Abstract
Plants can show long-term effects of environmental stresses and in some cases a stress “memory” has been reported to persist across generations, potentially mediated by epigenetic mechanisms. However, few documented cases exist of transgenerational effects that persist for multiple generations and it remains unclear if or how epigenetic mechanisms are involved. Here, we show that the composition of small regulatory RNAs in apomictic dandelion lineages reveals a footprint of drought stress and salicylic acid treatment experienced two generations ago. Overall proportions of 21 and 24 nt RNA pools were shifted due to grandparental treatments. While individual genes did not show strong up- or downregulation of associated sRNAs, the subset of genes that showed the strongest shifts in sRNA abundance was significantly enriched for several GO terms including stress-specific functions. This suggests that a stress-induced signal was transmitted across multiple unexposed generations leading to persistent changes in epigenetic gene regulation.
Keywords: small RNA, transgenerational effects, epigenetic inheritance, Taraxacum officinale, drought stress, salicylic acid
Stress exposure triggers responses that are mediated by changes in gene regulation (Heil 2002; Shao et al. 2008; Cramer et al. 2011). In plants, some responses to environmental stresses are long-lived. For instance, upon mild pathogen infection, plants can enter a “primed” state which is expressed as a quicker or more vigorous defense response upon a second infection later in life (Conrath et al. 2002). Similar defense-related induced effects, and also responses to other environmental triggers, have been demonstrated to persist into offspring generations in some cases (Agrawal 2002; Mandal et al. 2012; Slaughter et al. 2012; Wang et al. 2016).
Although several different mechanisms can underlie inherited environmental effects in plants (Crisp et al. 2016), epigenetic mechanisms are considered prime candidates because of their potential for environmental sensitivity (Dowen et al. 2012) and transgenerational stability (Cortijo et al. 2014). Especially DNA methylation can be transgenerationally stable in plants and this mechanism is often proposed to mediate environmental effects that persist for multiple generations (Boyko et al. 2007, 2010; Verhoeven et al. 2010; Ou et al. 2012; Bilichak et al. 2015). However, empirical support for this hypothesis remains scarce (Pecinka and Scheid 2012).
Accumulating evidence indicates that regulatory small RNAs (sRNAs) also have a role in plant transgenerational effects. Indeed, sRNA biogenesis mutants in A. thaliana show compromised transgenerational herbivore resistance (Rasmann et al. 2012), suggesting that sRNAs are required to sustain induced defense responses across generations. Changes in sRNA composition have been demonstrated in a number of species in response to heat (Ito et al. 2011; Bilichak et al. 2015; Song et al. 2016), drought (Matsui et al. 2008; Tricker et al. 2012), salinity (Borsani et al. 2005; Matsui et al. 2008; Ding et al. 2009; Song et al. 2016), cold, and osmotic stress (Song et al. 2016). In some cases, these sRNA alternations have been shown to persist in the offspring of stressed plants (Bilichak et al. 2015). The mechanisms that maintain changes of sRNAs across generations remain largely unclear but may be the result of feed-back loops involving (transiently) heritable DNA methylation changes (Wibowo et al. 2016).
Here, we used apomictic dandelion (Taraxacum officinale) to test the impact of environmental stress on sRNA composition in unexposed offspring two generations after stress treatment. Due to apomictic (clonal seed) reproduction, dandelion offspring are considered genetic copies allowing for multi-generation experiments without confounding effects of genetic differences between samples. Apomixis in triploid dandelion involves formation of unreduced egg cells that develop parthenogenetically into embryos (Bicknell and Koltunow 2004). It is possible that the absence of fertilization in apomicts promotes the transgenerational stability of novel epigenetic variants, as has been observed under vegetative propagation (Ong-Abdullah et al. 2015), because nonsexual reproduction may partly bypass the extensive reprogramming that occurs during male gametogenesis and early embryogenesis (Kawashima and Berger 2014). In apomictic dandelion, first-generation offspring of stress-exposed plants have previously been demonstrated to show modified phenotypes and DNA methylation patterns, suggesting potential for environment-induced transgenerational epigenetic inheritance (Verhoeven et al. 2010; Verhoeven and van Gurp 2012).
