Abstract
Objectives
To analyse the influence of genetic alterations and differential expression of transcription intermediary factor 1 (TIF1) genes in the pathophysiology of cancer-associated myositis (CAM).
Methods
Paired blood and tumour DNA samples from patients with anti-TIF1γ-positive CAM and from controls were analysed by whole-exome sequencing for the presence of somatic mutations and loss of heterozygosity (LOH) in their TIF1 genes. The genesis and maintenance of the autoimmune process were investigated immunohistochemically by studying TIF1γ expression in the different tissues involved in CAM (skin, muscle and tumour) based on the immunohistochemical H-score.
Results
From seven patients with anti-TIF1γ-positive CAM, we detected one somatic mutation and five cases of LOH in one or more of the four TIF1 genes compared with just one case of LOH in tumours from TIF1γ-negative myositis patients (86% vs 17%; P = 0.03). Compared with type-matched control tumours from non-myositis patients, TIF1γ staining was more intense in tumours from anti-TIF1γ-positive patients (H-score 255 vs 196; P = 0.01). Also, TIF1γ staining in muscle was slightly more intense in anti-TIF1γ-positive than in anti-TIF1γ-negative myositis (H-score 22 vs 5; P = 0.03). In contrast, intense TIF1γ staining was detected in the skin of both myositis and control patients.
Conclusion
Tumours from paraneoplastic anti-TIF1γ-positive patients showed an increased number of genetic alterations, such as mutations and LOH, in TIF1 genes. These genetic alterations, in the context of a high expression of TIF1γ in the tumour, muscle and skin of these patients may be key to understanding the genesis of paraneoplastic myositis.
Keywords: myositis, dermatomyositis, paraneoplastic diseases, autoantibodies, genetics
Rheumatology key messages
Genetic alterations in TIF1 genes are increased in tumours from anti-TIF1γ-positive cancer-associated myositis patients.
TIF1γ is highly expressed in the tumour, muscle and skin of anti-TIF1γ-positive patients.
TIF1 gene mutations in tumours with high expression of TIFγ may trigger myositis.
Introduction
Both genetic and environmental factors increase the risk of developing specific types of myositis, but the underlying cause of this disease remains unknown [1]. Nevertheless, in nearly one-third of the patients diagnosed with DM, a specific form of myositis with characteristic skin findings, disease occurrence shows a close temporal relationship with that of malignancy. For these patients with cancer-associated myositis (CAM), the association has both clinical and mechanistic implications. From a clinical standpoint, this relationship highlights the importance of screening newly diagnosed myositis patients for the presence of an occult neoplasm. From a mechanistic perspective, it strongly suggests cancer as one of the causes of myositis.
However, the risk of developing CAM is not the same in all myositis patients. Recently a significant association between the presence of circulating serum autoantibodies against human transcription intermediary factor 1 (TIF1) family proteins and cancer development in myositis patients was demonstrated [2]. Moreover, while the most common target in anti-TIF1-positive CAM is TIF1γ, other proteins of the TIF1 family may also be simultaneously targeted by the immune system. Thus anti-TIF1α is detected in >65% of these patients and anti-TIF1β is found in ∼10%. However, when anti-TIF1α or anti-TIF1β are positive, usually anti-TIF1γ is also positive, consequently, testing for anti-TIF1γ is more efficient that testing for all the TIF1 proteins separately [3]. Yet, it is still unknown why these autoantibodies are more often detected in CAM.
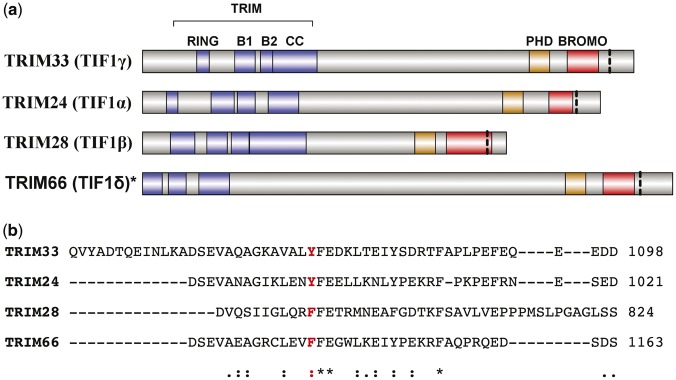
TIF1 proteins belong to the tripartite motif (TRIM)-containing family and participate in several biological processes involved in transcriptional regulation, cellular proliferation and apoptosis. These functions are also important for cancer development and the over-/underexpression of TIF1 proteins is associated with different types of tumours [4]. In addition, all TIF1 proteins share a C-terminal chromatin reading unit consisting of a plant homeodomain finger and a bromodomain (BROMO) that is highly conserved among TIF1 family members but which is not present in any of the other TRIM proteins (Fig. 1A) [5, 6].
