the optional RNase step. DNA purity and concentration were calculated using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE).
Library Preparation and Sequencing
Purified samples were shipped to the University of Utah Health Science Center Genomics Core Facility to generate paired-end libraries with 350-bp mean inserts using the TruSeq DNA PCR-Free Preparation Kit (Illumina, San Diego, CA). Multiplexed samples were sequenced using an Illumina HiSeq sequencer with 125-cycle paired-end sequencing using version-4 chemistry.
Mapping and Variant Discovery
Raw reads were aligned to the D. melanogaster genome (Flybase, r6.06) using the Burrows-Wheeler Aligner program (BWA, version 0.7.12) with default settings. Unmapped, low quality (<20), and singleton reads were removed from the Binary Alignment Map (BAM) file, which was then sorted by genomic coordinate using SAMtools (version 1.1). Duplicate reads were removed and the resulting de-duplicated BAM files were merged using Picard (version 1.123). Reads were locally realigned around putative indels using tools in the Genome Analysis Toolkit (GATK, version 3.3) from the Broad Institute. Variants were identified using the Unified Genotyper tool from GATK and annotated with SnpEff (version 4.1) using a custom database built through SnpEff, based on the latest Gene Transfer Format (GTF, r6.06) release from FlyBase (Attrill et al. 2016).
Filtering for Quality Control and Sampling Error
A series of quality control measures were performed using the VariantFiltration tool from GATK. To start, SNPs within five nucleotides of a putative insertion or deletion (indel) were discarded to control for mapping errors that occur frequently near indels. In addition we filtered closely clustered SNPs (3 SNPs within a window of 10 nucleotides), SNPs with poor phred-based quality scores (QUAL <30), high strand bias (FS >60) or greater than four mapping quality scores of zero across all samples (MQ >4). Variant Call Format (VCF) files were converted into a customized SNP table with reference and alternate allele counts for each population and ordered by genomic position for chromosomes X, 2L, 2R, 3L, and 3R using command line text editors. During this step SNPs with less than 30× coverage across all six populations or with more than one alternate allele were discarded. Using custom R scripts (R Core Team 2015), SNPs with a minor allele frequency of <0.05 across all six populations were filtered as well.
After quality control filters were applied, the SNP table contained 1,046,373 SNPs with 72.6 ± 5.9 (S.E.M.)-fold coverage across all six populations. To support that our SNPs were largely natural polymorphisms and not due to misalignments, we compared them with those available in the Drosophila Genetic Resource Panel (DGRP; Mackay et al. 2012). To do this we downloaded the VCF file for the DGRP Freeze 2.0 calls from dgrp2.gnets.ncsu.edu/data.html. Genomic location was extracted from the dgrp2.vcf file using custom command line text editors to generate a file with the chromosome and position of each SNP. The genomic positions of the DGRP calls and our set of SNPs were based on different FlyBase genome assemblies, resulting in discordant genomic coordinates. To match our coordinates we used the Drosophila Sequence Coordinates Converter from FlyBase (Attrill et al. 2016) to switch our SNPs from the version 6 assembly to the version 5 assembly. The converted positions were uploaded into R and compared with the DGRP SNP locations. In total we found 94.6% overlap between our SNPs and naturally occurring polymorphisms in the DGRP lines (data not shown).
In order to filter SNPs that could be explained by sampling error we used a scaled Chi-Squared test (Huang et al. 2012), which was applied independently to each replicate pair (i.e. FA compared with SA; Supplemental Code). To correct for multiple comparisons we used a Bonferonni correction with a 0.01 adjusted P-value threshold. This step filtered ∼88% of the SNPs with 120,825, 115,000 and 168,100 SNPs remaining for replicates A, B, and C, respectively.
While it is certainly possible that chromosomal rearrangements such as copy number variants, indels, and inversions have contributed to the evolution of these populations, it is challenging to find evidence of structural variants from Pool-SEQ data. Kapun et al. (2014) provide a useful resource in this regard—a set of SNP markers in seven inversions known to segregate in D. melanogaster populations. We cross-referenced this published list with our SNP data and identify only three matches: 1/73 of the In(3L)P alleles, 1/150 of the In(3R)Mo alleles, and 1/144 of the In(3R)C alleles. The observation of single SNPs, rather than suites of SNPs, in our data, suggests that these inversions are either not present in our lines, or are segregating at frequencies too low to be detected. Other labs using D. melanogaster lines that have been laboratory domesticated for many years have also failed to find concrete evidence of these inversion alleles in Pool-SEQ data (Graves et al. 2017), which is consistent with the idea that the expansion of some of these inversions has been recent (Langley et al. 2012).