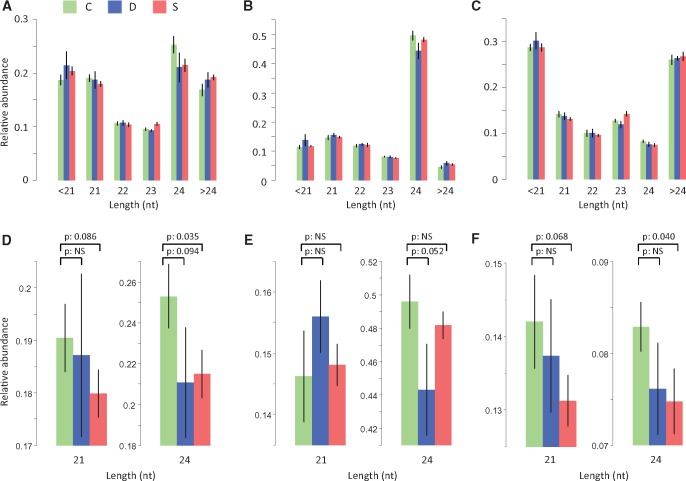
We grew first-generation (G1) plants under either drought stress, salicylic acid exposure (SA; a plant hormone that is involved in several processes including defense signaling in response to pathogens; Vicente and Plasencia 2011), or under control conditions. Second (G2) and third (G3) generation apomictic progenies were obtained by single-seed descent for four replicate lineages per experimental group and were grown under common control conditions (supplementary text S1, Supplementary Material online). sRNAs were sequenced at generation G3 in four individual plants per experimental group (supplementary text S1 and S2, Supplementary Material online). As no reference assembly currently exists for dandelion, we first assessed differences in sRNA composition between treatment groups and control using the total sRNA libraries. Using permutation tests based on random reshuffling of sample labels when comparing G3 control samples to either G3 drought or SA samples (supplementary text S3, Supplementary Material online), we found a significant reduction in the proportion of 24 nt sRNAs after grandparental SA treatment (bootstrap test, P = 0.035), and also marginally significant shifts in 24 nt sRNAs after grandparental drought treatment (bootstrap test, P = 0.094) and in 21 nt sRNAs after grandparental SA treatment (bootstrap test, P = 0.086) (fig. 1A and D). The most pronounced changes occurred for sRNAs of size 24 nt whose relative abundance in the total sRNA population was reduced in both of the stress conditions compared with the control. Relative loss of TE-associated 24 nt sRNA has been reported for a variety of biotic and abiotic stressors (Dowen et al. 2012; Lunardon et al. 2016; McCue et al. 2012). These changes appear to be mainly the result of hypomethylation and loss of RdDM targeting of transposable element (TE) sequences (Tran et al. 2005; McCue et al. 2013).
Fig. 1.
Length composition for the read libraries: all sRNAs (A and D), mapped to annotated TEs (B and E) and mapped to gene-annotated transcripts (C and F) (mean ± SE). Bottom panels (D, E, and F) are enlargements of top panels showing P values from permutation tests performed for 21 and 24 nt sRNA size classes. P values larger than 0.1 are labeled “NS” (not significant). Treatment groups (C: control; D: drought; S: salicylic acid) refer to grandparental treatments.
To understand the stress-induced shifts in 21 and 24 nt sRNA composition in more detail, we took advantage of a recent TE database that was generated based on de novo clustering of repetitive sequences from the T. officinale genome (Ferreira de Carvalho et al. 2016a; supplementary text S2, Supplementary Material online). We aligned sRNAs to these TE sequences, and compared the relative size abundance across conditions. A loss of 24 nt sRNAs was also observed in these TE-annotated sequences, at least after drought stress (P = 0.052, fig. 1B and E). Loss of 24 nt TE-associated sRNAs is typically accompanied by gains in 21 nt sRNAs due to an increase in the transcription of precursors for this class of sRNA (Dowen et al. 2012; McCue et al. 2012). Consistent with this, the loss of 24 nt TE-mapping sRNAs after drought stress cooccurred with an increase in 21 nt sRNAs, although this increase was not statistically significant (P = 0.139, fig. 1E).
