Abstract
Diatoms (Bacillariophyta) are a species-rich group of eukaryotic microbes diverse in morphology, ecology, and metabolism. Previous reconstructions of the diatom phylogeny based on one or a few genes have resulted in inconsistent resolution or low support for critical nodes. We applied phylogenetic paralog pruning techniques to a data set of 94 diatom genomes and transcriptomes to infer perennially difficult species relationships, using concatenation and summary-coalescent methods to reconstruct species trees from data sets spanning a wide range of thresholds for taxon and column occupancy in gene alignments. Conflicts between gene and species trees decreased with both increasing taxon occupancy and bootstrap cutoffs applied to gene trees. Concordance between gene and species trees was lowest for short internodes and increased logarithmically with increasing edge length, suggesting that incomplete lineage sorting disproportionately affects species tree inference at short internodes, which are a common feature of the diatom phylogeny. Although species tree topologies were largely consistent across many data treatments, concatenation methods appeared to outperform summary-coalescent methods for sparse alignments. Our results underscore that approaches to species-tree inference based on few loci are likely to be misled by unrepresentative sampling of gene histories, particularly in lineages that may have diversified rapidly. In addition, phylogenomic studies of diatoms, and potentially other hyperdiverse groups, should maximize the number of gene trees with high taxon occupancy, though there is clearly a limit to how many of these genes will be available.
Keywords: diatoms, Bacillariophyta, phylogenomics, phylotranscriptomics, incomplete lineage sorting
Introduction
Diatoms are a hyperdiverse lineage of microbial eukaryotes that form the base of marine food webs and produce roughly 20% of Earth’s oxygen (Field et al. 1998). Despite their importance, progress in understanding diatom evolutionary relationships has not kept pace with other similarly diverse groups. Illustrative of this, the first molecular phylogeny of diatoms, based on SSU rDNA sequences from 11 taxa (Medlin et al. 1993), was published the same year as a landmark rbcL phylogeny of 499 seed plants (Chase et al. 1993). Nearly 25 years later, plant relationships are now supported by hundreds of plastid genomes (Ruhfel et al. 2014), hundreds of nuclear genes (Wickett et al. 2014; Zeng et al. 2014), and a combined data set of 17 genes from 640 taxa (Soltis et al. 2011). Although phylogenetic studies of diatoms have seen substantial gains in taxon sampling, reliance on the SSU rDNA gene persists (Medlin 2016) despite its known limitations (Soltis et al. 1999; Theriot et al. 2009). Plastid loci have also proven to be informative (Theriot et al. 2015), but many diatom relationships remain uncertain. For example: 1) support among the major clades, including the earliest splits in the tree, is often low (Theriot et al. 2010, 2015; Li et al. 2015); 2) monophyly of the three diatom classes (Coscinodiscophyceae [radial centric diatoms], Mediophyceae [polar centric diatoms], and Bacillariophyceae [pennate diatoms]) can be sensitive to locus selection, alignment strategy, and outgroup choice (Medlin 2016); 3) relationships within the major classes can vary depending on the locus and method of phylogenetic inference (Theriot et al. 2010, 2015); and 4) the sister lineage to pennate diatoms (Bacillariophyceae), and within that clade, the sister to raphid pennates, are still uncertain.
Transcriptome (RNA-seq) data have become a widely used source of data for phylogenetic studies (Wen et al. 2015), providing hundreds to thousands of informative markers for resolving relationships across a broad range of evolutionary scales (Johnson et al. 2013; Wickett et al. 2014; Shen et al. 2016). Methodological advances have made it possible to extend these approaches to nonmodel organisms with few genomic resources. For example, identification of orthologous loci has been improved for data sets with incomplete or uneven phylogenetic sampling (Emms and Kelly 2015), and flexible strategies for orthology determination can accommodate the complex nature of transcriptome-based phylogenetic data sets (Yang and Smith 2014). Transcriptome data are, however, inherently noisy. Genes expressed at low levels may be partly assembled or absent for some taxa, leading to biases in ortholog clustering or multiple sequence alignments with incomplete data (Grabherr et al. 2011a; Boussau et al. 2013). Genes that are exclusive to smaller subclades within a large, species-rich group will also result in “missing” data for taxa in other parts of the tree. Although the impacts of variation in alignment column and taxon occupancy have been explored to different degrees in phylotranscriptomic studies (Andrade et al. 2014; Gonzalez et al. 2015; Lemer et al. 2015), the levels below which an individual gene matrix shifts from informing to impeding species-tree inferences are not always clear and will likely vary depending on the size and depth of the phylogeny. Identifying the optimal balance between phylogenetic signal and matrix occupancy (i.e., including the maximum possible number of alignment characters for the largest number of taxa) is useful both for interpreting phylogenetic results and guiding future studies.
We sampled the growing number of sequenced diatom genomes and transcriptomes and systematically evaluated the impacts of column and taxon occupancy on patterns of conflict and concordance among gene trees. We found that gene trees with low taxon occupancy exhibited high levels of discordance with the species phylogeny. Although much of the discordance reflected uncertainty in gene trees, strongly supported discordance was concentrated on short internal branches, which are relatively common across the diatom phylogeny. Nevertheless, we were able to recover a set of consistent and strongly supported species relationships across much of the phylogeny, suggesting that many challenging evolutionary relationships within diatoms can be resolved despite a highly variable and complex set of nuclear gene histories.
Results
Data Sampling and Species Tree Reconstructions
Total numbers of assembled transcripts ranged from 17,155 to 124,647 (x̄=39,112 ± 17,993) for the 94 diatom taxa in our study. After pruning and filtering orthologous clusters into single-copy alignments, we generated data subsets to reflect varying levels of both site and taxon occupancies prior to estimating gene trees. Alignments were trimmed at three occupancy cutoffs (0.2, 0.5, and 0.8) at each site in a gene alignment (i.e., column occupancy), and each of these three data treatments was further segregated into six exclusive or partially overlapping data sets based on the proportion of ingroup taxa present in a gene alignment (i.e., taxon occupancy). The 18 resulting data subsets ranged in size from 32 to 3,622 loci and 8 to 94 diatom taxa, with total combined alignment lengths ranging from 7,696 to 1.96 million amino acids (table 1). Average bootstrap support across gene trees ranged from 70.7 ± 12.1% to 69.8 ± 6.7% for gene trees in the 0.2 and 0.5 alignment column occupancy data subsets, respectively. Average gene tree bootstrap support decreased slightly with increasing taxon occupancy (all gene trees at 0.2 alignment column occupancy: y=−0.196x + 77.349, R2=0.168; all gene trees at 0.5 alignment column occupancy: y=−0.191 + 76.293, R2=0.159; all gene trees at 0.8 alignment column occupancy: y=−0.124 + 68.545, R2=0.059).
Table 1.
Alignment Counts for Data Partitions Used in Gene and Species Tree Analyses.
| Data Partition | Minimum Alignment Column Occupancy Proportion |
||
|---|---|---|---|
| 0.2 | 0.5 | 0.8 | |
| All | 3,622 (1,961,524) | 3,614 (1,446,913) | 3,324 (760,500) |
| 100% taxon occupancy | 32 (8,685) | 32 (8,077) | 32 (7,696) |
| 80–100% taxon occupancy | 517 (178,996) | 517 (154,571) | 512 (132,435) |
| 40–60% taxon occupancy | 390 (194,673) | 389 (150,355) | 366 (84,676) |
| 10–20% taxon occupancy | 1,219 (763,552) | 1,223 (509,180) | 1,105 (219,918) |
| Combined 10–20%, 40–60%, 80–100% taxon occupancy | 2,126 (1,137,221) | 2,129 (814,106) | 1,983 (437,029) |
Note.—Numbers in parentheses represent lengths of concatenated amino acid alignments. The “all” data partition represents all orthologous clusters, with taxon occupancy ranging from 8.4% to 100% (i.e., 8–94 diatom taxa).
Summary-coalescent and concatenation analyses resulted in species tree topologies that were largely consistent across data subsets, with the exception of data subsets with the lowest (10–20%) taxon occupancy (fig. 1). In the lowest taxon-occupancy data sets, topological resolution of deeper nodes was highly variable among genes and often lacked consensus support (supplementary file 1, Supplementary Material online). High levels of missing data can be problematic for summary-coalescent analyses (Vachaspati and Warnow 2015), which produced outlier tree topologies for low-taxon-occupancy data sets (fig. 1, clusters 2 and 3). Concatenation-based analyses appeared to be less sensitive to taxon occupancy (fig. 1). The following results and discussion are mostly limited to data subsets with high taxon occupancy (table 1), as these produced a more consistent set of topologies. Throughout this article, we assess species-tree support based on the combined ASTRAL, ASTRAL-mlbs, IQ-TREE SH-aLRT, and IQ-TREE ultrafast bootstrapping analyses.