Fig. 1.
Functional domains of the TIF1 proteins and alignment of the region corresponding to the TIF1γ mutation
(A) Functional domains of the tripartite motif proteins [5]. (B) Alignment of the region corresponding to the TIF1γ mutation. The mutation is marked with a vertical dashed lined in (A) and is displayed in bold in (B). aPositions with conserved residues; residues with strongly similar properties; residues with weakly similar properties.
Taken together, the association of TIF1 myositis with cancer, the coexistence of different autoantibodies against different TIF1 proteins in the same patient and the presence of common TIF1 protein domains with a high level of identity suggest that a mutated form of a common area in TIF1 proteins is the original antigen in TIF1 paraneoplastic myositis. A similar mechanism was recently identified in scleroderma, in which mutations in the gene encoding RNA III polypeptide A were detected in patients with autoantibodies against these antigenic peptides, which have been shown to trigger disease occurrence. Interestingly, in this study, while just three of six patients showed mutations, a greater number (five of six) of patients presented a deletion of one allele in the genetic region of interest [loss of heterozygosity (LOH)] [7]. LOH is the most frequent way to lose a mutant allele in human cancer and this is key to tumour immunoediting, since tumour cells with mutations producing a neoantigen may be eliminated by the immune system and replaced by tumour cells with LOH in that region (without the antigenic mutation) [7, 8].
To test our hypothesis, we analysed tissue samples from patients diagnosed with CAM for the presence of somatic mutations and an LOH of the TIF1 genes. In addition, the genesis and maintenance of the autoimmune process were studied by examining the expression of TIF1γ (TRIM33) in tissues involved in CAM (skin, muscle and tumour).
Methods
Patient population
All DM patients in the historical myositis cohorts of the Vall d’Hebron hospital and the Hospital Clinic (Barcelona, Spain) from 2008 to 2016 who had available skin, muscle and/or tumour biopsies were included in this study. The corresponding tumour, muscle and skin control samples were collected from patients in the Vall d’Hebron Pathology Department and the Hospital Clinic’s Muscle Diseases Unit.
The diagnosis of DM was based on the criteria of Bohan and Peter [9]. Only patients with definite or probable myositis were included in the study. Most cases of cancer associated with myositis occur within 3 years of myositis onset [10]. This is why we defined CAM, in accordance with Troyanov’s modified Bohan and Peter criteria, as myositis occurring within 3 years of cancer diagnosis [11]. Myositis that developed >3 years before or after cancer onset was considered as non-CAM.
Standard protocol approvals and patient consents
Informed consent was obtained from participating patients for the use of their biologic samples for research purposes. The study was approved by the Institutional Review Boards of the Vall d’Hebron and Clinic hospitals.
Serological assays and HLA typing
Anti-TIF1γ antibodies were detected using an in-house enzyme-linked immunosorbent assay and confirmed by immunoblot, as reported previously [12]. High-resolution class I and II HLAs were detected by PCR followed by sequence-based typing.
Normal/tumour paired exome sequencing
Tumour tissue was manually macrodissected from 5 × 10 mm formalin-fixed, paraffin-embedded slices and DNA was extracted using the Maxwell 16 formalin-fixed, paraffin-embedded tissue low elution volume DNA purification kit with the Maxwell 16 instrument (Promega, Fitchburg, WI, USA). Genomic DNA was isolated from blood samples using the QIAamp DNA Blood kit. Paired-end whole-exome sequencing libraries were prepared using the SureSelect XT Human All Exon V5 Target Enrichment System exome purification kit (Agilent, Santa Clara, CA, USA) and sequenced using an Illumina HiSeq 2000 sequencing system according to the manufacturer’s instructions.
Bioinformatic analysis
Paired-end 100-base sequences were aligned with human reference build 37 using the Burrows-Wheeler aligner. Somatic variants and LOH in TIF1 genes [TIF1α (TRIM24), TIF1β (TRIM28), TIF1γ (TRIM33) and TIF1δ (TRIM66)] were identified using VarScan2 [13]; allelic ratio graphs were generated using TITAN [14]. Somatic mutations and LOH were included in the statistical analysis if the number of reads was >10 and the P-values in Fisher’s exact test was <0.05. Somatic mutations were confirmed by Sanger sequencing and their impact on protein structure was determined using SIFT [15] and PolyPhen2 [16]. Mutated regions among TIF1 proteins were aligned using Clustal Omega [17]. Immune Epitope Database Analysis Resource consensus tools were used to determine whether peptides containing the specific mutations were likely to bind with high affinity to the patient’s HLA alleles [18].