PCA and Pearson Product-Moment Correlation
We used the PCA (prcomp) function of the stats package (version 3.1.3) in R to visualize differences in reference allele frequency across populations. In R, we plotted the pairwise coordinates of the first 3 components which explained 88% of the variation (supplementary table S1, Supplementary Material online). Also using the stats package in R we performed the Test for Association/Correlation Between Paired Samples (cor.test) to calculate the Pearson Product-Moment Correlation Coefficient (r) for pairwise associations of reference allele frequencies between populations. P-values for all correlations were <2.0×10−16 (supplementary fig. S1B, Supplementary Material online).
Starvation Resistance
Flies were removed from selection for one generation to control for parental effects. The progeny were aged to 4 days post-eclosion, briefly anesthetized with CO2, then separated into vials containing non-caloric 1% agar. Each vial contained either five males or females from the three selected populations (SA, SB, or SC), with a total of 10 vials per sex, resulting in a total of 100 flies (50 males, 50 females) per population. All vials were maintained on a 24-h light cycle at 25°C and scored for mortality every 8 h until the completion of the experiment. Every 3 days flies were manually transferred (without anesthetic) to fresh vials to replenish water resources. Data were split for each sex and processed in R, using a one-way ANOVA with population as the categorical factor for each sex followed by a Tukey post hoc test.
Heterozygosity and FST
Heterozygosity for each SNP was calculated as 2pq with the alternate alleles divided by total coverage for that SNP, per population. Custom functions in R were used to calculate the average heterozygosity of SNPs within 100 kb windows with a step size of 2 kb across the genome. Mean block heterozygosity was calculated from the total number of 100 kb windows for each population, with a one-way ANOVA and Tukey’s post hoc test used to find statistical differences. We calculated the variance in the mean heterozygosity for all 100 kB blocks within each population and used the “F Test to Compare Two Variances” (var.test) function in the stats package in R to test for pairwise differences between populations. We found contiguous blocks of low heterozygosity by locating overlapping 100 kb blocks whose mean heterozygosity was less than 0.05 using the Intervals (version 0.15.0) package in R. Contiguous blocks were analyzed for count, size and genomic location. Overlapping regions of contiguous blocks from populations SA, SB, and SC were found and the coordinates of these overlaps were used to find genes using a custom gene database, built from the r6.06 FlyBase release.
To estimate average differentiation between the replicate populations of each treatment, we calculated genome-wide Fst between the replicate populations of the three F and three S populations. Fst was calculated as (Htotal−Hwith)/Htotal where Htotal is the expected heterozygosity between three populations of a selection treatment under panmixia and Hwith is the heterozygosity averaged over the three populations. Plotted values are rolling averages of these FST estimates taken over every 1,000 SNPs.
Permutation Testing
We designed a custom permutation script in R to test whether the set of 1,796 overlapping candidate SNPs were higher than would be expected under a random model. To do this we randomly sampled SNPs from our total set of 1,046,373 for 3 populations of sizes 37,062, 45,506 and 61,370 to match the number of candidate SNPs found in SA–C. We then calculated the number of SNPs that overlapped between these three sets. This procedure was iterated 106 times to generate a distribution of expected 3-way SNP overlap under a random model. Our observed value of 1,796 was well outside the boundary of this distribution (P < 1.0×10−6, fig. 8C).