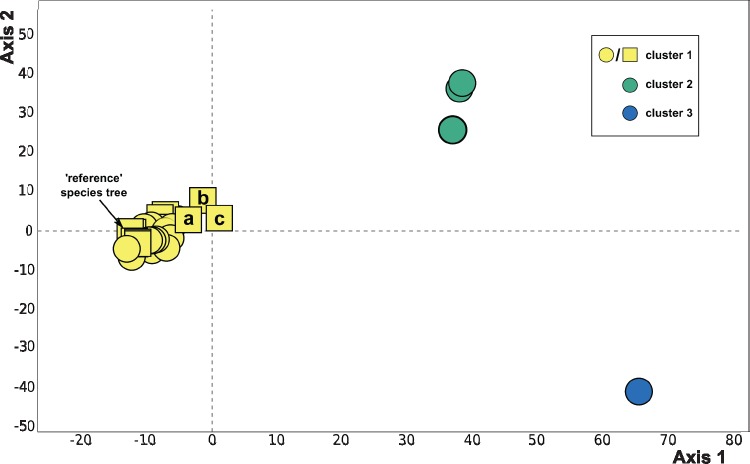
Fig. 1.
Clustering of species trees based on Robinson–Foulds symmetric distance for all data treatments (see table 1) using ASTRAL and ASTRAL-MLBS (circles) or IQ-TREE analysis of a concatenated matrix (squares). Most species trees fall within cluster 1, including the tree shown in figure 2. Cluster 1 also includes concatenation-based species trees with 10–20% taxon occupancy at alignment column occupancy cutoffs of 0.8 (a), 0.5 (b), and 0.2 (c). Outlying clusters 2 and 3 represent ASTRAL and ASTRAL-mlbs species trees for 10–20% taxon occupancy partitions and alignment column occupancy cutoffs of 0.2 and 0.5 (cluster 2; four trees total, two are topologically identical) and 0.8 (cluster 3; two topologically identical trees).
For high-taxon-occupancy data sets, most relationships across the diatom phylogeny were consistent and strongly supported (fig. 2 and supplementary file 1, Supplementary Material online). Differences in branching orders between species trees generated from these data sets were few (supplementary file 1, Supplementary Material online) and involved only ca. 10% of nodes in the species tree, including the branching order of major polar centric clades, the position of Staurosira, and minor variations within Skeletonema, Chaetoceros, and Cyclotella/Thalassiosira. We found consistently strong support for monophyly of pennate diatoms (Bacillariophyceae) and weak to strong support for monophyly of polar centric diatoms (Mediophyceae), excluding Attheya (discussed below). Mediophytes were recovered as paraphyletic by phylogenetic analyses of one data set (0.8 alignment column occupancy, 40–60% taxon occupancy; supplementary file 1, Supplementary Material online), but support for this result was low. No analysis rooted with outgroups (see Materials and Methods) recovered radial centrics (Coscinodiscophyceae) as monophyletic, and relationships among the radial centric lineages were consistent across high-taxon-occupancy data treatments and analyses. Araphid pennates were consistently paraphyletic, and branching order of the araphid pennate lineages was generally consistent, with the exception that the position of Staurosira varied slightly depending on the data set and type of analysis (supplementary file 2, Supplementary Material online). The biddulphioid genus Attheya was sister to pennate diatoms with weak to strong support in all but two analyses (ASTRAL and ASTRAL-mlbs for ≥0.5 alignment column occupancy, 100% taxon occupancy), both of which recovered (Attheya + Mediophyceae) with low support.
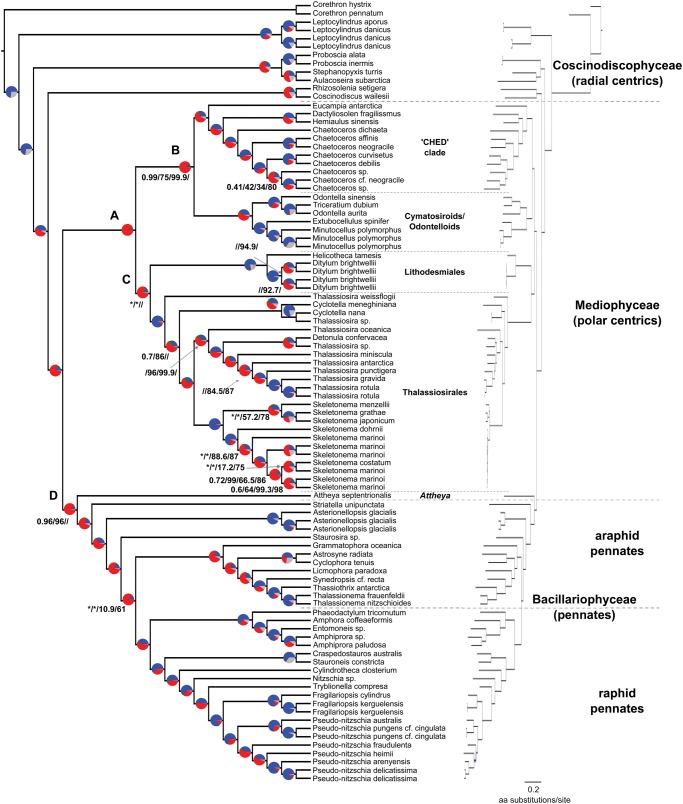
Fig. 2.
Concatenation-based cladogram with gene-tree concordance pie charts (left) and phylogram (right) using the 80–100% taxon occupancy and 0.8 alignment column occupancy data subset. Pie chart color coding: blue—fraction of gene trees supporting the shown split; green—fraction of gene trees supporting the second most common split; red—fraction of gene trees supporting all other alternative partitions; gray—fraction of gene trees with <33% bootstrap support at that node. Support values are only shown for nodes with less than full support from ASTRAL/ASTRAL-MLBS/IQ-TREE SH-aLRT/IQ-TREE ultrafast bootstrapping analyses. Asterisks (*) identify splits not supported by ASTRAL or ASTRAL-MLBS analyses. Nodes labeled A, B, C, and D varied topologically among data treatments and analyses and are discussed throughout the main text. Clades that showed variable phylogenetic placements are also identified.
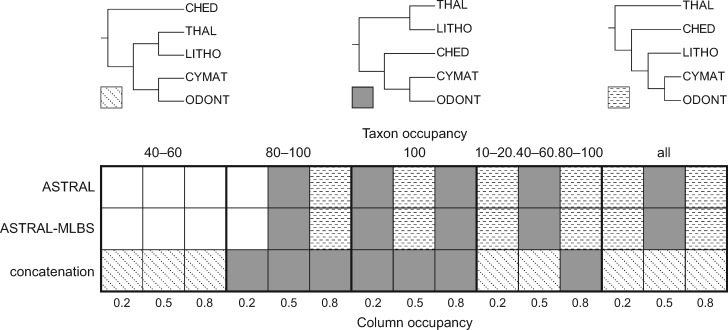
Mediophytes (excluding Attheya) were consistently split into four strongly supported clades: 1) Lithodesmiales [Ditylum and Helicotheca]; 2) Thalassiosirales [Cyclotella, Skeletonema, and Thalassiosira]; 3) cymatosiroids + odontelloids [Extubocellulus, Minutocellus, Odontella and Triceratium]; and 4) a “CHED” clade [Chaetoceros, Hemiaulus, Eucampia, and Dactyliosolen]. Relationships among these clades, however, varied considerably depending on data treatment and phylogenetic method (fig. 3). Species-tree topologies from concatenation analyses supported two topologies that differed only in whether CHED was sister to (Lithodesmiales + Thalassiosirales) or all other mediophytes (fig. 3). Concatenation analyses were more sensitive to taxon occupancy than column occupancy threshold, with the 40–60% taxon occupancy data set driving support for (CHED + other mediophytes) (fig. 3). In contrast, the (CHED + (Lithodesmiales + Thalassiosirales)) topology was supported by seven data subsets, all of which had high taxon occupancy (≥80%) (fig. 3). The latter topology was also supported by 10 ASTRAL and ASTRAL-mlbs analyses, making it the most commonly recovered topology in our analyses. The dominant topology recovered by ASTRAL and ASTRAL-mlbs analyses shared no sister relationships with either of the two concatenation topologies (fig. 3). Additional information about topological variation within araphid pennates and polar centrics is available in supplementary file 2, Supplementary Material online.
Fig. 3.
The three most commonly recovered topologies for major polar centric diatom clades. The four large boxes correspond to the four different taxon occupancy treatments, and each column represents a different column occupancy treatment (table 1). Each data treatment was analyzed with ASTRAL (top row), ASTRAL-MLBS (middle row), or IQ-TREE with a concatenated matrix (bottom row). The recovered topology for each analysis is identified by the shading or stippling according to the top panel. Empty boxes correspond to three other minority topologies (see supplementary file 2, Supplementary Material online, for full results).