Immunohistochemical studies
Skin and tumour samples were fixed in 5% buffered formalin, embedded in paraffin blocks and cut into 4 µm tissue sections. Sections were deparaffinized with EZ Prep (Ventana/Roche, Tucson, AZ, USA). Cell conditioning solution 1 (Ventana/Roche) was used for antigen retrieval and 3% hydrogen peroxide was used to inhibit endogenous peroxidase. The sections were subsequently incubated for 1 h with a 1:100 dilution of anti-TIF1γ antibody (rabbit polyclonal, IHC-00216; Bethyl Laboratories, Montgomery, TX, USA) and stained using the ultraVIEW universal diaminobenzidine detection kit. All steps were automated using the BenchMark XT automatic slide stainer (Ventana/Roche). Paired samples were processed simultaneously to avoid possible day-to-day variations in staining performance. The results of the immunohistochemistry study were interpreted independently by two experienced pathologists (B.F. and S.D.) who were blinded to the patient groups. The average of both readers’ scores were used for analysis.
Frozen muscle biopsies were cut in 8 μm sections, fixed with acetone for 10 min, blocked in peroxidase-blocking reagent (SM801; Dako, Santa Clara, CA, USA) for 10 min and then incubated for 30 min with a 1:100 dilution of the aforementioned polyclonal rabbit anti-TIF1γ antibody. The sections were subsequently incubated with a 1:100 dilution of anti-rabbit IgG horseradish peroxidase–linked secondary antibody for 30 min at 37 °C and stained using diaminobenzidine as the substrate. The results of the immunohistochemistry study were interpreted by three experienced neuromuscular experts (J.G.J., A.S.O. and I.P.F.) blinded to the patient groups.
The specificity of the polyclonal anti-TIF1γ antibody was confirmed using four non-myositis muscle biopsies as the negative control and myositis skin biopsies incubated with a monoclonal anti-TIF1γ antibody (mouse monoclonal, H00051592-M01; Abnova, Taipei City, Taiwan) as the positive control.
Immunohistochemistry extent and intensity in the skin, tumour and muscle biopsies were quantified using the H-score, as reported previously [19]. This score ranges from 0 to 300, with 300 corresponding to strong staining in 100% of the tumour cells.
Statistical analysis
H-scores between independent groups were compared using Student’s t-test and paired samples using a paired t-test. The percentage of patients with genomic changes in the genes of interest was compared with that of the control group using Fisher’s exact test. Logistic regression was used to calculate the odds of cancer in patients with anti-TIF1γ antibodies. All statistical analyses were performed using Stata/MP 14.0 (StataCorp, College Station, TX, USA). Two-sided P-values ⩽0.05 were considered to indicate statistical significance.
Results
Patient population
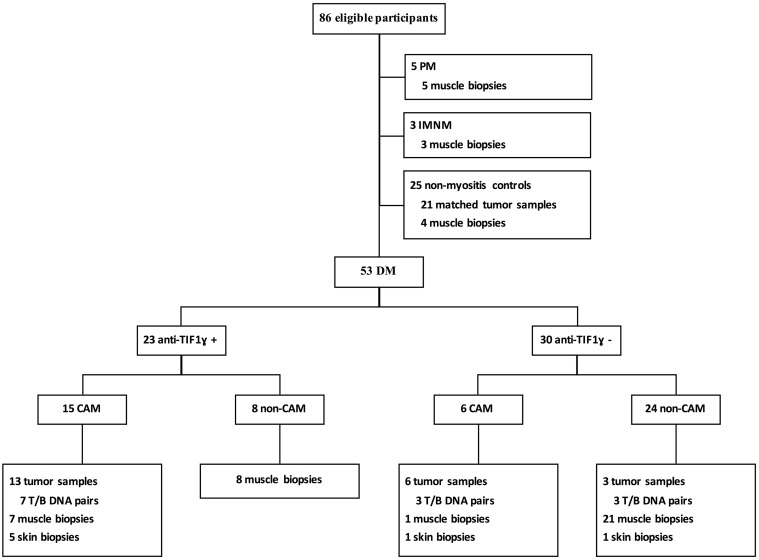
The study included samples from 61 adult patients (47 females) with myositis (21 patients with CAM) and 25 controls (21 type-matched tumours from non-myositis patients and 4 histologically normal muscle biopsies) (Fig. 2). Twenty-three of the 53 DM patients (43%) were anti-TIF1γ-positive. Of those, 15 (65%) had CAM (11 adenocarcinoma, 2 squamous cell carcinoma and 2 lymphoma). Only 6 of the 30 (20%) anti-TIF1γ-negative patients had cancer and myositis synchronously (1 sarcoma, 3 adenocarcinoma, 1 squamous cell carcinoma and 1 lymphoma; 2 had anti-Mi2, 1 had anti-Jo1 and 3 had no detectable myositis-specific autoantibodies). The odds ratio for cancer in the anti-TIF1γ-positive vs the anti-TIF1γ-negative patients was 11.25 (95% CI 3.3, 38). There were no significant age or sex differences between patients who were anti-TIF1γ-positive (56 years old, 87% female) and those who were anti-TIF1γ-negative (55 years old, 68% female) (Table 1 and Fig. 2).