It is unclear if and how such sRNA shifts impact gene expression. Previous studies have reported that TEs proximal to or overlapping genes can affect transcription under stress, probably as a result of RdDM-mediated DNA methylation loss (Lister et al. 2008; Hollister and Gaut 2009; Dowen et al. 2012; Wang et al. 2013; Quadrana et al. 2016). Another mechanism by which sRNAs can affect gene expression is through trans-acting posttranscriptional modifications (Borsani et al. 2005). We recently assembled the complete transcriptome of dandelion (Ferreira de Carvalho et al. 2016b). Over 13,500 genes could be annotated by homology with A. thaliana. We aligned our sRNA libraries to these transcriptomes (supplementary text S2, Supplementary Material online). On average about 53% of the reads from each library met our quality control criteria and could be successfully aligned. Similar to TE-aligned sRNAs, the relative abundance of transcriptome-aligned 24 nt sRNAs was reduced after grandparental stress treatments (most clearly after grandparental SA treatment: bootstrap test, P = 0.04; see fig. 1C and F), but now also a reduction in the relative abundance of 21 nt sRNAs was indicated (bootstrap test, P = 0.068; fig. 1F).
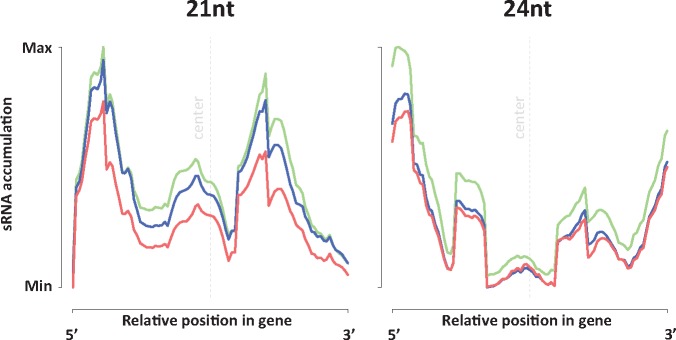
The observed loss of transcriptome-aligned 24 nt sRNAs was enigmatic, as 24 nt sRNA are typically depleted in genic sequences. To explore this issue in more detail, we studied the density distribution of 24 nt sRNA along our annotated transcripts. Our analysis shows that 24 nt sRNAs mostly mapped towards the 5′ and 3′ flanks of the genes, suggesting vestiges of a sRNA signal that originates from sequences outside of gene bodies, such as promoters or intergenic regions (fig. 2). In contrast, 21 nt sRNA showed a peak density more toward the center of gene bodies. The distributional patterns reported here resemble previously reported genic signatures of sRNA abundance in well-annotated genomes such as maize (Gent et al. 2013; Lunardon et al. 2016), A. thaliana (Dowen et al. 2012), and rice (Li et al. 2012). The relative decrease in transcript-associated 24 nt sRNA after stress exposure may suggest a loss of methylation in gene flanking regions (and possibly also in TE sequences within genes) and consequent gene expression upregulation. However, a quality genome reference assembly will be required to test this hypothesis. Together, the specific changes in sRNA profiles that we observed are in line with previous observations in stress-exposed plants, but our results indicate the stress-associated sRNA footprint is maintained transgenerationally for at least two unexposed generations after the stress treatment.
Fig. 2.
Spatial accumulation of 21 and 24 nt sRNA reads in gene-mapping transcripts. Lines represent density distributions of sRNA mapping location along the transcript. Each gene-mapping transcript was scaled to a length of 1000 bp and sRNA mapping positions (pooled replicates) inside each transcript were transformed accordingly. The counts for each transcript were afterwards collapsed into a single transcript model by calculating the averaged number of sRNA hits for each transcript coordinate across all length-normalized transcripts. Color code for treatment groups: control (green), drought (blue), and SA (red).