Concordance and Conflict between Gene Trees and Species Trees
We examined gene-tree/species-tree conflict and concordance across data subsets, with a particular focus on relationships among the four polar centric clades described earlier and the placement of Attheya, as these have been inconsistently resolved in past studies (Medlin 2016). For these analyses, we used as the reference species tree the concatenation-based tree with 0.8 alignment column occupancy and 80–100% taxon occupancy (fig. 2), and 0.8 column-occupancy cutoff data sets. To verify the robustness of our results to data subset choice, reference species-tree topology, and phylogenetic strategy, gene-tree concordance analyses were also performed using the ASTRAL species tree from the 0.5 column occupancy, 40–60% taxon occupancy data subset as a reference species tree, chosen because it showed the greatest RF distance to the reference concatenation species tree of all high taxon occupancy species trees within the primary cluster of RF analysis (cluster 1, fig. 1).
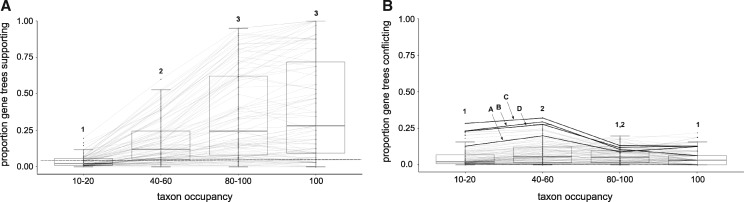
In general, gene tree discordance was common along the backbone of the species tree (e.g., deep splits within the polar centrics and araphid pennates) (fig. 2 and supplementary file 3, Supplementary Material online) as well as in shallow within-genus and within-species clades. Discordant loci typically were not dominated by a single alternative topology but rather many different topologies. In addition, discordant nodes in gene trees had disproportionately low bootstrap support compared with concordant nodes (fig. 4a and supplementary file 4, Supplementary Material online). This result highlights relatively low levels of phylogenetic signal overall in many of the gene trees (fig. 4a and supplementary file 4, Supplementary Material online). We also found consistently strong positive correlations between internal branch length (from the concatenated IQ-TREE analysis) and strongly supported nodes that were concordant between gene and species trees (fig. 4b and supplementary file 4, Supplementary Material online). Consistent with this, short branches subtending the four polar centric clades and Attheya had greater numbers of discordant than concordant topologies (fig. 4b, inset). Taxon occupancy impacted gene-tree concordance most strongly, with the proportion of concordant gene trees increasing between the 10–20%, 40–60%, and 80–100% taxon-occupancy data sets; taxon occupancy had, by contrast, relatively little impact on the observed proportion of discordant gene trees (fig. 5 and supplementary file 4, Supplementary Material online). The proportion of gene trees with strong support for the species tree was consistently low across taxon occupancy subsets for nodes subtending polar centric clades and Attheya (fig. 5a), whereas the proportion of gene trees with strongly supported conflict was relatively high at these nodes (fig. 5b). These trends were robust with respect to choice of reference tree (supplementary files 3 and 4, Supplementary Material online).
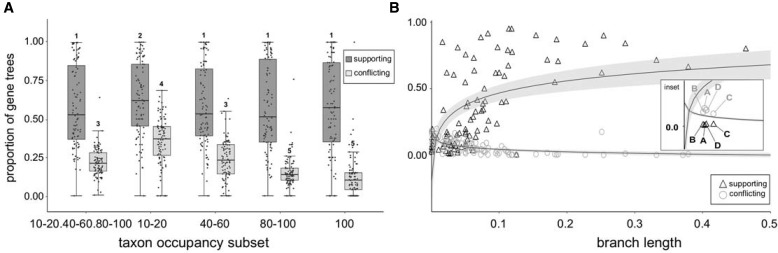
Fig. 4.
Gene tree concordance and discordance across all nodes in the species tree depicted in figure 2 in relation to bootstrap support and branch length. (a) Box plots summarizing the proportion of gene trees still resolved as supporting or conflicting nodes within the species tree, shown as points, when bootstrap support cutoff for the gene tree is increased from 33% to 70% (results shown for 0.8 column occupancy data sets). Shared numbers above whiskers indicate no significant difference at P < 0.05. (b) IQ-TREE branch length versus proportion of gene trees that support or conflict with a node (results shown for 80–100% taxon occupancy and 0.8 column occupancy data set). Each pair of points (triangle + circle) represents the proportion of gene trees supporting or conflicting, respectively, with a node on the species tree. A split in a gene tree was considered concordant if it was shared with the species tree and had ≥70% bootstrap support in the gene tree. Splits in a gene tree were considered discordant if they had ≥70% bootstrap support and were not shared with the species tree. The inset shows the nodes labeled in figure 2, with other data points removed for clarity; shaded areas delimit 95% confidence intervals.
Fig. 5.
Relationships between taxon occupancy and proportion of gene trees that were identified as concordant or discordant with the species tree shown in figure 2. (a) High-taxon-occupancy data sets have a greater proportion of gene trees that are concordant with the species tree. (b) The proportion of gene trees in conflict with the species tree is relatively invariable across varying levels of taxon occupancy. For both panels, each line corresponds to a split in the species tree at different levels of taxon occupancy. The four focal nodes identified in figure 2 are likewise identified in panel (b), but all of them have uniformly low gene-tree support and so fall below the dashed line in panel (a). A split in a gene tree was considered concordant if it was shared with the species tree and had ≥70% bootstrap support in the gene tree. Splits in a gene tree were considered discordant if they had ≥70% bootstrap support and were not shared with the species tree. Shared numbers above whiskers indicate no significant difference in mean values at P < 0.05.
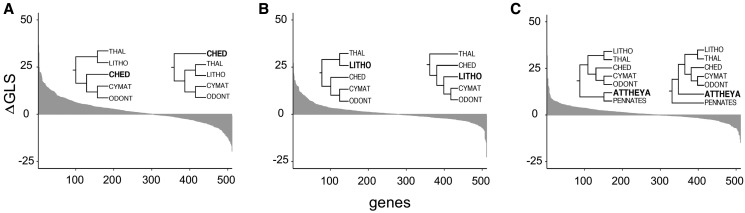
Following Shen et al. (2017), comparison of genewise log-likelihood scores between the most commonly recovered polar centric topology and the most commonly recovered alternative topologies revealed substantial, albeit minority, support for the alternative topologies (fig. 6 and supplementary file 5, Supplementary Material online). Similarly, a strong minority of loci favored placement of the genus Attheya as sister to the polar centric clade rather than sister to the pennate clade (fig. 6). For each pair of contrasting topologies shown in figure 6, removal of loci with the highest or lowest difference in log-likelihood scores did not change the topology of the optimal phylogenetic reconstruction.
Fig. 6.
Genewise log-likelihood differences for competing relationships of polar centric diatom clades and for the placement of Attheya. (a) Most commonly recovered versus the second most recovered polar centric topology; (b) most commonly recovered versus the third most recovered polar centric topology; and (c) (pennates + Attheya) versus (polar centrics + Attheya).
Discussion
Considerable progress toward reconstructing a large, comprehensively sampled phylogenetic hypothesis for diatoms has been made since publication of the first SSU rDNA tree (Medlin et al. 1993). Most of this effort has focused on increasing the number and diversity of sampled species, which is often a principal bottleneck for phylogenetic studies of highly diverse microbial lineages. The development of new phylogenetic markers represents another major hurdle to establishing a robust phylogenetic hypothesis for diatoms (Theriot et al. 2015).
The goal of this study was to determine whether large nuclear data sets can resolve some historically difficult relationships within diatoms, a lineage of microbial eukaryotes comparable in age and species richness to angiosperms. We applied summary-coalescent and concatenation-based approaches of phylogenetic inference to a range of taxon occupancy thresholds for inclusion of individual gene alignments and, within those alignments, individual columns. These experiments allowed us to determine the sensitivity of species-tree inferences to basic parameters related to data set size and composition. The largest data sets were also the sparsest, consisting of up to 3,622 genes and 1.96 million total aligned amino acids, but with individual gene alignments containing as few as 8 of the 94 total ingroup taxa and alignment columns lacking data for as many as 92 of 94 ingroup taxa. At the other extreme, the smallest data set was the most complete, consisting of just 32 genes with at least 80% column occupancy in all 94 ingroup taxa. Our results showed that occupancy thresholds can be relaxed considerably for concatenation-based and, to a lesser extent, summary-coalescent methods with relatively minor effects on the species tree topology (figs. 1 and3).
Phylotranscriptomics Resolves Some Challenging Relationships and Underscores the Recalcitrance of Others
Comparisons of our results to the most comprehensive phylogenetic analysis of diatoms to date are limited by lack of overlap in taxon sampling (i.e., just 32 of the 43 diatom genera in our study were included among the 131 genera sampled by Theriot et al. 2015). In addition, critical relationships in previous studies varied by data set and optimality criterion, confounding cross-study comparisons and also underscoring the need for increased character sampling (Theriot et al. 2015). A definitive reconstruction of the entire diatom phylogeny was not the central goal of this study, so we focused on historically challenging parts of the tree. We also focused on results from the high taxon- and column-occupancy cutoffs, which exhibited the greatest levels of topological congruence.