Fig. 2.
Flow chart of the experiment
IMNM: immune-mediated necrotizing myopathy; T/B: tumour/blood.
Table 1.
Patients and samples
| Anti-TIF1γ-positive | Anti-TIF1γ-negative | Total | |||
|---|---|---|---|---|---|
| CAM | Non-CAM | CAM | Non-CAM | ||
| Total number of patients, n (%) | 15 (65) | 8 (35) | 6 (20) | 24 (80) | 53 |
| Paired tumour/blood DNA, n | 7 | – | 3 | 3 | 13 |
| Mean germline TIF1 events | 7.9 | – | 8.3 | 6.7 | |
| Somatic mutations (SM) of TIF1, n (%) | 1 (14) | – | 0 (0) | 0 (0) | 1 |
| Patients with LOHa, n (%) | 5 (71) | – | 1 (33) | 0 (0) | 6 |
| TIF1γ (TRIM33), n (%) | 1 (14) | – | 1 (33) | 0 (0) | 2 |
| TIF1α (TRIM24), n (%) | 2 (29) | – | 0 (0) | 0 (0) | 2 |
| TIF1β (TRIM28), n (%) | 1 (14) | – | 0 (0) | 0 (0) | 1 |
| TIF1δ (TRIM66), n (%) | 2 (29) | – | 0 (0) | 0 (0) | 2 |
| Patients with SM or LOH, n (%) | 6 (86) | – | 1 (33) | 0 (0) | 7 |
| TIF1γ staining, mean (s.d.) | |||||
| Myositis patient tumour | 255 (56) | – | 251 (49) | 140 (113) | 243 (66) |
| Paired control tumour | 196 (33) | – | 152 (99) | 190 (0) | 183 (59) |
| Muscle | 37.3 (15.9) | 9.5 (23.3) | – | 4.9 (7.3) | 10 (26) |
| Skin | 300 (0) | – | 300 (0) | – | 300 (0) |
LOH in two TIF1 genes (TIF1α and TIF1β) was detected in the samples of one patient.
Normal/tumour paired exome sequencing
Tumour DNA samples from seven patients (54 years old, all female) with anti-TIF1γ-positive CAM and six who were anti-TIF1γ-negative (three CAM and three non-CAM; 58 years old, 83% female) were pair-sequenced with their respective whole-blood DNA samples (supplementary Figs. S1 and S2, available at Rheumatology Online). Exome sequencing yielded a median depth of coverage of the TIF1 genes between 139x and 231x, with >91% of the exonic regions of these genes having at least 10 reads.
The total number of germline single-nucleotide variants detected in the TIF1 genes of patients with TIF1γ-positive CAM vs those with TIF1γ-negative CAM was not significantly different (Table 1).
A somatic mutation in the BROMO region of the TIF1γ gene (c.3299T>C) was detected in the tumour tissue of one of the anti-TIF1γ-positive patients. This mutation led to an amino acid change (p.1072Tyr>Cys) in a C-terminal region close to the BROMO domain and highly similar between TIF1 members (Fig. 1B; supplementary Table S1, available at Rheumatology Online). Both SIFT and PolyPhen2 predicted that the change would significantly modify protein structure. In silico analysis of the binding strength of the normal peptide and of the mutated peptides encoded by the mutated region to the patient’s HLA proteins revealed that both bound with high affinity to HLA-A and HLA-B. In fact, the mutation did not modify the binding affinity of these peptides for any of the patient’s HLA alleles (Table 2).