Gene Mapping and Ontology Analyses
We used custom scripts in R to identify candidate SNPs that fell within the gene regions defined by the FlyBase r6.06 release. Genes containing multiple candidate SNPs were counted once, resulting in a list of 382 candidate genes (supplementary table S3, Supplementary Material online). We then looked for enriched GO terms using Gowinda, using our set of 1,796 candidate SNPs as input and the 1,046,373 quality control filtered SNPs as background (Kofler and Schlotterer 2012). GO terms and associated genes were obtained from FuncAssociate (Version 3.0; Berriz et al. 2009). We set Gowinda to run 106 simulations with the gene-definition and mode parameters set to “gene.” This resulted in a set of 279 GO terms with P < 0.05. We corrected this set for hierarchical clustering using GO-Module and filtered for GO categories containing five or more reference genes (Yang et al. 2011). These procedures resulted in a set of 77 enriched GO terms (supplementary table S4, Supplementary Material online).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We would like to thank the editor of the manuscript and the anonymous reviewers for their insightful comments. This study was supported by the National Institutes of Health (R15-GM100395 to A.G.G.); the National Science Foundation (IOS-1355210 to A.G.G.); the University of Nevada Las Vegas Faculty Opportunity Award (to A.G.G.); experiments performed in the University of Nevada Genomics Core facility were supported by the National Institute of General Medical Sciences (P20GM103440); and sequencing for the project described was performed at the Huntsman Cancer Institute at the University of Utah and supported by the National Cancer Institute (P30CA042014). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Author Contributions
C.M.H. and A.G.G. planned experiments and wrote the paper with assistance from all other authors. K.M.L. and C.M.H. prepared genomic DNA, oversaw library prep and maintained data storage. M.H. and C.M.H. designed and executed alignment pipeline. M.B. and C.M.H. performed intra-replicate genetic correlations and designed sliding window analysis to measure and plot genome-wide heterozygosity. L.E. and C.M.H. designed drift filter and statistical analysis. All other experiments were performed by C.M.H. Each author contributed to editing the final manuscript.
References
- Aldrich JC, Maggert KA.. 2015. Transgenerational inheritance of diet-induced genome rearrangements in Drosophila. PLoS Genet. 11(4):e1005148.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attrill H, Falls K, Goodman JL, Millburn GH, Antonazzo G, Rey AJ, Marygold SJ; The FlyBase Consortium. 2016. FlyBase: establishing a Gene Group resource for Drosophila melanogaster. Nucleic Acids Res. 44(D1):D786–D792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin-Brown JG, Long AD, Thornton KR.. 2014. The power to detect quantitative trait loci using resequenced, experimentally evolved populations of diploid, sexual organisms. Mol Biol Evol. 31(4):1040–1055.http://dx.doi.org/10.1093/molbev/msu048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard JWO, Melvin RG, Simpson SJ.. 2008. Starvation resistance is positively correlated with body lipid proportion in five wild caught Drosophila simulans populations. J Insect Physiol. 54(9):1371–1376.http://dx.doi.org/10.1016/j.jinsphys.2008.07.009 [DOI] [PubMed] [Google Scholar]
- Banerjee KK, Ayyub C, Sengupta S, Kolthur-Seetharam U.. 2012. dSir2 deficiency in the fatbody, but not muscles, affects systemic insulin signaling, fat mobilization and starvation survival in flies. Aging 4(3):206–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen HJ, Vaessin H, Bier E, Kolodkin A, D'Evelyn D, Kooyer S, Jan YN.. 1992. The Drosophila couch potato gene: an essential gene required for normal adult behavior. Genetics 131(2):365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beller M, Bulankina AV, Hsiao H-H, Urlaub H, Jäckle H, Kühnlein RP.. 2010. PERILIPIN-dependent control of lipid droplet structure and fat storage in Drosophila. Cell Metab. 12(5):521–532. [DOI] [PubMed] [Google Scholar]
- Bennett AF. 2003. Experimental evolution and the Krogh principle: generating biological novelty for functional and genetic analyses1. Physiol Biochem Zool. 76(1):1–11.http://dx.doi.org/10.1086/374275 [DOI] [PubMed] [Google Scholar]
- Bergland AO, Behrman EL, O’Brien KR, Schmidt PS, Petrov DA.. 2014. Genomic evidence of rapid and stable adaptive oscillations over seasonal time scales in Drosophila PLoS Genet. 10:e1004775.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriz GF, Beaver JE, Cenik C, Tasan M, Roth FP.. 2009. Next generation software for functional trend analysis. Bioinformatics 25(22):3043–3044.http://dx.doi.org/10.1093/bioinformatics/btp498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse RT, Choi J, Reardon K, Rodriguez J, Graham S, Diop S, Ocorr K, Bodmer R, Oldham S.. 2010. High-fat-diet-induced obesity and heart dysfunction are regulated by the TOR pathway in Drosophila. Cell Metab. 12(5):533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer ML. 2013. Kinematic analysis of axial rotations and the effects of stress selection on takeoff flight performance. UNLV Theses, Dissertations, Professional Papers, and Capstones. 1806. http://digitalscholarship.unlv.edu/thesesdissertations/1806.
- Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E.. 2001. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol. 11(4):213–221. [DOI] [PubMed] [Google Scholar]
- Buescher JL, Musselman LP, Wilson CA, Lang T, Keleher M, Baranski TJ, Duncan JG.. 2013. Evidence for transgenerational metabolic programming in Drosophila. Dis Model Mech. 6(5):1123–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke MK. 2012. How does adaptation sweep through the genome? Insights from long-term selection experiments. Proc R Soc B Biol Sci. 279(1749):5029–5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke MK, Dunham JP, Shahrestani P, Thornton KR, Rose MR, Long AD.. 2010. Genome-wide analysis of a long-term evolution experiment with Drosophila. Nature 467(7315):587–590. [DOI] [PubMed] [Google Scholar]
- Burke MK, Long AD.. 2012. What paths do advantageous alleles take during short-term evolutionary change? Mol Ecol. 21(20):4913–4916. [DOI] [PubMed] [Google Scholar]
- Carneiro M, Rubin C-J, Di Palma F, Albert FW, Alfoldi J, Barrio AM, Pielberg G, Rafati N, Sayyab S, Turner-Maier J, et al. 2014. Rabbit genome analysis reveals a polygenic basis for phenotypic change during domestication. Science 345(6200):1074–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien A, Edgar DB, Trela JM.. 1976. Deoxyribonucleic acid polymerase from the extreme thermophile Thermus aquaticus. J Bacteriol. 127(3):1550–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, et al. 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450(7167):203–218. [DOI] [PubMed] [Google Scholar]
- Cogni R, Kuczynski C, Koury S, Lavington E, Behrman EL, O'Brien KR, Schmidt PS, Eanes WF.. 2014. The intensity of selection acting on the couch potato gene-spatial-temporal variation in a diapause cline. Evolution 68(2):538–548 [DOI] [PubMed] [Google Scholar]
- Coyle S, Kroll E.. 2008. Starvation induces genomic rearrangements and starvation-resilient phenotypes in yeast. Mol Biol Evol. 25(2):310–318.http://dx.doi.org/10.1093/molbev/msm256 [DOI] [PubMed] [Google Scholar]
- Franssen SU, Nolte V, Tobler R, Schlötterer C.. 2015. Patterns of linkage disequilibrium and long range hitchhiking in evolving experimental Drosophila melanogaster populations. Mol Biol Evol. 32(2):495–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland T. 2003. Selection experiments: an under-utilized tool in biomechanics and organismal biology In: Bels VL, Gasc JP, Casinos A, editors. Vertebrate biomechanics and evolution. Oxford: BIOS Scientific Publishers Limited; p. 23–56. [Google Scholar]
- Garland T, Morgan MT, Swallow JG, Rhodes JS, Girard I, Belter JG, Carter PA.. 2002. Evolution of a small-muscle polymorphism in lines of house mice selected for high activity levels. Evolution 56(6):1267.. [DOI] [PubMed] [Google Scholar]
- Garland T, Rose MR, editors. 2009. Experimental evolution: concepts, methods, and applications of selection experiments. Berkeley (CA: ): University of California Press. [Google Scholar]
- Garza-Garcia A, Patel DS, Gems D, Driscoll PC.. 2007. RILM: a web-based resource to aid comparative and functional analysis of the insulin and IGF-1 receptor family. Hum Mutat. 28(7):660–668.http://dx.doi.org/10.1002/humu.20491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gefen E, Gibbs AG.. 2009. Interactions between enviornmental stress and male mating success may enhance evolutionary divergence of stress-resistant Drosophila populations. Evolution 63(6):1653–1659.http://dx.doi.org/10.1111/j.1558-5646.2009.00648.x [DOI] [PubMed] [Google Scholar]
- Genné-Bacon EA. 2014. Thinking evolutionarily about obesity. Yale J Biol Med. 87(2):99–112. [PMC free article] [PubMed] [Google Scholar]
- Gibbs AG, Reynolds LA.. 2012. Drosophila as a model for starvation: evolution, physiology, and genetics In: McCue MD, editor. Comparative physiology of fasting, starvation, and food limitation. Berlin, Heidelberg: Springer; p. 37–51.Available from: http://link.springer.com/10.1007/978-3-642-29056-5_4. [Google Scholar]