In our analyses, the backbone of the phylogeny was fully resolved, strongly supported (based on ASTRAL, ASTRAL-mlbs, and IQ-TREE support metrics), and relatively robust to inference method and taxon and column occupancy thresholds. Radial centric diatoms were decidedly not monophyletic, similar to results of other multi-gene studies (Ashworth et al. 2012; Theriot et al. 2015). Support for monophyly of the Mediophyceae has been relatively consistent, with the caveat that Attheya moves in and out of mediophytes in different analyses (Theriot et al. 2010, 2015; Medlin 2016). Our results further strengthened support for monophyly of Mediophyceae to the exclusion of Attheya. Relationships among four strongly supported clades within mediophytes were, however, among the most highly variable across data sets and analyses (fig. 3). The four subclades resolved into six different configurations across data partitions and analyses, and although a plurality of them (17/45) supported one topology (also recovered by Theriot et al. 2015), the five clades were separated by short branches, possibly indicative of rapid diversification or frequent hybridization early on in mediophyte evolution. As a result, lack of phylogenetic signal and the effects of incomplete lineage sorting on short internodes may make it challenging to resolve these nodes with sequence-based analyses (Shen et al. 2017).
The evolution of axial symmetry, isogamy, and active motility in the pennate diatoms (Bacillariophyceae) represent key transitions in the evolution of diatoms, and reconstructing these transitions requires knowledge of the sister lineage to the pennate and raphid pennate clades. Our analyses placed the araphid pennate genus Striatella, which has been notoriously difficult to place phylogenetically (Sato et al. 2008; Theriot et al. 2010; Medlin 2016), as sister to all other pennate diatoms. Many studies that have recovered Attheya in this position did not include Striatella in their analyses (Rampen et al. 2009; Theriot et al. 2009; Sorhannus and Fox 2012). In our analyses, Attheya was recovered consistently, and generally with strong support, as sister to Striatella + pennates, supporting the prediction that its highly variable placement in previous analyses was the result of too few characters (Theriot et al. 2015). Nevertheless, the phylogenetic position of Attheya inferred here should still be considered provisional because of: 1) the short branch separating it from its sister taxon (fig. 2), 2) considerable support for an alternative placement (fig. 6), and 3) evidence that inclusion of Biddulphia, which is missing from our data set, may be necessary to resolve the placement of Attheya with greater certainty (Theriot et al. 2015).
Trends in Gene-Tree Concordance and Conflict
We analyzed topological congruence between gene trees and species trees using a concatenation-based species tree as a point of reference (IQ-TREE analysis of the 0.8 alignment column occupancy cutoff, 80–100% taxon occupancy data subset). A reference tree is required for comparative purposes here, and is not meant to represent the true species tree, which is unknown. Additional analyses (supplemental files 3 and 4, Supplementary Material online) show that the topological comparisons presented here are robust to the choice of reference. Here, the concatenation-based tree is utilized for several reasons, including: 1) size (512 loci) and completeness (80–100% taxon occupancy, ≥80% alignment column occupancy) of the underlying data set; 2) relatively low levels of bootstrap support across many gene trees, possibly suggestive of moderate to high levels of gene tree error (Anisimova et al. 2011), a case in which concatenation-based methods have been shown to outperform summary-coalescent methods (Gatesy and Springer 2014; Mirarab and Warnow 2015); 3) recovery of the most commonly recovered relationships among the major mediophyte clades (fig. 3); and 4) a lack of consistent alternative topologies supported by the underlying gene trees (fig. 2 and supplementary file 3, Supplementary Material online). Importantly, the trends we report in gene-tree/species-tree concordance and discordance were also recovered when using a different tree topology inferred using a summary-coalescent approach and different data subset (supplementary files 3 and 4, Supplementary Material online), indicating that our results are largely robust to phylogenetic method and data set characteristics. Our gene-tree and concatenation-based species-tree estimates were based on amino acid alignments, which we chose based on the size and depth of the diatom phylogeny (200 My, Sorhannus 2007) as well as the high reported rates of sequence evolution in diatoms (Bowler et al. 2008). It is nonetheless possible that the application of nucleotide alignments could affect gene-tree inferences (Hall 2005) and, consequently, species-tree reconstructions (Mirarab and Warnow 2015) and patterns of gene-tree congruence. Future analyses may benefit from an exploration of the relative strengths and weaknesses of amino acid versus nucleotide alignments in phylogenomic analyses at this scale.
Relatively high levels of discordance among gene trees are common in phylogenomic data sets of large, diverse groups (Degnan and Rosenberg 2009). Across our analyses, gene tree discordance was typically lowest at low taxonomic levels, with the exception of some shallow within-genus (e.g., Skeletonema) and within-species (e.g., S. marinoi) nodes, which may exhibit high levels of hybridization and incomplete lineage sorting (Harrison and Larson 2014; Edwards et al. 2016). Conversely, discordance was relatively higher at deeper levels within the phylogeny, similar to findings for other groups (Smith et al. 2015). This was particularly true for deeper nodes within the polar centric and pennate diatom clades (fig. 2 and supplementary file 3, Supplementary Material online). This general pattern could reflect a combination of methodological and biological factors, including misaligned or incorrectly identified orthologs spanning large phylogenetic distances (Emms and Kelly 2015) or lineage-specific gains or losses of genes leading to nonrandomly distributed data (Xi et al. 2016). Evidence for the latter is found in genome comparisons between the model diatoms, Phaeodactylum tricornutum and Cyclotella nana (formerly Thalassiosira pseudonana), which only share roughly 44% of their gene families (Bowler et al. 2008). Considering the age (ca. 200 My; Sorhannus 2007) and diversity (ca. 100,000 species; Mann and Vanormelingen 2013) of diatoms, these are likely to remain as persistent challenges in efforts to reconstruct the species phylogeny of diatoms.
A number of salient trends emerged when comparing gene- and species-tree topologies. First, for gene tree topologies with high bootstrap support, our analyses showed that gene-tree/species-tree concordance was much greater than discordance for longer internal branches. As branch lengths decreased, the number of discordant gene trees can surpass the number of concordant ones. These trends are consistent with predictions based on both coalescent theory (Pamilo and Nei 1988; Maddison and Wiens 1997) and empirical phylogenomic studies (Lambert et al. 2015; Streicher et al. 2016; Blom et al. 2017). The high levels of well-supported gene-tree discordance concentrated on short internal branches suggests that incomplete lineage sorting is likely to be common across many parts of the diatom species tree, a challenge that may be overcome using summary-coalescent methods with high-taxon-occupancy gene matrices.
We also found that levels of gene-tree concordance increased with increasing taxon occupancy of the gene trees. Topological conflicts between gene and species trees tended to have low bootstrap support in gene trees—a trend that was most evident in the highest taxon-occupancy data subsets (fig. 4a). This suggests that a substantial portion of perceived conflict in our data set, and potentially other phylotranscriptomic data sets, reflects lack of phylogenetic signal in gene trees (Blom et al. 2017). Our results suggest that studies aimed at producing resolved species trees might benefit from filtering out low-signal genes by applying higher taxon-occupancy and bootstrap-support cutoffs to individual gene trees. Bootstrap cutoffs, or metrics that incorporate bootstrap support values across a gene tree, have previously been shown to increase the efficiency, support, or robustness of species-tree inferences (Salichos and Rokas 2013; Salichos et al. 2014; Streicher et al. 2016; Blom et al. 2017), and a more thorough exploration of their impacts, specifically in coalescent-based phylogenetic analyses, is warranted (Mirarab and Warnow 2015; Sayyari and Mirarab 2016). In our analyses, we found that average bootstrap support of individual gene trees decreased slightly with increasing taxon occupancy. However, the patterns of support seen for the four focal nodes in this study (fig. 5) do not completely follow the overall patterns of bootstrap support for all gene trees in relation to taxon occupancy. In fact, we do see (fig. 5b) a reflection of the negative correlation between bootstrap support and taxon occupancy for nodes A, B, C, and D in that the proportion of gene trees in conflict with the reference tree (at ≥70%) increases most strongly when taxon occupancy decreases from 80–100% to 40–60%. Variability between overall trends and patterns at our focal nodes underscores that careful dissection of gene-tree support for individual branches is necessary in many cases.
Although summary-coalescent methods benefit from large numbers of loci (Streicher et al. 2016), even ones with weak phylogenetic signal (Blom et al. 2017), these methods produced outlier tree topologies in our analyses of data sets with very low taxon occupancy (fig. 1). Although this impact may be less of a problem for smaller trees (Lambert et al. 2015), we recovered numerous implausible topologies (e.g., nonmonophyly of Thalassiosirales, pennates, or raphid pennates) with generally weak support (supplementary file 1, Supplementary Material online). Considering the challenging nature of phylotranscriptomic data sets, some of the correlations we found between gene-tree concordance and taxon occupancy, conflict, and uncertainty may also reflect technical challenges in fully capturing the phylogenetic signal in our data set (Sistrom et al. 2014). Improved tools for transcriptome-based orthology assessment, and a fuller understanding of the impacts of incorrect orthology assessment on phylogenetic inference (Yang and Smith 2014; Smith et al. 2015), will undoubtedly move phylotranscriptomic studies of nonmodel organisms forward in this regard.