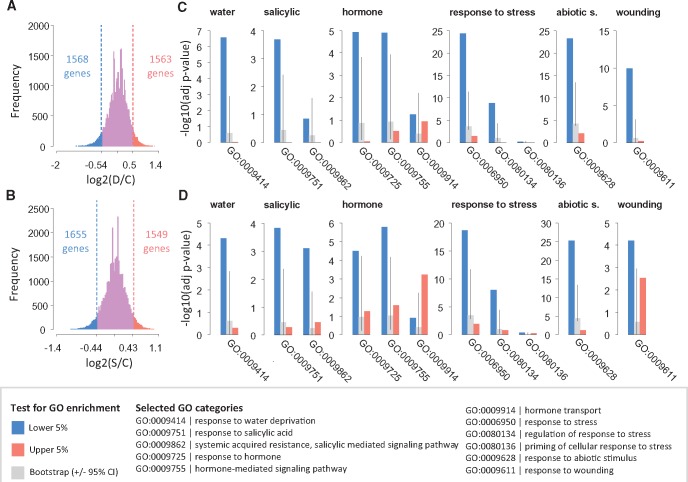
In order to identify specific genes that show different sRNA abundance comparing control and stress treatments, we performed differential analysis using DESeq2. DESeq2 uses negative binomial generalized linear models to statistically test each gene for a difference between experimental groups in the number of (sRNA) reads mapping to that gene (Love et al. 2014). We applied DESeq2 to different sRNA lengths: 21 nt, 24 nt sRNAs and for all length classes combined (18–30 nt). After adjusting for multiple testing (FDR = 0.10), our results showed virtually no significant sRNA enrichment or depletion at individual genes (supplementary table S4, Supplementary Material online). This is consistent with an observed overall high similarity between individual sRNA libraries, both within and between experimental groups (supplementary text S2, Supplementary Material online). We argue that the induced and transgenerationally inherited sRNA effects are subtle and may not be readily detectable using our approach that involved sRNA sequencing of individual plants, not pooled samples. We therefore focused, instead, on sets of genes that were either most depleted or enriched for sRNA in the stress groups and we tested if these gene sets were overrepresented for specific GO-terms. Using Fisher’s Exact tests our analysis revealed a significant overrepresentation for hundreds of GO categories, depending on grandparental treatment and sRNA length class (supplementary text S5, Supplementary Material online; our significance testing included evaluation against a bootstrapping-derived null distribution of enrichment to account for potential baseline biases in the dandelion transcriptome when compared with the Arabidopsis reference gene set). A large fraction of these significant GO-categories overlapped between the two stress treatments, suggesting a generalized stress response. The 5% of genes that were most depleted or enriched for 21 nt sRNAs were significantly enriched for about 400–500 GO categories in both the control-drought comparison and in the control-SA comparison. The 5% of genes that were most depleted for 24 nt sRNAs were enriched for many more GO terms than the 5% of genes that showed the strongest increase in associated 24 nt sRNAs, suggesting a strong biological signal in the relaxation of 24 nt-based gene silencing after grandparental stress.
We searched the list of significantly enriched GO terms for specific keywords that are associated with the grandparental stresses: “water” and “drought” for drought treatment, “salicylic” and “hormone” for SA treatment and “response to stress”, “abiotic stimulus” and “wounding” for stress treatments in general (see fig. 3). For instance, the GO term 0006950 (“response to stress”) showed strong statistical evidence for enrichment both after drought and after SA treatment, pointing to an active stress memory. For all key words, except “drought” for which no significantly enriched GO terms were detected, significantly enriched GO terms were found in both stress treatments, suggesting that these GO terms reflect a general stress response rather than a treatment-specific response. However, two SA-related GO categories (GO term 0009862: systemic acquired resistance, salicylic-mediated signaling pathway; and GO term 0009914: hormone transport) were affected only in the SA set, indicating a more treatment-specific pattern.
Fig. 3.