Table 2.
Affinity of the mutated TIF1γ peptide for the different HLA proteins of the patient
| Mutation | HLA allele | Wild-type peptide | Percentile ranka | Mutated peptide | Percentile ranka | Prediction method |
|---|---|---|---|---|---|---|
| p.1072Tyr>Cys | HLA-A*03: 01 | AVALYFEDK | 1.5 | AVALCFEDK | 1.4 | Consensus (ann/smm) |
| HLA-A*11: 01 | AVALYFEDK | 0.5 | AVALCFEDK | 0.55 | Consensus (ann/smm) | |
| HLA-B*44: 03 | SEVAQAGKAVALYF | 0.1 | SEVAQAGKAVALCF | 0.1 | ann | |
| HLA-C*06: 02 | LYFEDKLTEI | 1.6 | LCFEDKLTEI | 4.7 | ann | |
| HLA-C*16: 01 | VAQAGKAVALYF | 1.8 | VAQAGKAVALCF | 3.5 | netmhcpan | |
| HLA-DRB1*07: 01 | DSEVAQAGKAVALYF | 16.39 | DSEVAQAGKAVALCF | 14.02 | Consensus (comb.lib./smm/nn) | |
| HLA-DRB1*04: 05 | YFEDKLTEIYSDRTF | 19.49 | CFEDKLTEIYSDRTF | 19.49 | Consensus (smm/ann/sturniolo) |
The mutated TIF1γ peptide is underlined. The Immune Epitope Database Analysis Resource did not contain sufficient information about HLA-B*47:01, the other HLA-B allele of the patient, to allow predictions regarding affinity. aPercentiles <1% (in bold) were considered indicative of strong binding of the peptide with the HLA molecule. ann: neural network; comb.lib: scoring matrices derived from combinatorial peptide libraries; smm: stabilized matrix method.
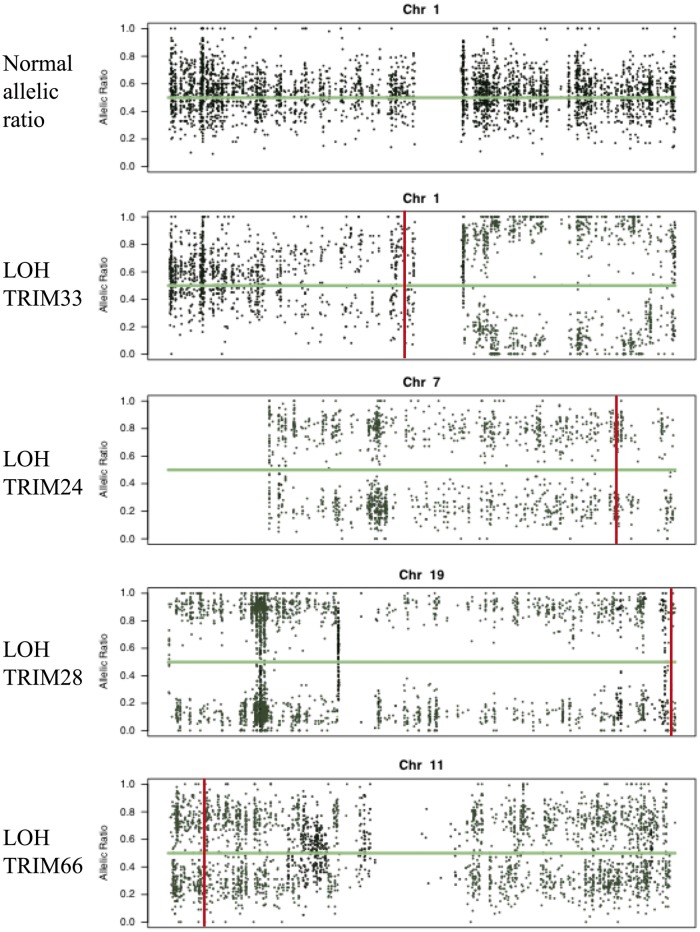
In addition to this mutation, the tumours of five other TIF1γ-positive patients (71.4%) showed evidence of a LOH for one or more of the TIF1 genes. Conversely, just one of the six tumours from TIF1γ-negative patients (16.7%) showed significant LOH in these regions (supplementary Table S1, available at Rheumatology Online). In samples from four of the five patients with TIF1γ-positive CAM and a LOH, all the informative nucleotides showed this genetic alteration. Also, in the tumours of four of these five patients, the LOH in TRIM genes was one of the widespread chromosomal events leading to the loss of important genetic information contained in one of the copies of the chromosome. No tumour showed LOH in the HLA class I region of chromosome 6 (Table 1 and Fig. 3; supplementary Table S1 and Supplementary Figs. S1 and S2, available at Rheumatology Online).