The focal nodes that varied across our analyses (e.g., relationships among mediophyte clades and the placement of Attheya) are especially challenging with respect to all of the trends just described. The branches subtending these nodes are short, and the proportion of gene trees in conflict with the inferred species tree exceeds the proportion of gene trees congruent with the species tree, even at higher bootstrap cutoffs for gene trees (fig. 4). Moreover, none of these nodes showed a strong correlation between taxon occupancy and gene tree support, instead maintaining low levels of gene tree support and high levels of conflict across taxon occupancy cutoffs (fig. 5). Finally, pairwise comparisons between competing topologies revealed substantial support for alternatives to the best inferred species tree (fig. 6). The relatively high rates of gene tree error or discordance at these nodes may argue for increased reliance on concatenation-based phylogenetic strategies in determining a singular species tree topology (Mirarab and Warnow 2015), though high levels of incomplete lineage sorting may offset gains in this regard (Mirarab et al. 2014). Further, identifying the underlying source of these high levels of gene-tree conflict will shed new light on processes shaping the macroevolutionary history of diatoms.
Taken together, the patterns of discordance in our analyses provide insights into previous and future efforts in reconstructing the phylogeny of both diatoms and perhaps other similarly diverse groups. For example, widespread discordance among gene trees cautions against overinterpreting phylogenetic trees based on one or a few genes (Medlin 2016). Short internal branches, which may be the result of rapid species radiations and/or high rates of historical hybridization, are a common feature of the diatom phylogeny. Resolution of these nodes is especially challenging for analyses based on a small number of genes, as discordance between gene trees was more common than concordance for some of these nodes. Across diatoms, the lack of dominant alternative topologies at nodes with relatively high levels of gene tree conflict, together with the relatively short observed branch lengths at these nodes, suggests that uncertainty introduced by incomplete lineage sorting—as opposed to a consistent set of subordinate signals introduced by hybridization events—is the more likely underlying cause of the discordance (Galtier and Daubin 2008; Smith et al. 2015). This may be alleviated to some extent by increasing within-species sampling in future studies (Maddison and Knowles 2006). Hybridization as an alternative explanation for gene tree discordance should not be ruled out, however, especially at lower taxonomic levels (Casteleyn et al. 2009).
Conclusions
While phylogenomic data sets have generally delivered on the promise of providing fully resolved species tree, they have likewise delivered—repeatedly and for a diverse range of groups—on predictions made decades ago that the complex and highly varied histories of genes within species will challenge our model of species phylogenies as simple bifurcating trees (Maddison and Wiens 1997). Moreover, alternative topologies for historically recalcitrant nodes often find substantial support across phylogenomic data sets, and may be disproportionately impacted by a small number of genes, or even sites, in the genome (Shen et al. 2017). The diatom phylogeny is not immune to these challenges, and will likely remain difficult to fully resolve even with increased taxonomic sampling. A “winner-take-all democracy” approach to phylogenetic reconstruction (Maddison and Wiens 1997) clearly will not capture the complex underlying history of diatom genomes (Huson and Bryant 2006) and may not, in the end, provide the appropriate comparative framework for studies of trait and genome evolution (Hahn and Nakhleh 2016) in this important lineage.
Materials and Methods
Data Sources
The primary data for this study was generated by The Gordon and Betty Moore Foundation’s Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP), which provided RNA-seq data for 92 diatom accessions from 40 genera (Keeling et al. 2014). We also compiled predicted proteins from the sequenced genomes of three diatoms (Phaeodactylum tricornutum [Bowler et al. 2008], Fragilariopsis cylindrus [Mock et al. 2017], and Cyclotella nana [Armbrust et al. 2004]) and three heterokont outgroups (Ectocarpus siliculosus [Cock et al. 2010], Nannochloropsis gaditana [Radakovits et al. 2012], and Aureococcus anophagefferens [Gobler et al. 2011]). In addition, we used high-depth RNA-seq data for an additional outgroup, Triparma pacifica, and two diatoms, Leptocylindrus danicus and Hemiaulus sinensis (Kessenich et al. 2014). In total, this resulted in an initial data set consisting of 97 diatoms and four heterokont outgroup accessions.
Processing and Assembly of RNA-Seq Reads
Raw RNA-seq data were downloaded from the Sequence Read Archive (SRA) hosted by The National Center for Biotechnology Information (NCBI). SRA-formatted files were converted to fastq format using the fastq-dump utility (ver. 2.7.0) (SRA Toolkit Development Team 2016). Reads from separate experimental treatments of identical strains were concatenated into single data sets.
RNA-seq reads were processed and assembled following recommendations outlined in the Oyster River Protocol (MacManes 2015). Briefly, predicted errors in raw reads were corrected using Rcorrector with options “-k 31 -t 15” (Song and Florea 2015). Corrected reads were then trimmed using Trimmomatic (ver. 0.32) with options “ILLUMINACLIP: 2: 40: 15 LEADING: 2 TRAILING: 2 SLIDINGWINDOW: 4: 2 MINLEN: 20” (Bolger et al. 2014). Sequences representing common laboratory vectors and diatom rRNA genes and organelle genomes were filtered using bowtie2 (ver. 2.2.3) with default settings (Langmead and Salzberg 2012). Overlapping forward and reverse reads were merged using BBMerge (ver. 8.8) with the option “strict = t” (Bushnell 2014). Merged nuclear reads were assembled using Trinity (ver. 2.2.0) with default settings (Grabherr et al. 2011b). Assembled nuclear transcripts were translated into amino acid sequences using TransDecoder (ver. 2.0.1) with default settings. Translation predictions were informed by BLASTP searches of the longest ORFs to the Swiss-Prot database and HMMER searches to the Pfam database (Eddy 2011; Finn et al. 2016). Quality of RNA-seq assemblies was assessed with TransRate (ver. 1.01) scoring (Smith-Unna et al. 2016), using P. tricornutum as a reference, and through identification of conserved eukaryotic orthologs in the assembly with BUSCO (ver. 1.2) (Simão et al. 2015). Each assembly was filtered for redundancy with CD-HIT (-c 0.99 -n 5) (Fu et al. 2012) prior to ortholog clustering. Three MMETSP accessions were discarded due to low-sequence read counts and poor assembly, resulting in a total of 94 diatom accessions and four outgroup taxa. All subsequent analyses used conceptual amino acid translations. Example commands for RNA-seq read processing and assembly steps described earlier can be found at https://github.com/mparkscbg/MMETSP-analyses.
Ortholog Selection, Gene Alignment, and Tree Building, and Species Tree Reconstructions
In selecting and applying orthologous loci for alignment and gene-tree reconstruction, our aim was to capture a large initial data set for interrogation of the diatom phylogeny, and to explore the impact of data completeness on species tree inference. We applied two explicit types of data filtering during the processes of sequence alignment and gene- and species-tree reconstruction: 1) trimming of orthologous alignments by minimal alignment column occupancy cutoffs prior to gene-tree estimation, and 2) filtering of ortholog trees and alignments used in species-tree building by taxon occupancy. We define column occupancy as the fraction of species with data for a particular column in a given ortholog alignment; the fraction is calculated based on the total number of species in the orthogroup. For example, an alignment column in which 10 of the 20 total species in an alignment had a gap (“-”) character would have a column occupancy of 0.5. We define taxon occupancy as the fraction of taxa present in an alignment based on the total number of taxa included in our analysis. An ortholog alignment that included 47 of the 94 total diatom taxa considered in this study would have a taxon occupancy of 0.5.
Ortholog Selection
We performed an all-versus-all BLASTP search of the filtered transcripts using NCBI-BLAST (ver. 2.3.0+) (Camacho et al. 2009) with an e-value cutoff of 0.001. These searches were parallelized using GNU Parallel (Tange 2011). BLASTP results were subsequently used for clustering of orthologous groups with Orthofinder (ver. 0.4.0) using default settings (Emms and Kelly 2015). The resulting orthologous clusters were filtered to include only clusters with at least one nondiatom outgroup, at least 20 unique taxa, and maximum taxon redundancy of 1,000 transcripts per cluster (i.e., the number of transcripts per taxon in an orthologous cluster).
Gene Alignments and Gene Tree Estimation
Ortholog alignments and trees were constructed with a modified version of the phylogenomic_data set_construction pipeline, using the “rooted ingroups” (RT) strategy for paralog pruning (Yang and Smith 2014). This strategy is recommended for data sets with multiple high-quality outgroups and can be used when the genome duplication history of the ingroup is unknown, as is the case for diatoms. For this pipeline, we used UPP (ver. 2.0) (Nguyen, Mirarab et al. 2015) to create multiple sequence alignments and FastTree (ver. 2.1) (Price et al. 2010) to construct an initial phylogenetic tree for each orthologous cluster.