Distribution for sRNA fold change in gene-mapping transcripts after grandparental drought stress (A) and salicylic acid (B) against control. Bar plots show p values of GO term enrichment tests (blue: enrichment test in set of genes with reduced sRNAs after stress; orange: enrichment test in set of genes with increased sRNAs after stress), in the case of the drought (C) and the salicylic acid (D) sets. Grey bars indicate P values obtained from random bootstrapping (absence of enrichment), which can be affected by biases in the dandelion reference transcriptome in comparison to the Arabidopsis reference gene set. Error bars indicate 95% bootstrap confidence intervals.
In summary, it is well known that stress responses can be mediated by changes in sRNA-associated gene silencing. Our results suggest that this regulation may persist for several generations after stress. sRNA-based multi-generational inheritance of environmental stress has been previously demonstrated in some animal systems (e.g., Gapp et al. 2014; Rechavi et al. 2014) where underlying mechanisms of sRNA inheritance are at least partly different from plants. Although effects on gene expression remain to be evaluated, our study is to our knowledge the first demonstration in plants of modified sRNAs two generations removed from the stress trigger. Our results show no clear statistically significant effects on individual genes, which may be due to low sequencing depth of the libraries, or lack of sensitivity of our differential analysis. However, we were able to uncover a sRNA signal among genes involved in stress-related functions. This illustrates that an epigenetic signal travelled between generations preserving footprints of grandparental stress, and that this memory implicates genes that are known to be involved in stress responses. Although we did not explore the nature of the transgenerationally inherited epigenetic signal, this signal could be a stress-induced change in TE-associated DNA methylation, which in plants can be stably inherited and can trigger RNA-mediated gene expression changes in offspring (Wibowo et al. 2016).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
Keygene N.V., Wageningen, kindly provided T. officinale BAC sequences that we used for preliminary analysis of TE-mapping sRNAs. Seeds from the T. officinale hemicyclum lineage were kindly provided by Jan Kirschner from the CAS Institute of Botany, Pruhonice. This work was supported by the Netherlands Organisation for Scientific Research (grant numbers 864.10.008 and 884.10.003); and an ERC starting grant to O.R. (grant number 335624). F.J. acknowledges support from the Technical University of Munich-Institute for Advanced Study funded by the German Excellence Initiative and the European Union Seventh Framework Programme under grant agreement #291763. sRNA data generated for this study are deposited in the NCBI Sequence Read Archive (SRA study SRP096310, BioProject accession number PRJNA360587).
References
- Agrawal AA. 2002. Herbivory and maternal effects: mechanisms and consequences of transgenerational induced plant resistance. Ecology 8312:3408–3415. [Google Scholar]
- Bicknell RA, Koltunow AM.. 2004. Understanding apomixis: recent advances and remaining conundrums. Plant Cell 16:S228–S245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilichak A, Ilnytskyy Y, Woycicki R, Kepeshchuk N, Fogen D, Kovalchuk I.. 2015. The elucidation of stress memory inheritance in Brassica rapa plants. Front Plant Sci. 6:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O, Zhu JH, Verslues PE, Sunkar R, Zhu JK.. 2005. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 1237:1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko A, Blevins T, Yao YL, Golubov A, Bilichak A, Ilnytskyy Y, Hollander J, Meins F, Kovalchuk I.. 2010. Transgenerational adaptation of arabidopsis to stress requires DNA methylation and the function of dicer-like proteins. PLoS One 53:e9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko A, Kathiria P, Zemp FJ, Yao YL, Pogribny I, Kovalchuk I.. 2007. Transgenerational changes in the genome stability and methylation in pathogen-infected plants (virus-induced plant genome instability). Nucleic Acids Res. 355:1714–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrath U, Pieterse CMJ, Mauch-Mani B.. 2002. Priming in plant–pathogen interactions. Trends Plant Sci. 75:210–216. [DOI] [PubMed] [Google Scholar]