Fig. 3.
Allelic ratio of the paraneoplastic anti-TIFγ-positive patients with loss of heterozygosity in any of the TIF1 proteins
Vertical lines indicate the positions of the TIF1 genes. The normal allelic ratio is provided for comparison.
Taken together, the results demonstrated a significant increase in somatic mutations or a LOH in the TIF1 genes of tumours from patients with TIF1γ-positive CAM than in those from the tumours of anti-TIF1γ-negative patients (86% vs 16.7%; P = 0.03) (Table 1).
Immunohistochemical studies
TIF1γ staining in both anti-TIF1γ-positive (H-score 255 vs 196; P = 0.01) and anti-TIF1γ-negative tumour samples (H-score 251 vs 152; P = 0.03) was more intense than in the paired same tumours from patients without myositis. The differences between anti-TIF1γ-positive and negative patients were not significant (supplementary Fig. S3, available at Rheumatology Online). Specifically, the H-score of the patient with the TIF1γ mutation was 280 (H-score of the corresponding non-myositis tumour = 210) and the mean H-score of the anti-TIF1γ-positive patients with an LOH in their TIF1 genes was 242 (mean H-score of the corresponding non-myositis tumours = 180). TIF1γ skin staining was extremely intense in both myositis patients and the controls (all H-scores = 300) (supplementary Fig. S3, available at Rheumatology Online).
Staining for TIF1γ was faint in muscle compared with that in tumour and skin, but it was slightly more intense in muscle biopsies from patients with anti-TIF1γ-positive vs anti-TIF1γ negative disease (H-score = 22 vs 5; P = 0.03). Among anti-TIF1γ-positive patients, staining was more intense in the muscle samples from those with than without CAM (H-score = 37 vs 10; P = 0.2), but the difference was not statistically significant. In samples from all of the non-DM controls, the H-score for TIF1γ staining was <20 and the four non-myositis muscle biopsies tested were negative for TIF1γ (supplementary Fig. S3, available at Rheumatology Online).
Discussion
In this study we showed an increased number of genetic modifications, such as mutations and LOH, in TIF1 genes of tumours from patients with anti-TIF1γ-positive myositis. We also found a high expression of TIF1γ in the tumour, muscle and skin of these patients.
Our findings support the hypothesis that the co-occurrence of mutations in peptide regions of TIF1 with high affinity for HLA class I and tumours with high-level TIF1 protein expression may initiate a strong adaptive immune response against neoplastic cells with the mutation. This would lead to two very different scenarios: either the tumour escapes immune attack by hiding or eliminating the targeted antigen or all of the tumour cells, and thus the tumour, are eliminated. While there is the possibility that the presence of anti-TIF1γ antibodies abolish the tumour-suppressive role of TIF1γ [20] and is thus the cause (rather than the consequence) of CAM, this mechanism would not explain the genetic lesions found in the tumours of our patients with anti-TIF1γ-positive CAM.
LOH in the TIF1 genes was detected with high frequency in the tumours of patients with anti-TIF1γ-positive CAM. Based on previous reports about the role of immunoediting in cancer [8], in these neoplasms, LOH may be the most common method to escape the immune system attack. As immune pressure on the mutated cancer cells increases, those with widespread genomic changes that effectively delete the mutated neoantigen may have a survival benefit, such that they are able to expand and eventually replace the entire tumour area. Joseph et al. [7] demonstrated the existence of a similar mechanism in SSc. Based on our findings and Joseph et al.’s results [7], LOH of the mutated gene may be a faster or more efficient way for tumours to escape immunosurveillance than other methods, such as hiding the antigen from the cell surface by reducing HLA class I expression through LOH on chromosome 6 [21]. Paradoxically, by selecting tumour cells with widespread chromosomal genomic modifications, the immune response would favour the survival of the most undifferentiated and malignant neoplastic cells.
Interestingly, 25–50% of adult patients positive for anti-TIF1γ have no history of malignancy. According to the hypothesis proposed in our study, these patients may have developed an immune response that effectively eradicated the tumour. This would suggest that TIF1 immunization, whether passively by anti-TIF1 immunoglobulin injection or by vaccination with TIF1 antigens, could theoretically result in therapeutic activity against tumours characterized by mutations and high-level expression of TIF1 proteins. However, either approach could also trigger myositis, thus limiting their clinical applicability. Moreover, the lack of an association between anti-TIF1γ autoantibodies and cancer in children [22] suggests an alternative cause of myositis in paediatric patients: either children are immunized against TIF1γ from a different antigenic source (e.g. infectious) or the antigen recognized by children with anti-p155/140 antibodies corresponds to a different set of proteins with the same molecular weight.