- Graves JL, Hertweck KL, Phillips MA, Han MV, Cabral LG, Barter TT, Greer LF, Burke MK, Mueller LD, Rose MR.. 2017. Genomics of parallel experimental evolution in Drosophila. Mol Biol Evol. 34(4):831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönke S, Müller G, Hirsch J, Fellert S, Andreou A, Haase T, Jäckle H, Kühnlein RP.. 2007. Dual lipolytic control of body fat storage and mobilization in Drosophila. PLoS Biol. 5(6):e137.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirao-Rico S, Aguade M.. 2009. Positive selection has driven the evolution of the Drosophila insulin-like receptor (InR) at different timescales. Mol Biol Evol. 26(8):1723–1732.http://dx.doi.org/10.1093/molbev/msp088 [DOI] [PubMed] [Google Scholar]
- Hardy CM, Birse RT, Wolf MJ, Yu L, Bodmer R, Gibbs AG.. 2015. Obesity-associated cardiac dysfunction in starvation-selected Drosophila melanogaster. Am J Physiol Regul Integr Comp Physiol. 309(6):R658–R667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshman LG, Schmid JL.. 1998. Evolution of starvation resistance in Drosophila melanogaster: aspects of metabolism and counter-impact selection. Evolution 52(6):1679.. [DOI] [PubMed] [Google Scholar]
- Hodgkin AL, Huxley AF.. 1952. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 117(4):500–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Hallas R, Sinclair C, Mitrovski P.. 2001. Levels of variation in stress resistance in drosophila among strains, local populations, and geographic regions: patterns for desiccation, starvation, cold resistance, and associated traits. Evol Int J Org Evol. 55:1621–1630. [DOI] [PubMed] [Google Scholar]
- Huang W, Richards S, Carbone MA, Zhu D, Anholt RRH, Ayroles JF, Duncan L, Jordan KW, Lawrence F, Magwire MM, et al. 2012. Epistasis dominates the genetic architecture of Drosophila quantitative traits. Proc Natl Acad Sci U S A. 109(39):15553–15559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalvingh KM, Chang PL, Nuzhdin SV, Wertheim B.. 2014. Genomic changes under rapid evolution: selection for parasitoid resistance. Proc R Soc B Biol Sci. 281(1779):20132303–20132303.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumbo-Lucioni P, Ayroles JF, Chambers M, Jordan KW, Leips J, Mackay TF, De Luca M.. 2010. Systems genetics analysis of body weight and energy metabolism traits in Drosophila melanogaster. BMC Genomics 11:297.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang L, Aggarwal DD, Rashkovetsky E, Korol AB, Michalak P.. 2016. Rapid genomic changes in Drosophila melanogaster adapting to desiccation stress in an experimental evolution system. BMC Genomics 17(1). Available from: http://www.biomedcentral.com/1471-2164/17/233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapun M, van Schalkwyk H, McAllister B, Flatt T, Schlötterer C.. 2014. Inference of chromosomal inversion dynamics from Pool-Seq data in natural and laboratory populations of Drosophila melanogaster. Mol Ecol. 23(7):1813–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawecki TJ, Lenski RE, Ebert D, Hollis B, Olivieri I, Whitlock MC.. 2012. Experimental evolution. Trends Ecol Evol. 27(10):547–560. [DOI] [PubMed] [Google Scholar]
- Kayashima Y, Sato A, Kumazawa S, Yamakawa-Kobayashi K.. 2013. A heteroallelic Drosophila insulin-like receptor mutant and its use in validating physiological activities of food constituents. Biochem Biophys Res Commun. 434(2):258–262. [DOI] [PubMed] [Google Scholar]
- Kessner D, Novembre J.. 2015. Power analysis of artificial selection experiments using efficient whole genome simulation of quantitative traits. Genetics 199(4):991–1005.http://dx.doi.org/10.1534/genetics.115.175075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. 1955. Solution of a process of random genetic drift with a continuous model. Proc Natl Acad Sci U S A. 41(3):144–150.http://dx.doi.org/10.1073/pnas.41.3.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Schlotterer C.. 2012. Gowinda: unbiased analysis of gene set enrichment for genome-wide association studies. Bioinformatics 28(15):2084–2085.http://dx.doi.org/10.1093/bioinformatics/bts315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Schlotterer C.. 2014. A guide for the design of evolve and resequencing studies. Mol Biol Evol. 31(2):474–483.http://dx.doi.org/10.1093/molbev/mst221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs HA, Johnson WA.. 1980. The role of citric acid in intermediate metabolism in animal tissues. FEBS Lett. 117(S1):K2–K10. [DOI] [PubMed] [Google Scholar]