- Cortijo S, Wardenaar R, Colome-Tatche M, Gilly A, Etcheverry M, Labadie K, Caillieux E, Hospital F, Aury J-M, Wincker P, et al. 2014. Mapping the epigenetic basis of complex traits. Science 3436175:1145–1148. [DOI] [PubMed] [Google Scholar]
- Cramer GR, Urano K, Delrot S, Pezzotti M, Shinozaki K.. 2011. Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol. 11:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp PA, Ganguly D, Eichten SR, Borevitz JO, Pogson BJ.. 2016. Reconsidering plant memory: intersections between stress recovery, RNA turnover, and epigenetics. Sci Adv. 22:e1501340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D, Zhang LF, Wang H, Liu ZJ, Zhang ZX, Zheng YL.. 2009. Differential expression of miRNAs in response to salt stress in maize roots. Ann Bot. 1031:29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen RH, Pelizzola M, Schmitz RJ, Lister R, Dowen JM, Nery JR, Dixon JE, Ecker JR.. 2012. Widespread dynamic DNA methylation in response to biotic stress. Proc Natl Acad Sci U S A. 10932:E2183–E2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira de Carvalho J, de Jager VCL, Van Gurp TP, Wagenmaker N, Verhoeven KJF.. 2016a. Recent and dynamic transposable elements contribute to genomic divergence under asexuality. BMC Genomics 17884:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira de Carvalho J, Oplaat C, Pappas N, Derks M, de Ridder D, Verhoeven KJF.. 2016b. Heritable gene expression differences between apomictic clone members in Taraxacum officinale: insights into early stages of evolutionary divergence in asexual plants. BMC Genomics 17203:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapp K, Jawaid A, Sarkies P, Bohacek J, Pelczar P, Prados J, Farinelli L, Miska E, Mansuy IM.. 2014. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci. 175:667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Ellis NA, Guo L, Harkess AE, Yao YY, Zhang XY, Dawe RK.. 2013. CHH islands: de novo DNA methylation in near-gene chromatin regulation in maize. Genome Res. 234:628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil M. 2002. Ecological costs of induced resistance. Curr Opin Plant Biol. 54:345–350. [DOI] [PubMed] [Google Scholar]
- Hollister JD, Gaut BS.. 2009. Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 198:1419–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Gaubert H, Bucher E, Mirouze M, Vaillant I, Paszkowski J.. 2011. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 4727341:115. [DOI] [PubMed] [Google Scholar]
- Kawashima T, Berger F.. 2014. Epigenetic reprogramming in plant sexual reproduction. Nat Rev. Genet. 159:613–624. [DOI] [PubMed] [Google Scholar]
- Li X, Zhu JD, Hu FY, Ge S, Ye MZ, Xiang H, Zhang GJ, Zheng XM, Zhang HY, Zhang SL, et al. 2012. Single-base resolution maps of cultivated and wild rice methylomes and regulatory roles of DNA methylation in plant gene expression. BMC Genomics 13:300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR.. 2008. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 1333:523–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 1512:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunardon A, Forestan C, Farinati S, Axtell MJ, Varotto S.. 2016. Genome-wide characterization of maize small RNA loci and their regulation in the required to maintain repression6-1 (rmr6-1) mutant and long-term abiotic stresses. Plant Physiol. 1703:1535–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal R, Kathiria P, Psychogios N, Bouatra S, Krishnumarthy R, Wishart D, Kovalchuk I.. 2012. Progeny of tobacco mosaic virus-infected Nicotiana tabacum plants exhibit trans-generational changes in metabolic profiles. Biocatal Agric Biotechnol. 12:115–123. [Google Scholar]
- Matsui A, Ishida J, Morosawa T, Mochizuki Y, Kaminuma E, Endo TA, Okamoto M, Nambara E, Nakajima M, Kawashima M, et al. 2008. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 498:1135–1149. [DOI] [PubMed] [Google Scholar]