After initial contact with the antigen, maturing B and T cells undergo somatic hypermutation, which increases their specificity for the antigen. The 105- to 106-fold increase in the mutation rate of regions recognizing the antigen (e.g. the B cell receptor locus) stochastically generates cells with a range of affinities for the antigen [23]. By increasing the survival and replication rate of those cells with the highest antigen affinity, the immune system progressively selects an ‘elite’ set of memory cells that perpetuate the immune response even in the absence of the original antigen.
Our results show that the peptides resulting from the region of TIF1γ yielding the mutation bound strongly to HLA class I alleles, regardless of the presence of the mutation, and that TIF1γ was overexpressed in the tumours of patients with CAM. We therefore hypothesize that the increased expression of a neoantigen with very strong binding to HLA class I may generate an intense immune response able to induce a sudden drop in tumour neoantigen availability during the early phases of affinity maturation by eliminating all tumour cells containing the mutated peptide. Consequently, TIF1 antigens expressed in muscle and skin, the two largest tissues in the body and thus the source of an unlimited amount of antigen, would exert pressure on maturing immune cells to increase their affinity for the wild-type form of TIF1, thereby inducing the autoimmune disease. Given previous reports suggesting that regenerating muscle fibres express higher concentrations of autoantigens [24], the initial attack on muscle would enhance the expression of TIF1 proteins by increasing the number of regenerating cells. This may create a positive feedback loop able to perpetuate the disease indefinitely. The increased levels of TIF1γ detected in the muscle samples of our patients with anti-TIF1γ-positive CAM were likely due to this positive feedback loop. The overall low expression of TIF1γ in muscle is consistent with the clinical observation that patients with anti-TIF1γ usually present with mild forms of myositis characterized predominantly by skin involvement [3, 25].
Our study has several limitations. First, because CAM is a rare condition, the number of patients was limited and the availability of samples from different tissues differed among the participating patients. However, the number of samples in each study group was sufficient to confirm our main hypothesis. Second, whole-exome sequencing was performed rather than using an exome panel focused on TIF1 proteins. Although the chosen approach reduced the sequencing depth in these regions and thus the likelihood of finding mutations present in small number of tumour cells, it allowed us to demonstrate the absence of LOH in the HLA class I region and to gain a broad understanding of the genetic modifications in the tumours included in the study. In the future, collaborative international efforts to gather a greater number of paired blood/tumour samples from myositis patients and a careful longitudinal analysis of the genetic modifications of tumours from CAM patients over time (through tumour relapses or newly developed metastasis) may shed further light to the exciting relationship between cancer and myositis.
In conclusion, our study demonstrates that tumours from paraneoplastic anti-TIF1γ-positive patients show an increased number of genetic alterations, such as mutations and LOH, in TIF1 genes. Also, these patients present high expression of TIF1γ in the tumour, muscle and skin. These two facts may be key to understanding why these patients develop myositis and how the disease is sustained over time.
Supplementary Material
Acknowledgements
We thank Dr S. Diaz for her assistance in interpreting the skin and tumour immunohistochemical staining. We also thank Dr A.L. Mammen, Dr M. Casal-Dominguez, Dr K. Pak, Dr A. Derfoul, R. Yeker, C. Parks and D. Amici for critical reading of the manuscript and suggestions for its improvement. The research of Dr Pinal-Fernandez is supported by the National Institutes of Health Intramural Research Program and by a fellowship from the Myositis Association.
Funding: This work was funded by the Instituto de Salud Carlos III (grants PI12-01320 and PI15-02100), co-financed by the European Regional Development Fund.
Disclosure statement: The authors have declared no conflicts of interest.
Supplementary data
Supplementary data are available at Rheumatology Online.