For each orthologous group, the alignment representing the largest identified orthologous clade with at least eight taxa was trimmed using the phyutility_wrapper.py script from Yang and Smith (2014) for minimal column occupancy at three cutoff values prior to reconstructing the final ortholog tree: 0.2, 0.5, and 0.8. Final tree-building and bootstrapping of gene trees (100 bootstrap pseudoreplicates per alignment) were performed with RAxML (ver. 8.2.9) (Stamatakis 2014) using the PROTCATJTT model for all alignments at each column-occupancy cutoff. SumTrees (Sukumaran and Holder 2010) was used to summarize bootstrap results onto the best-scoring maximum likelihood trees and to collapse nodes with <33% bootstrap support in order to minimize potential impacts of gene-tree estimation error on species-tree reconstructions (Mirarab and Warnow 2015; Sayyari and Mirarab 2016). However, we acknowledge that the interaction of branch contraction and bootstrap threshold may reduce accuracy in some cases and that further explorations of this using either simulated data or lineages with well-established relationships is warranted. An example of the full set of commands used to run the phylogenomic_data set_construction pipeline can be found at https://github.com/mparkscbg/MMETSP-analyses.
Species Tree Reconstructions
For each remaining locus at 0.2, 0.5, and 0.8 column occupancy cutoffs, gene alignments (for concatenation analyses, described below) and gene trees (for summary-coalescent analyses, described below) were further segregated into six exclusive or partially overlapping pools prior to species tree estimation, as follows: 1) all alignments/gene trees, in which ingroup taxon occupancy ranged from 8.4% to 100%; 2) alignments/gene trees with 100% taxon occupancy; 3) 80–100% taxon occupancy; 4) 40–60% taxon occupancy; 5) 10–20% taxon occupancy; and 6) the combination of alignments/gene trees with 10–20%, 40–60%, or 80–100% taxon occupancy. This resulted in pruned and filtered ortholog alignments and gene trees residing in 1 or more of 18 different data treatments (six taxon occupancy categories for each of three alignment column occupancy cutoffs).
We applied both a summary-coalescent approach with two measures of topological support, and a concatenation-based approach to phylogenetic inference of gene trees and alignments for each of the 18 data treatments, resulting in 54 total species tree estimates. We used ASTRAL (ver. 4.10.8) (Mirarab and Warnow 2015) with default settings for summary-coalescent species tree estimation, with species tree topology and node support estimated with standard ASTRAL support values (i.e., local posterior probability) and multilocus bootstrapping (hereafter referred to as ASTRAL and ASTRAL-mlbs, respectively). For concatenation-based analyses, we used ProtTest (ver. 3.4.2) (Guindon et al. 2003; Darriba et al. 2011) to determine the best-fitting model of protein evolution for each gene alignment in a data subset, based on the AICc selection criterion; the resulting models were dominated by LG + I+G and LG + G (69.7% ± 11.6%), with the models WAG + I+G, JTT + I+G, and VT + I+G accounting for either the majority or a substantial portion of remaining gene alignments (14.6% ± 9.8%). Loci for each column and taxon occupancy combination category were concatenated with AMAS (Borowiec 2016). Concatenation-based species trees were estimated using IQ-TREE with ultrafast bootstrapping and SH-aLRT testing (1,000 replicates each) to evaluate support (Guindon et al. 2010; Minh et al. 2013; Nguyen, Schmidt et al. 2015; Chernomor et al. 2016).
The RT strategy for ortholog detection returns alignments and unrooted ortholog trees without outgroup sequences. We used Yang and Smith’s (2014) “monophyletic outgroups” (MO) pipeline, which does not remove outgroup taxa, on a subset of the original orthogroups to determine whether pruning of outgroup taxa affected the branching order or resolution of the earliest splits in the diatom phylogeny. These analyses were based on a total of 306 orthologous clusters that met the following criteria: 80–100% taxon occupancy, fewer than 1,000 transcripts per taxon, and outgroup sampling that included, 1) Triparma and Aureococcus or Aureococcus alone (91 clusters), 2) Triparma and Ectocarpus or Ectocarpus alone (154 clusters), or 3) Triparma and Nannochloropsis or Nannochloropsis alone (61 clusters). This allowed use of the highly curated, genome-based stramenopile proteomes (Aureococcus, Ectocarpus, and Nannochloropsis) as outgroups, as recommended for the MO pipeline (Yang and Smith 2014), while avoiding assumptions about relationships among Aureococcus, Ectocarpus, and Nannochloropsis, which are inconsistently ordered in other phylogenetic studies (Brown et al. 2010; Gomez et al. 2011). Column occupancy cutoff was set at 0.8, and other parameter settings were specified as described previously. The resulting gene trees were summarized in ASTRAL using default settings. For this analysis, the two representatives of the genus Corethron were supported as monophyletic and sister to all other diatoms with high (0.99) ASTRAL support. Based on these results, Corethron was used for subsequent rooting of RT trees.
Gene-Tree and Species-Tree Concordance and Conflict
We applied several strategies to investigate levels of support and conflict among species trees generated from different data subsets and analysis strategies, and between gene trees and species trees, using the concatenation-based species tree with 0.8 alignment column occupancy and the 80–100% taxon occupancy data set, as a reference species tree. To verify that our results concerning the impact of bootstrap cutoff, internal branch length, and taxon occupancy (all described below) were robust to choice of reference species tree, the analyses were also run using the high taxon occupancy, summary-coalescent species tree with the greatest RF distance from the reference species tree (ASTRAL species tree estimated with 0.5 alignment column occupancy and 40–60% taxon occupancy) and the associated 0.5 alignment column occupancy cutoff gene trees. These analyses are described in full in the following sections.
Topological Concordance of Species Trees
Topological concordance between species trees for all data subsets and phylogenetic analyses (54 species trees in total) was estimated by Robinson–Foulds symmetric differences and the Ward clustering method using TreeScape (ver. 1.10.18) (Jombart et al. 2017).
Gene tree concordance was analyzed for 45 data treatments (three column occupancy classes each for all taxon occupancy classes except “all,” which failed to complete on our computing clusters), by evaluating gene tree concordance against the reference species tree. Concordance was quantified using the PhyParts software package (Smith et al. 2015) and the ETE3 Python toolkit (Huerta-Cepas et al. 2016) as implemented in PhyPartsPieCharts (https://github.com/mossmatters/MJPythonNotebooks; last accessed January 12, 2017). PhyParts requires rooted trees, so gene trees for each partition were rooted using radial centric taxa as present based on order of divergence as supported by the reference species tree; gene trees that did not include radial centric taxa were not used in PhyParts analyses. Gene-tree bootstrap cutoffs were kept at 33% as applied through SumTrees during gene-tree reconstruction.
Impact of Gene Tree Bootstrap Cutoff, Species Tree Branch Lengths and Taxon Occupancy
PhyParts analyses were repeated as described earlier but with bootstrap support cutoffs increased from 33% to 70%. For each species tree, we determined the average proportion of support and conflict persisting following increased bootstrap cutoff as follows:
where n = total node count in species tree, x = number of gene trees supporting or conflicting with a species-tree node with bootstrap support cutoff of 70%, and y = number of gene trees supporting or conflicting with a species tree node with bootstrap support cutoff of 33%. Significant differences between resultant mean values was determined through ANOVA (standard weighted means analysis for correlated samples) followed by Tukey’s HSD test (www.vassarstats.net).
We also investigated the impact of branch length and taxon occupancy on the proportion of gene trees supporting or conflicting with species tree nodes. For each node of the reference species tree, the proportion of gene trees supporting or conflicting that node (with 70% bootstrap cutoff applied to gene trees) was recorded for each data subset, along with the associated branch length at that node. The correlation between proportions of gene trees and branch lengths was estimated through regression analysis using the ggplot2 package (Wickham 2009) for R (R Development Core Team 2015). The impact of taxon occupancy was estimated by determining the proportion of gene trees supporting or conflicting with the reference species tree for each of the 10–20%, 40–60%, 80–100%, and 100% column occupancy data subsets at each alignment column occupancy cutoff. Statistical analyses followed those described for analyses of bootstrap cutoffs.