- McCue AD, Nuthikattu S, Reeder SH, Slotkin RK.. 2012. Gene expression and stress response mediated by the epigenetic regulation of a transposable element small RNA. PLoS Genet. 82: e1002474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCue AD, Nuthikattu S, Slotkin RK.. 2013. Genome-wide identification of genes regulated in trans by transposable element small interfering RNAs. RNA Biol. 108:1379–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong-Abdullah M, Ordway JM, Jiang N, Ooi SE, Kok SY, Sarpan N, Azimi N, Hashim AT, Ishak Z, Rosli SK, et al. 2015. Loss of Karma transposon methylation underlies the mantled somaclonal variant of oil palm. Nature 5257570:533–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou XF, Zhang YH, Xu CM, Lin XY, Zang Q, Zhuang TT, Jiang LL, von Wettstein D, Liu B.. 2012. Transgenerational inheritance of modified DNA methylation patterns and enhanced tolerance induced by heavy metal stress in rice (Oryza sativa L.). PLoS One 79:e41143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecinka A, Scheid OM.. 2012. Stress-induced chromatin changes: a critical view on their heritability. Plant Cell Physiol. 535:801–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrana L, Silveira AB, Mayhew GF, LeBlanc C, Martienssen RA, Jeddeloh JA, Colot V.. 2016. The Arabidopsis thaliana mobilome and its impact at the species level. Elife 5:e15716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmann S, De Vos M, Casteel CL, Tian DL, Halitschke R, Sun JY, Agrawal AA, Felton GW, Jander G.. 2012. Herbivory in the previous generation primes plants for enhanced insect resistance. Plant Physiol. 1582:854–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechavi O, Houri-Ze'evi L, Anava S, Goh WSS, Kerk SY, Hannon GJ, Hobert O.. 2014. Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell 1582:277–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao HB, Chu LY, Jaleel CA, Zhao CX.. 2008. Water-deficit stress-induced anatomical changes in higher plants. C R Biol. 3313:215–225. [DOI] [PubMed] [Google Scholar]
- Slaughter A, Daniel X, Flors V, Luna E, Hohn B, Mauch-Mani B.. 2012. Descendants of primed Arabidopsis plants exhibit resistance to biotic stress. Plant Physiol. 1582:835–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YP, Ci D, Tian M, Zhang DQ.. 2016. Stable methylation of a non-coding RNA gene regulates gene expression in response to abiotic stress in Populus simonii. J Exp Bot. 675:1477–1492. [DOI] [PubMed] [Google Scholar]
- Tran RK, Zilberman D, de Bustos C, Ditt RF, Henikoff JG, Lindroth AM, Delrow J, Boyle T, Kwong S, Bryson TD, et al. 2005. Chromatin and siRNA pathways cooperate to maintain DNA methylation of small transposable elements in Arabidopsis. Genome Biol. 611:R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tricker PJ, Gibbings JG, Lopez CMR, Hadley P, Wilkinson MJ.. 2012. Low relative humidity triggers RNA-directed de novo DNA methylation and suppression of genes controlling stomatal development. J Exp Bot. 6310:3799–3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven KJF, Jansen JJ, van Dijk PJ, Biere A.. 2010. Stress-induced DNA methylation changes and their heritability in asexual dandelions. New Phytol. 1854:1108–1118. [DOI] [PubMed] [Google Scholar]
- Verhoeven KJF, van Gurp TP.. 2012. Transgenerational effects of stress exposure on offspring phenotypes in apomictic dandelion. PLoS One 76:e38605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente MRS, Plasencia J.. 2011. Salicylic acid beyond defence: its role in plant growth and development. J Exp Bot. 6210:3321–3338. [DOI] [PubMed] [Google Scholar]
- Wang X, Weigel D, Smith LM.. 2013. Transposon variants and their effects on gene expression in Arabidopsis. PLoS Genet. 92:e1003255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Xin CY, Cai J, Zhou Q, Dai TB, Cao WX, Jiang D.. 2016. Heat priming induces trans-generational tolerance to high temperature stress in wheat. Front Plant Sci. 7:501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibowo A, Becker C, Marconi G, Durr J, Price J, Hagmann J, Papareddy R, Putra H, Kageyama J, Becker J, et al. 2016. Hyperosmotic stress memory in Arabidopsis is mediated by distinct epigenetically labile sites in the genome and is restricted in the male germline by DNA glycosylase activity. Elife 5:e13546. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.