- Langley CH, Stevens K, Cardeno C, Lee YCG, Schrider DR, Pool JE, Langley SA, Suarez C, Corbett-Detig RB, Kolaczkowski B, et al. 2012. Genomic variation in natural populations of Drosophila melanogaster. Genetics 192(2):533–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long A, Liti G, Luptak A, Tenaillon O.. 2015. Elucidating the molecular architecture of adaptation via evolve and resequence experiments. Nat Rev Genet. 16(10):567–582.http://dx.doi.org/10.1038/nrg3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TFC, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han Y, Magwire MM, Cridland JM, et al. 2012. The Drosophila melanogaster genetic reference panel. Nature 482(7384):173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins NE, Faria VG, Nolte V, Schlotterer C, Teixeira L, Sucena E, Magalhaes S.. 2014. Host adaptation to viruses relies on few genes with different cross-resistance properties. Proc Natl Acad Sci U S A. 111(16):5938–5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masek P, Reynolds LA, Bollinger WL, Moody C, Mehta A, Murakami K, Yoshizawa M, Gibbs AG, Keene AC.. 2014. Altered regulation of sleep and feeding contributes to starvation resistance in Drosophila melanogaster. J Exp Biol. 217(Pt 17):3122–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na J, Musselman LP, Pendse J, Baranski TJ, Bodmer R, Ocorr K, Cagan R, Rulifson E.. 2013. A Drosophila model of high sugar diet-induced cardiomyopathy. PLoS Genet. 9(1): e1003175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel JV. 1962. Diabetes mellitus: a “Thrifty” genotype rendered detrimental by “Progress”? Am J Hum Genet. 14:353–362. [PMC free article] [PubMed] [Google Scholar]
- Olsen KM, Caicedo AL, Polato N, McClung A, McCouch S, Purugganan MD.. 2006. Selection under domestication: evidence for a sweep in the rice waxy genomic region. Genetics 173(2):975–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco-terWengel P, Kapun M, Nolte V, Kofler R, Flatt T, SchlöTterer C.. 2012. Adaptation of Drosophila to a novel laboratory environment reveals temporally heterogeneous trajectories of selected alleles: genomic signatures of adaptation to new enviornment. Mol Ecol. 21(20):4931–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öst A, Lempradl A, Casas E, Weigert M, Tiko T, Deniz M, Pantano L, Boenisch U, Itskov PM, Stoeckius M, et al. 2014. Paternal diet defines offspring chromatin state and intergenerational obesity. Cell 159(6):1352–1364. [DOI] [PubMed] [Google Scholar]
- Paaby AB, Bergland AO, Behrman EL, Schmidt PS.. 2014. A highly pleiotropic amino acid polymorphism in the Drosophila insulin receptor contributes to life-history adaptation: adaptive polymorphism at InR. Evolution 68(12):3395–3409.http://dx.doi.org/10.1111/evo.12546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paaby AB, Blacket MJ, Hoffmann AA, Schmidt PS.. 2010. Identification of a candidate adaptive polymorphism for Drosophila life history by parallel independent clines on two continents: ALLELIC VARIATION AT INR. Mol Ecol. 19(4):760–774.http://dx.doi.org/10.1111/j.1365-294X.2009.04508.x [DOI] [PubMed] [Google Scholar]
- Palu RAS, Thummel CS.. 2016. Sir2 Acts through hepatocyte nuclear factor 4 to maintain insulin signaling and metabolic homeostasis in Drosophila. PLOS Genet. 12:e1005978.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2015. R: A language and enviornment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing. Available from: http://www.R-project.org/
- Reiter LT, Potocki L, Chien S, Gribskov M, Bier E.. 2001. A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res. 11(6):1114–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remolina SC, Chang PL, Leips J, Nuzhdin SV, Hughes KA.. 2012. Genomic basis of aging and life-history evolution in Drosophila melanogaster: genomics of life-history evolution. Evolution 66(11):3390–3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds LA. 2013. The effects of starvation selection on Drosophila melanogaster life history and development. UNLV Theses, Dissertations Professional Paper and Capstones. 1876. http://digitalscholarship.unlv.edu/thesesdissertations/1876.