References
- 1. Dalakas MC. Inflammatory muscle diseases. N Engl J Med 2015;373:393–4. [DOI] [PubMed] [Google Scholar]
- 2. Trallero-Araguas E, Rodrigo-Pendas JA, Selva-O’Callaghan A. et al. Usefulness of anti-p155 autoantibody for diagnosing cancer-associated dermatomyositis: a systematic review and meta-analysis. Arthritis Rheum 2012;64:523–32. [DOI] [PubMed] [Google Scholar]
- 3. Fujimoto M, Hamaguchi Y, Kaji K. et al. Myositis-specific anti-155/140 autoantibodies target transcription intermediary factor 1 family proteins. Arthritis Rheum 2012;64:513–22. [DOI] [PubMed] [Google Scholar]
- 4. Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer 2011;11:792–804. [DOI] [PubMed] [Google Scholar]
- 5. Khetchoumian K, Teletin M, Mark M. et al. TIF1δ, a novel HP1-interacting member of the transcriptional intermediary factor 1 (TIF1) family expressed by elongating spermatids. J Biol Chem 2004;279:48329–41. [DOI] [PubMed] [Google Scholar]
- 6. Venturini L, You J, Stadler M. et al. TIF1γ, a novel member of the transcriptional intermediary factor 1 family. Oncogene 1999;18:1209–17. [DOI] [PubMed] [Google Scholar]
- 7. Joseph CG, Darrah E, Shah AA. et al. Association of the autoimmune disease scleroderma with an immunologic response to cancer. Science 2014;343:152–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schreiber RD, Old LJ, Smyth MJ.. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science 2011;331:1565–70. [DOI] [PubMed] [Google Scholar]
- 9. Bohan A, Peter JB.. Polymyositis and dermatomyositis (second of two parts). N Engl J Med 1975;292:403–7. [DOI] [PubMed] [Google Scholar]
- 10. Hida A, Yamashita T, Hosono Y. et al. Anti-TIF1-γ antibody and cancer-associated myositis: a clinicohistopathologic study. Neurology 2016;87:299–308. [DOI] [PubMed] [Google Scholar]
- 11. Troyanov Y, Targoff IN, Tremblay JL. et al. Novel classification of idiopathic inflammatory myopathies based on overlap syndrome features and autoantibodies: analysis of 100 French Canadian patients. Medicine 2005;84:231. [DOI] [PubMed] [Google Scholar]
- 12. Labrador-Horrillo M, Martinez MA, Selva-O’Callaghan A. et al. Anti-TIF1γ antibodies (anti-p155) in adult patients with dermatomyositis: comparison of different diagnostic assays. Ann Rheum Dis 2012;71:993–6. [DOI] [PubMed] [Google Scholar]
- 13. Koboldt DC, Zhang Q, Larson DE. et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012;22:568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ha G, Roth A, Khattra J. et al. TITAN: inference of copy number architectures in clonal cell populations from tumor whole-genome sequence data. Genome Res 2014;24:1881–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ng PC, Henikoff S.. Predicting deleterious amino acid substitutions. Genome Res 2001;11:863–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Adzhubei IA, Schmidt S, Peshkin L. et al. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sievers F, Wilm A, Dineen D. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 2014;7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moutaftsi M, Peters B, Pasquetto V. et al. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat Biotechnol 2006;24:817–9. [DOI] [PubMed] [Google Scholar]
- 19. McClelland RA, Finlay P, Walker KJ. et al. Automated quantitation of immunocytochemically localized estrogen receptors in human breast cancer. Cancer Res 1990;50:3545–50. [PubMed] [Google Scholar]
- 20. Pommier RM, Gout J, Vincent DF. et al. TIF1γ suppresses tumor progression by regulating mitotic checkpoints and chromosomal stability. Cancer Res 2015;75:4335–50. [DOI] [PubMed] [Google Scholar]
- 21. Jimenez P, Canton J, Collado A. et al. Chromosome loss is the most frequent mechanism contributing to HLA haplotype loss in human tumors. Int J Cancer 1999;83:91–7. [DOI] [PubMed] [Google Scholar]
- 22. Gunawardena H, Wedderburn LR, North J. et al. Clinical associations of autoantibodies to a p155/140 kDa doublet protein in juvenile dermatomyositis. Rheumatology 2007;47:324–8. [DOI] [PubMed] [Google Scholar]
- 23. William J, Euler C, Christensen S, Shlomchik MJ.. Evolution of autoantibody responses via somatic hypermutation outside of germinal centers. Science 2002;297:2066–70. [DOI] [PubMed] [Google Scholar]
- 24. Casciola-Rosen L, Nagaraju K, Plotz P. et al. Enhanced autoantigen expression in regenerating muscle cells in idiopathic inflammatory myopathy. J Exp Med 2005;201:591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Targoff IN, Mamyrova G, Trieu EP. et al. A novel autoantibody to a 155-kd protein is associated with dermatomyositis. Arthritis Rheum 2006;54:3682–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.