Quantification of Support for Alternative Species Tree Topologies
The distribution of support for three contrasting pairs of topologies was measured through comparisons of genewise log-likelihood support (Shen et al. 2017). These pairs captured the majority of recovered species tree topological discrepancies, and represented the most common topologies recovered in species trees for polar centric clades and for the biddulphioid genus Attheya. For each comparison, the reference species tree was adjusted in Mesquite (ver. 3.2) (Maddison and Maddison 2017) to reflect the appropriate topology while maintaining branch lengths. Sitewise log-likelihood scores were obtained for each topology using the –f G and the PROTGAMMAAUTO settings of RAxML (ver. 8.2.10) (Stamatakis 2014) and summed across each gene using the aligned supermatrix for the 80–100% taxon occupancy and ≥0.8 alignment column occupancy data set to obtain genewise log-likelihood scores. For each contrasting pair of topologies, the gene partition with the highest and lowest difference in log-likelihood scores were removed from the alignment matrix, and the species tree was recalculated with IQ-Tree as described previously. These analyses were also performed at the 0.2 and 0.5 alignment column occupancy cutoffs (80–100% taxon occupancy), and with the 80–100% taxon occupancy data subset with all gap-containing positions removed.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank members of the Alverson and Wickett labs and three anonymous reviewers for critical comments on earlier versions of the article. We thank David Chafin, Jeff Pummill, and Pawel Wolinski of the Arkansas High Performance Computing Center (AHPCC), and the Chicago Botanic Garden, for computational resources and support. We also thank those who contributed diatom samples to the MMETSP project. This work was supported by the National Science Foundation (NSF) (grant no. DEB-1353131 to A.J.A. and DEB-1353152 to N.J.W.) and multiple awards from the Arkansas Biosciences Institute to A.J.A. Computational resources through the AHPCC were funded through multiple NSF grants and the Arkansas Economic Development Commission; resources available at the Chicago Botanic Garden were funded by NSF (DEB-1239992 and DEB-1342873 to N.J.W.).
References
- Andrade SCS, Montenegro H, Strand M, Schwartz ML, Kajihara H, Norenburg JL, Turbeville JM, Sundberg P, Giribet G.. 2014. A transcriptomic approach to ribbon worm systematics (Nemertea): resolving the Pilidiophora problem. Mol Biol Evol. 31(12):3206–3215. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Gil M, Dufayard JF, Dessimoz C, Gascuel O.. 2011. Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst Biol. 60(5):685–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou S, Allen AE, Apt KE, Bechner M, et al. 2004. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306(5693):79–86. [DOI] [PubMed] [Google Scholar]
- Ashworth MP, Ruck EC, Lobban CS, Romanovicz DK, Theriot EC.. 2012. A revision of the genus Cyclophora and description of Astrosyne gen. nov. (Bacillariophyta), two genera with the pyrenoids contained within pseudosepta. Phycologia 51(6):684–699. [Google Scholar]
- Blom MPK, Bragg JG, Potter S, Moritz C.. 2017. Accounting for uncertainty in gene tree estimation: summary-coalescent species tree inference in a challenging radiation of Australian lizards. Syst Biol. 66:352–366. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina Sequence Data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiec ML. 2016. AMAS: a fast tool for alignment manipulation and computing of summary statistics. PeerJ 4:e1660.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussau B, Szollosi GJ, Duret L, Gouy M, Tannier E, Daubin V.. 2013. Genome-scale coestimation of species and gene trees. Genome Res. 23(2):323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, Maheswari U, Martens C, Maumus F, Otillar RP, et al. 2008. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456(7219):239–244. [DOI] [PubMed] [Google Scholar]
- Brown JW, Sorhannus U, Gilbert MTP.. 2010. A molecular genetic timescale for the diversification of autotrophic stramenopiles (Ochrophyta): substantive underestimation of putative fossil ages. PLoS One 5(9):e12759.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell B. 2014. BBMerge. Walnut Creek (CA: ): Joint Genome Institute. [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL.. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casteleyn G, Adams NG, Vanormelingen P, Debeer AE, Sabbe K, Vyverman W.. 2009. Natural hybrids in the marine diatom Pseudo-nitzschia pungens (Bacillariophyceae): genetic and morphological evidence. Protist 160(2):343–354. [DOI] [PubMed] [Google Scholar]
- Chase MW, Soltis DE, Olmstead RG, Morgan D, Les DH, Mishler BD, Duvall MR, Price RA, Hills HG, Qiu Y-L, et al. 1993. Phylogenetics of seed plants: an analysis of nucleotide sequences from the plastid gene rbcL. Ann Mo Bot Garden 80(3):528–580. [Google Scholar]
- Chernomor O, von Haeseler A, Minh BQ.. 2016. Terrace aware data structure for phylogenomic inference from supermatrices. Syst Biol. 65(6):997–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock JM, Sterck L, Rouze P, Scornet D, Allen AE, Amoutzias G, Anthouard V, Artiguenave F, Aury JM, Badger JH, et al. 2010. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 465(7298):617–621. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27(8):1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan JH, Rosenberg NA.. 2009. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol Evol. 24(6):332–340. [DOI] [PubMed] [Google Scholar]
- Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol. 7(10):e1002195.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SV, Potter S, Schmitt CJ, Bragg JG, Moritz C.. 2016. Reticulation, divergence, and the phylogeography-phylogenetics continuum. Proc Natl Acad Sci U S A. 113(29):8025–8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S.. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16:157.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field CB, Behrenfeld MJ, Randerson JT, Falkowski P.. 1998. Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281(5374):237–240. [DOI] [PubMed] [Google Scholar]
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, et al. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44(D1):D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W.. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28(23):3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Daubin V.. 2008. Dealing with incongruence in phylogenomic analyses. Philos Trans R Soc Lond B Biol Sci. 363(1512):4023–4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatesy J, Springer MS.. 2014. Phylogenetic analysis at deep timescales: unreliable gene trees, bypassed hidden support, and the coalescence/concatalescence conundrum. Mol Phylogenet Evol. 80:231–266. [DOI] [PubMed] [Google Scholar]
- Gobler CJ, Berry DL, Dyhrman ST, Wilhelm SW, Salamov A, Lobanov AV, Zhang Y, Collier JL, Wurch LL, Kustka AB, et al. 2011. Niche of harmful alga Aureococcus anophagefferens revealed through ecogenomics. Proc Natl Acad Sci U S A. 108(11):4352–4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez F, Moreira D, Benzerara K, Lopez-Garcia P.. 2011. Solenicola setigera is the first characterized member of the abundant and cosmopolitan uncultured marine stramenopile group MAST-3. Environ Microbiol. 13(1):193–202. [DOI] [PubMed] [Google Scholar]
- Gonzalez VL, Andrade SCS, Bieler R, Collins TM, Dunn CW, Mikkelsen PM, Taylor JD, Giribet G.. 2015. A phylogenetic backbone for Bivalvia: an RNA-seq approach. Proc R Soc B Biol Sci. 282(1801):20142332.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011a. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011b. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat Biotechnol. 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O.. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O, Rannala B.. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52(5):696–704. [DOI] [PubMed] [Google Scholar]
- Hahn MW, Nakhleh L.. 2016. Irrational exuberance for resolved species trees. Evolution 70(1):7–17. [DOI] [PubMed] [Google Scholar]
- Hall BG. 2005. Comparison of the accuracies of several phylogenetic methods using protein and DNA sequences. Mol Biol Evol. 22(3):792–802. [DOI] [PubMed] [Google Scholar]
- Harrison RG, Larson EL.. 2014. Hybridization, introgression, and the nature of species boundaries. J Hered. 105(S1):795–809. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J, Serra F, Bork P.. 2016. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol. 33(6):1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D.. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 23:254–267. [DOI] [PubMed] [Google Scholar]
- Johnson BR, Borowiec ML, Chiu JC, Lee EK, Atallah J, Ward PS.. 2013. Phylogenomics resolves evolutionary relationships among ants, bees, and wasps. Curr Biol. 23(20):2058–2062. [DOI] [PubMed] [Google Scholar]
- Jombart T, Kendall M, Almagro-Garcia J, Colijn C.. 2017. treescape: statistical exploration of landscapes of phylogenetic trees. Mol Ecology Resources. doi: 10.1111/1755-0998.12676. [DOI] [PMC free article] [PubMed]
- Keeling PJ, Burki F, Wilcox HM, Allam B, Allen EE, Amaral-Zettler LA, Armbrust EV, Archibald JM, Bharti AK, Bell CJ, et al. 2014. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 12(6):e1001889.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessenich CR, Ruck EC, Schurko AM, Wickett NJ, Alverson AJ.. 2014. Transcriptomic insights into the life history of bolidophytes, the sister lineage to diatoms. J Phycol. 50(6):977–983. [DOI] [PubMed] [Google Scholar]
- Lambert SM, Reeder TW, Wiens JJ.. 2015. When do species-tree and concatenated estimates disagree? An empirical analysis with higher-level scincid lizard phylogeny. Mol Phylogenet Evol. 82:146–155. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemer S, Kawauchi GY, Andrade SCS, Gonzalez VL, Boyle MJ, Giribet G.. 2015. Re-evaluating the phylogeny of Sipuncula through transcriptomics. Mol Phylogenet Evol. 83:174–183. [DOI] [PubMed] [Google Scholar]
- Li CL, Ashworth MP, Witkowski A, Dąbek P, Medlin LK, Kooistra WHCF, Sato S, Zgłobicka I, Kurzydłowski KJ, Theriot EC, et al. 2015. New insights into Plagiogrammaceae (Bacillariophyta) based on multigene phylogenies and morphological characteristics with the description of a new genus and three new species. PLoS One 10(10):e0139300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacManes MD. 2015. An opinionated guide to the proper care and feeding of your transcriptome. bioRxiv. 10.1101/035642.