- Rion S, Kawecki TJ.. 2007. Evolutionary biology of starvation resistance: what we have learned from Drosophila: starvation resistance in Drosophila. J Evol Biol. 20(5):1655–1664.http://dx.doi.org/10.1111/j.1420-9101.2007.01405.x [DOI] [PubMed] [Google Scholar]
- Rose MR, Graves JL, Hutchinson EW.. 1990. The use of selection to probe patterns of pleiotropy in fitness characters In: Gilbert F, editor. Insect life cycles: genetics, evolution and co-ordination. London: Springer; p. 29–42. Available from: http://dx.doi.org/10.1007/978-1-4471-3464-0_3. [Google Scholar]
- Sajid W, Kulahin N, Schluckebier G, Ribel U, Henderson HR, Tatar M, Hansen BF, Svendsen AM, Kiselyov VV, Norgaard P, et al. 2011. Structural and biological properties of the Drosophila Insulin-like peptide 5 show evolutionary conservation. J Biol Chem. 286(1):661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt PS, Duvernell DD, Eanes WF.. 2000. Adaptive evolution of a candidate gene for aging in Drosophila. Proc Natl Acad Sci U S A. 97(20):10861–10865.http://dx.doi.org/10.1073/pnas.190338897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt PS, Zhu C-T, Das J, Batavia M, Yang L, Eanes WF.. 2008. An amino acid polymorphism in the couch potato gene forms the basis for climatic adaptation in Drosophila melanogaster. Proc Natl Acad Sci U S A. 105(42):16207–16211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwasinger-Schmidt TE, Kachman SD, Harshman LG.. 2012. Evolution of starvation resistance in Drosophila melanogaster: measurement of direct and correlated responses to artificial selection: evolution of starvation resistance. J Evol Biol. 25(2):378–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V, Swaminathan A, Bao R, Pile LA.. 2008. Drosophila SIN3 is required at multiple stages of development. Dev Dyn. 237(10):3040–3050. [DOI] [PubMed] [Google Scholar]
- Speakman JR. 2008. Thrifty genes for obesity, an attractive but flawed idea, and an alternative perspective: the “drifty gene” hypothesis. Int J Obes. 32(11):1611–1617. [DOI] [PubMed] [Google Scholar]
- Stemshorn KC, Reed FA, Nolte AW, Tautz D.. 2011. Rapid formation of distinct hybrid lineages after secondary contact of two fish species (Cottus sp.). Mol Ecol. 20:1475–1491. [DOI] [PubMed] [Google Scholar]
- Stöger R. 2008. The thrifty epigenotype: an acquired and heritable predisposition for obesity and diabetes? BioEssays 30(2):156–166. [DOI] [PubMed] [Google Scholar]
- Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS.. 2001. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 292(5514):107–110. [DOI] [PubMed] [Google Scholar]
- Templeton AR. 2006. Population genetics and microevolutionary theory. Hoboken (NJ: ): Wiley-Liss. [Google Scholar]
- Tenaillon O, Rodriguez-Verdugo A, Gaut RL, McDonald P, Bennett AF, Long AD, Gaut BS.. 2012. The molecular diversity of adaptive convergence. Science 335(6067):457–461. [DOI] [PubMed] [Google Scholar]
- Teotónio H, Chelo IM, Bradić M, Rose MR, Long AD.. 2009. Experimental evolution reveals natural selection on standing genetic variation. Nat Genet. 41(2):251–257. [DOI] [PubMed] [Google Scholar]
- Teotonio H, Rose MR.. 2000. Variation in the reversibility of evolution. Nature 408(6811):463–466.http://dx.doi.org/10.1038/35044070 [DOI] [PubMed] [Google Scholar]
- Turner TL, Miller PM.. 2012. Investigating natural variation in drosophila courtship song by the evolve and resequence approach. Genetics 191(2):633–642.http://dx.doi.org/10.1534/genetics.112.139337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TL, Stewart AD, Fields AT, Rice WR, Tarone AM.. 2011. Population-based resequencing of experimentally evolved populations reveals the genetic basis of body size variation in Drosophila melanogaster. PLoS Genet. 7:e1001336.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Li J, Lee Y, Lussier YA.. 2011. GO-Module: functional synthesis and improved interpretation of Gene Ontology patterns. Bioinformatics 27(10):1444–1446.http://dx.doi.org/10.1093/bioinformatics/btr142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Udpa N, Gersten M, Visk DAnn. W, Bashir A, Xue J, Frazer KA, Posakony JW, Subramaniam S, Bafna V, Haddad GG.. 2011. Experimental selection of hypoxia-tolerant Drosophila melanogaster. Proc Natl Acad Sci U S A. 108(6):2349–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.