- Maddison WP, Knowles LL.. 2006. Inferring phylogeny despite incomplete lineage sorting. Syst Biol. 55(1):21–30. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR.. 2017. Mesquite: a modular system for evolutionary analysis. Version 3.2. http://mesquiteproject.org
- Maddison WP, Wiens JJ.. 1997. Gene trees in species trees. Syst Biol. 46(3):523–536. [Google Scholar]
- Mann DG, Vanormelingen P.. 2013. An inordinate fondness? The number, distributions, and origins of diatom species. J Eukaryot Microbiol. 60(4):414–420. [DOI] [PubMed] [Google Scholar]
- Medlin LK. 2016. Evolution of the diatoms: major steps in their evolution and a review of the supporting molecular and morphological evidence. Phycologia 55(1):79–103. [Google Scholar]
- Medlin LK, Williams DM, Sims PA.. 1993. The evolution of the diatoms (Bacillariophyta). I. Origin of the group and assessment of the monophyly of its major divisions. Eur J Phycol. 28(4):261–275. [Google Scholar]
- Minh BQ, Nguyen MA, von Haeseler A.. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirarab S, Reaz R, Bayzid MS, Zimmermann T, Swenson MS, Warnow T.. 2014. ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics 30(17):i541–i548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirarab S, Warnow T.. 2015. ASTRAL-II: coalescent-based species tree estimation with many hundreds of taxa and thousands of genes. Bioinformatics 31(12):i44–i52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mock T, Otillar RP, Strauss J, McMullan M, Paajanen P, Schmutz J, Salamov A, Sanges R, Toseland A, Ward BJ, et al. 2017. Evolutionary genomics of the cold-adapted diatom Fragilariopsis cylindrus. Nature 541(7638):536–540. [DOI] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen NP, Mirarab S, Kumar K, Warnow T.. 2015. Ultra-large alignments using phylogeny-aware profiles. Genome Biol. 16:124.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamilo P, Nei M.. 1988. Relationships between gene trees and species trees. Mol Biol Evol. 5(5):568–583. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP.. 2010. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5(3):e9490.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2015. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing.
- Radakovits R, Jinkerson RE, Fuerstenberg SI, Tae H, Settlage RE, Boore JL, Posewitz MC.. 2012. Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropsis gaditana. Nat Commun. 3:686.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampen SW, Schouten S, Elda Panoto F, Brink M, Andersen RA, Muyzer G, Abbas B, Sinninghe Damsté JS.. 2009. Phylogenetic position of Attheya longicornis and Attheya septentrionalis (Bacillariophyta). J Phycol. 45(2):444–453. [DOI] [PubMed] [Google Scholar]
- Ruhfel BR, Gitzendanner MA, Soltis PS, Soltis DE, Burleigh JG.. 2014. From algae to angiosperms–inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol Biol. 14:23.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salichos L, Rokas A.. 2013. Inferring ancient divergences requires genes with strong phylogenetic signals. Nature 497(7449):327–331. [DOI] [PubMed] [Google Scholar]
- Salichos L, Stamatakis A, Rokas A.. 2014. Novel information theory-based measures for quantifying incongruence among phylogenetic trees. Mol Biol Evol. 31(5):1261–1271. [DOI] [PubMed] [Google Scholar]
- Sato S, Mann DG, Matsumoto S, Medlin LK.. 2008. Pseudostriatella (Bacillariophyta): a description of a new araphid diatom genus based on observations of frustule and auxospore structure and 18S rDNA phylogeny. Phycologia 47(4):371–391. [Google Scholar]
- Sayyari E, Mirarab S.. 2016. Fast coalescent-based computation of local branch support from quartet frequencies. Mol Biol Evol. 33(7):1654–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X-X, Hittinger CT, Rokas A.. 2017. Contentious relationships in phylogenomic studies can be driven by a handful of genes. Nat Ecol Evol. 1(5):0126.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen XX, Zhou XF, Kominek J, Kurtzman CP, Hittinger CT, Rokas A.. 2016. Reconstructing the backbone of the Saccharomycotina yeast phylogeny using genome-scale data. G3 Genes Genomes Genet. 6:3927–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Sistrom M, Hutchinson M, Bertozzi T, Donnellan S.. 2014. Evaluating evolutionary history in the face of high gene tree discordance in Australian Gehyra (Reptilia: Gekkonidae). Heredity 113(1):52–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, Moore MJ, Brown JW, Yang Y.. 2015. Analysis of phylogenomic datasets reveals conflict, concordance, and gene duplications with examples from animals and plants. BMC Evol Biol. 15:150.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Unna R, Boursnell C, Patro R, Hibberd J, Kelly S.. 2016. TransRate: reference free quality assessment of de novo transcriptome assemblies. Genome Res. 26:1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Smith SA, Cellinese N, Wurdack KJ, Tank DC, Brockington SF, Refulio-Rodriguez NF, Walker JB, Moore MJ, Carlsward BS, et al. 2011. Angiosperm phylogeny: 17 genes, 640 taxa. Am J Bot. 98(4):704–730. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE, Wolf PG, Nickrent DL, Chaw S, Chapman RL.. 1999. The phylogeny of land plants inferred from 18S rDNA sequences: pushing the limits of rDNA signal? Mol Biol Evol. 16(12):1774–1784. [DOI] [PubMed] [Google Scholar]
- Song L, Florea L. (Song2015 co-authors). 2015. Rcorrector: efficient and accurate error correction for Illumina RNA-seq reads. GigaScience 4(1):48.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorhannus U. 2007. A nuclear-encoded small-subunit ribosomal RNA timescale for diatom evolution. Mar Micropaleontol. 65(1-2):1–12. [Google Scholar]
- Sorhannus U, Fox MG.. 2012. Phylogenetic analyses of a combined data set suggest that the Attheya lineage is the closest living relative of the pennate diatoms (Bacillariophyceae). Protist 163(2):252–262. [DOI] [PubMed] [Google Scholar]
- SRA-Tools [Internet]. 2016. [downloaded 2016 26 August 2016]. http://ncbi.github.io/sra-tools/
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streicher JW, Schulte JA, Wiens JJ.. 2016. How should genes and taxa be sampled for phylogenomic analyses with missing data? An empirical study in iguanian lizards. Syst Biol. 65(1):128–145. [DOI] [PubMed] [Google Scholar]
- Sukumaran J, Holder MT.. 2010. DendroPy: a Python library for phylogenetic computing. Bioinformatics 26(12):1569–1571. [DOI] [PubMed] [Google Scholar]
- Tange O. 2011. GNU Parallel: the command-line power tool. USENIX Mag. 36:42–47. [Google Scholar]
- Theriot EC, Ashworth M, Ruck E, Nakov T, Jansen RK.. 2010. A preliminary multigene phylogeny of the diatoms (Bacillariophyta): challenges for future research. Plant Ecol Evol. 143(3):278–296. [Google Scholar]
- Theriot EC, Ashworth MP, Nakov T, Ruck E, Jansen RK.. 2015. Dissecting signal and noise in diatom chloroplast protein encoding genes with phylogenetic information profiling. Mol Phylogenet Evol. 89:28–36. [DOI] [PubMed] [Google Scholar]
- Theriot EC, Cannone JJ, Gutell RR, Alverson AJ.. 2009. The limits of nuclear-encoded SSU rDNA for resolving the diatom phylogeny. Eur J Phycol. 44(3):277–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vachaspati P, Warnow T.. 2015. ASTRID: Accurate Species TRees from Internode Distances. BMC Genomics 16(Suppl 10):S3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J, Egan AN, Dikow RB, Zimmer EA.. 2015. Utility of transcriptome sequencing for phylogenetic inference and character evolution. In: Hörandl E, Appelhans MS, editors. Next-generation sequencing in plant systematics. International Association for Plant Taxonomy. p. 1–41. Germany: Koeltz Scientific Books.
- Wickett NJ, Mirarab S, Nguyen N, Warnow T, Carpenter E, Matasci N, Ayyampalayam S, Barker MS, Burleigh JG, Gitzendanner MA, et al. 2014. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci U S A. 111:E4859–E4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2009. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag. [Google Scholar]
- Xi ZX, Liu L, Davis CC.. 2016. The impact of missing data on species tree estimation. Mol Biol Evol. 33(3):838–860. [DOI] [PubMed] [Google Scholar]
- Yang Y, Smith SA.. 2014. Orthology inference in nonmodel organisms using transcriptomes and low-coverage genomes: improving accuracy and matrix occupancy for phylogenomics. Mol Biol Evol. 31(11):3081–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Zhang Q, Sun R, Kong H, Zhang N, Ma H.. 2014. Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times. Nat Commun. 5:4956.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.